Abstract
The Wnt signaling pathway plays key roles in various developmental processes. Wnt5a, which activates the non-canonical pathway, has been shown to be particularly important for axon guidance and outgrowth as well as dendrite morphogenesis. However, the mechanism underlying the regulation of Wnt5a remains unclear. Here, through conditional disruption of Foxg1 in hippocampal progenitors and postmitotic neurons achieved by crossing Foxg1fl/fl with Emx1-Cre and Nex-Cre, respectively, we found that Wnt5a rather than Wnt3a/Wnt2b was markedly upregulated. Overexpression of Foxg1 had the opposite effects along with decreased dendritic complexity and reduced mossy fibers in the hippocampus. We further demonstrated that FOXG1 directly repressed Wnt5a by binding to its promoter and one enhancer site. These results expand our knowledge of the interaction between Foxg1 and Wnt signaling and help elucidate the mechanisms underlying hippocampal development.
Keywords: Wnt5a, Foxg1, hippocampus, granule cell, pyramidal neuron, dendritic arborization, mossy fiber, Wnt
Introduction
Members of the Wnt family are known to be crucial in numerous developmental processes, including telencephalic patterning, cell fate determination, axon guidance, and dendritic arborization, by triggering different signaling pathways [1–7]. In the developing telencephalon, Wnt members usually exhibit unique expression patterns and are involved in distinct developmental events. Disruption of Wnt3a, which is specifically expressed in the cortical hem, a signal center that plays key roles in the patterning of the telencephalon, results in the loss of the whole hippocampus, whereas forced Wnt3a expression facilitates neurogenesis, learning, and memory in the hippocampus [8–11]. In hippocampal neurons, Wnt3a is also involved in determining axonal position and outgrowth by controlling the remodeling of microtubules [12]. Wnt2b is involved in cell fate determination through activating the canonical Wnt signaling pathway [13, 14]. Wnt2b also controls the differentiation of neural progenitor cells [15, 16]. Wnt5a usually activates the non-canonical Wnt pathway and is highly conserved from Caenorhabditis elegans to humans [17–19]. Wnt5a is expressed in the medial pallium of the telencephalon at early developmental stages and is postnatally expressed in postmitotic neurons in the developing hippocampus [20, 21]. Previous studies have shown that Wnt5a is required for axon guidance and outgrowth [22–26]. Moreover, recently, Wnt5a was reported to be essential for dendritic branching in hippocampal neurons [21, 27]. Despite these critical roles, the upstream regulators of Wnt5a during telencephalic development remain elusive.
Foxg1, one of the Forkhead box transcription factors, has been reported to mainly act as a transcriptional repressor [28–30]. Foxg1-null mutants exhibit significant expansion of the cortical hem, where multiple Wnts are enriched [31]. The expression of Wnt3a, 2b and 5a spreads to the neocortex. FOXG1 has also been reported to direct optic development through transcriptional suppression of Wnt8b [32]. Previous studies have shown that during early facial and forebrain development, the canonical Wnt/ß-catenin signaling pathway transcriptionally targets Fgf8 signaling, which subsequently triggers Foxg1 expression [33, 34]. Moreover, Foxg1 is strongly reduced in the truncated medial wall neuroepithelium in Wnt3a mutants [9]. To further elucidate the mutual interaction between Foxg1 and Wnts and considering the role of Wnt5a in the development of dendrites, we conditionally disrupted Foxg1 in the medial pallium and postmitotic neurons of the hippocampus and found that Wnt5a was significantly upregulated accompanied by impairments in dendritic complexity and mossy fibers. Overexpression of Foxg1 had the opposite effects. Moreover, combined with chromatin immunoprecipitation (ChIP) analysis, these results indicated that FOXG1 directly suppresses Wnt5a. These results expand our knowledge of the interaction between Foxg1 and Wnt signaling and the mechanisms underlying the development of the hippocampus.
Materials and Methods Animals
The Foxg1fl/fl and the CAG-loxp-stop-loxp-Foxg1-IRES-EGFP mouse lines were generated as previously reported [35, 36]. Emx1-Cre mice (stock number: 005628) were purchased from the Jackson Laboratory. NEX-Cre mice were generated as previously described [37]. Conditional disruption of Foxg1 was achieved by crossing Foxg1fl/fl mice with Emx1-Cre or NEX-Cre mice. For overexpression of Foxg1 in the progenitors and postmitotic neurons, the CAG-loxp-stop-loxp-Foxg1-IRES-EGFP line was crossed with the Emx1-Cre and NEX-Cre lines, respectively. The day of vaginal plug detection was considered to be embryonic day 0.5 (E0.5), and the day of birth was designated postnatal day P0. All mice were bred in the animal facility at Southeast University. Gender was not considered in this study. All experiments were performed according to the approved guidelines of Southeast University.
Immunostaining and In Situ Hybridization
Brains were fixed by transcardial perfusion with cold 4% paraformaldehyde (PFA) in PBS, postfixed at 4°C overnight, cryoprotected in 30% sucrose, embedded in OCT, and stored at –70°C until use. The brains were cryosectioned at 16 μm on a Leica CM 3050S cryostat. Immunostaining was then performed as previously described [35]. The following antibodies were used: guinea pig anti-Calbindin (Millipore, AB1778, 1:250), rabbit anti-Calretinin (Millipore, AB5054, 1:500), rat anti-Ctip2 (Abcam, ab18465, 1:2000), rabbit anti-FOXG1 (Abcam, AB18259, 1:500), rabbit anti-Prox1 (Millipore, AB5475, 1:1000), and mouse anti-Synaptoporin (SCBT, sc376761). The secondary antibodies were: Alexa Fluor 568 goat anti-guinea-pig IgG (Molecular Probes, A11075, 1:500), Alexa Fluor 555 donkey anti-mouse IgG (Invitrogen, A31570), Alexa Fluor 546 donkey anti-rabbit IgG (Life, A10040, 1:500), Alexa Fluor 647 donkey anti-rabbit IgG (Life, A31573, 1:500), and Alexa Fluor 546 goat anti-rat IgG (Molecular Probes, A11081, 1:500). DAPI was from Sigma-Aldrich (D9564).
In situ hybridization was performed as previously described [38]. The probes were obtained by PCR amplification.
Real-Time PCR
Total RNA was isolated from the E14.5 dorsal telencephalon using the RNeasy Plus Mini Kit (Qiagen, 74104) according to the manufacturer’s instructions, and each sample was reverse-transcribed into cDNA using the PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa, RR047A). Tissue from at least three brains of each genotype were used. Quantitative real-time PCR was performed as previously described [35]. The following primers were used for the analyses: Wnt3a-F: 5′-GCAGCTGTGAAGTGAAGAC-3′ and Wnt3a-R: 5′-GGTGTTTCTCTACCACCATCTC-3′; Wnt2b-F: 5′-GCCAAAGAGAAGAGGCTTAA-3′ and Wnt2b-R: 5′-TCAGTCCGGGTGGCGTGGCG-3′; Wnt5a-F: 5′-CTCGGGTGGCGACTTCCTCTCCG-3′ and Wnt5a-R: 5′-CTATAACAACCTGGGCGAAGGAG-3′. Each sample was analyzed in triplicate reactions. The relative gene expression of the samples was normalized to the most reliable endogenous gene (glyceraldehyde 3-phosphate dehydrogenase, GAPDH).
