Abstract
Background:
Molecular diagnostics such as pathogen-directed PCRs have transformed testing for ocular infections since the late 1990s. Although these assays remain important diagnostic tools for samples with low biomass, the lack of diagnostic range motivates alternative molecular approaches for ocular infections. The aim of this study was to determine the performance of a high-throughput RNA sequencing approach, RNA-seq, to detect infectious agents in ocular samples from patients with presumed ocular infections.
Methods:
We compared the performance of RNA-seq to pathogen-directed PCRs using remnant nucleic acids from 41 aqueous or vitreous samples of patients with presumed ocular infections. Pathogen-directed PCRs were performed at the CLIA-certified Stanford Clinical Virology Laboratory. RNA-seq was performed in a masked manner at the Proctor Foundation at the University of California San Francisco. Percent positive and negative agreement between the two testing approaches were calculated. Discordant results were subjected to orthogonal testing.
Results:
The positive percent agreement between RNA-seq and pathogen-directed PCRs was 100% (95% confidence interval (CI): 78.5% to 100%). The negative percent agreement was 92.6% (95% CI: 76.6% to 97.9%). RNA-seq identified pathogens not on the differential diagnosis for 9.7% (4/41) of the samples. Two pathogens solely identified with RNA-seq were confirmed with orthogonal testing.
Conclusions:
RNA-seq can accurately identify common and rare pathogens in aqueous and vitreous samples of patients with presumed ocular infections. Such an unbiased approach to testing has the potential to improve diagnostics although practical clinical utility warrants additional studies.
Introduction
Non-specific testing may be of low clinical utility and can be wasteful of healthcare resources.[1] High-throughput sequencing, with its hypothesis-free approach to testing that can potentially diagnose any infectious etiology with a single test, is transforming clinical molecular diagnostics.[2–4] The cost of sequencing has dramatically decreased over the past decade, but remains significantly more expensive than available targeted molecular diagnostics. From a clinical standpoint, it is also unclear if such an approach is more informative than the current laboratory testing practice of ophthalmologists.[3, 5, 6] In this study, we compared the performance of metagenomic RNA sequencing (RNA-seq) to pathogen-directed polymerase chain reaction (PCRs) assays for diagnosing ocular infections.
Materials and methods
Ethical approval was obtained from the University of California San Francisco (UCSF) and the Stanford University Institutional Review Boards. This study adhered to the tenets of the Declaration of Helsinki. From June 2018 to December 2019, the CLIA-certified Stanford Clinical Virology laboratory received 41 aqueous or vitreous samples for pathogen-directed PCR testing from multiple medical centers across the United States. Total nucleic acids (DNA and RNA) were extracted as previously described.[7] PCR tests at Stanford include CMV, HSV-1, HSV-2, VZV, and Toxoplasma gondii PCRs, although the number of pathogens tested is at the discretion of the ordering physician. The remaining extracted total nucleic acids of all samples with sufficient residual volume were then sent to the Proctor Foundation at UCSF for RNA-seq which was performed as previously described.[8] Briefly, 5 μL of extracted total nucleic acids of each sample was first converted to cDNA and sequencing libraries were prepared using the NEBNext RNA ULTRA II kit (New England Biolabs) according to the manufacturer’s instructions and then amplified with 16 PCR cycles. Samples were sequenced on the NovaSeq system (NovaSeq 6000; Illumina) using 150-nucleotide paired-end sequencing. Analysis of the sequenced data was made using a rapid computational pipeline to identify pathogens.[8] Paired-end reads were subjected to two rounds of human sequencing read removal. In the initial step, all paired-end reads were aligned to the human reference genome 38 (hg38) and the Pantroglodytes genome (panTro4, 2011, UCSC), using the Spliced Transcripts Alignment to a Reference (STAR) aligner (v25.4b).[9] Unaligned reads were quality filtered using PriceSeqFilter (v1.2) and reads that were at least 95% identical were compressed by cd-hit-dup (cd-hit v4.7).[10] Another round of human reads removal was performed using Bowtie2 (v2.3.4.1). The remaining non-host reads were subjected to taxonomic classification using the Centrifuge Taxonomic Classifier engine using an index created from the NCBI nucleotide non-redundant sequences (3/3/2018).[11] The pre-specified criteria for positive pathogen are 1) it is known to be a human pathogen and represented the most abundant reads after background subtraction or 2) two or more unique reads covering separate regions in the HSV, VZV, or CMV genomes. All samples were de-identified and all laboratory personnel at the Proctor Foundation were masked to the results obtained at the Stanford Clinical Virology laboratory.
