Abstract
Proton therapy allows to avoid excess radiation dose on normal tissues. However, there are some limitations. Indeed, passive delivery of proton beams results in an increase in the lateral dose upstream of the tumor and active scanning leads to strong differences in dose delivery. This study aims to assess possible differences in the transcriptomic response of skin in C57BL/6 mice after TBI irradiation by active or passive proton beams at the dose of 6 Gy compared to unirradiated mice. In that purpose, total RNA was extracted from skin samples 3 months after irradiation and RNA-Seq was performed. Results showed that active and passive delivery lead to completely different transcription profiles. Indeed, 140 and 167 genes were differentially expressed after active and passive scanning compared to unirradiated, respectively, with only one common gene corresponding to RIKEN cDNA 9930021J03. Moreover, protein–protein interactions performed by STRING analysis showed that 31 and 25 genes are functionally related after active and passive delivery, respectively, with no common gene between both types of proton delivery. Analysis showed that active scanning led to the regulation of genes involved in skin development which was not the case with passive delivery. Moreover, 14 ncRNA were differentially regulated after active scanning against none for passive delivery. Active scanning led to 49 potential mRNA-ncRNA pairs with one ncRNA mainly involved, Gm44383 which is a miRNA. The 43 genes potentially regulated by the miRNA Gm44393 confirmed an important role of active scanning on skin keratin pathway. Our results demonstrated that there are differences in skin gene expression still 3 months after proton irradiation versus unirradiated mouse skin. And strong differences do exist in late skin gene expression between scattered or scanned proton beams. Further investigations are strongly needed to understand this discrepancy and to improve treatments by proton therapy.
Subject terms: Gene expression analysis, Non-coding RNAs, Transcriptomics, Cancer therapy
Introduction
Radiotherapy is a double-edge sword as it can be efficient on tumors but it can also alter normal tissues crossed by the radiations. Proton therapy (PT) should allow to avoid side effects on healthy tissues as the dose deposit is maximal at the end of the beam course under the form of a Bragg peak. Moreover, PT is often indicated for children cancers when organ at risk (OAR) are at proximity of the tumors. However, toxicities are still encountered after treatment by PT. Indeed, several studies on pediatric patients treated by PT for medulloblastoma1, brain tumors2, head and neck cancers3 and lung tumors4 have shown an increase in side effects compared to conventional radiotherapy, including Intensity-modulated radiotherapy.
These toxicities could be linked to the limitations of PT. Indeed, heterogeneity can occur in the lateral penumbra due to the variety of tissues encountered5. Moreover, to treat the entire volume of a tumor, Bragg peaks have to be added (SOBP, Spread-Out Bragg Peak) thus resulting in an increase in the dose received upstream of the tumor where plateau phases are also added6. In addition, PT can be delivered by two different techniques: passive scattering or active scanning. Passive beam delivery is the older technique: dose is delivered by a scattered proton beam which conforms to the tumor by collimators and compensator. This results in an increase in the lateral dose upstream of the tumor in normal tissues7 as shown in patients treated for breast cancer8. Active beam delivery allows a better conformation of the dose to the tumor7 but it leads to strong disparities in dose delivery in normal and tumor tissues in terms of: (1) dose rate (up to several Gy/s in the distal layer of the tumor); (2) duration (some seconds in the distal zone compared to several minutes in the proximal layer); and (3) fractionation (dose delivered at once for the most distal part compared to a lot of times in the proximal part)9. This can result in differences in biological responses in OAR localized near the tumor.
In the literature, there are few studies concerning effects of irradiation in the plateau phase before the Bragg peak of proton beams on transcription in normal tissues. Gridley et al. have shown an increase in gene expression of Prdx6 and Sod3 only after total body irradiation (TBI) proton irradiation in mice10. In the same manner, an increase in the transcription of NOX4 was observed by Chang et al. in hematopoietic stem cells after TBI proton mouse irradiation11. In rat eye exposed to proton beams, Mao et al. have shown an increase in the expression of genes involved in apoptosis and oxidative stress response12. In mouse brain, the expression of genes related to oxidative stress was also changed after protons compared to photons13. In an in vitro model of primary dermal fibroblasts, Nielsen et al. have shown that transcription of genes involved in inflammation and malignant transformation were differentially regulated by protons compared to photons14. And there are even less studies on gene expression after Pencil Beam Scanning (PBS) versus Double Scattering (DS) proton beams. Gridley et al. assessed 152 genes related to p53 and DNA damage pathways in human lung epithelial cells, only 2 genes were differentially expressed by a factor more than 2 in the plateau phase before the Bragg peak : BTG2 and SIAH1 concerning passive delivery, and only one : WT1 for active scanning15.