Western Blot
The dentate gyrus (DG) was collected at P0 after hypothermic anesthesia and prepared as described previously [35]. The primary antibodies used were rabbit anti-FOXG1 (Abcam, AB18259, 1:3000) and rabbit anti-β-actin (Cell Signaling Technology, 1:5000). HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7074S, 1:3000) was used as the secondary antibody.
Microscopy and Image Analysis
Sections were viewed under a confocal microscope (Olympus FV1000), and images were collected and analyzed using FV10-ASW image analysis software. The images were optimized for size, color, and contrast using Adobe Illustrator.
Golgi Staining and Morphometric Analysis
P17 brains were stained using an FD Rapid GolgiStain kit according to the manufacturer’s instructions and as previously described [39, 40]. Briefly, brains were impregnated with Golgi solution (a mixture of solutions A and B) at room temperature in the dark for 6 days, transferred to solution C for 3 days, and sectioned at 120 μm on a vibrating microtome (VT1000; Leica Microsystems). Five brains per genotype from different parents (n > 3) were impregnated. Images of the dendritic arbors of pyramidal neurons and granule cells were captured under Z-stack mode using an EVOS FL Auto microscope (Life Technology) with a 40× objective lens. The dendrite of each neuron was manually traced using ImageJ software (NIH). Sholl analysis in ImageJ was used to assess the dendritic complexity by examining the number of dendritic intersections per 20-μm concentric radial distance from the soma. Significant differences in dendritic complexity were determined by two-way ANOVA (genotype and circle radius as factors) in GraphPad Prism software. Values were considered significant at P <0.05.
ChIP-Seq Analyses and qPCR
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed using E16.5 wild-type telencephalons with an anti-FOXG1 antibody as described previously [29]. The sequencing reads were mapped to the mouse reference genome (mm9 on the UCSC genome browser). The gene sequences for Wnt5a (NC_000080.6) were obtained using NCBI. The qPCR primers spanned three FOXG1 putative binding sites: the promoter (a) and two enhancer regions (b, c): Wnt5a-a-F: 5′-AGAGAGGAGGAGCTGACAAT-3′ and Wnt5a-a-R: 5′-AATCCCTGTGCCCAAGATG-3′; Wnt5a-b-F: 5′-GCGCAGTCAATCAACAGTAAAC-3′ and Wnt5a-b-R 5′-GGCTTCTGGAGAGGAAAGAAAG-3′; Wnt5a-c-F: 5′-CTGGCTGGCTCTGGTTT-3′ and Wnt5a-c-R: 5′-TTTCTACGCCAGAGTTTAAGAATTG-3′. The fold enrichment from three independent experiments were compared using GraphPad software to determine the statistical significance. Values are expressed as the mean ± SEM.
Luciferase assay
The mouse Wnt5a-luciferase reporter (pmrt) contains nucleotides from − 1273 to + 621 relative to the transcription start site of the mouse Wnt5a gene (NC_000080.6). The Wnt5a-enhancer 1 (E1) and 2 (E2) regions contain nucleotides from + 5241 bp to + 6441 bp and + 194.2 kb to + 195.2 kb of the transcription start site, respectively. The reporter vectors were constructed by cloning pmrt or the E1 and E2 fragments into the pGL3-basic or pGL3-promoter reporter plasmids. The primer sequences were as follows: Wnt5a-prmt-F: 5′- GCGTGCTAGCCCGGGCTCGAGGGCGCAAGGAGACATCCC-3′ and Wnt5a-prmt-R: 5′- ACTTAGATCGCAGATCTCGAGGGCACGGAGAGGAAGTCGC-3′; Wnt5a-E1-F: 5′- AATCGATAAGGATCCGTCGACCTGCTGATGCTTGGGAGCTG-3′ and Wnt5a-E1-R: 5′- CTCTCAAGGGCATCGGTCGACGAAAGAAAGGAGCAGATGTTTATTGC-3′; Wnt5a-E2-F: 5′- AATCGATAAGGATCCGTCGACAACATCTCTTTATATAGTAAGGCTCTAGGATG-3′ and Wnt5a-E2-R: 5′-CTCTCAAGGGCATCGGTCGACACAAGTCCAACCAATCAACTAAAATAA-3′. A luciferase assay was performed using Neuro2A cells. For transfections, 1 × 105 cells per well were seeded onto 24-well plates and incubated with DMEM/F12 complex medium overnight. The second day, pGL3-basic-prmt, pGL3-promoter-E1, and pGL3-promoter-E2 were added with either pCAG EN (control) or pCAG-Foxg1 (500 ng/well for each construct) together with the SV40 plasmid containing the Renilla luciferase gene (50 ng/well) as an internal plasmid control. The transfection medium was replaced by complete medium containing antibiotics after 6 h of transfection. The luciferase activities were assayed 48 h after transfection. The firefly luciferase readout was normalized to the respective Renilla luciferase readout from the same cell lysate. The fold change in response to FOXG1 activity was calculated with respect to the controls. All the values are expressed as the mean ± SEM of three biological replicates.
Results
Abnormal Expression of Wnt5a After Deletion of Foxg1 in the Hippocampal Primordium
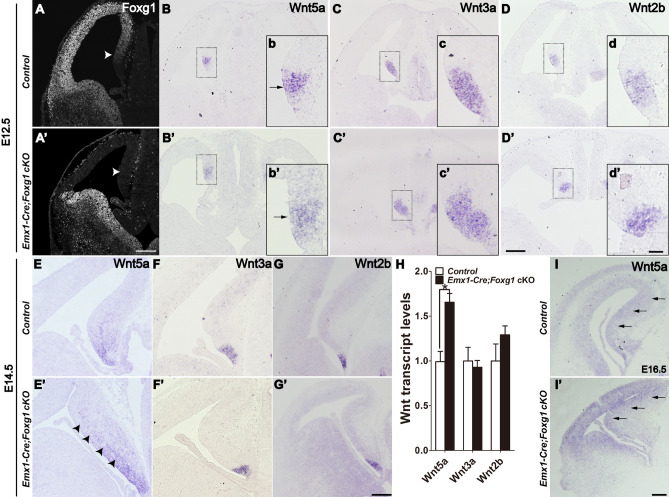
To gain an in-depth understanding of the regulation of Wnt5a by Foxg1 in the developing telencephalon, we first conditionally inactivated Foxg1 in the dorsal telencephalic progenitors from E10.5 onwards by crossing Emx1-Cre mice with Foxg1fl/fl mice (referred to as Emx1-Cre;Foxg1 cKO). The deletion efficiency of Foxg1 was assessed by immunostaining for FOXG1. Compared to that of the control, FOXG1 was completely undetectable in the dorsal telencephalon, including the cortical hem, where multiple Wnts are enriched (Fig. 1A, A’). The Foxg1-/- mice in which Foxg1 was deleted as early as E8.5 exhibited severe expansion of the cortical hem with the expression of Wnt5a and Wnt3a/Wnt2b, two specific markers for the cortical hem, which spread to the whole cortex [31]. We then examined the expression of the three Wnt members at E12.5 in Emx1-Cre; Foxg1 cKO mice. As shown in Fig. 1B and 1B’, there was a slight expansion of Wnt5a in the medial pallium compared to that of controls, but no differences in the expression of Wnt3a and Wnt2b (Fig. 1C–D’), consistent with our previous report [41]. At E14.5, Wnt5a was expressed not only in the cortical hem but also in the hippocampal primordium in controls (Fig. 1E). A marked upregulation was detected in the Emx1-Cre; Foxg1 cKO mice (Fig. 1E’). Similarly, there were no changes in Wnt3a and Wnt2b (Fig. 1F–G’). Moreover, qPCR analysis revealed that Wnt3a and Wnt2b were expressed at comparable levels in control and the Emx1-Cre; Foxg1 cKO mice, while Wnt5a was increased by ~50% (Fig. 1H). Upregulation of Wnt5a was also evident at E16.5 in Emx1-Cre;Foxg1 cKO mice (Fig. 1I, I’). These results suggest that FOXG1 is involved in the regulation of Wnt5a rather than Wnt3a/Wnt2b from E10.5 onwards.
