Abstract
Chemotherapy resistance is the main impediment in the treatment of acute myeloid leukaemia (AML). Despite rapid advances, the various mechanisms inducing resistance development remain to be defined in detail. Here we report that loss-of-function mutations (LOF) in the histone methyltransferase EZH2 have the potential to confer resistance against the chemotherapeutic agent cytarabine. We identify seven distinct EZH2 mutations leading to loss of H3K27 trimethylation via multiple mechanisms. Analysis of matched diagnosis and relapse samples reveal a heterogenous regulation of EZH2 and a loss of EZH2 in 50% of patients. We confirm that loss of EZH2 induces resistance against cytarabine in the cell lines HEK293T and K562 as well as in a patient-derived xenograft model. Proteomics and transcriptomics analysis reveal that resistance is conferred by upregulation of multiple direct and indirect EZH2 target genes that are involved in apoptosis evasion, augmentation of proliferation and alteration of transmembrane transporter function. Our data indicate that loss of EZH2 results in upregulation of its target genes, providing the cell with a selective growth advantage, which mediates chemotherapy resistance.
Subject terms: Experimental models of disease, Acute myeloid leukaemia
Introduction
Acute myeloid leukaemia (AML) is a heterogeneous haematological malignancy, characterised by clonal expansion of abnormal, undifferentiated myeloid precursor cells. Even though many patients with AML respond well to induction chemotherapy, relapse and refractory disease are common, representing the major cause of treatment failure. Treatment with cytarabine (AraC) and daunorubicin (DNR) remains the standard care for AML patients, although several new therapeutic strategies have been implemented within the last years1–3. Epigenetic dysregulation of DNA methylation or histone modifications has been identified in many malignant tumors4,5 and can be considered as a cause of cancer development and progression6,7. Since considerable insight concerning those epigenetic changes has been gained in recent years, many therapy concepts targeting the involved regulatory factors have been proposed and hold promise for novel treatment approaches8,9.
Enhancer of zeste homolog 2 (EZH2) is a lysine methyltransferase found as the central core protein of the polycomb repressive complex 2 (PRC2)10. Comprising four subunits (SUZ12, EED, EZH2/EZH1 and RbAp46), this complex mediates transcriptional repression by catalysing the trimethylation of histone H3 at lysine 27 (H3K27me3)11. EZH2 has been found to serve a dual purpose, as either tumour suppressor or oncogene, depending on the type of cancer12–17. In leukaemia, overexpression of EZH2 has been observed in CLL18, paediatric T-ALL19 and CML20, while other studies reported EZH2 levels to be decreased in CMML21 as well as ALL18,19,22. A recent study of Basheer et al. suggests opposing roles of EZH2 in initiation and maintenance of AML23.
EZH1, an EZH2 homolog capable of partially compensating EZH2 function, holds an essential role in preserving pathological stem cells24. Therefore, it might contribute to the already complex role of EZH2 in hematopoietic malignancies25–27. Although EZH2 loss-of-function mutations seem to be rare in AML28, loss of EZH2 by other mechanisms have been frequently reported and appear to play a major role in disease progression29,30. Absence of EZH2 in leukaemia cells was recently found to aberrantly activate BCAT1, resulting in enhanced mTOR signaling27 and activation of the oncogene Hmga2 by causing an epigenetic switch from H3K27 trimethylation to H3K27 acetylation31. Furthermore, reduced disease-free survival was found to be associated with EZH2 mutations in myeloid malignancies28,32,33 including AML23. In addition, chemoresistance was found in a recent study on AML patients with poor prognosis and downregulated EZH234.
In our previous study35, examining diagnosis/relapse pairs of 50 cytogenetically normal (CN) AML patients, we found mutations in epigenetic modifiers, including EZH2, frequently gained at relapse, suggesting epigenetic mechanisms to be involved in disease progression in a subset of patients. The current study aims to evaluate the importance of chemotherapy resistance in AML. We investigated EZH2 mutations and their functional loss of methyltransferase activity using patient samples, in vivo and in vitro patient-derived xenografts (PDX), and haematopoietic cell lines. We found EZH2 loss-of-function mutations to be involved in the development of resistance against cytarabine and observed upregulation of EZH2 target genes due to loss of H3K27 trimethylation.
Results
Recurrent EZH2 mutations at diagnosis
In our previous work, we analysed 664 AML patients to study recurrently mutated genes, including EZH236. In this cohort, 25 patients (4 %) carried an EZH2 mutation at the time of diagnosis (27 mutations in total, Fig. 1a). Most of these mutations (n = 20, 74%) were located in the SET ([Su(var)3-9, Enhancer-of-zeste and Trithorax]) or CXC (cysteine-rich region, sometimes referred to as pre-SET) domain at the C-terminus of the protein and are responsible for the catalytic activity of the methyltransferase. Furthermore, 41% (11) of mutations cause a stop-gain or frameshift, resulting in a truncated protein. An additional two frameshift mutations result in an elongated protein variant. Mutations most frequently co-occurring with mutated EZH2 were found in RUNX1, ASXL1, DNMT3A and TET2 (44%, 40%, 20% and 20%, Supplementary Fig. 1a). Additionally, RUNX1 and ASXL1 mutations were found to occur more often in EZH2 mutated patients (44% and 40%) than in EZH2 wild type patients (14% and 10%, p = 4.6e−04, and p = 9.3 e−05, Fisher's Exact Test). In contrast, NPM1, the most frequently mutated gene in our cohort, was found to be mutated less often in EZH2 mutated (12%) than in EZH2 wild type patients (34%) (p = 2.8 e−02, Fisher's Exact Test). Interestingly, KDM6A and EZH2 mutations were found to be mutually exclusive. Most patients with EZH2 mutations (76%, n = 19) can be assigned to the adverse risk group (Supplementary Fig. 1a), according to the recent ELN classification37.
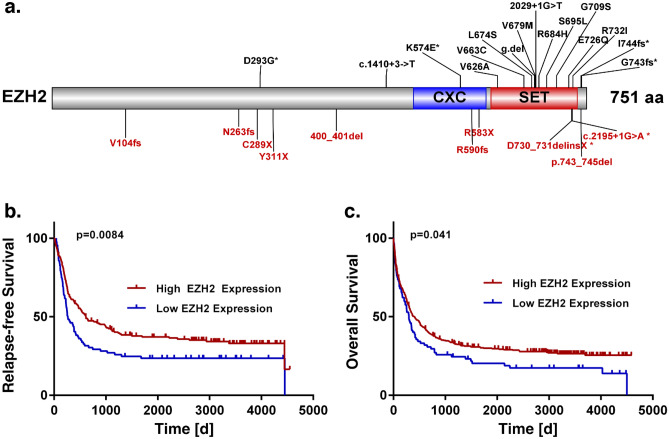
Figure 1.
Recurrent EZH2 mutations. (a) Schematic overview of EZH2 protein structure (NM_004456.4) and identified mutations (27 in total, c.2195+1G>A appeared twice) in a cohort of 664 AML patients at diagnosis. Functional domains are indicated at distinct locations and truncating mutations are displayed in red. Patients from Metzeler et al. 2016 (AMLCG-1999, AMLCG-2008). (b–c) Survival analysis of patients with low or high EZH2 mRNA expression at the time point of diagnosis. EZH2 high and low groups defined by the upper and lower quartile of EZH2 mRNA expression, independent of mutation status. (b) Relapse-free survival (RFS). (c) Overall survival (OS). Patients from AMLCG 1999 (GSE37642), n = 517. 21 patients harboured an EZH2 mutation. P-value calculated by log-rank test.