Results
Of the 5 available pathogen-directed PCRs, the clinicians ordered 4 ±1 (mean±standard deviation) assays per sample with Toxoplasma gondii PCR being the least likely to be ordered (10/41). Infectious etiologies were identified in 12 out of the 41 samples (29%) based on clinician testing practices. RNA-seq correctly identified all 12 pathogens identified by pathogen-directed PCRs. Additionally, RNA-seq identified pathogens in 4 other samples for a total of 16 samples (39%) (Figure 1A and 1B). Those pathogens were Toxoplasma gondii, CMV, rubella, and Pithomyces chartarum (Figure 1C). Toxoplasma gondii and CMV were not ordered by the treating clinician. These results were later confirmed at Stanford via pathogen-directed PCR. For the sample positive for rubella, the clinician ordered only CMV PCR. For the case in which Pithomyces chartarum was detected, HSV-1/HSV-2/CMV/VZV PCRs were ordered on the aqueous fluid, but no bacterial or fungal cultures were ordered on the aqueous fluid or cornea. The presence of rubella and Pithomyces chartarum RNA were confirmed with pathogen-directed reverse transcription (RT)- PCR at the Proctor Foundation. Therefore, the positive and negative percent agreement between RNA-seq and pathogen-directed PCRs were 100% (95% confidence interval (CI): 78.5% to 100%) and 92.6% (95% CI: 76.6% to 97.9%), respectively. The overall agreement was 95.1% (95% CI: 83.9% to 98.7%). The kappa coefficient was 0.90 (95% CI: 0.75 to 1.00).
Figure 1: Performance of RNA-seq compared to pathogen-directed PCRs.
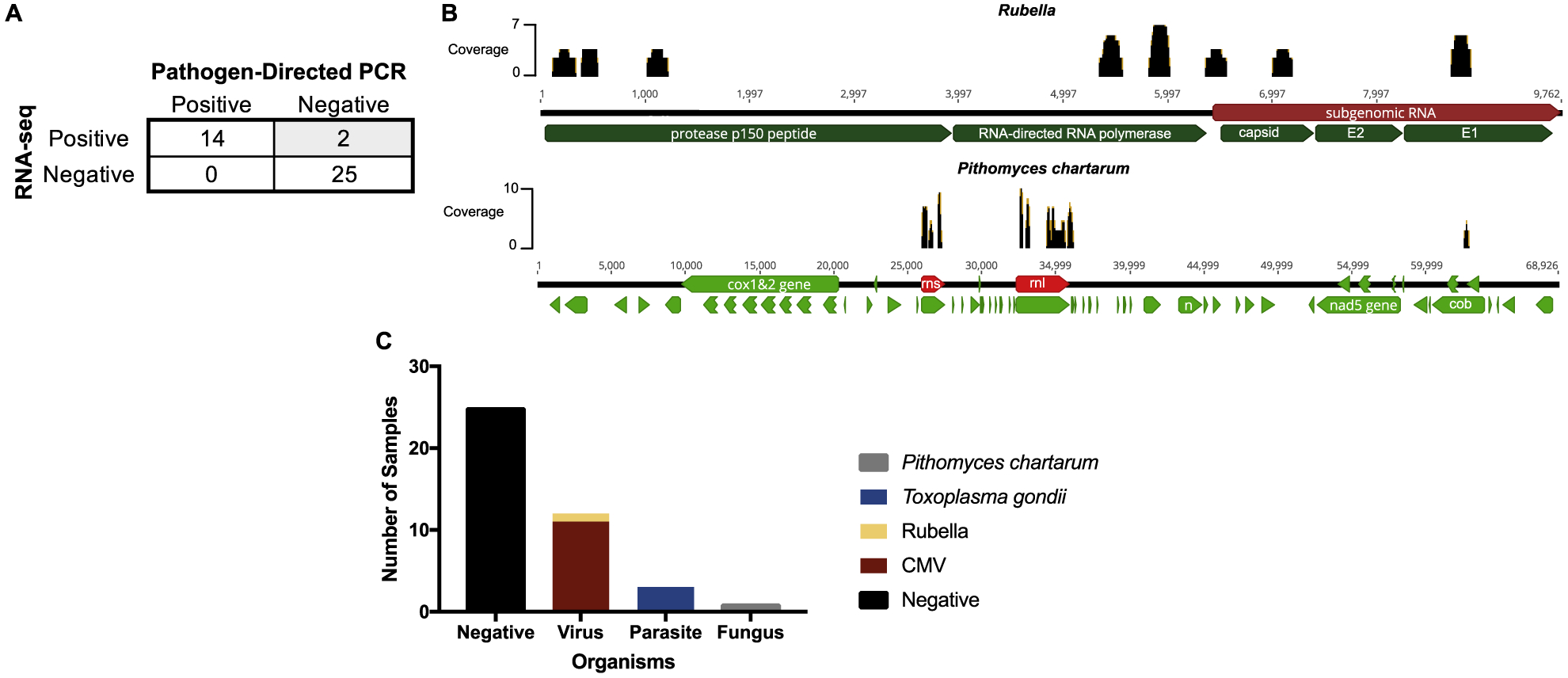
A, 2 × 2 table comparing the performance of RNA-seq compared to pathogen-directed PCRs. B, Genome coverage maps for rubella and Pithomyces chartarum, the two samples highlighted in grey in panel A. C, Pathogens detected by RNA-seq. Abbreviations: cytomegalovirus, CMV.
Discussion
Previously, we showed that DNA-seq for intraocular fluid samples from patients with presumed ocular infections had excellent positive and negative concordance to pathogen-directed PCRs.[12] Of the samples negative by conventional diagnostics, an additional 22% of the samples were positive by DNA-seq for pathogens and were subsequently confirmed with orthogonal testing. This suggests that the clinicians’ suspicion for infectious etiologies exceeds the capabilities of conventional diagnostics, indicating a need to expand our testing armamentarium. In this study, the sample that tested positive for rubella on RNA-seq came from a patient carrying the presumed diagnosis of Fuchs’ heterochromic iridocyclitis. Rubella and CMV are pathogens known to be associated with Fuchs’ heterochromic iridocyclitis.