Fig. 1.
Upregulation of Wnt5a after deletion of Foxg1 in the hippocampal primordium. A, A’ Immunostaining showing that FOXG1 is efficiently disrupted in the dorsal telencephalon, including the hippocampal primordium (arrowhead). B–D’ In situ analysis of Wnt5a, Wnt3a, and Wnt2b. b–d’ Magnified views of the boxed areas in B–D’. E–G’ Representative in situ images of Wnt5a, Wnt3a, and Wnt2b in the cortical hem at E14.5. Arrows in b, b’ and arrowheads in E, E’ indicate the excessive expression of Wnt5a in the medial pallium. H qPCR analysis of Wnt5a, Wnt3a, and Wnt2b in the telencephalon. I, I’ In situ analysis of Wnt5a at E16.5. Values are expressed as the mean ± SEM, n = 3, *P < 0.05. Scale bars, (A–I’) 200 µm; (b–d’) 50 µm.
Downregulation of Wnt5a After Overexpression of Foxg1 in the Hippocampal Primordium
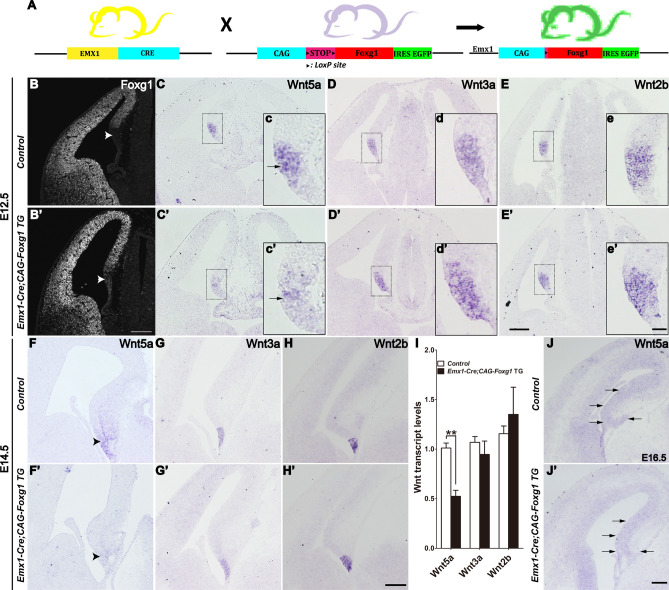
To confirm the regulation of Wnt5a by FOXG1, we overexpressed Foxg1 in the dorsal telencephalon by crossing the Emx1-Cre line with the CAG-loxp-stop-loxp-Foxg1-IRES-EGFP line (referred to as Emx1-Cre; CAG-Foxg1 TG) (Fig. 2A). The FOXG1 expression in the dorsal telencephalon was stronger than that of the controls (Fig. 2B and 2B’). As shown in Fig. 2B’, notable ectopic expression of FOXG1 was also detected in the cortical hem where FOXG1 was normally absent. Interestingly, Wnt5a was clearly downregulated in the cortical hem at E12.5 and almost undetectable at E14.5 (Fig. 2C, C’ and 2F, F’), while Wnt3a and Wnt2b remained unchanged in the Emx1-Cre;CAG-Foxg1 TG mice (Fig. 2D–E’ and 2G–H’). qPCR analysis further demonstrated the downregulation of Wnt5a in the Emx1-Cre;CAG-Foxg1 TG mice with both Wnt3a and Wnt2b unaltered in either case (Fig. 2I). The reduction of Wnt5a was detected as well at E16.5 in the Emx1-Cre; CAG-Foxg1 TG mice (Fig. 2J, J’). Thus, FOXG1 functions upstream of Wnt5a in the developing telencephalon.
Fig. 2.
Downregulation of Wnt5a after Foxg1 overexpression in the hippocampal primordium. A Schematic of the strategy for overexpression of Foxg1. B, B’ FOXG1 expression assessed by immunofluorescence. Arrowheads indicate ectopic expression of FOXG1 in the TG cortical hem (B’), which is normally undetectable (B). C–E’ In situ hybridization of Wnt5a (C, C’), Wnt3a (D, D’), and Wnt2b (E, E’) in E12.5 controls (C–E) and Emx1-cre;Foxg1 TG mice (C’–E’) showing downregulation of Wnt5a (arrows) and comparable levels of Wnt3a and Wnt2b. c–e’ Magnified views of the boxed areas in (C–E’). F–H’ In situ hybridization for Wnt5a (F, F’), Wnt3a (G, G’), and Wnt2b (H, H’) at E14.5. Wnt5a is almost undetectable in the hippocampal primordium (arrowheads). No changes occur in Wnt3a and Wnt2b. I qPCR showing a significant decrease in Wnt5a. J, J’ In situ analysis of Wnt5a at E16.5. Values are expressed as the mean ± SEM, n = 3, **P < 0.01. Scale bars, (B–J’) 200 µm; (c–e’) 50 µm.
Foxg1 Regulates Wnt5a in Postmitotic Neurons
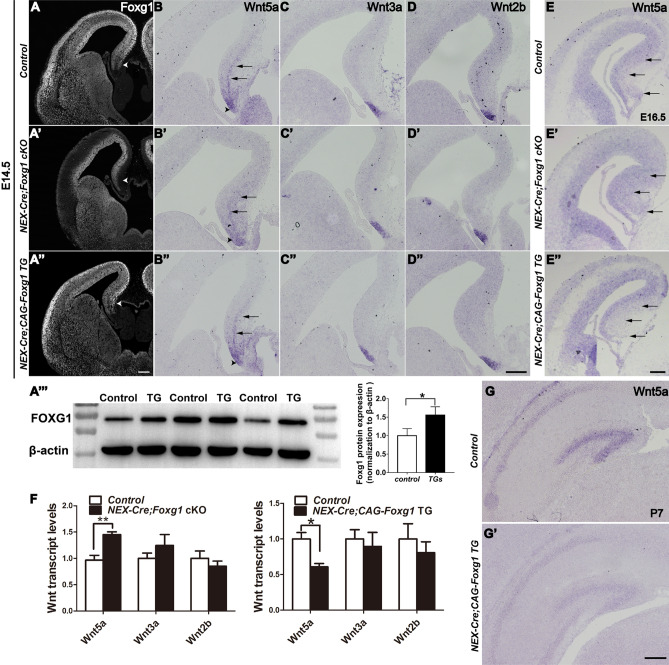
To further determine whether Foxg1 regulates Wnt5a in postmitotic neurons, we conditionally inactivated Foxg1 by crossing NEX-Cre with Foxg1fl/fl mice (referred to as NEX-Cre; Foxg1 cKO). As expected, Foxg1 expression was eliminated in the postmitotic neurons but remained in the ventricular zone and the subventricular zone in the NEX-Cre; Foxg1 cKO mice (Fig. 3A–A’) when examined at E14.5. In the controls, apart from the shrinking cortical hem, Wnt5a was also expressed in the postmitotic hippocampal neurons aligned along the medial pallium (Fig. 3B, E). However, in the NEX-Cre; Foxg1 cKO mice, the expression of Wnt5a was expanded more widely at E14.5 and E16.5 (Fig. 3B’, E’). The expression of Wnt3a and Wnt2b was comparable in control and NEX-Cre; Foxg1 cKO mice (Fig. 3C–D’). Next, Foxg1 was overexpressed in postmitotic neurons by crossing NEX-Cre mice with CAG-loxp-stop-loxp-Foxg1-IRES-EGFP mice (referred to as NEX-Cre;CAG- Foxg1 TG) (Fig. 3A’’). At the protein level, FOXG1 was strongly increased by ~45% at P0 in the NEX-Cre; CAG-Foxg1 TG mice (Fig. 3A’’’). Downregulation of Wnt5a was evident in the postmitotic neurons in the medial pallium at E14.5 and E16.5, while Wnt3a and Wnt2b were unchanged (Fig. 3B’’–E’’). qPCR showed a 1.4-fold increase in Wnt5a in the NEX-Cre;Foxg1 cKO mice and a 0.4-fold decrease in the NEX-Cre;CAG-Foxg1 TG mice, and that the levels of Wnt3a and Wnt2b were unchanged was also confirmed (Fig. 3F). Wnt5a has been shown to be highly expressed in the DG and CA1 region in postnatal stages (Fig. 3G). Due to postnatal lethality, examination of Wnt5a in the NEX-Cre; Foxg1fl/fl cKOs was unachievable. We then assessed Wnt5a in the NEX-Cre;CAG- Foxg1 TG mice at P7 and found that it was significantly reduced (Fig. 3G’). Taken together, these data indicate that Foxg1 regulates Wnt5a in postmitotic hippocampal neurons.
