Abstract
Glutamate is the most abundant excitatory neurotransmitter in the central nervous system, therefore its precise control is vital for maintaining normal brain function and preventing excitotoxicity1. Removal of extracellular glutamate is achieved by plasma membrane-bound transporters, which couple glutamate transport to sodium, potassium and pH gradients using an elevator mechanism2–5. Glutamate transporters also conduct chloride ions via a channel-like process that is thermodynamically uncoupled from transport6–8. However, the molecular mechanisms that allow these dual-function transporters to carry out two seemingly contradictory roles are unknown. Here we report the cryo-electron microscopy structure of a glutamate transporter homologue in an open-channel state, revealing an aqueous cavity that is formed during the transport cycle. By studying functional properties combined with molecular dynamics simulations, we show that this cavity is an aqueous-accessible chloride permeation pathway gated by two hydrophobic regions and is conserved across mammalian and archaeal glutamate transporters. Our findings provide insight into the mechanism by which glutamate transporters support their dual function and add a crucial piece of information to aid mapping of the complete transport cycle shared by the SLC1A transporter family.
Excitatory amino acid transporters (EAATs) are glutamate transporters in the solute carrier 1A (SLC1A) family. EAATs couple glutamate transport to the co-transport of three sodium (Na+) ions and one proton (H+) and the counter-transport of one potassium (K+) ion4. In addition to coupled transport, binding of substrate and Na+ ions to EAATs activates a reversible and thermodynamically uncoupled chloride (Cl−) conductance6–8. Mutations that alter channel properties have been linked to episodic ataxia type 6 (EA6)9,10.
Structures of EAAT111, the neutral amino acid transporter 2 (ASCT2)12–14, and archaeal homologs GltPh2,15–23 and GltTk24–26 reveal SLC1A family members have a similar trimeric structure wherein each protomer functions independently26–28. They utilize a twisting elevator mechanism of transport, in which the transport domain, containing the substrate and coupled ions, shuttles across the membrane during transport while the scaffold domain makes inter-subunit contacts and remains anchored to the membrane2,13. This results in sequential transitions between an “outward-facing” state (OFS), an “intermediate outward-facing” state (iOFS), and an “inward-facing” state (IFS) (Fig. 1a). A clear pathway for the uncoupled Cl− conductance has not been observed in the available structures and the molecular determinants required for activation remain unclear. Electrophysiological studies have identified several residues in EAAT1 that likely line the channel based on their ability to exclusively alter Cl− conductance properties when mutated7,29–35. Some of these residues line an aqueous cavity at the interface of the transport and scaffold domain (domain interface) in the iOFS, suggesting the cavity may be a partially formed Cl− channel that fully opens in a different conformational state20,29.
Fig. 1 |. GltPh utilizes an elevator mechanism that can be probed by cross-links between the transport and scaffold domains.
a, GltPh protomer viewed parallel to the membrane in the OFS (PDB:2NWX), iOFS (PDB:3V8G) and IFS (PDB:3KBC) with the scaffold domain (salmon), the transport domain (blue) and bound aspartate (black). The three cysteine pairs, XL1, XL2 and XL3, are in purple, green and pink, respectively. b, Cα-Cα distances of cysteine pairs in the known crystal structures of GltPh.
Here we investigate the relationship between the elevator mechanism of transport and Cl− channel opening and present a cryo-EM structure of an open-channel conformation. We present molecular dynamics (MD) simulations of GltPh and functional analysis of EAAT1 to confirm the cavity observed at the domain interface is an aqueous-accessible Cl− permeation pathway conserved across SLC1A transporters.
GltPh can be trapped in an open-channel conformation
To uncover how glutamate transporters enter an ion-conducting state during substrate translocation, we utilized a disulfide cross-linking strategy to isolate different stages of the transport cycle in GltPh. Three cysteine pairs were introduced into a fully functional cysteine-less GltPh36 such that one cysteine residue was located on the transport domain and the other on the scaffold domain (Fig. 1a). Cysteine pairs were chosen for their proximity in the OFS (XL1), iOFS (XL2) and IFS (XL3) stages of the transport cycle (Fig. 1b). The homobifunctional cross-linking reagent, mercury, was used to promote complete cross-linking to ensure sample homogeneity and was verified using an SDS-PAGE gel-shift assay (Extended Data Fig. 1a).
GltPh-XL1, XL2 and XL3, were crystallized for structural determination. GltPh-XL1 and GltPh-XL3 were solved at 3.7Å and 3.5Å resolutions, respectively (Extended Data Table 1). The structure of GltPh-XL1 was similar to the OFS (Cα RMSD = 0.5Å) and GltPh-XL3 similar to the IFS (Cα RMSD = 0.6Å) (Extended Data Fig. 1b). GltPh-XL1 was cross-linked spontaneously with an average Cα−Cα of 6.5Å, while GltPh-XL3 was cross-linked in the presence of HgCl2 with a Cα−Cα of 8.8Å. A continuous pore was not observed in either of these structures, in agreement with previous strudies2,16. Despite extensive screening and crystal optimization, the structure of GltPh-XL2 could not be determined. Instead, purified protein was reconstituted into nanodiscs and examined using single-particle cryo-electron microscopy (cryo-EM). Samples were prepared using physiologically relevant concentrations of NaCl and aspartate and full cross-linking of all protomers was ensured by the addition of HgCl2 and confirmed by SDS-PAGE (Extended Data Fig. 1c). 3D classification was performed by imposing a C3 symmetry and initial rounds of refinement revealed all three protomers in a conformation similar to the iOFS20 (Extended Data Fig. 2a, Extended Data Table 2).
Symmetry expansion was used to identify any structural variation between protomers within the GltPh-XL2 trimer. Particles representing individual protomers were subject to focused 3D classification, revealing ten classes representing two distinct entities (Extended Data Fig. 2a). Nine classes adopted a conformation resembling the iOFS, whereas one class exhibited considerable differences. The unique class and also the clearest class resembling the iOFS were further refined and post-processed to 4.0Å and 3.7Å resolutions, respectively (Extended Data Fig. 2a-c, Extended Data Table 2). The iOFS structure was essentially indistinguishable from that previously reported20 (Cα RMSD = 0.9Å) (Extended Data Fig. 1d, 2). However, the unique class revealed a novel structure with substantial conformational differences to the iOFS, including an aqueous-accessible pathway (Fig. 2, Extended Data Fig. 2). We therefore refer to this novel conformation as the Cl−-conducting state (ClCS).
Fig. 2 |. GltPh can be trapped in an open-channel conformation.
a, Surface representation of GltPh-XL1 in the OFS, GltPh-XL2 in the iOFS and ClCS, and GltPh-XL3 in the IFS, viewed from the membrane plane. The color scheme is the same as in Fig. 1. The Cα positions of L152C and G351C are shown as spheres in red and blue, respectively. The dashed line through L152C highlights its unchanging position throughout the transport cycle as G351C moves down. b, Cross-sections of (a) through R276. Residues lining the domain interface are labelled. Close-up of the “constriction zone” viewed from the extracellular space in the iOFS (contour level 12.9σ) (c) and the ClCS (contour level 10.3σ) (d) fitted in their respective cryo-EM maps. e, Close-up of the narrowest point of the cavity in the ClCS (contour level 10.3σ). Noise removed for clarity; contour levels calculated using Chimera.