Conventional paradigm is that ionizing radiations mainly act via DNA damage16. More recent observations led to rethink this theory. Actually, epigenetic effects are one of the most relevant pathways with non-targeted effects (for review,17). Among epigenetic modifications, DNA methylation, histone modifications and non-coding RNAs (ncRNAs) modulations are the most important. Indeed, human genome code for 98% for ncDNA. Resulting ncRNAs, like lncRNAs and miRNA, can regulate mRNAs by inhibiting their translation or by inducing their degradation18,19. miRNAs levels were shown to change after irradiation in in vitro models20–23 and also in vivo24–26. They could play an essential role in cell death and cycle arrest27. Therefore, miRNAs are interesting targets to study as they could be implicated in radiation sensitivity28–32. Khan et al. showed that 2 Gy TBI proton irradiation of mice led to specific patterns of miRNAs according to the organ in brain, testis and liver33.
Our study aims at evaluating transcriptional responses after PBS versus DS proton beams. In this purpose, C57BL/6 which are able to develop side effects after irradiation, were total body irradiated in the plateau phase before the Bragg peak and skin samples were taken 3 months after irradiation to perform transcriptional analysis of total RNA, i.e. mRNA and ncRNA. Sequencing led to the differentially expressed genes (DEGs), STRING analysis revealed potential protein–protein interactions and analysis of GO-terms enrichments allowed to point out implicated pathways. Potential interactions between mRNAs of regulated genes and ncRNAs were also investigated.
Materials and methods
Animals
Ten weeks-old C57BL/6 mice were purchased at Charles River Laboratories (L’Arbresle, France). C57BL/6 mice had been specifically selected for genetic background. C57BL/6 are known to allow the development of late side effects after irradiation unlike other genetic backgrounds that preferentially develop early effects. To overcome the impact of the ovarian cycle and thus avoid a known sex effect in radiation-induced carcinogenesis, experiments were performed on male animals34. Mice were housed in a continuously controlled environment, with enrichment adapted to their species. Seven days were observed between the arrival of the mice and the application of the experimental procedures to allow them to acclimatize to their new environment and thus limit their stress.
Ethics statement
All experimental procedures involving mice were conducted in compliance with the experimental research protocol approved by the ethics committee of the Institut Curie CEEA-IC #118 (authorization APAFiS# 27721-2020101612316744-v3 given by National Authority) and in accordance with the European Union Council (2010/63/UE) and ARRIVE guidelines for the use of laboratory animals.