Fig. 3.
Abnormal expression of Wnt5a after deletion or overexpression of Foxg1 in postmitotic neurons. A–A’’’ Immunostaining of FOXG1 in control, NEX-cre;Foxg1 cKO, and NEX-cre;Foxg1 TG brains at E14.5. The arrowhead in A’ indicates the loss of Foxg1 in postmitotic neurons. A’’, A’’’ Immunofluorescence and Western blot analysis of FOXG1. Foxg1 is overexpressed in the NEX-cre;Foxg1 TG brain. B–D’’ In situ hybridization for Wnt5a (B–B’’), Wnt3a (C–C’’), and Wnt2b (D–D’’) in E14.5 control, NEX-cre;Foxg1 cKO, and NEX-cre;Foxg1 TG brains showing Wnt5a expression in the cortical hem (arrowheads) and the postmitotic hippocampal neurons (arrows). E–E’’ In situ hybridization for Wnt5a at E16.5. F qPCR showing significant changes in Wnt5a. G, G’ Photomicrographs of in situ Wnt5a in control and NEX-cre;Foxg1 TG brains at P7. Values are expressed as the mean ± SEM, n ≥ 3, *P < 0.05, **P < 0.01. Scale bars, 200 µm.
Abnormal Development of the Mossy Fibers and Dendrites of Granule Cells in NEX-Cre; CAG-Foxg1 TG Mice
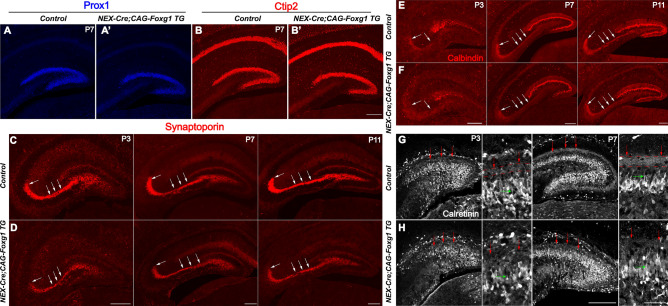
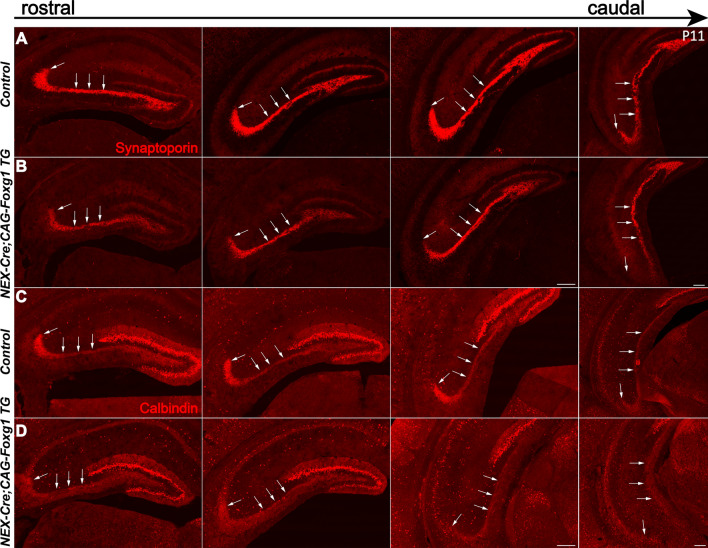
Wnt5a is involved in axon guidance and outgrowth and was recently shown to be required for dendritic branching [25, 26, 42–44]. Here, we examined the development of mossy fibers and the dendrites of granule cells in the DG of NEX-Cre;CAG-Foxg1 TG mice. The morphology of the DG was grossly normal at P7, as viewed by immunostaining with anti-Prox1 and Ctip2, specific markers for granule cells (Fig. 4). Next, immunostaining with anti-Synaptoporin and anti-Calbindin were used to examine the mossy fibers at P3, P7, and P11. In controls, long and thick Synaptoporin+ fibers were observed to project to the CA3 region (Fig. 4C). However, in the NEX-Cre; CAG-Foxg1 TG mice, although mossy fibers properly projected to the CA3 region, they were substantially thinner than those of controls (Fig. 4D). Similar defects were showed by Calbindin immunostaining (Fig. 4E, F). To examine the dendritic development of granule cells, we immunostained with anti-Calretinin at P3 and P7. In the control DGs, Calretinin+ proximal dendrites of granule cells formed a weak band at P3 and a clear band at P7 on the inner side of the molecular layer (Fig. 4G). In the NEX-Cre;CAG-Foxg1 TG mice, the Calretinin+ band was almost undetectable at P3, and much weaker at P7 than that in controls (Fig. 4H), suggesting a developmental defect in the dendrites of granule cells. To further confirm these defects, we immunostained with anti-Synaptoporin and anti-Calbindin in serial sections of P11 brains. As shown in Figure 5, compared to the control DG, from the rostral to the caudal levels the mossy fibers were thinner in the NEX-Cre; CAG-Foxg1 TG mice, further demonstrating that Foxg1 is required for the development of mossy fibers.
Fig. 4.
Impaired development of granule cell dendrites in the NEX-cre;Foxg1 TG brain. A–B’ Representative images of P7 mouse brain sections stained with antibodies against Prox1 and Ctip2 to mark granule cells in the DG. C–F Immunostaining of anti-Synaptoporin (C, D) and Calbindin (E, F) showing the mossy fibers (white arrows) at P3, P7, and P11. G, H Immunostaining of Calretinin labeling developing dendrites of granule cells at P3 and P7. Red arrows indicate the band formed by the proximal dendrites of granule cells in the molecular layer, while green arrows indicate the dendrites of granule cells. DG, dentate gyrus. Scale bars in magnified views, 50 µm; in A–H, 200 µm.