In order to evaluate the prognostic importance of EZH2, we examined the survival of patients dependent on their EZH2 mutation and expression status. The overall survival (OS) of patients harbouring EZH2 mutations did not differ significantly from patients without mutation (Supplementary Fig. 1b). However, low EZH2 mRNA expression was significantly associated with poor relapse-free survival (RFS) and OS in publicly available independent data sets of the AMLCG 1999 trial (GSE37642, Fig. 1b-c) and HOVON (GSE14468, Supplementary Fig. 1) study groups38–40. Additionally, monosomy 7, resulting in reduced EZH2 expression, was associated with poor overall survival (Supplementary Fig. 1c).
Relevance of EZH2 status in AML relapse
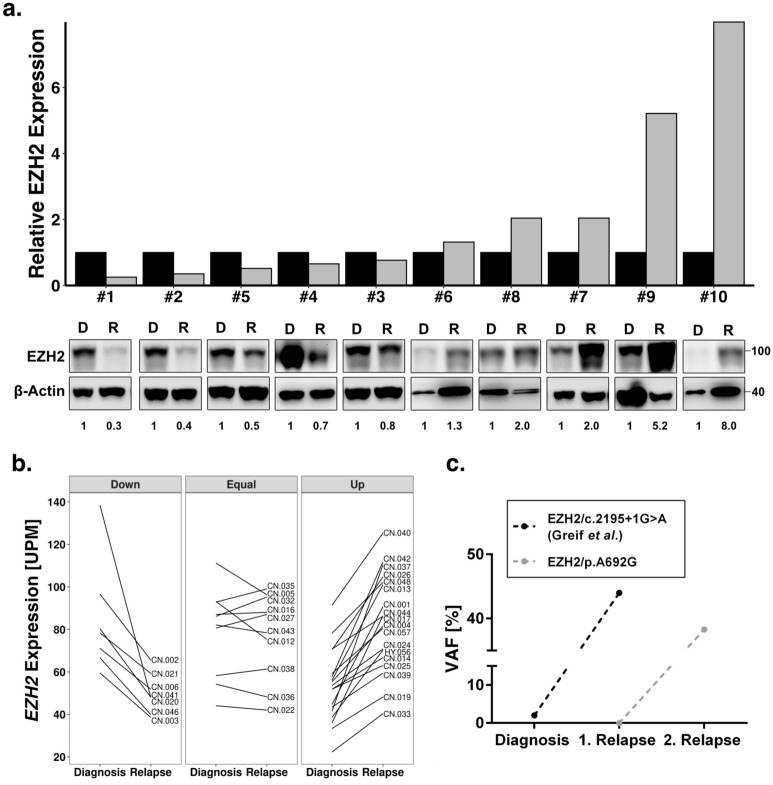
To further investigate the poor survival in patients with low EZH2 mRNA expression, we compared protein expression in a set of matched diagnosis and relapse pairs of ten AML patients without EZH2 mutations (Supplementary Table 2). In 50% of patients, we observed decreased levels of EZH2 protein expression, whereas the other half revealed increased protein expression levels in relapse (Fig 2a). An increase of at least 2-fold in protein expression was found in four patients, whereas a strong decrease (2-fold or more) in protein expression was observed in three patients. An additional analysis of EZH2 mRNA expression in 32 CN-AML patients revealed a similar heterogenous picture. Downregulation of EZH2 was found in 22% of patients, while upregulation was found in 53% (Fig. 2b). Additionally, we identified two relapse-associated EZH2 mutations. EZH2/p.A692G found in the second relapse of patient CN-021 from the Greif et al. cohort35 and EZH2/Y733LfsX6 found in the first relapse of a patient from the AML-CG cohort. Both mutations revealed subclonal outgrowth during the course of treatment and increasing variant allele frequencies (VAFs) in relapsed patients (Fig. 2c). Additionally, we found an increase of VAFs in the relapse of three other EZH2 mutations found in the Greif et al. cohort35 (Supplementary Fig. 2).
Figure 2.
Relevance of EZH2 status in AML relapse. (a) Immunoblot for EZH2 protein expression in 10 AML patients at diagnosis and relapse. MW, molecular weight; β-actin, loading control. The ratio of EZH2 to β-actin expression is indicated below and presented in the histogram above. Each relapse value was normalized to the corresponding diagnosis sample. None of the patients carried an EZH2 mutation. (b) EZH2 mRNA expression between diagnosis and relapse of 32 CN-AML patients from Greif et al. cohort35. Up and down are defined as a change in mRNA expression of at least 20%. Three patients carried an EZH2 mutation. (c) Variant allele frequency of the two relapse-associated EZH2 mutations with outgrowth in first and second relapse.
Functional characterisation of EZH2 mutations
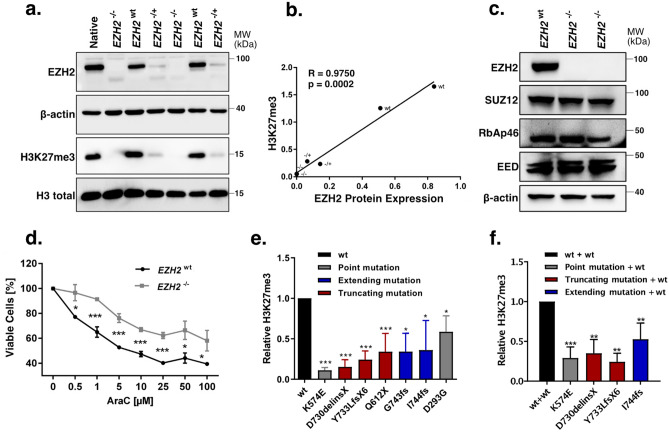
To evaluate the biochemical activity of the EZH2 variants, we measured global H3K27 trimethylation levels in a 293T/EZH2−/− model (Fig. 3a), which was established through CRISPR/Cas9 mediated genome editing, targeting exon 3 of EZH2. We found EZH2 protein expression levels to be strongly correlated with global H3K27me3 levels (Fig. 3b). In fact, global H3K27me3 was not detectable in any of the tested EZH2−/− clones, whereas EZH2+/− clones showed decreased EZH2 expression as well as reduced global H3K27me3 levels (Fig. 3a). Since EZH2 is only one part of the PRC2 complex, we additionally analysed protein expression of the remaining components SUZ12, RBAP46 and EED. We could not detect aberrant expression of these subunits in both 293T/EZH2−/− clones (Fig. 3c). Interestingly, both clones showed an increased resistance against AraC compared to the wild type clones (Fig. 3d, Supplementary Fig. 3a) and a slightly reduced colony count was observed in a colony formation assay (Supplementary Fig. 3c). Re-expression of seven different EZH2 variants, (Supplementary Fig. 3b) found in the AML-CG-1999 and AML-CG-2008 studies, could only partially rescue global H3K27me3 levels, indicating a LOF phenotype, while the re-expressed wildtype protein was able to restore complete activity (Fig. 3e).