Total body irradiation with PBS or DS proton beams
Irradiations were performed the same day for both cohorts at the Proton Therapy Center (CPO, Institut Curie, Orsay, France) using a C230 IBA accelerator providing proton beams either DS or PBS. Mice (n = 9 per condition) received a single sub-lethal dose of 6 Gy (physical dose) at an energy of 190.6 MeV in the plateau phase of the Bragg peak. For the IBA DS clinical machine, nozzle-specific settings are automatically chosen in order to achieve the desired field size, range, and modulation width. The range of the DS mode was then set to 23.9 g/cm2 and the modulation to the smallest value: this un-modulated beam was achieved by irradiating without rotating the modulator and placing the beam on the first thickness of the modulation wheel. The PBS energy (190.6 MeV) has been selected in order to ensure that both modes had the same range. The spot size at isocenter was approximately 4.5 mm, and a 5 × 5 cm2 field was created by scanning a single, narrow, monoenergetic proton-beam incident on the central axis with spots arranged in a uniform grid (same weight for all spots) with center-to-center spacing of 3 mm. Several studies have already made comparisons between these two modes35,36, and no significant differences were found for the LETd (dose-averaged linear energy transfer) values between the two modes. In this way, radiation field was the smallest possible to ensure the whole body irradiation and radiation time and linear energy transfer were equivalent for both delivery modes. The mean dose rate used was the standard rate in conventional treatment settings: 6 Gy in 11 s for PBS (32 Gy/min) and 6 Gy in 2.5 min for DS (2.4 Gy/min). Each mouse was irradiated individually. The absolute dose per monitor unit was determined at the center of the field in the plateau region of the pseudo-monoenergetic 190.6 MeV pristine Bragg peak (at a measurement depth of 3 g cm−2) according to IAEA TRS-398 recommendations37. A plane parallel ionization chamber (PPC05, IBA dosimetry, Belgium), with a 9.9 mm diameter sensitive volume was used for absorbed dose-to-water measurements. The chamber was cross-calibrated under reference conditions at the isocenter against a Semiflex-type chamber calibrated under reference conditions in a 60Co beam at the French national metrology institute (CEA-LNHB), in terms of absorbed dose-to-water37,38. Polarity and recombination effects lower than 0.3% were found with this chamber for both modes, and a beam quality correction factor kQ of 1.028 was used. Dose distribution in the mouse is shown in Fig. 1 with a dose color wash using a CT (computed tomography) of a mouse with a 5 cm collimator and a 190.6 MeV Bragg peak (ISOGRAY, DOSISOFT, Cachan, France). Dose inhomogeneity is estimated at ± 5% by dose calculation (pencil beam scanning algorithm) within the mouse. Control mice followed the same course as irradiated mice. Mice were anesthetized with 4% isoflurane before being placed in prone position on a stage positioned in the irradiation field. The legs and tail of the mouse were taped with a flexible adhesive tape to prevent movement. The gas anesthesia was maintained with the mask throughout the irradiation, which will last a maximum of 15 min. Isoflurane was increased to 1.5–2% in 50% air room/50% oxygen. Mouse is detached after irradiation and placed in a clean cage during the awakening time.
Figure 1.
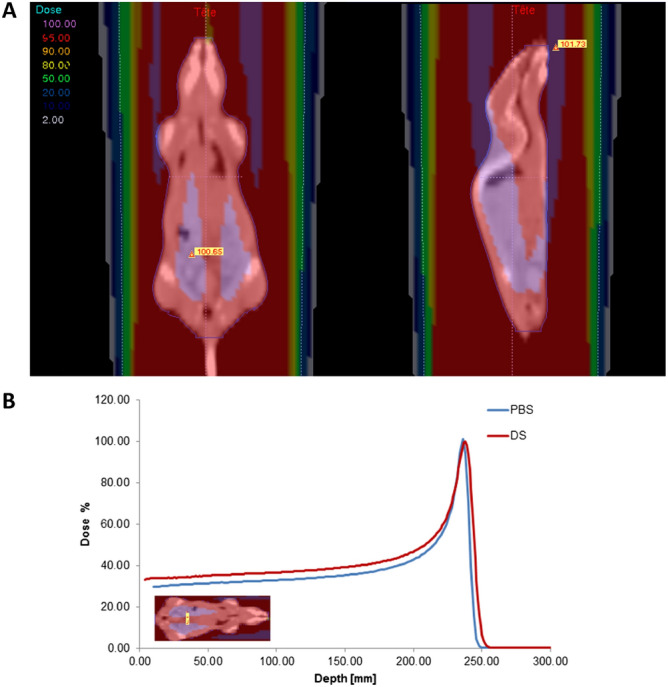
Dose distribution for murine experiments: (A) dose color wash using computed tomography of a mouse for the 5 cm diameter irradiation and double-scattered 190.6 MeV proton beam and (B) the corresponding depth dose distribution is also shown for both irradiation modes.