Fig. 5.
Impaired development of mossy fibers in the NEX-cre; Foxg1 TG brain at P11. A–D Immunostaining of anti-Synaptoporin (A, B) and Calbindin (C, D) showing the mossy fibers (white arrows), from the rostral to the caudal levels. Scale bars, 200 µm.
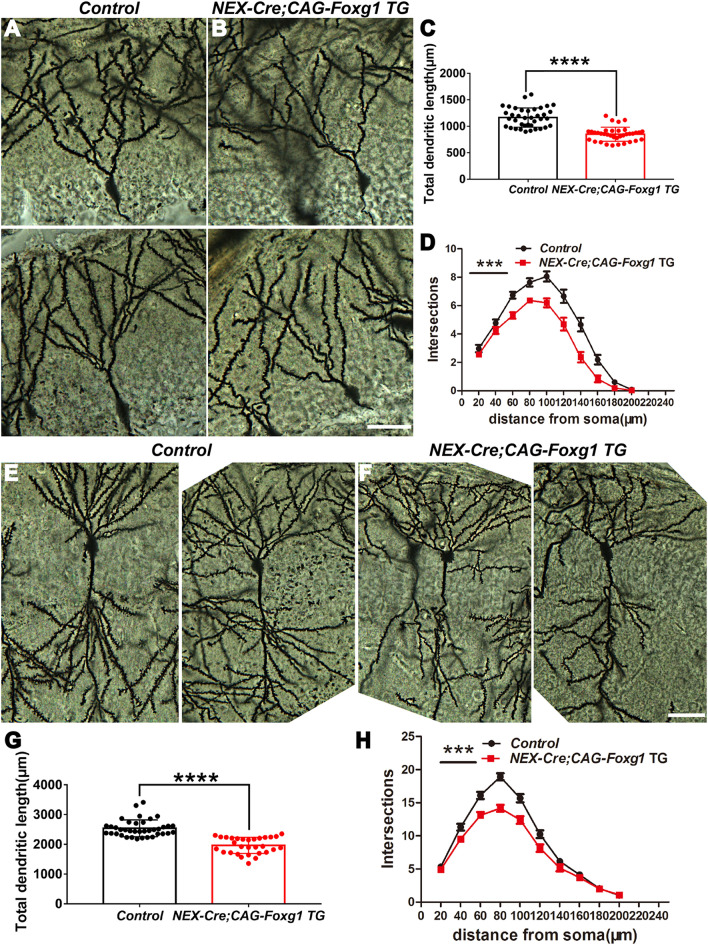
We then used Golgi-Cox staining to further explore the developmental dendritic defects. At P17 in controls, the dendritic tree of granule cells was well developed with numerous and complicated arbors (Fig. 6A). In contrast, we observed a substantial reduction in the dendritic complexity in the NEX-Cre;CAG- Foxg1 TG mice (Fig. 6B), and the total dendritic length was significantly reduced (Fig. 6C). We next quantified the dendritic complexity by counting the intersections where the dendrites crossed the concentric circles drawn at 20-μm intervals around a granule cell body. Statistical analysis showed a strong decrease in the number of intersections compared with the control neurons (Fig. 6D). Similar deficits were observed in CA1 pyramidal neurons (Fig. 6E, F). The total dendritic length was reduced by ~24% (Fig. 6G), and the number of intersections was also significantly decreased in the NEX-Cre; CAG-Foxg1 TG mice (Fig. 6H). Together, these results suggest that the Foxg1-Wnt5a signaling pathway is required for the development of neurites in the DG as well as the CA region.
Fig. 6.
Reduced dendritic complexity in CA1 and DG after Foxg1 overexpression. A, B Representative images showing the morphology of granule cells at P17 in control and TG mice by Golgi staining. C Total dendritic length is significantly reduced in NEX-cre;Foxg1 TG mice (control, n = 38 cells from 5 mice; TG, n = 36 cells from 5 mice, ****P < 0.0001; t-test). D Sholl analysis of the dendritic complexity of granule cells (control, n = 38 cells from 5 mice; TG, n = 36 cells from 5 mice, ***P < 0.0001; two-way ANOVA). E, F Representative images of Golgi-stained CA1 pyramidal neurons from control (E) and TG (F) mice. G Total dendritic length of CA1 pyramidal neurons is also reduced in TG mice (control, n = 38 cells from 5 mice; TG, n = 30 cells from 5 mice, ****P < 0.0001; t-test). H Sholl analysis of the dendritic complexity of hippocampal CA1 pyramidal neurons (control, n = 38 cells from 5 mice; TG, n = 30 cells from 5 mice; ***P < 0.0001; two-way ANOVA). CA, cornu ammonis; scale bars, 20 μm.
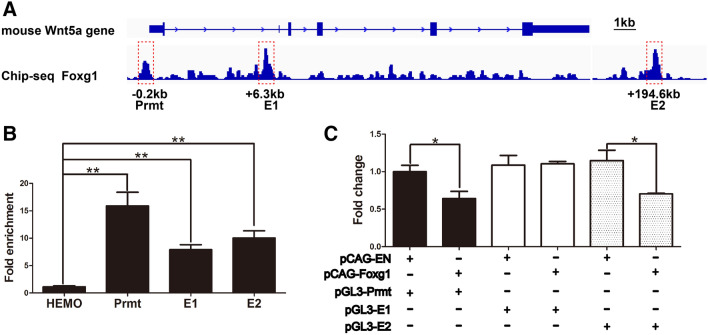
FOXG1 Directly Represses Wnt5a
To investigate whether FOXG1 directly regulates Wnt5a, we analyzed the Wnt5a locus and found three putative FOXG1 consensus binding motifs: one was located at the promotor domain (0.2 kb upstream of the transcription start site, RefSeq accession number: NM_ 009524), and the other two were 6.3 kb and 194.6 kb downstream enhancer sites (Fig. 7A, referred to as Prmt, E1, and E2, respectively). ChIP-qPCR was then carried out, and showed 15.89 ± 2.49-fold, 7.89 ± 0.91-fold, and 10.02 ± 1.32-fold enrichment of FOXG1 on Prmt, E1, and E2, respectively (Fig. 7B). The luciferase assay showed that when PGL3-Prmt was co-transfected with PCAG-Foxg1, the activity was reduced by ~36.0 ± 9.4% (P < 0.05). No significant inhibition was detected when PGL3-E1 was co-transfected with PCAG-Foxg1. The luciferase activity was reduced by ~30.4 ± 6.8% (P < 0.05) when PGL3-E2 was co-transfected (Fig. 7C). Thus, Foxg1 directly binds to the promotor and E2 enhancer domains to repress Wnt5a expression.
Fig. 7.
FOXG1 represses Wnt5a by directly binding its promoter and E2 enhancer sites. A Three FOXG1 consensus binding sites denoted Prmt, E1, and E2 at the Wnt5a gene locus. B ChIP-QPCR exhibits high enrichment of FOXG1 on the three hypothetical loci (n = 3, **P < 0.01). C Luciferase assays showing significant inhibition of Wnt5a by FOXG1 through the promoter and E2 sites (n = 4, *P < 0.05; Student’s t-test).