Figure 3.
Evaluation of EZH2 mutations found in AML patients at diagnosis. (a) Comparison of EZH2 expression and global H3K27me3 between EZH2−/−, EZH2+/− and EZH2wt sc clones in 293T cells. MW, molecular weight. β-actin and H3 total, loading controls. (b) Correlation between EZH2 protein expression and global H3K27me3 in 293T sc clones. Pearson's correlation. (c) Immunoblot for EZH2, SUZ12, RbAP46 and EED expression in 293T/EZH2−/− sc clones. MW, molecular weight; β-actin, loading control. (d) AraC resistance in one 293T/EZH2−/− and one 293T/EZH2wt sc clone. Cells were treated for 72 h with different concentrations of AraC. Viable cells relative to untreated control. (e) H3K27me3 levels after re-expression of seven EZH2 mutations, detected in patient diagnosis samples. Colours referring to protein structural changes caused by the mutation. 293T/EZH2−/− cells were transfected transiently with EZH2 constructs 72 h before protein isolation, and global H3K27me3 was evaluated by immunoblot. Values relative to the wild type. (f) H3K27me3 levels after re-expression of four EZH2 mutants in combination with wild type EZH2. 293T/EZH2−/− cells were transfected transiently with EZH2 wildtype and EZH2 mutant constructs 72 h before protein isolation. Values relative to the wild type. Unpaired, two-tailed Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001. Error bars indicate mean ± s.d of at least three independent experiments.
In addition, co-expression of these variants with wild type EZH2 led to a reduction of H3K27me3 levels in four mutations, suggesting a dominant-negative effect (Fig. 3f).
To validate the robustness of our 293T/EZH2−/− model, we performed the rescue experiment with two previously described EZH2 variants. EZH2/p.Y646N, a gain-of-function mutation found in lymphomas16,17 and EZH2/Y731, a LOF mutation41. We were able to verify the functions of both mutations with our model (Supplementary Fig. 3d and e).
EZH2 depletion promotes resistance in K562 cells
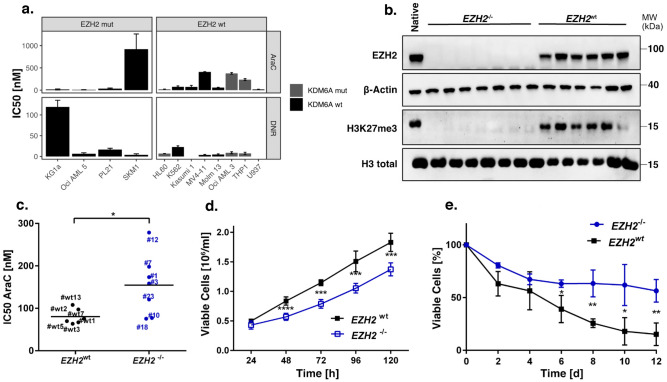
In order to study the impact of EZH2 mutations on chemoresistance in a hematopoietic context, we screened 12 AML cell lines with EZH2 mutant or wild type background (Supplementary Table 1). We identified two EZH2 mutated cell lines, SKM-1 and KG-1a, that seem to be more resistant against cytarabine and daunorubicin, respectively (Fig. 4a). Notably, three of the EZH2wt cell lines were harbouring KDM6A mutations, which can also affect drug resistance42. Furthermore, we found a strong positive correlation between H3K27me3 levels and EZH2 protein expression (Supplementary Fig. 3a). Next, we established seven EZH2−/− single cell (sc) knockout clones in the myeloid cell line K562, using CRISPR Cas9 genome editing (Fig. 4b, Supplementary Fig. 4e). Both knockout and control cells were treated for 72 h with either AraC or DNR. In K562/EZH2−/− clones, increased chemoresistance was found against AraC, while sensitivity against DNR was not affected (Fig. 4c, Supplementary Fig. 4c). Additionally, we observed reduced proliferation in K562/EZH2−/− clones compared to K562/EZH2+/+ clones (Fig. 4d). Furthermore, the response towards AraC treatment was studied in a long-term proliferation assay, consequently treating single cell clones for 12 days with a low dose of AraC. In accordance with the short-term assay, also the K562/EZH2−/− sc clones of the long-term assay displayed higher resistance against AraC (Fig. 4e, Supplementary Fig. 4d).
Figure 4.
EZH2 depletion promotes resistance in the myeloid cell line K562. (a) Comparison of IC50 values for DNR and AraC in twelve haematopoietic cell lines. Cells were treated with AraC/DNR for 72 h. (b) Immunoblot for EZH2 expression and global H3K27me3 of seven EZH2−/− and six EZH2wt sc clones in K562 cells. MW, molecular weight; β-actin and H3 total, loading controls. (c) Comparison of AraC IC50 values in EZH2wt (n = 6) and EZH2−/− (n = 7) clones. Cells were treated with AraC/DMSO for 72 h. Each value represents the mean of three independent experiments. (d) Proliferation of EZH2wt (n = 4) and EZH2−/− (n = 7) clones for 5 d. Medium was changed every 48 h. (e), Long-term low dose AraC treatment in EZH2wt (n = 3) and EZH2−/− (n = 3) clones. Cells were treated with 30 nM AraC/DMSO for 12 d. Viable cells relative to untreated control. Unpaired, two-tailed Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001. Error bars indicate mean ± s.d of three independent experiments.
EZH2 re-expression sensitises to AraC treatment in K562 cells
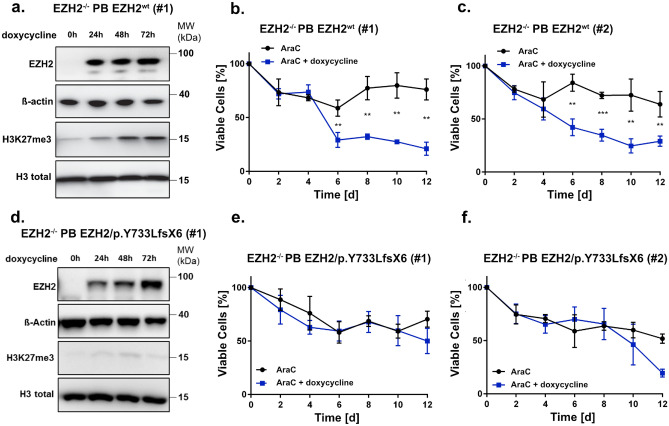
To investigate if re-expression of EZH2 can reconstitute baseline H3K27me3 levels and therefore sensitise cells to AraC treatment, we established a stable, doxycycline-inducible EZH2 expression system via the PiggyBac transposon system (Supplementary Fig. 5a–d). For this reason, DNA coding for EZH2wt (AA1-751) was introduced into K562/EZH2−/− cells (clone #7). Re-expression of wildtype EZH2 was able to restore global H3K27me3 levels after 48 h (Fig. 5a and Supplementary Fig. 5e). Furthermore, sensitivity against AraC could be restored in a long-term low dose AraC treatment experiment (Fig. 5b,c). Additionally, we introduced DNA coding for the relapse-associated mutation EZH2/Y733LfsX6 (AA1-737, Supplementary Fig. 5c,d). Re-expression of this mutant after doxycycline induction did not result in restoration of H3K27me3 levels (Fig. 5d and Supplementary Fig. 5f). Likewise, the mutation was not able to restore sensitivity against AraC treatment, indicating an involvement of H3K27 trimethylation in the phenotype of chemoresistance (Fig. 5e,f). Doxycycline alone had no effect on the sensitivity of either EZH2wt or EZH2−/− cells towards AraC treatment (Supplementary Fig. 5g,h).