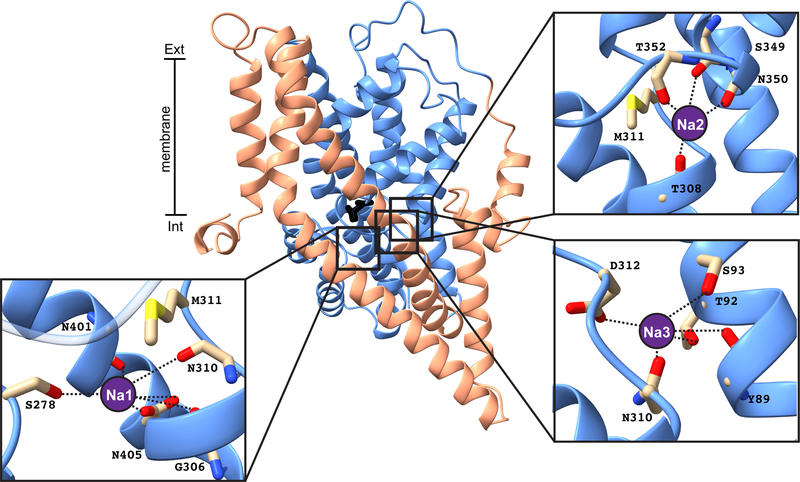
The scaffold domain (TM1, TM2, TM4 and TM5) in the iOFS and ClCS structures is relatively similar (Cα RMSD = 0.7Å). However, the transport domain in ClCS (TM3, TM6, TM7, TM8, HP1 and HP2) is further towards the IFS, following a ~5.7Å translation and ~17.7° rotation (Extended Data Fig. 3c). In this novel conformation, the position of G351C, located on the tip of HP2, is ~8.4Å closer to the cytoplasm compared to its position in the iOFS (Fig. 2a). In both states, HP2 is closed over the bound aspartate. While densities corresponding to Na+ ions were not observed, the orientation of residues involved in coordinating the three Na+ ions suggests they are present, with the side chain of M311 pointing towards Na1 and Na216,25 (Extended Data Fig. 4). Interestingly, the Cα-Cα distances between the cysteine residues at positions 152 and 351 are similar in both the iOFS and the ClCS (6.5Å and 6.6Å, respectively), yet the absolute positions of C152 and C351 varies where C351 is above C152 in the iOFS, and below it in the ClCS (Fig. 2a, Extended Data Fig. 3d, e). As the Cα-Cα distances are within the range required for a disulfide bond and the energetically favorable OFS was not captured, we infer that the cysteine residues in both structures are cross-linked. An additional ~5.6Å downward movement of the transport domain and a further ~8.7° rotation are required to reach the IFS, placing the ClCS approximately two-thirds of the way along the substrate translocation pathway (Extended Data Fig. 3c).
The existence of two distinct cross-linked states (iOFS and ClCS) in this sample likely explains our inability to obtain well diffracting crystals. Using cryo-EM, these two states were able to be separated and the nanodisc surrounding the trimer visualized. Of the 220,938 trimer particles used for analysis, 63.9% contained all three protomers in the iOFS, 28.7% contained one protomer in the ClCS, 6.5% contained two protomers in the ClCS and only 0.9% had all protomers in the ClCS simultaneously (Extended Data Fig. 5a). In a trimer containing one ClCS protomer, a slight bending of the nanodisc was observed around the ClCS transport domain compared to the iOFS (Extended Data Fig. 5b) and non-protein densities were observed between TM1, TM2, TM5 of one protomer and TM4 of the neighboring protomer (Extended Data 5c[dummy]). While we cannot unambiguously assign a molecule into these densities, their branched nature suggests they may be bound lipids participating in lipid-protein interactions.
Previously, Verdon and Boudker20 observed a cavity lined by hydrophobic residues in TM2 and TM5 at the domain interface of the iOFS, which permitted solvent accessibility from the cytoplasmic side. The further twisted and downward position of the transport domain in the ClCS increases the gap between it and the scaffold domain, dismantling the ‘constriction zone’ formed by S65, Y195, M286 and P304 at the intracellular end of the cavity in the iOFS37 (Fig. 2b-d). As the transport domain transitions between the iOFS and ClCS, TM7 pulls away from the scaffold domain to extend the cavity and reveal a cluster of hydrophobic residues in TM2 (F50 and V51) and TM5 (L212) that are now accessible to the extracellular solvent (Fig. 2b). This results in an aqueous pathway connecting the extracellular and intracellular milieu with the narrowest point of the pore occurring at a cluster of solvent-exposed hydrophobic residues, L13, I16, M202, and P356 together with R276 – the proposed anion selectivity filter for the Cl− channel in the SLC1A family30,38 (Fig. 2e). Thus, our ClCS structure reveals a putative Cl− permeation pathway in GltPh.
GltPh-ClCS is in an open-channel conformation
We sought atomic-level evidence that the water-accessible pathway in the ClCS functions as an ion-permeable pore by employing MD simulations combined with free-energy calculations. We performed a 200-ns MD simulation of our cryo-EM structure of ClCS after embedding it into a lipid bilayer, during which we observed near-complete hydration through the domain interface, followed by a 100-ns MD simulation in the presence of an external electric field during which a continuous hydration pathway was formed at the interface (Fig. 3a, Extended Data Fig. 6a). This suggests that the captured ClCS conformation can facilitate the movement of water and potentially ions via a continuous water pathway. The solvent accessible surface area (SASA) values calculated for residues lining the permeation pathway in the ClCS are higher than those in the OFS (Extended Data Fig. 6b), supporting the involvement of the domain interface in the conduction pathway.
Fig. 3 |. Energetic landscape for the movement of Cl− through GltPh-ClCS.
a, The ClCS structure was embedded into a lipid bilayer and simulated for 300 ns with and without an external electric field during which a continuous water pathway (red surface) formed at the domain interface. b, The inter-domain water molecules were used to place individual Cl− ions (purple spheres) to seed the windows for US simulations that resulted in the underlying free-energy profile for Cl− conduction. c, Occupancy of Cl− captured during 1.2-μs US simulation is shown in light red isosurface. Residues which form the Cl− pathway (polar (green), hydrophobic (cyan), basic (blue), acidic (red)) are labeled. The ClCS protomer is shown in cartoon, scaffold domain (salmon) and transport domain (blue). d, The free energy profile for Cl− conduction through ClCS along the pore axis, obtained from the US simulations, shows favorable interactions (minima) between the Cl− ion and R276/R52/K55, and energy barriers formed by hydrophobic residues as indicated. Overall, the largest barrier against the movement of Cl− observed is 1.9 kcal/mol. e, Residues interacting with the permeating Cl− ion.
To gauge ion permeation through this pathway quantitatively, we used the position of water molecules hydrating the inter-domain space to place Cl− ions along the putative ion permeation pathway (Fig. 3b) to seed a set of Umbrella Sampling (US) simulations for the ClCS structure, amounting to 1.2 μs of sampling (Fig. 3c). The converged free-energy profiles captured from these simulations revealed the presence of hydrophobic residues (V12, I16, F50, V51, M202, A205, and L212) in the pathway that provide an energy barrier against Cl− movement (Fig. 3d, Extended Data Table 3, Extended Data Fig. 6c, d). The free-energy profile also revealed energy minima corresponding to the interaction of Cl− with the basic residues R276, R52, and K55 (Fig. 3d, Extended Data Table 3). Additionally, the permeating Cl− ion interacts with several residues lining the domain interface during its translocation, including S65, which plays an important role in anion permeation7, 35 (Fig. 3c, e, Extended Data Table 3). The estimated diameter of the narrowest part of the pore is ~5Å, as calculated by Hole39. This is sufficient to accommodate a dehydrated thiocyanate ion40, the largest and most permeant anion carried by EAATs8,41. Together, these results show that GltPh can transition into a water- and Cl−-conducting conformation, and that hydrophobic residues potentially gate either end of this channel.
The EAAT1 open-channel conformation conducts Cl−
To explore the functional relevance of the different structural states isolated in GltPh, we introduced the same three cysteine pairs (XL1, XL2 and XL3) into a fully functional cysteine-less EAAT1 (E1)42 and expressed the constructs in Xenopus laevis oocytes. Uptake of L-[3H]glutamate increased upon incubation of oocytes with the reducing agent dithiothreitol (DTT), suggesting that all cysteine pairs are able to form a spontaneous disulfide bond. Incubation with the oxidizing agent copper phenanthroline (CuPh), which promotes disulfide formation, resulted in reduced L-[3H]glutamate uptake (Extended Data Fig. 7a). These effects were only observed when the two cysteines were expressed within an individual protomer (Extended Data Fig. 7f), indicating that the disulfide bond restricts the elevator movement of the transport domain, thus inhibiting transport.