Discussion
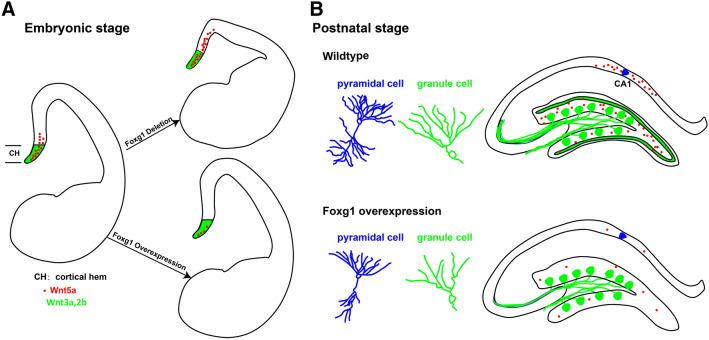
Wnt signaling plays crucial roles in various developmental processes [1, 5, 6, 45–52]. Wnt5a has been demonstrated to be required for axon outgrowth, dendrite morphogenesis, and synaptic plasticity [25, 27, 42, 44, 53, 54]. However, how Wnt5a is regulated remains unclear. In this study, we report that Foxg1 suppressed Wnt5a directly during the development of the hippocampus at both embryonic and postnatal stages (Fig. 8).
Fig. 8.
Schematic representation of the role of Foxg1 in hippocampal development via Wnt5a. A During the embryonic stage (E12.5–E14.5), deletion of Foxg1 leads to expansion of the Wnt5a-expressing region, while Foxg1-overexpression represses Wnt5a expression in the hippocampal primordium. B During postnatal development, forced expression of Foxg1 in postmitotic pyramidal and granule cells leads to downregulation of wnt5a and disturbs the development of dendrites and mossy fibers.
FOXG1 has been shown to interact with Wnt signaling in a complicated way. FOXG1 transcriptionally suppresses Wnt8b during optic development [32]. During early facial and forebrain development, the canonical Wnt/ß-catenin pathway targets Fgf8 signaling, which subsequently triggers Foxg1 expression [33, 34]. Interestingly, in Wnt3a mutants, Foxg1 is severely reduced in the truncated medial neuroepithelium [9]. On the other hand, in Foxg1-null mutants in which Foxg1 is disrupted as early as E8.5, the cortical hem markedly expands into the neocortex with the expression of Wnt3a, Wnt2b and Wnt5a spread throughout the whole cortical field [31]. Here, we showed that deletion of Foxg1 from E10.5 onwards did not cause expansion of the cortical hem, while Wnt3a and Wnt2b exhibited a normal expression pattern. Interestingly, we found that Wnt5a was upregulated. Moreover, Wnt5a expression was significantly increased when FOXG1 was deleted in postmitotic neurons in the medial pallium from which the hippocampus and the DG are derived. Overexpression of Foxg1 reduced Wnt5a but had no effects on Wnt3a/Wnt2b. We further demonstrated that FOXG1 directly suppressed Wnt5a by binding to its promoter and enhancer domains. Our results suggest that FOXG1 regulates Wnt5a but not Wnt3a/Wnt2b in the medial pallium during telencephalic development and expand our knowledge of the interaction between Foxg1 and Wnt signaling.
In mammals, the hippocampus plays a crucial role in learning and memory [55–60]. During mnemonic functions, the hippocampus coordinates the interactions among brain areas including the medial prefrontal cortex, medial entorhinal cortex, and amygdala, to which the CA1 pyramidal neurons project [61–64]. The DG receives information from the entorhinal cortex and serves as the primary afferent pathway into the hippocampus [65–67]. Dysfunction of the neural circuits is closely associated with a variety of neurological disorders [68–71]. Patients suffering from FOXG1 syndrome have severe cognitive defects [72–75]. Duplications of FOXG1 are associated with cognitive defects and mental retardation [76–78]. Studies using mouse models have shown that Foxg1 is involved in hippocampal formation as well as neurite development [35, 40]. Recently, Wnt5a was reported to be required for dendrite branching and growth in hippocampal neurons [21]. Wnt5a also regulates axon guidance and outgrowth [25, 26, 44, 79]. In the present study, loss of Foxg1 led to evident upregulation of Wnt5a in the hippocampal primordium, while overexpression of Foxg1 resulted in dramatic downregulation of Wnt5a in the granule cells and pyramidal neurons, accompanied by reduced dendritic complexity and mossy fibers. Our study showed that FOXG1 acts as a direct repressor of Wnt5a. FOXG1 suppresses Wnt5a not only at early developmental stages but also in postnatal hippocampal development. Our results help to elucidate the mechanisms underlying the interaction between Foxg1 and Wnt signaling during telencephalic development.
Acknowledgements
We thank Yiquan Wei and Li Liu for their assistance in the laboratory and with animal care, and other members of the laboratory for helpful discussions. This work was supported by grants from the National Natural Science Foundation of China (31930045 and 81870899) and the National Key R&D Program of China (2016YFA0501001).
Conflict of interest
The authors have no conflicts of interest to declare.
References
- 1.Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 2.Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396:370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 3.Kiecker C, Niehrs C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development. 2001;128:4189–4201. doi: 10.1242/dev.128.21.4189. [DOI] [PubMed] [Google Scholar]
- 4.Houart C, Caneparo L, Heisenberg C, Barth K, Take-Uchi M, Wilson S. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron. 2002;35:255–265. doi: 10.1016/S0896-6273(02)00751-1. [DOI] [PubMed] [Google Scholar]
- 5.Lyuksyutova AI, Lu CC, Milanesio N, King LA, Guo N, Wang Y, et al. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 2003;302:1984–1988. doi: 10.1126/science.1089610. [DOI] [PubMed] [Google Scholar]
- 6.Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 7.Vallee A, Lecarpentier Y, Guillevin R, Vallee JN. Opposite interplay between the canonical WNT/beta-catenin pathway and PPAR gamma: A potential therapeutic target in gliomas. Neurosci Bull. 2018;34:573–588. doi: 10.1007/s12264-018-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 9.Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. [Google Scholar]
- 10.Shruster A, Offen D. Targeting neurogenesis ameliorates danger assessment in a mouse model of Alzheimer’s disease. Behav Brain Res. 2014;261:193–201. doi: 10.1016/j.bbr.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Xu N, Zhou WJ, Wang Y, Huang SH, Li X, Chen ZY. Hippocampal Wnt3a is necessary and sufficient for contextual fear memory acquisition and consolidation. Cereb Cortex. 2015;25:4062–4075. doi: 10.1093/cercor/bhu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanganello E, Zahavi EE, Burute M, Smits J, Jordens I, Maurice MM, et al. Wnt signaling directs neuronal polarity and axonal growth. iScience. 2019;13:318–327. doi: 10.1016/j.isci.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh M. WNT2B: comparative integromics and clinical applications (Review) Int J Mol Med. 2005;16:1103–1108. [PubMed] [Google Scholar]
- 14.Katoh M, Kirikoshi H, Terasaki H, Shiokawa K. WNT2B2 mRNA, up-regulated in primary gastric cancer, is a positive regulator of the WNT- beta-catenin-TCF signaling pathway. Biochem Biophys Res Commun. 2001;289:1093–1098. doi: 10.1006/bbrc.2001.6076. [DOI] [PubMed] [Google Scholar]
- 15.Kubo F, Takeichi M, Nakagawa S. Wnt2b inhibits differentiation of retinal progenitor cells in the absence of Notch activity by downregulating the expression of proneural genes. Development. 2005;132:2759–2770. doi: 10.1242/dev.01856. [DOI] [PubMed] [Google Scholar]