[13, 14] While the clinician was able to order CMV-directed PCR, rubella testing was not available. Here, DNA-seq would not have been useful as it cannot detect RNA viruses such as rubella. RNA-seq is required.
The identification of Pithomyces chartarum in an aqueous sample with RNA-seq is also notable. Pithomyces species are dematiaceous fungi commonly found in plants and decaying vegetation and can cause diseases in animals and humans.[15, 16] While Pithomyces species have been isolated from human corneal specimens in the United States[16], little clinical description of ocular infections due to Pithomyces has been documented in the literature. The aqueous sample was obtained from a non-Caucasian man undergoing a Descemet stripping automated endothelial keratoplasty (DSAEK) with corneal edema and filamentary keratitis. He had undergone several prior intraocular surgeries, including a complex cataract surgery, pars plana vitrectomy, and anterior chamber intraocular lens implantation that was subsequently exchanged for a scleral fixated intraocular lens. Here, it is unclear if the presence of Pithomyces chartarum RNA in the aqueous fluid was truly pathogenic or simply a bystander.
In this study of 41 intraocular fluid samples from patients with presumed ocular infections, we found that RNA-seq was highly concordant with pathogen-directed PCRs. It also has the potential to improve upon DNA-seq because of its ability to detect RNA viruses. A major concern with unbiased testing is the potential for false positives. In this study, the pathogens identified solely with RNA-seq were confirmed with orthogonal testing. As with any diagnostic assay interpretation, clinical correlation is needed. It is also worthwhile to note that 25 out of 41 samples (61%) were negative by both RNA-seq and pathogen-directed PCRs. While it is possible that RNA-seq still needs optimization for clinical samples, it does highlight the challenge of distinguishing intraocular inflammation associated with an infectious etiology versus non-infectious causes of intraocular inflammation. A negative result with RNA-seq is of clinical importance as it may prompt the investigation for malignancy or the transition to immunosuppressive therapy.