Glutamate application to oocytes expressing EAAT1 generates an overall current composed of two components: the coupled transport current and the uncoupled Cl− current (Extended Data Fig. 7b). The net conductance is inwardly-rectifying with the current reversing direction at +33 ± 3 mV and the bulk of the current is generated at −100 mV due to glutamate transport. In contrast, a linear conductance with a current reversal potential of −24 mV is characteristic of the isolated Cl− channel component in oocytes6,43. Thus, changes in both reversal potential and current amplitude at positive membrane potentials are indicative of alterations to the contribution of Cl− to the total conductance observed. Consistent with uptake experiments, incubation of E1-XL1 and E1-XL3 with DTT resulted in a larger glutamate-induced current at −100 mV and incubation with CuPh resulted in a smaller current (Extended Data Fig. 7c, e). However, the conductance of E1-XL2 was markedly different to E1; the reversal potential was negative (−25.9 ± 2.3 mV) with large currents at positive membrane potentials under all conditions, characteristic of enhanced Cl− channel activity (Extended Data Fig. 7d). These features were only observed when the two cysteine residues were within the same protomer (Extended Data Fig. 7g), confirming that an intra-protomer cross-link traps the transporter in the ClCS.
To investigate the role of the hydrophobic residues identified (Fig. 3d, Extended Data Table 3) on gating the uncoupled Cl− conductance, the equivalent residues were mutated in EAAT1. Hydrophobic residues (F50, T54, L88, M89, M286, L269) were mutated to alanine to reduce side chain hydrophobicity, while A289, was converted to phenylalanine to introduce hydrophobicity (Extended Data Fig. 7h). Compared to wild-type EAAT1, the reversal potential of the conductance of the alanine mutants shifts to more hyperpolarized potentials, demonstrating an increase in the proportion of current carried by Cl− (Extended Data Fig. 7i). Conversely, A289F resulted in a significant positive shift of the reversal potential to +76 ± 2 mV, virtually eliminating any contribution from Cl− to the net conductance. Collectively, these data confirm that the permeation pathway identified in ClCS is conserved in EAAT1 and forms a functional Cl− channel gated by hydrophobic residues at either end.
Discussion
Glutamate transporters are enigmatic membrane proteins with an unusual structure. Each of their three protomers forms a twisting elevator that supports a dual transport and channel function. It is thought that the physiological role of the Cl− channel is to reconcile ionic homeostasis and charge balance disrupted by glutamate transport1,44. In the retina, the Cl− conductance of EAAT5 contributes to a negative feedback loop that reduces glutamatergic excitatory transmission45,46. In cerebellar astrocytes, EAAT1/2 channel activity influences the resting intracellular Cl− concentration47. Disruption of EAAT1 Cl− channel activity underlies the pathogenesis of EA69,10, a disorder characterized by periods of imbalance and incoordination and often associated with neurological symptoms, including seizures and migraine. The P290R mutation results in reduced glutamate transport activity and an elevated Cl− conductance, the latter property causing an episodic ataxia-like phenotype and affecting glial cell morphology in a drosophila model9,10.
Here, we have mapped the conformational transitions required for dual function glutamate transporters to enter a Cl− conducting state (ClCS). The cryo-EM structure of GltPh-ClCS reveals that the transport domain undergoes a twisting vertical movement as it moves along the translocation pathway from the iOFS to the IFS with a continuous aqueous pore at the domain interface, which permits solvent accessibility from either side of the membrane and is lined by residues proposed to line the Cl− permeation pathway (Extended Data Fig. 7j)7,29–35, 37. Employing MD simulations combined with free-energy calculations, we characterize the conduction pathway and the energetics associated with Cl− conduction and functional experiments in EAAT1 identify a similar aqueous accessible pathway, suggesting a high degree of conservation across SLC1A family members. Our findings are in contrast to previous studies suggesting that channel opening requires a lateral movement of the transport domain and that transporters trapped in an outward-facing state (XL1) can enter a Cl−-conducting conformation38,48. We do not observe any evidence of a Cl− conducting state in XL1, either in electrophysiological recordings of E1-XL1 or in the structure of GltPh-XL1, where the Cα-Cα distance between V216 and A391 in the ClCS is 14.7Å, outside the range for cross-linking to occur.
We have further characterized molecular determinants for Cl− permeation and identified two clusters of hydrophobic residues at the intracellular and extracellular regions of the Cl− channel pathway. On the intracellular side of EAAT1, the hydrophobic pocket consists of F50, T54, M286, A289, whereas L88, M89 and L296 constitute the hydrophobic cluster on the extracellular side. Similar dual hydrophobic gating mechanisms are characteristic of ion channels with a lyotropic selectivity sequence38.These two hydrophobic regions act as gates that allow Cl− ions to enter the pathway before encountering the narrowest region near an arginine residue that determines anion selectivity of this uncoupled conductance (R477 in EAAT1 and R276 in GltPh)30,38. Residues that constitute the extracellular gate were previously identified as pore-lining residues using the substituted cysteine accessibility method37, however our study is the first to reveal their role as one of two hydrophobic gates. The hydrophobicity of residues that make up the two gates are conserved in other SLC1A members49,50; therefore, it is likely that anion permeation is enabled by common molecular determinants in the SLC1A family.
Solvent-accessible hydrophobic residues at either end of the permeation pathway renders ClCS an unstable conformation, consistent with predictions that channel opening occurs with low probability (~0.01) during the transport process41. Such a state may be stabilized by the membrane-like environment provided by the nanodisc, which could support movements of the transport domain by surrounding it with a compliant lipid bilayer and providing an additional network of protein-lipid interaction51. Indeed, a recent study also observed an intermediate state between the OFS and the IFS of membrane-reconstituted GltPh52.
In summary, this study has identified the conformational rearrangements and molecular determinants involved in Cl− channel activation of GltPh and EAAT1, providing crucial information to map the complete transport cycle shared by the SLC1A transporter family. Our data reveal how the dual transport and channel functions are achieved in a single membrane protein and may be relevant to understanding other dual function transporters such as SLC26A953 and SLC6 family transporters54,55. Furthermore, this work assists in understanding the functional roles the Cl− channel plays, notably, in maintaining cell excitability and osmotic balance45,46,56 and provides a framework for developing therapeutics to differentially modulate substrate transport or channel properties for the treatment of neurological disorders caused by EAAT dysfunction.
Methods
Data reporting.
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments.
Chemicals.
All chemicals were purchased from Sigma-Aldrich (Sydney, Australia) unless otherwise stated.
Site-directed mutagenesis.
Cysteine-less EAAT1 (E1) and cysteine-less GltPh (CLGltPh) were subcloned into the oocyte transcription vector (pOTV) and bacterial expression vector (pBAD24), respectively. The pBAD24 vectors encode a 6 x His tag at the C-terminal end of the cysteine-less GltPh cDNA. Mutagenesis was performed to using the Q5® Site-Directed Mutagenesis Kit (New England BioLabs® Inc.). The cDNA products were purified using the PureLink™ Quick Plasmid Miniprep Kit (Life Technologies) and sequences were verified by the Australian Genome Research Facility (Sydney, Australia). Residues for cysteine mutation were chosen due to their predicted proximity in the three known states of GltPh/EAAT1; the outward facing state (OFS), the intermediate outward facing state (iOFS) and the inward facing state (IFS). In addition to the three pairs reported in this study, several more cross-linking pairs were investigated. Most of these pairs resulted in states indistinguishable from the OFS (XL-1) and the IFS (XL-3). Others were non-functional in EAAT1 or could not be expressed in GltPh. XL-2 was the only double cysteine mutant that could be expressed in EAAT1, GltPh and whose structure was resolved in a novel state.
Protein expression and purification.