- 16.Cho SH, Cepko CL. Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development. 2006;133:3167–3177. doi: 10.1242/dev.02474. [DOI] [PubMed] [Google Scholar]
- 17.Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 18.Yoshikawa S, McKinnon RD, Kokel M, Thomas JB. Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature. 2003;422:583–588. doi: 10.1038/nature01522. [DOI] [PubMed] [Google Scholar]
- 19.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 20.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 21.Chen CM, Orefice LL, Chiu SL, LeGates TA, Hattar S, Huganir RL, et al. Wnt5a is essential for hippocampal dendritic maintenance and spatial learning and memory in adult mice. Proc Natl Acad Sci U S A. 2017;114:E619–E628. doi: 10.1073/pnas.1615792114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Zhu J, Yang GY, Wang QJ, Qian L, Chen YM, et al. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat Cell Biol. 2007;9:743–754. doi: 10.1038/ncb1603. [DOI] [PubMed] [Google Scholar]
- 23.Shafer B, Onishi K, Lo C, Colakoglu G, Zou Y. Vangl2 promotes Wnt/planar cell polarity-like signaling by antagonizing Dvl1-mediated feedback inhibition in growth cone guidance. Dev Cell. 2011;20:177–191. doi: 10.1016/j.devcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou Y, Lyuksyutova AI. Morphogens as conserved axon guidance cues. Curr Opin Neurobiol. 2007;17:22–28. doi: 10.1016/j.conb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Hutchins BI, Kalil K. Wnt5a induces simultaneous cortical axon outgrowth and repulsive axon guidance through distinct signaling mechanisms. J Neurosci. 2009;29:5873–5883. doi: 10.1523/JNEUROSCI.0183-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blakely BD, Bye CR, Fernando CV, Horne MK, Macheda ML, Stacker SA, et al. Wnt5a regulates midbrain dopaminergic axon growth and guidance. PLoS One. 2011;6:e18373. doi: 10.1371/journal.pone.0018373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bian WJ, Miao WY, He SJ, Wan ZF, Luo ZG, Yu X. A novel Wnt5a-Frizzled4 signaling pathway mediates activity-independent dendrite morphogenesis via the distal PDZ motif of Frizzled 4. Dev Neurobiol. 2015;75:805–822. doi: 10.1002/dneu.22250. [DOI] [PubMed] [Google Scholar]
- 28.Godbole G, Shetty AS, Roy A, D’Souza L, Chen B, Miyoshi G, et al. Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development 2018, 145. [DOI] [PMC free article] [PubMed]
- 29.Du A, Wu X, Chen H, Bai QR, Han X, Liu B, et al. Foxg1 directly represses Dbx1 to confine the POA and subsequently regulate ventral telencephalic patterning. Cereb Cortex. 2019;29:4968–4981. doi: 10.1093/cercor/bhz037. [DOI] [PubMed] [Google Scholar]
- 30.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 31.Muzio L, Mallamaci A. Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci. 2005;25:4435–4441. doi: 10.1523/JNEUROSCI.4804-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith R, Huang YT, Tian T, Vojtasova D, Mesalles-Naranjo O, Pollard SM, et al. The Transcription Factor Foxg1 Promotes Optic Fissure Closure in the Mouse by Suppressing Wnt8b in the Nasal Optic Stalk. J Neurosci. 2017;37:7975–7993. doi: 10.1523/JNEUROSCI.0286-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimamura K, Rubenstein JL. Inductive interactions direct early regionalization of the mouse forebrain. Development. 1997;124:2709–2718. doi: 10.1242/dev.124.14.2709. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Song L, Zhou CJ. The canonical Wnt/beta-catenin signaling pathway regulates Fgf signaling for early facial development. Dev Biol. 2011;349:250–260. doi: 10.1016/j.ydbio.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Tian C, Gong Y, Yang Y, Shen W, Wang K, Liu J, et al. Foxg1 has an essential role in postnatal development of the dentate gyrus. J Neurosci. 2012;32:2931–2949. doi: 10.1523/JNEUROSCI.5240-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Xiao H, Zhao C. Forced expression of Foxg1 in the cortical hem leads to the transformation of cajal-retzius cells into dentate granule neurons. J Dev Biol 2018, 6. [DOI] [PMC free article] [PubMed]
- 37.Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Gu X, Han X, Du A, Jiang Y, Zhang X, et al. A novel function for Foxm1 in interkinetic nuclear migration in the developing telencephalon and anxiety-related behavior. J Neurosci. 2014;34:1510–1522. doi: 10.1523/JNEUROSCI.2549-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Y, Liu J, Yu B, Ba R, Zhao C. Brpf1 Haploinsufficiency Impairs Dendritic Arborization and Spine Formation, Leading to Cognitive Deficits. Front Cell Neurosci. 2019;13:249. doi: 10.3389/fncel.2019.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu B, Liu J, Su M, Wang C, Chen H, Zhao C. Disruption of Foxg1 impairs neural plasticity leading to social and cognitive behavioral defects. Mol Brain. 2019;12:63. doi: 10.1186/s13041-019-0484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X, Gu X, Zhang Q, Wang Q, Cheng Y, Pleasure SJ, et al. FoxG1 Directly Represses Dentate Granule Cell Fate During Forebrain Development. Front Cell Neurosci. 2018;12:452. doi: 10.3389/fncel.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keeble TR, Halford MM, Seaman C, Kee N, Macheda M, Anderson RB, et al. The Wnt receptor Ryk is required for Wnt5a-mediated axon guidance on the contralateral side of the corpus callosum. J Neurosci. 2006;26:5840–5848. doi: 10.1523/JNEUROSCI.1175-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blakely BD, Bye CR, Fernando CV, Prasad AA, Pasterkamp RJ, Macheda ML, et al. Ryk, a receptor regulating Wnt5a-mediated neurogenesis and axon morphogenesis of ventral midbrain dopaminergic neurons. Stem Cells Dev. 2013;22:2132–2144. doi: 10.1089/scd.2013.0066. [DOI] [PubMed] [Google Scholar]
- 44.Clark CE, Richards LJ, Stacker SA, Cooper HM. Wnt5a induces Ryk-dependent and -independent effects on callosal axon and dendrite growth. Growth Factors. 2014;32:11–17. doi: 10.3109/08977194.2013.875544. [DOI] [PubMed] [Google Scholar]
- 45.Davis EK, Zou Y, Ghosh A. Wnts acting through canonical and noncanonical signaling pathways exert opposite effects on hippocampal synapse formation. Neural Dev. 2008;3:32. doi: 10.1186/1749-8104-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theil T, Aydin S, Koch S, Grotewold L, Ruther U. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129:3045–3054. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- 47.Nordin N, Li M, Mason JO. Expression profiles of Wnt genes during neural differentiation of mouse embryonic stem cells. Cloning Stem Cells. 2008;10:37–48. doi: 10.1089/clo.2007.0060. [DOI] [PubMed] [Google Scholar]