Follow-up and skin taking
Mice statute was monitored with caution during seven day after irradiation to detect any early signs of distress or ill-being. Mice were sacrificed 3 months after irradiation or when a weight loss greater than 20% of the initial weight was observed. When the mice were sacrificed, total exsanguination was performed to avoid the presence of blood in skin samples. In brief, mice were deeply anesthetized under 5% isoflurane throughout sampling. Rib cage was opened and then the blood was taken by direct puncture into the heart using a 25G needle followed by cervical dislocation after blood sampling. Then, skin from posterior legs (and other organs for another study) were collected.
mRNA and ncRNA sequencing
Total RNAs from posterior leg skin were extracted from 36 different mice (9 per condition). Skin samples have been ground using a cryogenic grinder (SPEX SamplePrep LLC, Methucen, NJ). Obtained powder was put in 1 mL of TRIzol (Ambion, Life Technologies, Carlsbad, CA, USA) before solvent extraction performed according to manufacturer recommendations. RNA concentrations and quality (A260/A280 ratio) were assessed by spectrophotometry (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA, USA) and RNA integrity was evaluated (denaturing gel electrophoresis). The 9 RNA samples per condition were then pooled randomly by three. A twelve-sample stranded library preparation was performed using a NEBNext rRNA-depleted (HMR) kit (New England BioLabs, Ipswich, MA, USA). The sequencing of 100-pb paired-end reads was performed on an Illumina Novaseq 6000 Sequencer (Illumina, San Diego, CA, USA).
RNAseq data analysis
The raw datasets for the twelve libraries were cleaned and trimmed with Trimmomatic v0.39: (using defaults parameters)39. Quantification for the abundances of transcripts was performed using pseudo-aligner Salmon v1.1.040 against cDNA or ncRNA of Mus musculus GRCm38 database. Differential gene expression analyses were performed using TMM41 normalization counts by DEseq2 [Love, 2014] and edgeR42. Significant (p-value adj. < 0.05) differentially expressed genes (DEGs) (fold change ≥ 2) were analyzed. A STRING analysis (v11.0, https://string-db.org) was performed (minimum interaction score > 0.7) to reveal potential protein interaction encoded by DEGs and corresponding GO-terms enrichments43. For identifying ncRNA-mRNA coexpression only pairs of DEGs mRNA-lncRNA with a Pearson correlation coefficient above 0.98 between TMM expression level were kept. LncTar software44 was used to predict potential mRNA-ncRNA interaction (using ndG < -0.20). Putative mRNA-ncRNA interaction network was drawn with CYTOSCAPE (v2.8.3, www.cytoscape.org).
Statistics
Body weights were analyzed by one-way ANOVA (normality and homogeneity of variances were previously checked). Differences were considered significant at P < 0.05. Differential expression analyses and other resulting analyses were conducted by comparing the expression levels of the transcribed between irradiated mice group and their respective non-irradiated control group.
Results
Follow-up of the mice after PBS or DS irradiation
After irradiation, mouse body weight tends to decrease after PBS or DS irradiation before returning to the same level as unirradiated mice several weeks after irradiation (Fig. 2). When comparing PBS and DS irradiation, there is a non-significant trend to a lower weight after DS than after PBS. No mice died or were euthanized due to weight loss greater than 20% of the initial weight. The two control groups of non-irradiated mice showed no significant differences. Moreover, all mice presented hair depigmentation.
Figure 2.
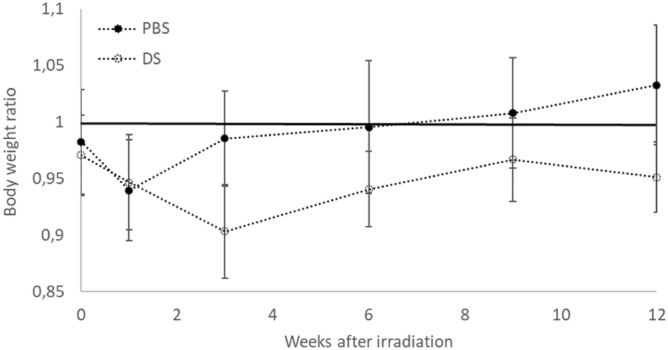
Mouse body weight ratio after PBS or DS 3 months after irradiation. Weight ratio was calculated as follows: irradiated mouse body weight divided by unirradiated mouse body weight. N = 9 in each group. Error bars represent standard error of the mean (SEM).