The routine implementation of high-throughput sequencing as diagnostic tests in the clinical laboratories will be inevitable. Reagent and sequencing costs are no longer prohibitive and many facilities now have access to various sequencing platforms. While reagent and sequencing costs for metagenomic deep sequencing are still more expensive than PCR reagents, the main barriers for the routine implementation are the labor intensive library sequencing workflow and the bioinformatics required for metagenomics. However, these challenges can be overcome with automation and the standardization of bioinformatic pipelines. From a clinical standpoint, a diagnostic test can be of the latest technology and is highly sensitive and specific, but may have little or no effect on patient clinical outcomes. Randomized controlled trials comparing clinical outcomes between standard testing approaches and RNA-seq could further guide clinical practice.
As in the case with retrospective studies, we cannot assess whether RNA-seq has an impact on patient clinical outcomes. In addition, the multiple freeze-thaws of the surplus nucleic acids in this study may underestimate the number of RNA viruses identified by RNA-seq given that RNA is highly susceptible to degradation.
In summary, RNA-seq can identify known and unknown pathogens from intraocular fluid samples of patients with presumed intraocular infections. Its clinical usefulness remains to be determined.
Highlights.
The testing performance of RNA-seq was compared against the available molecular diagnostics for ocular infections.
RNA-seq had high positive and negative concordances with pathogen-directed PCRs.
RNA-seq was able to detect pathogens not on the differential diagnosis.
RNA-seq outperforms conventional molecular diagnostics for ocular infections although clinical utility remains to be determined.
Funding:
Research reported in this manuscript was supported by the Research to Prevent Blindness Career Development Award (T.D.) and the National Eye Institute of the National Institutes of Health under Award Number K08EY026986 (T.D.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Bianchi MT, Alexander BM, Cash SS. Incorporating uncertainty into medical decision making: an approach to unexpected test results. Medical decision making : an international journal of the Society for Medical Decision Making 2009; 29(1): 116–24. [DOI] [PubMed] [Google Scholar]
- [2].Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nature reviews Genetics 2016; 17(5): 257–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Valdes L, Bispo P, Sobrin L. Application of Metagenomic Sequencing in the Diagnosis of Infectious Uveitis. Seminars in ophthalmology 2020; 35(5–6): 276–9. [DOI] [PubMed] [Google Scholar]
- [4].Muthappan V, Lee AY, Lamprecht TL, et al. Biome representational in silico karyotyping. Genome research 2011; 21(4): 626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Doan T, Wilson MR, Crawford ED, et al. Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome medicine 2016; 8(1): 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ung L, Bispo PJM, Doan T, et al. Clinical metagenomics for infectious corneal ulcers: Rags to riches? The ocular surface 2020; 18(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gomez CA, Sahoo MK, Kahn GY, et al. Dual-target, real-time PCR for the diagnosis of intraocular Toxoplasma gondii infections. Br J Ophthalmol 2019; 103(4): 569–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Doan T, Hinterwirth A, Worden L, et al. Gut microbiome alteration in MORDOR I: a community-randomized trial of mass azithromycin distribution. Nature medicine 2019. [DOI] [PubMed] [Google Scholar]
- [9].Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013; 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics (Oxford, England) 2012; 28(23): 3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome research 2016; 26(12): 1721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Doan T, Acharya NR, Pinsky BA, et al. Metagenomic DNA Sequencing for the Diagnosis of Intraocular Infections. Ophthalmology 2017; 124(8): 1247–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cunningham ET Jr, Baglivo E Fuchs heterochromic iridocyclitis--syndrome, disease, or both? American journal of ophthalmology 2009; 148(4): 479–81. [DOI] [PubMed] [Google Scholar]
- [14].Leveque TK, Goldstein DA. Moving From Syndromic Description to Etiologic Diagnosis for Uveitis. JAMA ophthalmology 2019; 137(4): 438–9. [DOI] [PubMed] [Google Scholar]
- [15].Shah H, Honeybul S, Tang S, Arthur I, McLaren S, Boan P. Mould meningitis associated with intravenous drug use. Medical mycology case reports 2018; 20: 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].da Cunha KC, Sutton DA, Gené J, et al. Pithomyces species (Montagnulaceae) from clinical specimens: identification and antifungal susceptibility profiles. Medical mycology 2014; 52(7): 748–57. [DOI] [PubMed] [Google Scholar]