Figure 5.
EZH2 re-expression sensitizes K562 cells to AraC treatment. (a,d) Immunoblot for EZH2 expression and global H3K27me3 in (a) EZH2−/− PB EZH2wt cells (clone #1) and (d) EZH2−/− PB EZH2/p.Y733LfsX6 (clone #1) after 0 h, 24 h, 48 h and 72 h of doxycycline induction. Cells were treated with 1 µg/ml doxycycline every 24 h. MW, molecular weight; β-actin and H3 total, loading controls. (b–c, e–f), AraC low dose long-term treatment in (b–c) EZH2−/− PB EZH2wt and (e–f) EZH2−/− PB EZH2/p.Y733LfsX6 cells. Cells were pre-treated for 3 d with doxycycline and then treated with 30 nM AraC/DMSO for 12 d. Cells were split and treated every 4 d and doxycycline was added every 48 h to ensure stable expression of EZH2. Error bars indicate mean ± s.d of three independent experiments. Unpaired, two-tailed Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001.
Upregulation of EZH2 target genes desensitises cells to AraC treatment
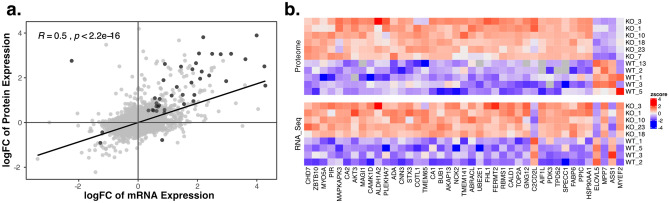
RNA sequencing and Proteome analysis was performed to uncover the molecular mechanism involved in EZH2-mediated chemoresistance. EZH2 knockout in K562 cells resulted in aberrant gene and protein expression (Supplementary Fig. 7), visible in transcriptional upregulation of 216 genes and downregulation of 42 genes as well as translational upregulation of 375 genes and downregulation of 205 genes (Supplementary Table 3 and Supplementary Table 4). The change in protein and RNA expression was found to be correlated (R = 0.5, p = 2.2e-16, Pearson’s correlation, Fig. 6a) and 41 genes showed differential expression of both mRNA and protein. Most of these genes (37) were upregulated, and only two downregulated in both measures (Fig. 6b). Amongst the upregulated genes, we identified FHL1 as well as UBE2E1, both involved in chemotherapy resistance and relapse in AML43,44. Additionally, upregulation of CA2, CNN3 and AKAP13 was found, which are suggested to be involved in chemotherapy resistance in glioblastoma, colon cancer and breast cancer, respectively. Furthermore, PDK3, TPD52, MYO5A, AKT3 and SPECC1 were upregulated, genes associated with poor prognosis in AML or involved in apoptosis. EZH2 ChIP-seq in K562 cells of a publicly available dataset (ENCSR000AQE, ENCSR000AKY) revealed peaks in the promoter region of FHL1, suggesting FHL1 to be a direct target of EZH2. In the promoter region of UBE2E1 no EZH2 peaks were found, but EZH2 binding was detected in a distal enhancer region (GeneHancer Accession: GH03J023748). Other potential direct targets of EZH2 are CNN3, AKAP13, TPD52, MYO5A, AKT3 and SPECC1 as EZH2 peaks were found in the respective promoter regions. Additionally, enhancers regulating FHL1 and TPD52 could be identified (GeneHancer Accession: GHOXJ136155 and GH08J080078). No peaks were assigned to the genes CA2 and PDK3.
Figure 6.
Upregulation of EZH2 target genes. (a) Correlation of protein and mRNA expression. Dark grey points representing genes differentially expressed in both measures (adj.P < 0.05). Pearson’s correlation. (b) Heatmap of the 41 genes differentially expressed (adj.P < 0.05) between EZH2−/− and wild type clones, in both protein and mRNA. The colour gradient from red to blue represents high to low expression of genes. White indicates no change.
Resistance of EZH2 mutated patient-derived xenografts (PDX)
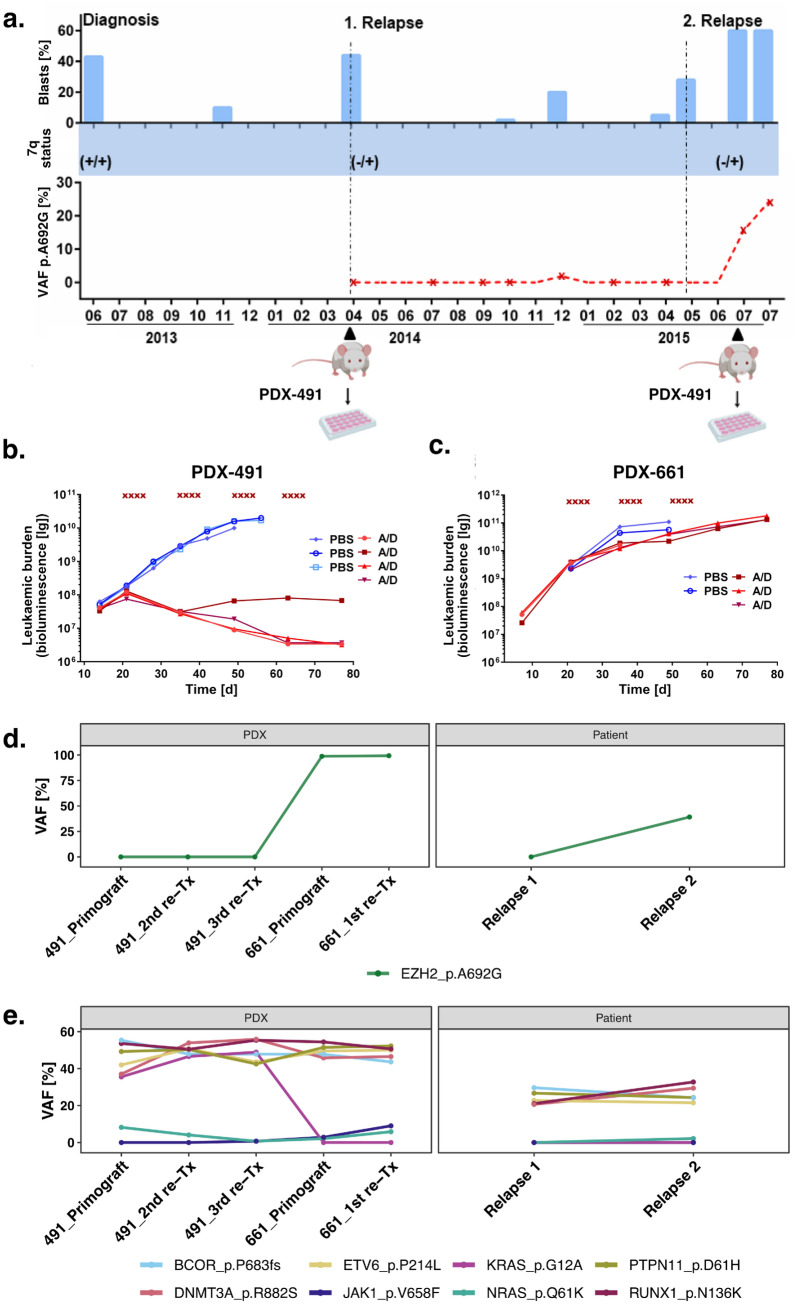
To extend our findings of EZH2 associated AraC resistance in vitro, we screened relapsed AML samples for clonal outgrowth of EZH2 mutated cells. We identified a 54-year-old patient who gained an EZH2 mutation (p.A692G) at second relapse. A summary of the patients’ course of disease including bone marrow blast counts from 9 time points is shown in Figure 7a. We established a sensitive, custom designed digital droplet PCR (ddPCR) assay to monitor the abundance of p.A692G during disease progression from first to second relapse. We detected the mutation only in the second relapse with a more than 20% increase within three months (Fig. 7a). Furthermore, the patient gained a heterozygous 7q deletion at first relapse, as analysed by MLPA (Fig. 7a).