Cysteine-less GltPh (CLGltPh) and double cysteine mutants were transformed into Escherichia coli (E. coli) Top10 cells (Invitrogen). Bacterial cells were inoculated in Luria Broth medium containing 100 μg/mL ampicillin at 37 °C until OD600 ~0.6–0.8. Protein expression was induced with 0.1% L-arabinose and harvested 4 h post-induction. All protein purification experiments were conducted at 4 °C. Membranes were first isolated and solubilized with 10 mM n-Dodecyl-β-D-maltoside (C12M; Anatrace) in binding buffer containing 20 mM HEPES-Tris (pH 7.5), 200 mM NaCl, and 5 mM Na-Asp for 1 h. Proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) and the histidine tag was subsequently removed by thrombin digestion (10 U/mg of protein). The proteins were further purified by size exclusion chromatography (SEC) using Superdex200 16/60 HiLoad gel filtration column (GE Healthcare). For CLGltPh, GltPh-XL1, the column was equilibrated with SEC running buffer containing 20 mM HEPES-Tris (pH 7.5), 25 mM NaCl, 25 mM KCl, 5 mM Na-Asp and 7 mM n-decyl-β-D-maltopyranoside (C10M; Anatrace). For GltPh-XL2 and GltPh-XL3, the SEC buffer contained 10 mM HEPES-KOH/NaOH (pH 7.5), 100 mM NaCl, 0.1 mM Na-Asp and 7 mM C10M. Proteins were concentrated to 6–7 mg/mL for gel analysis and crystallization.
Mercury-induced cross-linking and SDS-PAGE gel shift assay.
Purified proteins were diluted to 1 mg/mL and incubated for 1 h at room temperature in the presence or the absence of 10-fold molar excess HgCl2. Both HgCl2-treated and untreated protein samples were incubated with 500 μM 5 kDa methoxyl polyethylene glycol-maleimide (mPEG5K) and 0.5% SDS for 2 h at room temperature57. All samples were then re-suspended in 12% SDS loading buffer and run on a NuPAGE™ 4–12% Bis-Tris gel (Invitrogen) at 200 V for 45 min. Proteins were visualized via Coomassie blue staining. For gel source data, see Supplementary Figure 1.
Crystallography, data collection and analysis.
Protein solution at 6–7 mg/mL was incubated with 10-fold molar excess HgCl2 on ice for 1 h and dialyzed overnight at 4 °C against their respective SEC buffers to remove excess HgCl2. All crystals were grown using hanging-drop vapor diffusion. GltPh-XL1 was crystallized in condition containing 5–23% PEG 1000, 100 mM Li2SO4, 50 mM citric acid, 50 mM Na2HPO4 at 4 °C. GltPh-XL3 was crystallized in condition containing 30–35% PEG 400, 0.2 M MgCl2, 0.1 M KCl and 0.025 M Na-citrate pH 3.5–5 at 20 °C. Crystals of GltPh-XL1 was cryoprotected by soaking in a solution containing 20% PEG 1000, 50 mM citric acid, 50 mM Na2HPO4, 100 mM Li2SO4, 5% glycerol, 2 mM C10M followed by solution containing 30% PEG 1000, 50 mM citric acid, 50 mM Na2HPO4, 100 mM Li2SO4, 5% glycerol 2 mM C10M. All crystals were flash-frozen in liquid nitrogen.
All crystallographic data were collected on the Eiger 16M detector at the Australian Synchrotron (ACRF ANSTO) beamline MX2 at a wavelength of 0.976 Å58. All crystal diffraction datasets were indexed, integrated and scaled using XDS59 and the CCP4 software package60. One crystal per structure was used for analysis. Initial phases were obtained by molecular replacement with PHASER61 using published structures of GltPh in the OFS (PDB: 2NWX), the IFS (PDB: 3KBC) and the iOFS (PDB: 3V8G) as search models. The protein models were then manually built in COOT62 and further refined using REFMAC563 with TLS and three-fold non-crystallographic three-fold NCS restraints64. Phases were further improved by rounds of manual rebuilding followed by restrained refinement in REFMAC5 and PHENIX65 with tight three-fold NCS restraints. Unit cell parameters, data collection and refinement statistics are presented in Extended Data Table 1. Structural figures and animations were prepared using PyMOL66, Chimera67 or VMD68.
Cryo-EM sample preparation and imaging.
Membrane scaffold protein (MSP1E3) was expressed and purified in E. coli BL21(Gold) cells and purified GltPh proteins were reconstituted into nanodiscs as described previously69. Briefly, E.coli total lipid extract and 1-parmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) (Avanti) dissolved in chloroform were mixed in a ratio of 3:1 and dried under a gentle stream of nitrogen. The dried lipid film was resuspended in buffer containing 10 mM HEPES/KOH pH 7.5, 100 mM NaCl, 0.1 mM Na-Asp and 49 mM C10M by freeze-thaw cycles until the mixture was clear, which resulted in 33.5 mM lipid stock. GltPh proteins were mixed with MSP1E3 and the lipid stock in a molar ratio of 1:1:50 where the final lipid concentration was 4 mM and then incubated at 4 °C for 30 min. An equal volume of Biobeads SM-2 (Bio-Rad) was added to the nanodisc mixture for detergent removal, incubated at room temperature for 1 h on a rotator and then overnight at 4 °C upon replacing with fresh biobeads. The protein-embedded nanodiscs were isolated using a Superose 6 Increase 10/300 GL column (GE Lifesciences) in buffer containing 10 mM HEPES/KOH pH 7.5, 100 mM NaCl and 0.1 mM Na-Asp and concentrated to 4 mg/mL.
3.5 μL of protein-embedded nanodiscs supplemented with 1.5 mM Fos-Choline-8 (Anatrace) was applied to a glow-discharged holey carbon grid (Quantifoil R1.2/1.3, 200 mesh). Using Vitrobot Mark IV (FEI), grids were blotted for 3 s at 22 °C and 100% humidity, before flash-freezing in liquid ethane.
Data collection and processing.
Grids were imaged with a Talos Arctica transmission electron microscope (Thermo Fisher Scientific) at 200 kV equipped with a Falcon 3EC detector ((Thermo Fisher Scientific)) operating in counting mode. Data collection was performed at 150,000 x magnification, resulting in a physical pixel size of 0.986 Å. Each movie was exposed to a total dose of 40 electrons per Å over 29 frames with a total exposure time of 55 s. A total of 2,419 movie micrographs were collected over two sessions and merged together.
Data processing was performed in RELION 3.170. The frame stacks were aligned and corrected for local beam-induced motion using MotionCorr271 with 5 × 5 patches. Contrast transfer function (CTF) estimation was performed with CTFFIND72 and micrographs with poor statistics were discarded, yielding 1,843 micrographs that were used for further analysis. 1,199 particles were manually picked from multiple micrographs and subsequently subjected to 2D classification to create templates for autopicking. 731,422 particles were automatically picked, extracted and subjected to one round of 2D classification. An initial model was generated using 311,380 particles from the best twelve class averages from the 2D classification. 3D classification was performed using the initial model low pass filtered to 50 Å as reference, resulting in four 3D classes. Of those, 220,938 particles from two classes (that exhibited the indistinguishable trimeric assembly of GltPh and conformation) were combined, re-extracted and subjected to 3D refinement, post-process and Bayesian polishing. The resulting map of 3.9 Å resolution showed all protomers in a conformation similar to the iOFS (PDB: 3V8G). Symmetry expansion73 was performed on 662,814 protomer particles to probe for structural heterogeneity between the trimer protomers. The particles were then subjected to one round of 3D classification without image shifts using a regularization parameter of 40, yielding ten 3D classes. Of the ten 3D classes, nine classes closely resembled the iOFS conformation and one class consisting of 14.8% of the symmetry expanded particles was in the ClCS conformation. The best iOFS class and ClCS were selected for 3D refinement either without (unfocused refinement) or with (focused refinement) a local mask containing a single protomer excluding densities attributed to the nanodisc (Extended Data Fig. 2). The final resolution for the iOFS and the ClCS focused maps was 3.7 Å and 4.0 Å, respectively, using the gold-standard procedure74.
Model building.