Overview of the sequencing after PBS or DS irradiation
Table 1 provides an overview of sequencing metrics. Three replicates of pooled total RNAs from 3 different animal were sampled by experiment (DS and PBS) and by dose condition (0 and 6 Gy). In the end, 12 cDNA libraries were sequenced. Despite a lower number of reads in the 6 PBS proton beam samples, the remapping as well as the number of identifications was greater than DS proton beam samples testifying to a greater sample homogeneity within PBS proton beam experiment.
Table 1.
Overview of sequencing, assembly and analysis.
| DS | PBS | |
|---|---|---|
| Total number of bases sequenced | 93,995,555,464 | 85,127,017,962 |
| Reads | 465,324,532 | 421,420,881 |
| Remapping (mRNA + ncRNA) | 74.35% | 77.16% |
| mRNA transcripts identified | 34,093 | 36,747 |
| ncRNA transcripts identified | 19,746 | 19,738 |
| Number of coding DEGs | 167 | 140 |
| Number of non-coding DEGs | 0 | 14 |
| Number of putative interactions ncRNA-mRNA | 0 | 49 |
Quantitative analysis of the differentially expressed gene number after PBS or DS irradiation
Filtering of differentially expressed transcribed (|FC|> 2; padj < 0.05) showed that 140 and 167 genes were still regulated 3 months after the irradiation of mice with PBS and DS proton beam respectively (Fig. 3). Only ENSMUST0000175764.8 corresponding to RIKEN cDNA 9930021J03 gene is regulated by both types of irradiation.
Figure 3.
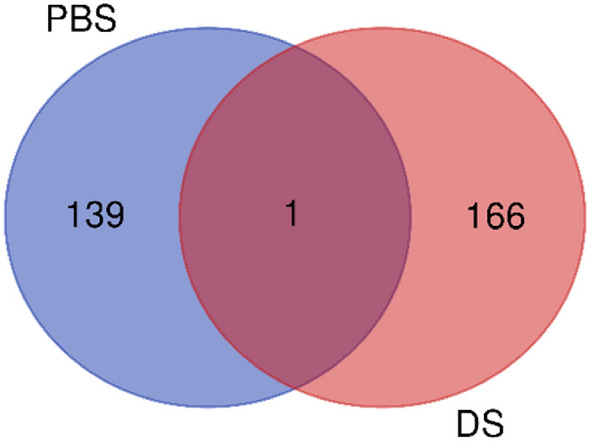
Distribution of differentially expressed genes between irradiated mice and their corresponding control group. Numbers corresponds to transcripts whose expression levels vary by more than a factor 2 and whose adjusted P value is less than 0.05.
List of differentially expressed genes after PBS or DS irradiation
Supplementary Table S1 presents the complete list of mouse differentially expressed transcripts 3 months after irradiation with PBS proton beam and compared to non-irradiated mice group. A 128 transcripts were founded to be overexpressed and 12 downregulated compared to unirradiated controls. Only 4 genes showed two isoforms: Cux1, Padi3, Taf5l and Slc25a37. Keratin-related genes were highly represented: 17 transcripts coding for keratin and 21 for keratin associated proteins.
Supplementary Table S2 presents the complete list of mouse differentially expressed transcripts 3 months after irradiation with DS proton beam and compared to non-irradiated mouse group. A 110 transcripts were founded to be overexpressed and 57 downregulated compared to unirradiated controls. Twelve genes showed two isoforms: Casr, Casc4, Evi2, Hdx, Marveld2, Olfr1258, Retreg2, Serpina3m, Sla, Ubp1, Vmn1r220 and Wdr61.
Supplementary Table S3 presents the list of the 14 mouse differentially expressed non-coding transcripts 3 months after irradiation with DS proton beam and compared to non-irradiated mouse group. Among these 14 non-coding RNAs, all are upregulated compared to control group. Gm44383, Gm44393 and Gm44460 belong to the class of miRNA. No non-coding RNAs passed filtering after irradiation with DS proton beam (|FC|> 2, p-adj < 0.05); ending further analysis.