Figure 7.
Resistance in an EZH2 mutated PDX model. (a) Course of disease of an AML patient suffering from two relapses (indicated by dashed vertical lines). 7q deletion was confirmed with MLPA. Variant allele frequency (VAF) of the p.A692G mutation was monitored by digital droplet PCR (time points of samples indicated by red stars). Blast count was measured from bone marrow at the indicated time points (blue bars). Samples used for PDX engraftment are indicated with black triangles. In July 2015 two samples were taken in the same month. (b–c) In vivo treatment of PDX mice. NSG mice were injected with patient material of relapse 1 and 2, establishing (b) PDX-491 and (c) PDX-661. 21 d after injection, mice were treated with AraC (100 mg/kg) and DaunoXome (1 mg/kg) (treatment days indicated with red x). Leukaemic burden was monitored in vivo by bioluminescence imaging. Control mice treated only with PBS are shown in blue. (d–e), Variant allele frequencies in the course of first to second relapse of (d) EZH2/p.A692G and (e) other mutations identified by targeted sequencing in Patient and PDX samples. PDX samples of first engraftment (primograft) as well as first (1st re-Tx) second (2nd re-TX) and third (3rd re-TX) re-transplantation.
Patient cells of the first and second relapse were serially transplanted into immune-deficient mice, establishing patient-derived xenografts PDX-AML-491 and PDX-AML-661 respectively (Fig. 7a)45. Leukaemic cells of the initial diagnosis did not engraft in the model. PDX cells were lentivirally transduced for transgenic expression of luciferase, enabling disease monitoring in vivo36. We treated the xenograft mice with a combination of cytarabine and daunorubicin and monitored the leukaemic burden over 80 days. We observed a drastic drop in leukaemic cells in the PDX-491 mice, with a complete cure of three out of four animals (Fig. 7b). The treatment of PDX-661 mice only had minimal effect (Fig. 7c).
Targeted sequencing of a panel of 68 recurrently mutated genes36 of patient and PDX samples revealed a strong increase of the clone harbouring the EZH2/p.A692G mutation in the PDX-661 samples (VAF: 98.8%) in comparison to the second relapse of the patient (VAF: 39.2%, Fig. 7d). The only other mutation illustrating an increase in variant allele frequency was a subclonal JAK1 mutation, detectable only in the PDX-661 cells. The majority of mutations (BCOR, DNMT3A, ETV6, PTPN11 and RUNX1) remained stable at all time points. Furthermore, two subclonal mutations in NRAS and KRAS were detected. Both were absent in the patient’s first relapse. KRAS was only detectable in the PDX-491 samples, while NRAS decreased during PDX-491 passaging but was detectable again in the PDX-661 as well as in the patient’s second relapse.
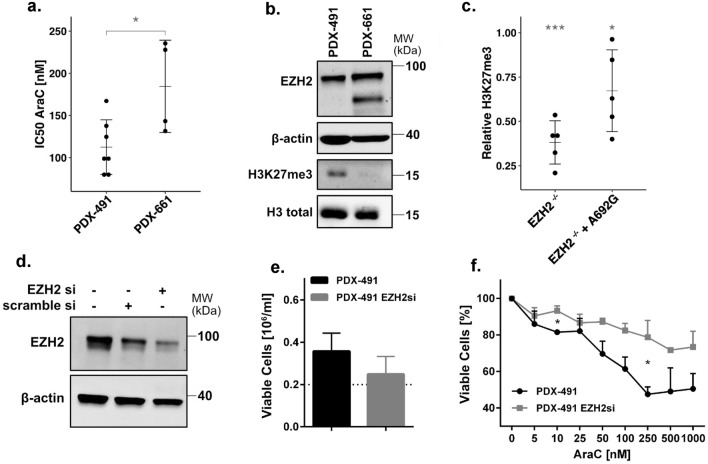
Dose-response analysis of PDX-491 and PDX-661 cells in vitro confirmed an increased resistance of PDX-661 towards AraC (Fig. 8a). Moreover, global H3K37me3 levels were completely depleted in PDX-661 cells, while EZH2 protein expression was stable (Fig. 8b). Transient transfection of the p.A692G mutation into our 293T/EZH2−/− model revealed decreased global H3K27me3 compared to the wild type, further confirming a LOF phenotype (Fig. 8c). To examine if the observed chemoresistance can be caused by EZH2 depletion, we established an siRNA knockdown (kd) targeting wild type EZH2 in the PDX-491 cells. EZH2 levels could thereby be reduced by approximately 40% (Fig. 8d). Treatment of these cells for 72 h with AraC resulted in lower proliferation (Fig. 8e) and an increased resistance (Fig. 8f).
Figure 8.
Knockdown of EZH2 in a patient-derived xenograft (PDX) model. (a) Comparison of IC50 AraC values for PDX-491 and PDX-661 in vitro. (b) Immunoblot of EZH2 expression and global H3K27me3 in PDX-491 and PDX-661. (c) H3K27me3 levels of EZH2/p.A692G in 293T/EZH2−/− cells. Values normalized to H3 loading control and relative to wild type. *indicates significant difference to the wild type. (d) Immunoblot of EZH2 expression in PDX-491 cells treated with 10 nM siRNA. Representative blot shown for two independent experiments. (e) Histogram showing the proliferation of PDX-491 cells with 10 nM siRNA. Cells were pre-treated for 2 d with siRNA and then incubated for another 3 d for the proliferation assay. (f) AraC treatment in PDX-491 cells with 10 nM siRNA. Cells were pre-treated for 2 d with siRNA and then treated for 72 h with AraC. Unpaired, two-tailed Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001. MW, molecular weight. β-actin and H3 total, loading controls. Error bars indicate mean ± s.d of three independent experiments.
Discussion
Development of resistance against standard chemotherapeutics is common in AML and can be induced through various mechanisms46. In this study we report that loss-of-function mutations in the histone methyltransferase EZH2 are associated with increased resistance against the antimetabolite cytarabine (AraC).