- 48.Fotaki V, Larralde O, Zeng S, McLaughlin D, Nichols J, Price DJ, et al. Loss of Wnt8b has no overt effect on hippocampus development but leads to altered Wnt gene expression levels in dorsomedial telencephalon. Dev Dyn. 2010;239:284–296. doi: 10.1002/dvdy.22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fotaki V, Price DJ, Mason JO. Wnt/beta-catenin signaling is disrupted in the extra-toes (Gli3(Xt/Xt)) mutant from early stages of forebrain development, concomitant with anterior neural plate patterning defects. J Comp Neurol. 2011;519:1640–1657. doi: 10.1002/cne.22592. [DOI] [PubMed] [Google Scholar]
- 50.Harrison-Uy SJ, Pleasure SJ. Wnt signaling and forebrain development. Cold Spring Harb Perspect Biol. 2012;4:a008094. doi: 10.1101/cshperspect.a008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- 52.Cuesta S, Funes A, Pacchioni AM. Social Isolation in Male Rats During Adolescence Inhibits the Wnt/beta-Catenin Pathway in the Prefrontal Cortex and Enhances Anxiety and Cocaine-Induced Plasticity in Adulthood. Neurosci Bull. 2020;36:611–624. doi: 10.1007/s12264-020-00466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farias GG, Alfaro IE, Cerpa W, Grabowski CP, Godoy JA, Bonansco C, et al. Wnt-5a/JNK signaling promotes the clustering of PSD-95 in hippocampal neurons. J Biol Chem. 2009;284:15857–15866. doi: 10.1074/jbc.M808986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varela-Nallar L, Alfaro IE, Serrano FG, Parodi J, Inestrosa NC. Wingless-type family member 5A (Wnt-5a) stimulates synaptic differentiation and function of glutamatergic synapses. Proc Natl Acad Sci U S A. 2010;107:21164–21169. doi: 10.1073/pnas.1010011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eichenbaum H. Hippocampus: cognitive processes and neural representations that underlie declarative memory. Neuron. 2004;44:109–120. doi: 10.1016/j.neuron.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 56.Lisman JE, Grace AA. The hippocampal-VTA loop: controlling the entry of information into long-term memory. Neuron. 2005;46:703–713. doi: 10.1016/j.neuron.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 57.Aimone JB, Wiles J, Gage FH. Potential role for adult neurogenesis in the encoding of time in new memories. Nat Neurosci. 2006;9:723–727. doi: 10.1038/nn1707. [DOI] [PubMed] [Google Scholar]
- 58.Surget A, Tanti A, Leonardo ED, Laugeray A, Rainer Q, Touma C, et al. Antidepressants recruit new neurons to improve stress response regulation. Mol Psychiatry. 2011;16:1177–1188. doi: 10.1038/mp.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Luo XP. Plasticity and metaplasticity of lateral perforant path in hippocampal dentate gyrus in a rat model of febrile seizure. Acta Physiol Sin. 2011;63:124–130. [PubMed] [Google Scholar]
- 60.Ji C, Zhu L, Chen C, Wang S, Zheng L, Li H. Volumetric Changes in Hippocampal Subregions and Memory Performance in Mesial Temporal Lobe Epilepsy with Hippocampal Sclerosis. Neurosci Bull. 2018;34:389–396. doi: 10.1007/s12264-017-0186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cenquizca LA, Swanson LW. Spatial organization of direct hippocampal field CA1 axonal projections to the rest of the cerebral cortex. Brain Res Rev. 2007;56:1–26. doi: 10.1016/j.brainresrev.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maren S, Quirk GJ. Neuronal signalling of fear memory. Nat Rev Neurosci. 2004;5:844–852. doi: 10.1038/nrn1535. [DOI] [PubMed] [Google Scholar]
- 63.Fanselow MS, Poulos AM. The neuroscience of mammalian associative learning. Annu Rev Psychol. 2005;56:207–234. doi: 10.1146/annurev.psych.56.091103.070213. [DOI] [PubMed] [Google Scholar]
- 64.Lee SH, Marchionni I, Bezaire M, Varga C, Danielson N, Lovett-Barron M, et al. Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron. 2014;82:1129–1144. doi: 10.1016/j.neuron.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blackstad TW. Commissural connections of the hippocampal region in the rat, with special reference to their mode of termination. J Comp Neurol. 1956;105:417–537. doi: 10.1002/cne.901050305. [DOI] [PubMed] [Google Scholar]
- 66.Blackstad TW. On the termination of some afferents to the hippocampus and fascia dentata; an experimental study in the rat. Acta Anat (Basel) 1958;35:202–214. doi: 10.1159/000141409. [DOI] [PubMed] [Google Scholar]
- 67.Forster E, Zhao S, Frotscher M. Laminating the hippocampus. Nat Rev Neurosci. 2006;7:259–267. doi: 10.1038/nrn1882. [DOI] [PubMed] [Google Scholar]
- 68.Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paluszkiewicz SM, Martin BS, Huntsman MM. Fragile X syndrome: the GABAergic system and circuit dysfunction. Dev Neurosci. 2011;33:349–364. doi: 10.1159/000329420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu W, Sudhof TC. A neural circuit for memory specificity and generalization. Science. 2013;339:1290–1295. doi: 10.1126/science.1229534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferhat AT, Halbedl S, Schmeisser MJ, Kas MJ, Bourgeron T, Ey E. Behavioural Phenotypes and Neural Circuit Dysfunctions in Mouse Models of Autism Spectrum Disorder. Adv Anat Embryol Cell Biol. 2017;224:85–101. doi: 10.1007/978-3-319-52498-6_5. [DOI] [PubMed] [Google Scholar]
- 72.Kortum F, Das S, Flindt M, Morris-Rosendahl DJ, Stefanova I, Goldstein A, et al. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet. 2011;48:396–406. doi: 10.1136/jmg.2010.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pratt DW, Warner JV, Williams MG. Genotyping FOXG1 Mutations in Patients with Clinical Evidence of the FOXG1 Syndrome. Mol Syndromol. 2013;3:284–287. doi: 10.1159/000345845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shoichet SA, Kunde SA, Viertel P, Schell-Apacik C, von Voss H, Tommerup N, et al. Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum Genet. 2005;117:536–544. doi: 10.1007/s00439-005-1310-3. [DOI] [PubMed] [Google Scholar]
- 75.Le Guen T, Bahi-Buisson N, Nectoux J, Boddaert N, Fichou Y, Diebold B, et al. A FOXG1 mutation in a boy with congenital variant of Rett syndrome. Neurogenetics. 2011;12:1–8. doi: 10.1007/s10048-010-0255-4. [DOI] [PubMed] [Google Scholar]
- 76.Brunetti-Pierri N, Paciorkowski AR, Ciccone R, Della Mina E, Bonaglia MC, Borgatti R, et al. Duplications of FOXG1 in 14q12 are associated with developmental epilepsy, mental retardation, and severe speech impairment. Eur J Hum Genet. 2011;19:102–107. doi: 10.1038/ejhg.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeung A, Bruno D, Scheffer IE, Carranza D, Burgess T, Slater HR, et al. 4.45 Mb microduplication in chromosome band 14q12 including FOXG1 in a girl with refractory epilepsy and intellectual impairment. Eur J Med Genet 2009, 52: 440–442. [DOI] [PubMed]
- 78.Tohyama J, Yamamoto T, Hosoki K, Nagasaki K, Akasaka N, Ohashi T, et al. West syndrome associated with mosaic duplication of FOXG1 in a patient with maternal uniparental disomy of chromosome 14. Am J Med Genet A. 2011;155A:2584–2588. doi: 10.1002/ajmg.a.34224. [DOI] [PubMed] [Google Scholar]
- 79.Coullery RP, Ferrari ME, Rosso SB. Neuronal development and axon growth are altered by glyphosate through a WNT non-canonical signaling pathway. Neurotoxicology. 2016;52:150–161. doi: 10.1016/j.neuro.2015.12.004. [DOI] [PubMed] [Google Scholar]