One subunit of GltPh in the iOFS (PDB: 3V8G, Chain C) and the IFS (PDB: 3KBC) were docked into the density map of the iOFS and the ClCS, respectively, using PHENIX75. For the trimer, three subunits of the iOFS were docked into the map. All models were refined by several rounds of real-space refinement in PHENIX76, and model building in coot62 and ISOLDE77.
Computational Methods.
Molecular dynamics (MD) simulations of the membrane-bound ClCS state of GltPh.
The simulation system was designed based on the experimental model of the ClCS state of GltPh. Protonation states of titratable residues were assigned using PROPKA78. The Membrane Builder module of CHARMM-GUI79 was used to embed the protein in a pre-equilibrated patch of membrane containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-Oleoyl-sn-glycero-3-phosphoglycerol (POPG), and 1-palmitoyl-2-oleoyl-sn-glycero- 3- phosphocholine (POPC) lipids at a 2:1:1 ratio. The protein-membrane system was solvated and neutralized with 150 mM NaCl. The final constructed system contained ~195,000 atoms with dimensions of 150 × 150 × 114 Å3 prior to any simulations.
All MD simulations were performed using NAMD280 with the CHARMM36 protein and lipid force fields81 and TIP3P water. The simulations were performed at a temperature of 310 K maintained using Langevin thermostat with a damping coefficient of γ=1.0 ps−1, and the Nosé-Hoover Langevin piston method82 was used to maintain a constant pressure of 1 bar. Long-range electrostatic forces were calculated using the particle mesh Ewald (PME) method83, and a 2 fs timestep was used for integration. Non-bonded interactions were calculated with a cutoff of 12 Å and a switching distance of 10 Å. The system was initially energy minimized and equilibrated for 5 ns with protein backbone atoms harmonically restrained to their initial positions with a force constant of k = 1 kcal/mol/Å2. These restraints were released during the following production run. All simulations were performed in the fully-bound state of the transporter, i.e., in the presence of the substrate and three Na+ ions16. The unresolved Na+ ions were modeled based on previous studies16, 84. The initial phase of the production run was performed for 200 ns in the absence of a membrane potential during which a near-complete hydration of the interface between the transport and scaffold domains was observed. Additional 100 ns of simulation was then performed in the presence of a uniform electric field (corresponding to a transmembrane potential of 800 mV) in the negative z direction (extracellular to intracellular; equivalent to positive at the extracellular side) to probe more efficiently the hydration pattern of the putative pore in the ClCS state. To prevent the disruption of the lipid bilayer under the applied external electric field, weak harmonic restraints were applied to the phosphorous atoms of the lipid molecules. Similarly, in order to maintain the bound state of Na2, which is known to have a low binding affinity25, a weak harmonic restraint was applied between Na2 and its binding pocket.
Free-energy calculations of chloride permeation through ClCS conformation of GltPh.
In order to probe the chloride (Cl−) conduction mechanism and to calculate the free-energy profile associated with the process, we performed umbrella sampling (US) simulations85. The initial positions of the permeant ion were seeded using the water molecules hydrating the putative pore in the CLCS state from the 300-ns simulation described above. For each umbrella, the position of the oxygen atom of a water was used to replace the water molecule with a Cl− ion, which was then restrained to that point using a harmonic potential (see below) to probe the energy landscape of ion movement in the respective portion of the reaction coordinate. US calculations were performed with distance (Z-component) between the Cl− ion and the membrane midplane as the reaction coordinate. In total, 60 windows were generated at a spacing of 1 Å, each simulated for 20 ns with accumulated sampling of 1.2 μs. US simulations were performed in the fully-bound state of GltPh, during which a weak harmonic restraint was applied to keep Na2 in its binding pocket (as described above). The potential of mean force (PMF) profile was then calculated using all the resulting umbrella trajectories with the WHAM implementation86.
Harvesting and preparing oocytes.
Female Xenopus laevis frogs were obtained from NASCO (Wisconsin, USA) and harvested as described previously87. Briefly, stage V oocytes were harvested following anaesthesia with 6.5 mM tricaine in 7.14 mM sodium bicarbonate, pH 7.5 and stored in OR-2 buffer (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2 5 mM hemosodium HEPES, pH 7.5). All surgical procedures have been approved by the University of Sydney Animal Ethics under the Australian Code of Practise for the Care and Use of Animals for Scientific Purposes. Oocytes were defolliculated by agitation with 2 mg/mL collagenase for 1 h. Following digestion, oocytes were injected with 4.6 ng of cRNA and stored with shaking at 16–18°C in standard frog Ringer’s solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM hemisodium HEPES, pH 7.5) supplemented with 50 μg/mL gentamycin, 50 μg/mL tetracycline, 2.5 mM sodium pyruvate and 0.5 mM theophylline.
Radiolabeled uptake in oocytes.
Uptake of L-[3H]glutamate (PerkinElmer Life Sciences) was measured in oocytes expressing E1, double cysteine derivatives and in uninjected oocytes (nudes). Oocytes were incubated in 10 μM L-[3H]glutamate for 10 min and uptake was terminated by washing oocytes three times in ice-cold frog Ringer’s solution. Oocytes were then lysed in 1 M NaOH and 1% SDS prior to adding scintillant (Optiphase HisSafe, PerkinElmer). Uptake of L-[3H]glutamate was measured using MicroBeta TriLux scintillation counter (PerkinElmer). For cross-linking studies, oocytes were treated with either 1 mM dithiothreitol (DTT) for 1 min or 400 μM copper phenanthroline for 5 min and washed three times in Ringer’s solution at room temperature prior to incubation with 10 μM L-[3H]glutamate.
Electrophysiology.
2–4 days after injections, currents were recorded using the two-electrode voltage clamp technique with a Geneclamp 500 amplifier (Axon Instruments, Foster City, CA, USA) interfaced with a PowerLab 2/20 chart recorder (ADInstruments, Sydney, Australia) and a Digidata 1322A (Axon Instruments, CA, USA), which was used in conjunction with Chart software (ADInstuments; Axon instruments). All recordings were made with a bath grounded via a 3 M KCl/ 1% agar bridge linking to a 3 M KCl reservoir containing a Ag/AgCl2 ground electrode to minimize offset potentials. Current-voltage relationships for substrate-elicited conductance were determined by measuring substrate-elicited currents during 245 ms voltage pulses between −100 mV and +60 mV at 10 mV steps. Background currents were eliminated by subtracting currents in the absence of substrate from substrate-elicited currents at corresponding membrane potentials. For cross-linking studies, current elicited by an initial control dose of 100 μM L-glutamate was recorded prior to 2 min incubation with 1 mM DTT. Currents were recorded following 1 mM DTT treatment and following subsequent 5 min incubation with 400 μM CuPh. For hydrophobic gate mutagenesis studies, current elicited was measured in Ringer’s solution containing 30 μM aspartate.
Extended Data
Extended Data Fig. 1 |. Cross-linking experiments on purified GltPh double cysteine transporters.
a, SDS-PAGE gel shift assay shows the extent of cross-linking in detergent-solubilized cysteine-less GltPh (CLGltPh) and the double cysteine GltPh transporters under untreated conditions, upon mPEG5K-maleimide treatment and following incubation with HgCl2 with arrows indicating the positions of differentially cross-linked protomers and mPEG5K-bound proteins. This is representative data from one experiment that was replicated at least two times from two separate protein purifications. For gel source data, see Supplementary Figure 1. b, Crystal structure of GltPh-XL1 (purple) and GltPh-XL3 (pink) superimposed on the OFS (PDB: 2NWX; left, grey) and the IFS (PDB: 3KBC; right, grey), respectively. c, SDS-PAGE analysis of purified GltPh-XL3 in nanodiscs. d, Cryo-EM structure of GltPh-XL2 (green) superimposed on the iOFS (PDB:3V8G, chain C; grey).