Protein–protein interactions of regulated genes after PBS or DS irradiation
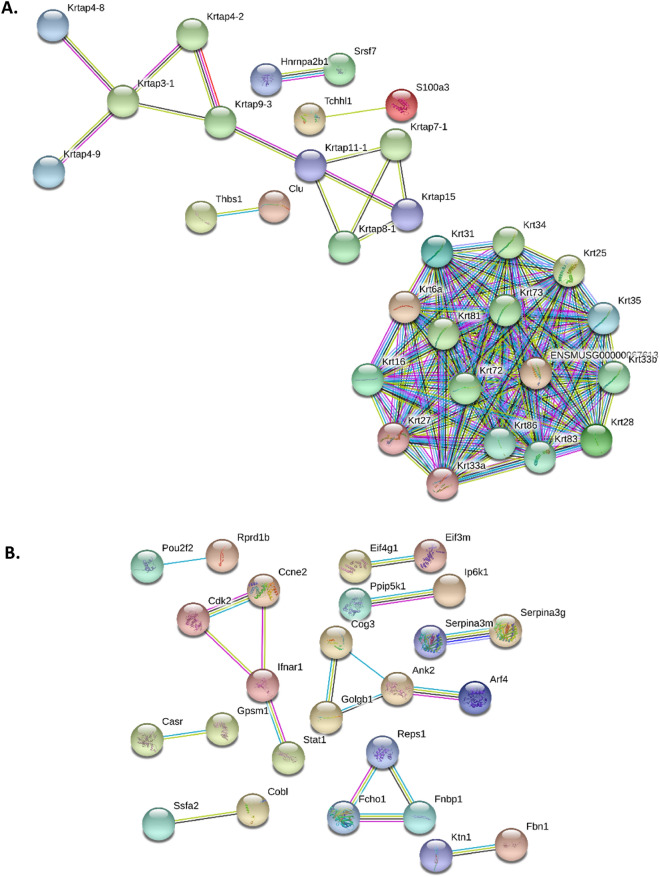
STRING protein–protein interaction analysis revealed that 31 of the 140 and 25 of the 167 genes regulated, after irradiation with PBS (Fig. 4A) and DS (Fig. 4B) proton beam respectively, were functionally related. However, none of the regulated genes were common between the two types of irradiation.
Figure 4.
Protein–protein interaction of mouse skin genes regulated 3 months after (A) PBS or (B) DS irradiation (STRING analysis).
GO-term enrichment of differentially expressed genes after PBS or DS irradiation
Analysis of GO-terms enrichments also confirmed the presence of two very different regulated gene profiles. While genes regulated after irradiation with PBS proton beam were centered on skin development and more precisely the formation of keratin (Table 2), those regulated after DS proton beam were less precise and localized mainly at the organelle level (Table 3).
Table 2.
GO-terms enrichment of differentially expressed genes after PBS irradiation.
| Biological process (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0042633 | Hair cycle | 7 of 95 | 0.0032 |
| GO:0008544 | Epidermis development | 10 of 237 | 0.0032 |
| GO:0043588 | Skin development | 9 of 220 | 0.0035 |
| Molecular function (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0005198 | structural molecule activity | 20 of 546 | 2.40e−08 |
| Cellular component (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0005882 | Intermediate filament | 26 of 120 | 1.65e−29 |
| GO:0045111 | Intermediate filament Cytoskeleton | 27 of 158 | 1.47e−28 |
| GO:0099513 | Polymeric cytoskeletal fiber | 29 of 581 | 5.32e−17 |
| GO:0099512 | Supramolecular fiber | 30 of 809 | 2.25e−14 |
| GO:0045095 | Keratin filament | 11 of 43 | 3.95e−13 |
| GO:0044430 | Cytoskeletal part | 31 of 1460 | 5.53e−09 |
| GO:0005856 | Cytoskeleton | 34 of 1933 | 6.68e−08 |
| GO:0043232 | Intracellular non-membrane-bounded organelle | 43 of 3809 | 9.94e−05 |
| GO:0005903 | Brush border | 6 of 133 | 0.0035 |
Table 3.