EZH2 mutations in AML are a very rare event. In fact, only 4% of our patients harboured these mutations at diagnosis, with the majority located in the catalytic SET domain, a known hotspot for EZH2 mutations28,47. Seemingly, the mutations induce loss of EZH2 function, independent of the type of mutation. (Fig. 3, Supplementary Fig. 3a). Apart from the SET domain (aa 605-725), also the post-SET domain (aa 725-746), which is essential for the formation of the cofactor S-adenosyl-L-methionine (SAM) binding pocket, was found crucial in maintaining enzymatic function48. Two of the mutations identified in our patients, D730_delinsX and Y733LfsX6, previously described in myelodysplastic syndromes (MDS)49, caused almost complete elimination of the post-SET domain, while two others, I744fs50,51 and G743fs, caused frame shifts that resulted in elongated protein variants, highlighting the importance of this domain. In contrast, the missense mutation K574E, located in the CXC domain, is likely to impair the domain's binding ability to the substrate nucleosome and thereby bringing the H3 tail out of reach50.
We found that complete loss of EZH2 promotes AraC resistance in HEK293T cells as well as the myeloid cell line K562 (Fig. 3, Fig. 4). Furthermore, increased resistance was observed in K562 cells expressing the LOF mutation Y733LfsX6 (Fig. 5d–f) and in a PDX model of a patient who gained the LOF mutation A692G at second relapse (Fig. 7). Additionally, low EZH2 mRNA expression correlated with poor overall and relapse-free survival (Fig. 1b–c). Our findings are therefore in concordance with the study of Göllner et al.34, who described AraC resistance in a shRNA knockdown of EZH2 in MV4-11 cells. However, elevated EZH2 expression has also been reported in AML patients52,53, and dual inhibition of EZH1/2 was found to eliminate quiescent leukaemic stem cells (LSCs) to prevent relapse25. These combined findings suggest a dual role of EZH2 as either tumour suppressor or oncogene. In our matched diagnosis/relapse pairs, EZH2 protein and mRNA expression levels were found to be highly patient specific, and in most cases, we observed up- or downregulation in relapse (Fig. 2). EZH2 therefore appears to bear an important function in disease progression, and close monitoring of expression and mutation status seems to be crucial in choosing the best treatment approach.
Interestingly, RUNX1 and ASXL1 mutations were significantly co-occurring with mutations in EZH2. Similar associations have been described before in myeloid malignancies including AML28,54,55. Therapy resistance was associated with frequent co-occurrence of EZH2 and RUNX1 LOF mutations56, suggesting a cooperative role of these mutations. ASXL1 LOF mutations on the other hand can establish an additive effect to EZH2 loss by additional reduction of H3K27 trimethylation through inhibition of PRC2 recruitment29.
Mutations in other PRC2 subunits (EZH1, EED, SUZ12 or RbAp48) are extremely rare in AML. In the cohort of 50 AML patients of Greif et al. (2018) none could be detected, while in a study of 165 AML patients from Faber et al. (2016), only EED mutations were found with a frequency of 1.8 %. Co-occurrence of EED and EZH2 mutations was found in only one of the patients. EZH2 requires direct interaction with EED to exert its enzymatic function57. Thus, also other mutations in the PRC2 complex like EED mutations harbour the potential to confer chemoresistance.
H3K27me3 levels can also be altered by the histone demethylase KDM6A. Loss-of-function mutations in KDM6A have been detected in AML and are associated with the development of chemoresistance42. Although we and other groups found mutations in both genes to be mutually exclusive58, expression levels of KDM6A and EZH2 have an antagonistic effect on global H3K27 trimethylation (Supplementary Fig. 4d). Further research is needed to investigate common and specific EZH2 and KDM6A target sites.
EZH2 is responsible for the trimethylation of H3K27 and therefore inactivation of its target genes. Knockout of EZH2 in K562 cells induced almost complete loss of H3K27me3 levels and resulted in the upregulation of 216 genes and 375 proteins (Fig. 6b). We identified FHL1 and UBE2E1 to be direct targets of EZH2. Overexpression of these genes has recently been described to be involved in resistance against cytarabine, and in relapse in AML patients43,44.
FHL1 might be involved in the transmembrane transport of chemotherapeutic agents. Fu et al.43 found upregulation of ABCC1 and ABCC4, encoding for the unidirectional efflux transporter proteins MRP1 and MRP4, in AML patients with high FHL1 expression. A slight upregulation of ABCC1 protein could also be detected in our data. Interestingly, Fu et al. also found expression of FHL1 to be negatively correlated to SLC29A1 (ENT1) expression. ENT1 is an influx transporter that mediates the uptake of chemotherapeutics and is downregulated upon loss of KDM6A42. Since EZH2 and KDM6A mutations were found to be mutually exclusive, those findings suggest an involvement of either EZH2 or KDM6A in the regulation of transmembrane transporter proteins, responsible for the release or uptake of chemotherapeutic agents. The ubiquitin-conjugating enzyme UBE2E1 can regulate the expression of HOX genes by its ability to ubiquitinate histones59. Although we did not detect any aberrant expression of HOX genes, upregulation of HOXA9 as well as HOXB7 was reported by Göllner et al. in a resistant EZH2 negative AML cell line model. We furthermore identified upregulation of the direct EZH2 target genes CNN3 and AKAP13, that are involved in chemotherapy resistance in colon cancer and breast cancer, respectively60,61, and the genes MYO5A, AKT3 and SPECC1, which are implicated in the evasion of apoptosis62–64. Additionally, upregulation of TPD52, involved in proliferation, migration, invasion and apoptosis, was found in many cancer types including AML65.
We conclude that loss-of-function mutations in the histone methyltransferase EZH2 have the potential to confer resistance against the chemotherapeutic agent cytarabine and suggest an involvement of upregulated EZH2 target genes in apoptosis, proliferation and transmembrane transport.
Materials and methods
Cell culture and patient samples
All cell lines (Supplementary Table 1) were acquired from DSMZ (Braunschweig, Germany) and cultured according to the supplier’s recommendations. Patient-derived xenograft (PDX) AML samples were serially passaged in NSG mice and re-isolated for in vitro cultivation as previously described23,45. Exclusion of mycoplasma contamination was performed continuously during cell culture using the MycoAlert Mycoplasma detection kit (Lonza, Basel, Switzerland). Analysis of patient samples was based on material of AML patients from the AMLCG-99 trial (NCT00266136), AMLCG-2008 trial (NCT01382147), and the Department of Medicine III, University Hospital, LMU. Mononuclear cells were enriched from bone marrow or peripheral blood by Ficoll density gradient centrifugation. Written informed consent for scientific use of sample material was obtained from all patients. The study was performed in accordance with the ethical standards of the responsible committee on human experimentation (written approval by the Research Ethics Boards of the medical faculty of Ludwig-Maximilians-Universität, Munich, number 068-08 and 222-10) and with the Helsinki Declaration of 1975, as revised in 2000. All animal trials were performed in accordance with the current ethical standards of the official committee on animal experimentation (Regierung von Oberbayern, number 55.2-1-54-2531-95-2010 and ROB-55.2Vet-143 2532.Vet_02-16-7).