Extended Data Fig. 2 |. Cryo-EM data processing protocol and refinement.
a, Data processing flow chart for GltPh reconstituted into nanodiscs in the presence of NaCl and aspartate. b, Fourier shell correlation (FSC) curves indicating the resolution at the 0.143 threshold of final masked (blue) and unmasked (red) maps for GltPh trimer iOFS (left), GltPh protomer ClCS (middle) and GltPh protomer iOFS (right). c, Final maps after Relion post-processing, colored according to local resolution estimation using Relion for GltPh trimer iOFS (left, 3.9Å resolution, contour level 7.2σ), GltPh protomer ClCS (middle, 4.0Å resolution, contour level 12.0σ) and GltPh protomer iOFS (right, 3.7Å resolution, contour level 9.5σ). Contour levels calculated using Chimera.
Extended Data Fig. 3 |. The conformational space of a GltPh protomer.
a, The front and b, top views of the cryo-EM map and atomic model of GltPh-XL2 in the iOFS (contour level 9.5σ) and ClCS (contour level 12.0σ). Density attributed to the scaffold domain, transport domain and HP2 are shown in salmon, blue and red, respectively. c, Conformational changes undertaken by a GltPh protomer during the substrate transport cycle viewed from the side and top. HP2 is colored for easier visualization of rotational changes observed in the transport domain. d, e, Close-up views of the L152C-G351C cross-link fitted in the iOFS (contour level 12.9σ) and ClCS (contour level 10.3σ) cryo-EM maps. Contour level calculated using Chimera.
Extended Data Fig. 4 |. Na+ coordination sites in the ClCS.
A close-up of the three Na+ coordination sites on the ClCS protomer are shown with residues interacting with Na+ ion (purple circle, modelled) shown in sticks. The scaffold and the transport domains are shown in salmon and blue, respectively, with the substrate in black sticks.
Extended Data Fig. 5 |. Nanodisc deformation supports transport domain movement by the ClCS and putative lipid binding sites.
a, Percentage of GltPh-XL2 trimers containing all three protomers in iOFS, in ClCS, or a mixture of both. Out of 220,938 trimers, 79,809 contained one or more protomers in the ClCS. Particle count within symmetry expanded data showed 63,470 trimers contained one ClCS, 14,394 trimers contained two ClCS and 1,945 trimers contained all protomers in the ClCS. b, Density map of GltPh-XL2 trimer (unfocused refinement) containing one ClCS protomer and embedded in nanodiscs (viewed from the membrane plane). The two iOFS protomers are shown in orange and the ClCS in blue. The nanodisc is shown in yellow. c, Putative lipid binding sites in the GltPh-XL2 trimer, where the transport domain is in blue and the scaffold domain in salmon. Identical lipid densities (green) were observed between protomers (contour level 7.2σ). TMs located within 5 Å of the putative lipid densities are labelled. Contour level calculated using Chimera.
Extended Data Fig. 6 |. Water conduction through GltPh-ClCS and setup of umbrella sampling simulations and convergence to capture Cl− movement through GltPh-ClCS.
a, The GltPh-ClCS structure was embedded into a lipid bilayer containing PE, PG, and PC lipids (mimicking experimental conditions). After an initial equilibration of 100 ns, the entire system was subjected to an external electric field of 800mV which resulted in a continuous water pathway through the interface of scaffold and transport domain. The GltPh-ClCS protomer is shown in cartoon, with the transport domain in blue and the scaffold domain in salmon. XL-2 residues L152/G351 are shown in red and blue spheres, respectively b, Residues lining the Cl− pathway have a higher solvent accessible surface area (SASA) in the GltPh-ClCS than in the OFS (calculated using the crystal structure of OFS PDB: 2NWX). c, Overlap between the corresponding windows used in US simulations. d, No significant changes between the free-energy profile obtained at 10ns (red), 15ns (blue), and 20 ns (green) were observed, highlighting convergence of US simulations.
Extended Data Fig. 7 |. The EAAT1 open-channel conformation conducts Cl−.
a, L-[3H]glutamate uptake into oocytes expressing cysteine-less EAAT1 and double cysteine transporter mutants in control conditions (grey), and following pre-incubation with DTT (cyan) or CuPh (orange). Number of cells (n) used for each condition is indicated in each graph and all measurements presented were taken across at least two batches of oocytes. b-e, L-glutamate elicited current-voltage relationships for cysteine-less E1 (b), E1-XL1 (c), E1-XL2 (d) and E1-XL3 (e) monitored in the same conditions as (a). To confirm cross-links E1-XL1, E1-XL2 and E1-XL3 were occurring within an individual protomer rather than between protomers of the trimeric complex, oocytes expressing single cysteine residues that make up E1-XL1 (K300C and W473C), E1-XL2 (L244C and G439C), and E1-XL3 (K300C and A470C) either alone or co-injected into an individual oocyte were also examined using the same approaches (f-g). Data represent standard error of the mean (mean ± SEM). h, hEAAT1 (PBD: 5LLU) highlighting residues forming the extracellular and intracellular hydrophobic gates. The scaffold domain is shown in grey and the transport domain in gold. The Cα atoms of the two introduced cysteine residues are shown as spheres (L244 in red and G439 in blue). i, Membrane reversal potentials (Erev) measured in oocytes expressing wild-type (n = 6) and mutant (n = 5) EAAT1 transporters. Each white circle represents a response from a single cell. Black bar represents mean ± SEM. Significance was determined using One-Way ANOVA with Bonferroni post-hoc test for multiple comparisons performed by GraphPad Prism 8, and exact P values are provided. j, Schematic representation of the substrate transport cycle. A single protomer is shown with the scaffold domain in salmon, transport domain in blue and substrate in black.
Extended Data Table 1 |.
Data collection and refinement statistics
| GltPh-XL1 (PDB 6X01) | GltPh-XL3 (PDB 6WZB) | |
|---|---|---|
| Data collection | ||
| Space group | P61 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 113.99, 113.99, 321.70 | 112.11, 204.57, 207.37 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 48.79 – 3.65 (3.90–3.65) | 49.31 – 3.45 (3.64–3.45) |
| Rsym or Rmerge | 0.15 (3.55) | 0.15 (1.52) |
| I / σI | 12.40 (1.20) | 10.30 (2.20) |
| CC1/2 (%) | 99.70 (85.10) | 99.7 (78.2) |
| Completeness (%) | 100 (100) | 100 (100) |
| Redundancy | 21.60 (20.50) | 13.80 (14.30) |
| Refinement | ||
| Resolution (Å) | 44.84 – 3.65 | 49.31– 3.45 |
| No. reflections | 26118 (2591) | 31734 (3120) |
| Rwork / Rfree | 27.23 (36.52) | 23.46 (30.58) |
| 30.86 (44.79) | 26.70 (34.37) | |
| No. atoms | 8862 | 9144 |
| Protein | 8856 | 9135 |
| Ligand/ion | 9 | 12 |
| Water | N/A | N/A |
| B-factors | 176.77 | 111.47 |
| Protein | 176.79 | 111.45 |
| Ligand/ion | 140.35 | 130.31 |
| Water | N/A | N/A |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.003 |
| Bond angles (°) | 0.60 | 0.62 |
| Ramachandran (%) | ||
| Favored | 95.96 | 96.66 |
| Allowed | 4.04 | 3.34 |
| Disallowed | 0.00 | 0.00 |
One crystal per structure was used for analysis and values in parentheses are for the highest-resolution shell.
Extended Data Table 2 |.