Proliferation assay
Suspension cells were treated with cytarabine (AraC, Selleck Chemicals, Houston, TX, USA), and daunorubicin (in-house). For short time assays, viable cells were treated once (d0) and counted after 72 h on Vi-Cell Cell Viability Analyzer (Beckman Coulter, Krefeld, Germany). For long-term proliferation assays, cells were treated three times (d0, d4, d8) and viable cells were counted every second day. Unpaired, two-tailed Student’s t-test and calculation of IC50 values were performed using GraphPad Prism version 6.07 (GraphPad Software, La Jolla, CA, USA). PiggyBac23,45,66 (PB)/EZH2 cells were pre-cultured with or without doxycycline (1μg/mL) for 72 h followed by treatment with AraC +/− doxycycline, which was added every 48 h. For knockdown experiments in PDX cells, siRNA targeting EZH2 (#s4918, Thermo Fisher Scientific, Waltham, USA) was transiently transfected (10 nM) via nucleofection (Supplementary Methods). Cells were pre-incubated for 48 h and then treated with AraC for 72 h.
Immunoblotting
Immunoblotting was performed as described before3. The following antibodies were used: anti-EZH2 (#5246, Cell Signaling Technology, Danvers, USA), anti-β-actin (A5441, Sigma Aldrich, St. Louis, USA), anti-H3 (ab1791, Abcam, Cambridge, UK), anti-H3K27me3 (#9733, Cell Signaling Technology, Danvers, USA), anti-SUZ12 (#3737, Cell Signaling Technology, Danvers, USA), anti-RbAP46 (#4522, Cell Signaling Technology, Danvers, USA), anit-EED (ab113911, Abcam, Cambridge, UK), anti-EZH1 (#42088, Cell Signaling Technology, Danvers, USA). Western blots were quantified using ImageJ version 1.50d and levels were normalized to the associated loading control (β-actin for EZH2, total H3 for H3K27me3).
In vivo therapy trial
Patient-derived xenograft (PDX) cells expressing enhanced firefly luciferase and mCherry were established as described previously45. For in vivo therapy trials, 1*105 PDX-AML-491 or 8*105 PDX-AML-661 luciferase-positive cells were injected intravenously into 11 or 16 week old male NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory, Bar Harbour, ME, USA), and tumour growth was regularly monitored by bioluminescence imaging (BLI) as described previously24. 21 days after transplantation, mice were treated with a combination of Cytarabine (AraC; 100 mg/kg, i.p., days 1-4 of therapy weeks) and liposomal daunorubicin (DaunoXome; 1mg/kg, i.v., days 1 and 4 of therapy weeks) every second week for three (AML-661, n = 3) or four (AML-491, n = 4) cycles. Tumour burden was regularly monitored by BLI and compared to untreated control mice. In total, 13 mice were included in this study; one AML-661 control mouse was sacrificed 14 days after injection due to leukaemia unrelated illness. End point of the study was end-stage leukaemia. All animal trials were performed in accordance with the current ethical standards of the official committee on animal experimentation (Regierung von Oberbayern, number 55.2-1-54-2531-95-2010 and ROB-55.2Vet-143 2532.Vet_02-16-7) and in compliance with the ARRIVE guidelines.
Ethics approval
We hereby confirm that all experimental protocols were approved by the Department of Medicine III, University Hospital, LMU Munich, the Department of Biology III and Center for Integrated Protein Science Munich (CIPSM); Human Biology and BioImaging, LMU Munich, Planegg Martinsried, Germany and the Helmholtz Zentrum München, Munich.
Supplementary Information
Acknowledgements
The authors thank all study participants. We thank M. Fritschle for animal handling. We thank Bianka Ksienzyk for cell sorting. We thank Karl-Peter Hopfner for the nice cooperation within mutation characterisation. We thank Dr. Igor Paron and Dr. Christian Deiml from the Proteomics and Signal Transduction Group and the Clinical Proteomics Group at the Max Planck Institute of Biochemistry, Martinsried, for their technical assistance and Michael Wierer und Matthias Mann for their support. K.H.M., P.A.G., G.S., H.L., I.J., and K.S. were supported by the German Research Council (DFG) within the SFB 1243 “Cancer Evolution”. J.M.K., R.M., K.V., L.E.W., M.R. and S.W. are members of the IRTG-1243 within the SFB 1243 of the DFG. TH was supported by the Wilhelm-Sander-Stiftung (Grant 2013.086.2) and the Physician Scientists Grant (G-509200-004) from the Helmholtz Zentrum München. E.U. is a fellow of the International Max Planck Research School for Molecular Life Sciences (IMPRS-LS) and is supported by the research training group 1721 (RTG 1721, DFG).
Author contributions
K.S., J.M.K. and S.W. conceived the study. J.M.K. and S.W. wrote the manuscript and analysed the data. J.M.K. performed all experiments which are not mentioned below. M.D.B. and S.B. supported and supervised CRISPR/Cas9 mediated genome editing experiments. R.M., K.V., M.R., M.F. and H.L. supported the experiments. L.E.W. performed RNA-seq experiments. Mass spectrometry-based proteomics experiments were designed by S.W., M.R. and E.U., performed by E.U. and analysed by S.W. and E.U. B.V. supported and supervised all mice and PDX experiments. K.H.M. performed mutation analysis in AML diagnosis. T.H. performed survival analysis and provided supplementary Fig. 1d. M.S. performed and evaluated MLPA analysis. K.S., I.J., G.S., W.H. and O.W. interpreted the data and supported the project by coordinating the teams and experiments. All authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The RNA-seq data generated for this study is available at GEO under the accession number: GSE162623 The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE67 partner repository with the dataset identifier PXD023139.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Julia M. Kempf and Sabrina Weser.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-84708-6.
References
- 1.Bradstock KF, et al. A randomized trial of high-versus conventional-dose cytarabine in consolidation chemotherapy for adult de novo acute myeloid leukemia in first remission after induction therapy containing high-dose cytarabine. Blood. 2005;105:481–488. doi: 10.1182/blood-2004-01-0326. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, et al. Meta-analysis of randomised clinical trials comparing idarubicin + cytarabine with daunorubicin + cytarabine as the induction chemotherapy in patients with newly diagnosed acute myeloid leukaemia. PLoS One. 2013;8:e60699. doi: 10.1371/journal.pone.0060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hann IM, et al. Randomized Comparison of DAT Versus ADE as Induction Chemotherapy in Children and Younger Adults With Acute Myeloid Leukemia Results of the Medical Research Council’s 10th AML Trial (MRC AML10) Blood. 1997;89:2311–2318. doi: 10.1182/blood.V89.7.2311. [DOI] [PubMed] [Google Scholar]
- 4.Marcucci G, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanwal, R., Gupta, K. & Gupta, S. Cancer Epigenetics: An Introduction. in Cancer Epigenetics: Risk Assessment, Diagnosis, Treatment, and Prognosis (ed. Verma, M.) 3–25 (Springer New York, 2015).
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Stahl M, et al. Hypomethylating agents in relapsed and refractory AML: outcomes and their predictors in a large international patient cohort. Blood Adv. 2018;2:923–932. doi: 10.1182/bloodadvances.2018016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cashen AF, Schiller GJ, O’Donnell MR, DiPersio JF. Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J. Clin. Oncol. 2010;28:556–561. doi: 10.1200/JCO.2009.23.9178. [DOI] [PubMed] [Google Scholar]
- 10.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 12.Chang C-J, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behrens C, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin. Cancer Res. 2013;19:6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 15.Zingg D, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 16.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bödör C, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013;122:3165–3168. doi: 10.1182/blood-2013-04-496893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabello D, et al. Overexpression of EZH2 associates with a poor prognosis in chronic lymphocytic leukemia. Blood Cells Mol. Dis. 2015;54:97–102. doi: 10.1016/j.bcmd.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 19.D’Angelo V, et al. EZH2 is increased in paediatric T-cell acute lymphoblastic leukemia and is a suitable molecular target in combination treatment approaches. J. Exp. Clin. Cancer Res. 2015;34:83. doi: 10.1186/s13046-015-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishioka C, Ikezoe T, Yang J, Yokoyama A. BCR/ABL increases EZH2 levels which regulates XIAP expression via miRNA-219 in chronic myeloid leukemia cells. Leuk. Res. 2016;45:24–32. doi: 10.1016/j.leukres.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon C, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basheer F, et al. Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J. Exp. Med. 2019;216:966–981. doi: 10.1084/jem.20181276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mochizuki-Kashio M, et al. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood. 2015;126:1172–1183. doi: 10.1182/blood-2015-03-634428. [DOI] [PubMed] [Google Scholar]
- 25.Fujita S, et al. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia. 2018;32:855–864. doi: 10.1038/leu.2017.300. [DOI] [PubMed] [Google Scholar]
- 26.Neff T, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu Z, et al. Loss of EZH2 reprograms BCAA metabolism to drive leukemic transformation. Cancer Discov. 2019;9:1228–1247. doi: 10.1158/2159-8290.CD-19-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 29.Abdel-Wahab O, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sashida G, et al. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J. Exp. Med. 2016;213:1459–1477. doi: 10.1084/jem.20151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, et al. TET2, ASXL1 and EZH2 mutations in Chinese with myelodysplastic syndromes. Leuk. Res. 2013;37:305–311. doi: 10.1016/j.leukres.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Grossmann V, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 34.Göllner S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat. Med. 2017;23:69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greif PA, et al. Evolution of cytogenetically normal acute myeloid leukemia during therapy and relapse: an exome sequencing study of 50 patients. Clin. Cancer Res. 2018;24:1716–1726. doi: 10.1158/1078-0432.CCR-17-2344. [DOI] [PubMed] [Google Scholar]
- 36.Metzeler KH, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 37.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J. Clin. Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wouters BJ, et al. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stief SM, et al. Loss of KDM6A confers drug resistance in acute myeloid leukemia. Leukemia. 2019 doi: 10.1038/s41375-019-0497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y, et al. Genome-wide identification of FHL1 as a powerful prognostic candidate and potential therapeutic target in acute myeloid leukaemia. EBioMedicine. 2020;52:102664. doi: 10.1016/j.ebiom.2020.102664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo H, et al. Microarray-based analysis and clinical validation identify ubiquitin-conjugating enzyme E2E1 (UBE2E1) as a prognostic factor in acute myeloid leukemia. J. Hematol. Oncol. 2016;9:125. doi: 10.1186/s13045-016-0356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vick B, et al. An advanced preclinical mouse model for acute myeloid leukemia using patients’ cells of various genetic subgroups and in vivo bioluminescence imaging. PLoS One. 2015;10:e0120925. doi: 10.1371/journal.pone.0120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Gu Y, Chen B. Mechanisms of drug resistance in acute myeloid leukemia. Onco. Targets. Ther. 2019;12:1937–1945. doi: 10.2147/OTT.S191621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guglielmelli P, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118:5227–5234. doi: 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One. 2013;8:e83737. doi: 10.1371/journal.pone.0083737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 50.Poepsel S, Kasinath V, Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 2018;25:154–162. doi: 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grubach L, et al. Gene expression profiling of Polycomb, Hox and Meis genes in patients with acute myeloid leukaemia. Eur. J. Haematol. 2008;81:112–122. doi: 10.1111/j.1600-0609.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- 53.Wen S, et al. Novel combination of histone methylation modulators with therapeutic synergy against acute myeloid leukemia in vitro and in vivo. Cancer Lett. 2018;413:35–45. doi: 10.1016/j.canlet.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 54.Gaidzik VI, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–2168. doi: 10.1038/leu.2016.126. [DOI] [PubMed] [Google Scholar]
- 55.Rinke J, et al. Molecular characterization of EZH2 mutant patients with myelodysplastic/myeloproliferative neoplasms. Leukemia. 2017;31:1936–1943. doi: 10.1038/leu.2017.190. [DOI] [PubMed] [Google Scholar]
- 56.Booth CAG, et al. Ezh2 and Runx1 mutations collaborate to initiate lympho-myeloid leukemia in early thymic progenitors. Cancer Cell. 2018;33:274–291.e8. doi: 10.1016/j.ccell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Ueda T, et al. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia. 2012;26:2557–2560. doi: 10.1038/leu.2012.146. [DOI] [PubMed] [Google Scholar]
- 58.Khan SN, et al. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia. 2013;27:1301–1309. doi: 10.1038/leu.2013.80. [DOI] [PubMed] [Google Scholar]
- 59.Zhu B, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 60.Nair VA, Al-Khayyal NA, Sivaperumal S, Abdel-Rahman WM. Calponin 3 promotes invasion and drug resistance of colon cancer cells. World J. Gastrointest. Oncol. 2019;11:971–982. doi: 10.4251/wjgo.v11.i11.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bentin Toaldo C, et al. Protein Kinase A-induced tamoxifen resistance is mediated by anchoring protein AKAP13. BMC Cancer. 2015;15:588. doi: 10.1186/s12885-015-1591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alves CP, et al. Myosin-Va contributes to manifestation of malignant-related properties in melanoma cells. J. Invest. Dermatol. 2013;133:2809–2812. doi: 10.1038/jid.2013.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hinz N, Jücker M. Distinct functions of AKT isoforms in breast cancer: a comprehensive review. Cell Commun. Signal. 2019;17:154. doi: 10.1186/s12964-019-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Agostino L, Giordano A. A novel dual signaling axis for NSP 5a3a induced apoptosis in head and neck carcinoma. Oncotarget. 2011;2:1055–1074. doi: 10.18632/oncotarget.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ha M, et al. Prognostic role of TPD52 in acute myeloid leukemia: a retrospective multicohort analysis. J. Cell. Biochem. 2019;120:3672–3678. doi: 10.1002/jcb.27645. [DOI] [PubMed] [Google Scholar]
- 66.Mulholland, C. B. et al. Recent evolution of a TET-controlled and DPPA3/STELLA-driven pathway of passive demethylation in mammals. bioRxiv 321604 (2020) 10.1101/321604.
- 67.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yılmaz Ş, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated for this study is available at GEO under the accession number: GSE162623 The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE67 partner repository with the dataset identifier PXD023139.