Cryo-EM data collection, refinement and validation statistics
| GltPh-XL2 Trimer (EMDB-21968) (PDB 6WYL) | GltPh-XL2 protomer iOFS (EMDB-21966) (PDB 6WYJ) | GltPh-XL2 protomer ClCS (EMDB-21967) (PDB 6WYK) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 150,000 | 150,000 | 150,000 |
| Voltage (kV) | 200 | 200 | 200 |
| Electron exposure (e−/Å2) | 40 | 40 | 40 |
| Defocus range (μm) | 0.5–3.5 | 05–3.5 | 0.5–3.5 |
| Pixel size (Å) | 0.986 | 0.986 | 0.986 |
| Symmetry imposed | C3 | C1 | C1 |
| Initial particle images (no.) | 713,422 | 662,814* | 662,814* |
| Final particle images (no.) | 220,938 | 105,135* | 98,093* |
| Map resolution (Å) | 3.9 | 3.7 | 4.0 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Refinement | |||
| Initial model used (PDB code) | 3V8G (Chain C) | 3V8G (Chain C) | 3KBC |
| Model resolution (Å)† | 3.8/3.8 | 3.5/3.5 | 3.8/3.9 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map sharpening B factor (Å2) | −226 | −199 | −189 |
| Model composition | |||
| Non-hydrogen atoms | 8847 | 2949 | 3083 |
| Protein residues | 1194 | 398 | 415 |
| Ligands | 3 | 1 | 1 |
| B factors (Å2) | |||
| Protein | 83.02 | 41.37 | 74.36 |
| Ligand | 81.47 | 40.77 | 73.83 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.012 | 0.010 | 0.007 |
| Bond angles (°) | 1.193 | 0.643 | 0.767 |
| Validation | |||
| MolProbity score | 1.26 | 1.64 | 1.53 |
| Clashscore | 3.39 | 5.08 | 5.65 |
| Poor rotamers (%) | 0.00 | 0.00 | 0.00 |
| Ramachandran plot | |||
| Favored (%) | 97.29 | 94.66 | 96.60 |
| Allowed (%) | 2.71 | 5.34 | 3.40 |
| Disallowed (%) | 0.00 | 0.0 | 0.00 |
Number of particles after symmetry expansion.
Resolution values are provided for masked/unmasked models.
Extended Data Table 3 |.
Residues that interact with Cl− in GltPh as captured with Umbrella Sampling simulations (and corresponding residues in EAAT1).
| GltPh | EAAT1 | Location | Publication |
|---|---|---|---|
| E9 | R47 | TM1 | N/A |
| V12 | F50 | TM1 | 38 |
| L13 | V51 | TM1 | 38 |
| I16 | T54 | TM1 | 38 |
| D37 | Y75 | TM2 | N/A |
| T41 | K79 | TM2 | N/A |
| F50 | L88 | TM2 | 37, 38 |
| V51 | M89 | TM2 | 37 |
| R52 | R90 | TM2 | 35 |
| L54 | L92 | TM2 | 38 |
| K55 | Q93 | TM2 | 35 |
| I61 | L99 | TM2 | 35 |
| S65 | S103 | TM2 | 20, 35 |
| Y195 | M279 | TM5 | 20, 37 |
| K196 | R280 | TM5 | 37 |
| N199 | A283 | TM5 | N/A |
| M202 | M286 | TM5 | 37, 38 |
| Q203 | W287 | TM5 | N/A |
| A205 | A289 | TM5 | 38 |
| V209 | I293 | TM5 | 37, 38 |
| L212 | L296 | TM5 | 37, 38 |
| E219 | E303 | TM5 | N/A |
| T271 | T358 | HP1 | 38 |
| V274 | G361 | HP1 | 38 |
| T275 | T362 | HP1 | 38 |
| R276 | R477* | HP1 | 30, 38 |
| S277 | S364 | HP1 | N/A |
| S279 | S366 | HP1 | 29 |
| G280 | A367 | HP1 | N/A |
| L282 | L369 | HP1 | 29 |
| P283 | P370 | HP1 | N/A |
| V284 | I371 | HP1 | 38 |
| M286 | F373 | HP1 | 20, 29, 37 |
| R287 | K374 | HP1 | N/A |
| P304 | P392 | TM7 | 29, 37 |
| A307 | A395 | TM7 | N/A |
| T308 | T396 | TM7 | 29 |
| P356 | P444 | HP2 | N/A |
| G357 | Q445 | HP2 | N/A |
| G359 | G447 | HP2 | N/A |
| A360 | L448 | HP2 | N/A |
Hydrophobic gate residues mutated in this study are in bold.
R477 of EAAT1 is found on TM8 and corresponds to R276, found in HP1 of GltPh.
Supplementary Material
Acknowledgements
This work was supported by the Australian National Health and Medical Research Council Project Grant APP1164494 (RR, RV, JF) and Fellowship APP1159347 (AGS), National Institutes of Health grants P41-GM104601 (ET) and R01-GM067887 (ET), computational resources provided by XSEDE (grant MCA06N060 to ET), NCSA Blue Waters (ET), and Microsoft Azure (ET), Research Training Program Scholarship (IC) and Beckman Institute Graduate Fellowship (SP).
We wish to thank and acknowledge the use of the Victor Chang Cardiac Research Institute Innovation Centre, funded by the NSW Government, and the Electron Microscope Unit at UNSW Sydney, funded in part by the NSW Government; and the use of the MX2 beamline at the Australian Synchrotron, part of ANSTO, and the Australian Cancer Research Foundation (ACRF) detector. We acknowledge the facilities and technical assistance of James Bouwer and Simon Brown from Cryo Electron Microscopy - Molecular Horizons, University of Wollongong and William Close from Microscopy Australia at the Australian Centre for Microscopy & Microanalysis, University of Sydney. We would also like to thank Douglas Chappell, Xiaoyu Wang and Olga Boudker for helpful discussions, Zhiyu Zhao for his assistance with simulations, and Cheryl Handford and those that support the Xenopus laevis colony at the University of Sydney.
Footnotes
Competing interest declaration
The authors declare no competing interests.
Data availability statement
All relevant crystallography, cryo-EM and MD data are available from the corresponding author upon request. The maps and the coordinates of the refined models have been deposited into the Protein Data Bank; GltPh-XL1 (PDB 6X01), GltPh-XL3 ( PDB 6WZB), GltPh-XL2 - iOFS (PDB 6WYJ, EMDB-21966), GltPh-XL2 - ClCS (PDB 6WYK, EMDB-21967), GltPh-XL2 trimer – iOFS (PDB 6WYL, EMDB-21968).
References
- 1.Vandenberg RJ, Ryan RM, Mechanisms of glutamate transport. Physiol Rev 93, 1621–1657 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Reyes N, Ginter C, Boudker O, Transport mechanism of a bacterial homologue of glutamate transporters. Nature 462, 880–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan RM, Vandenberg RJ, Elevating the alternating-access model. Nat Struct Mol Biol 23, 187–189 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Zerangue N, Kavanaugh MP, Flux coupling in a neuronal glutamate transporter. Nature 383, 634–637 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Wadiche JI, Arriza JL, Amara SG, Kavanaugh MP, Kinetics of a human glutamate transporter. Neuron 14, 1019–1027 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Fairman WA, Vandenberg RJ, Arriza JL, Kavanaugh MP, Amara SG, An excitatory amino-acid transporter with properties of a ligand-gated chloride channel. Nature 375, 599–603 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Ryan RM, Mindell JA, The uncoupled chloride conductance of a bacterial glutamate transporter homolog. Nat Struct Mol Biol 14, 365–371 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Wadiche JI, Amara SG, Kavanaugh MP, Ion fluxes associated with excitatory amino acid transport. Neuron 15, 721–728 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Parinejad N, Peco E, Ferreira T, Stacey SM, van Meyel DJ, Disruption of an EAAT-Mediated Chloride Channel in a Drosophila Model of Ataxia. J Neurosci 36, 7640–7647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter N, Kovermann P, Fahlke C, A point mutation associated with episodic ataxia 6 increases glutamate transporter anion currents. Brain 135, 3416–3425 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Canul-Tec JC et al. , Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544, 446–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garaeva AA et al. , Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat Struct Mol Biol 25, 515–521 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Garaeva AA, Guskov A, Slotboom DJ, Paulino C, A one-gate elevator mechanism for the human neutral amino acid transporter ASCT2. Nat Commun 10, 3427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X et al. , Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akyuz N et al. , Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature 518, 68–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E, Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature 445, 387–393 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Oh S, Boudker O, Kinetic mechanism of coupled binding in sodium-aspartate symporter GltPh. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes N, Oh S, Boudker O, Binding thermodynamics of a glutamate transporter homolog. Nat Struct Mol Biol 20, 634–640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scopelliti AJ, Font J, Vandenberg RJ, Boudker O, Ryan RM, Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5. Nat Commun 9, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdon G, Boudker O, Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nat Struct Mol Biol 19, 355–357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verdon G, Oh S, Serio RN, Boudker O, Coupled ion binding and structural transitions along the transport cycle of glutamate transporters. Elife 3, e02283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yernool D, Boudker O, Jin Y, Gouaux E, Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431, 811–818 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Boudker O, Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arkhipova V et al. , Binding and transport of D-aspartate by the glutamate transporter homolog GltTk. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guskov A, Jensen S, Faustino I, Marrink SJ, Slotboom DJ, Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nat Commun 7, 13420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arkhipova V, Guskov A, Slotboom DJ, Structural ensemble of a glutamate transporter homologue in lipid nanodisc environment. Nat Commun 11, 998 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan Y et al. , Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc Natl Acad Sci U S A 114, 1584–1588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erkens GB, Hanelt I, Goudsmits JM, Slotboom DJ, van Oijen AM, Unsynchronised subunit motion in single trimeric sodium-coupled aspartate transporters. Nature 502, 119–123 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Cater RJ, Vandenberg RJ, Ryan RM, The domain interface of the human glutamate transporter EAAT1 mediates chloride permeation. Biophys J 107, 621–629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cater RJ, Vandenberg RJ, Ryan RM, Tuning the ion selectivity of glutamate transporter-associated uncoupled conductances. J Gen Physiol 148, 13–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hotzy J, Schneider N, Kovermann P, Fahlke C, Mutating a conserved proline residue within the trimerization domain modifies Na+ binding to excitatory amino acid transporters and associated conformational changes. J Biol Chem 288, 36492–36501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Vandenberg RJ, Mutations in transmembrane domains 5 and 7 of the human excitatory amino acid transporter 1 affect the substrate-activated anion channel. Biochemistry 46, 9685–9692 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Kovermann P, Machtens JP, Ewers D, Fahlke C, A conserved aspartate determines pore properties of anion channels associated with excitatory amino acid transporter 4 (EAAT4). J Biol Chem 285, 23676–23686 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J et al. , Transient formation of water-conducting states in membrane transporters. Proc Natl Acad Sci U S A 110, 7696–7701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan RM, Mitrovic AD, Vandenberg RJ, The chloride permeation pathway of a glutamate transporter and its proximity to the glutamate translocation pathway. J Biol Chem 279, 20742–20751 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Ryan RM, Compton EL, Mindell JA, Functional characterization of a Na+-dependent aspartate transporter from Pyrococcus horikoshii. J Biol Chem 284, 17540–17548 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng MH, Torres-Salazar D, Gonzalez-Suarez AD, Amara SG, Bahar I, Substrate transport and anion permeation proceed through distinct pathways in glutamate transporters. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machtens JP et al. , Mechanisms of anion conduction by coupled glutamate transporters. Cell 160, 542–553 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS, HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Iwadate Y, Kawamura K, Igarashi K, Mochinaga J, Effective ionic radii of nitrite and thiocyanate estimated in terms of the Boettcher equation and the Lorentz-Lorenz equation. The Journal of Physical Chemistry 86, 5205–5208 (1982). [Google Scholar]
- 41.Wadiche JI, Kavanaugh MP, Macroscopic and microscopic properties of a cloned glutamate transporter/chloride channel. J Neurosci 18, 7650–7661 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seal RP, Amara SG, A reentrant loop domain in the glutamate carrier EAAT1 participates in substrate binding and translocation. Neuron 21, 1487–1498 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Barish ME, A transient calcium-dependent chloride current in the immature Xenopus oocyte. J Physiol 342, 309–325 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandenberg RJ, Huang S, Ryan RM, Slips, leaks and channels in glutamate transporters. Channels (Austin) 2, 51–58 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Veruki ML, Mørkve SH, Hartveit E, Activation of a presynaptic glutamate transporter regulates synaptic transmission through electrical signaling. Nat Neurosci 9, 1388–1396 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Wersinger E et al. , The glutamate transporter EAAT5 works as a presynaptic receptor in mouse rod bipolar cells. J Physiol 577, 221–234 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Untiet V et al. , Glutamate transporter-associated anion channels adjust intracellular chloride concentrations during glial maturation. Glia 65, 388–400 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Shabaneh M, Rosental N, Kanner BI, Disulfide cross-linking of transport and trimerization domains of a neuronal glutamate transporter restricts the role of the substrate to the gating of the anion conductance. J Biol Chem 289, 11175–11182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scopelliti AJ, Heinzelmann G, Kuyucak S, Ryan RM, Vandenberg RJ, Na+ interactions with the neutral amino acid transporter ASCT1. J Biol Chem 289, 17468–17479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zerangue N, Kavanaugh MP, ASCT-1 is a neutral amino acid exchanger with chloride channel activity. J Biol Chem 271, 27991–27994 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Zhou W et al. , Large-scale state-dependent membrane remodeling by a transporter protein. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matin TR, Heath GR, Huysmans GHM, Boudker O, Scheuring S, Millisecond dynamics of an unlabeled amino acid transporter. Nat Commun 11, 5016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walter JD, Sawicka M, Dutzler R, Cryo-EM structures and functional characterization of murine Slc26a9 reveal mechanism of uncoupled chloride transport. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galli A, Blakely RD, DeFelice LJ, Norepinephrine transporters have channel modes of conduction. Proc Natl Acad Sci U S A 93, 8671–8676 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingram SL, Prasad BM, Amara SG, Dopamine transporter-mediated conductances increase excitability of midbrain dopamine neurons. Nat Neurosci 5, 971–978 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Vandenberg RJ, Handford CA, Campbell EM, Ryan RM, Yool AJ, Water and urea permeation pathways of the human excitatory amino acid transporter EAAT1. Biochem J 439, 333–340 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Basilio D, Noack K, Picollo A, Accardi A, Conformational changes required for H(+)/Cl(−) exchange mediated by a CLC transporter. Nat Struct Mol Biol 21, 456–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aragao D et al. , MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J Synchrotron Radiat 25, 885–891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W, Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collaborative Computational Project N, The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 61.McCoy AJ et al. , Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Winn MD et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winn MD, Isupov MN, Murshudov GN, Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57, 122–133 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.The PyMOL Molecular Graphics System, Version 2.2.2 Schrödinger, LLC. [Google Scholar]
- 67.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Ritchie TK et al. , Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheres SH, Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Scheres SH, Chen S, Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liebschner D et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Croll TI, ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rostkowski M, Olsson MH, Sondergaard CR, Jensen JH, Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct Biol 11, 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Phillips JC et al. , Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Best RB et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martyna GJ, Tobias DJ, Klein ML, Constant pressure molecular dynamics algorithms. Journal of Chemical Physics 101, 4177 (1994). [Google Scholar]
- 83.Essmann U et al. , A smooth particle mesh Ewald method. Journal of Chemical Physics 103, 8577 (1995). [Google Scholar]
- 84.Huang Z, Tajkhorshid E, Identification of the third Na+ site and the sequence of extracellular binding events in the glutamate transporter. Biophys J 99, 1416–1425 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kästner J, Umbrella sampling. WIREs Computational Molecular Science 1, 932–942 (2011). [Google Scholar]
- 86.Grossfield A, WHAM: the weighted histogram analysis method. http://membrane.urmc.rochester.edu/wordpress/?page_id=126.
- 87.Poulsen MV, Vandenberg RJ, Niflumic acid modulates uncoupled substrate-gated conductances in the human glutamate transporter EAAT4. J Physiol 534, 159–167 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant crystallography, cryo-EM and MD data are available from the corresponding author upon request. The maps and the coordinates of the refined models have been deposited into the Protein Data Bank; GltPh-XL1 (PDB 6X01), GltPh-XL3 ( PDB 6WZB), GltPh-XL2 - iOFS (PDB 6WYJ, EMDB-21966), GltPh-XL2 - ClCS (PDB 6WYK, EMDB-21967), GltPh-XL2 trimer – iOFS (PDB 6WYL, EMDB-21968).