GO-terms enrichment of differentially expressed genes after DS irradiation.
| Biological process (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| – | |||
| Molecular function (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0005515 | Protein binding | 58 of 6456 | 0.0319 |
| GO:0005488 | Binding | 83 of 10,884 | 0.0469 |
| Cellular component (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0044464 | Cell part | 101 of 14,017 | 0.0043 |
| GO:0044424 | Intracellular part | 93 of 12,219 | 0.0043 |
| GO:0043231 | Intracellular membrane-bounded organelle | 76 of 9088 | 0.0043 |
| GO:0043229 | Intracellular organelle | 85 of 10,645 | 0.0043 |
| GO:0043227 | Membrane-bounded organelle | 80 of 9775 | 0.0043 |
| GO:0043226 | Organelle | 87 of 10,897 | 0.0043 |
| GO:0005623 | Cell | 101 of 14,044 | 0.0043 |
| GO:0005622 | Intracellular | 94 of 12,462 | 0.0043 |
| GO:0097135 | Cyclin E2-CDK2 complex | 2 of 2 | 0.0084 |
| GO:0005634 | Nucleus | 54 of 6086 | 0.0087 |
| GO:0005737 | Cytoplasm | 76 of 9909 | 0.0192 |
| GO:0044444 | Cytoplasmic part | 62 of 7673 | 0.0267 |
| GO:0044425 | Membrane part | 50 of 5857 | 0.0349 |
| GO:0016020 | Membrane | 60 of 7460 | 0.0349 |
Potential interactions between regulated gene mRNAs and non-coding RNAs after PBS irradiation
No non-coding RNAs passed filtering after irradiation with DS proton beam. After PBS irradiation, the combined analysis of the correlation of expression levels and theoretical required hybridization energy leaded to identify 49 potential mRNA-ncRNA pairs (Table 4).
Table 4.
Theoretical hybridization energy required to form ncRNA-mRNA pairs after PBS irradiation.
| Query | Length_Query | Target | Length_Target | dG | ndG |
|---|---|---|---|---|---|
| Gm44393 | 138 | Krtap1-4 | 593 | − 19.08 | − 0.2 |
| Gm44393 | 138 | Slc7a8 | 4084 | − 20.67 | − 0.2 |
| Gm44393 | 138 | Hoxc13 | 2461 | − 19.52 | − 0.2 |
| Gm44393 | 138 | Krtap1-5 | 1041 | − 18.17 | − 0.2 |
| Gm44393 | 138 | Krtap4-16 | 954 | − 17.41 | − 0.2 |
| Gm44393 | 138 | Krt31 | 1581 | − 19.17 | − 0.2 |
| Gm44393 | 138 | Krt34 | 1609 | − 19.17 | − 0.2 |
| Gm44393 | 138 | Krtap4-7 | 984 | − 17.81 | − 0.2 |
| Gm44393 | 138 | Krt28 | 1624 | − 20.46 | − 0.2 |
| Gm44393 | 138 | Krtap16-1 | 1888 | − 18.65 | − 0.2 |
| Gm44393 | 138 | Krtap4-9 | 1059 | − 18.71 | − 0.2 |
| Gm44393 | 138 | Capn12 | 3040 | − 21.05 | − 0.2 |
| Gm44393 | 138 | Dlx3 | 2607 | − 21.23 | − 0.2 |
| Gm44393 | 138 | Krtap4-2 | 974 | − 18.20 | − 0.2 |
| Gm44393 | 138 | Gm11938 | 663 | − 20.76 | − 0.2 |
| Gm44393 | 138 | Gm11562 | 817 | − 20.77 | − 0.2 |
| Gm44393 | 138 | Krtap2-4 | 828 | − 20.77 | − 0.2 |
| Gm44393 | 138 | Gm11937 | 396 | − 20.77 | − 0.2 |
| Gm44393 | 138 | Gprc5d | 1317 | − 14.67 | − 0.2 |
| Gm44393 | 138 | Gjb2 | 2406 | − 17.81 | − 0.2 |
| Gm44393 | 138 | Krtap9-3 | 759 | − 20.79 | − 0.2 |
| Gm44393 | 138 | Pinlyp | 843 | − 19.13 | − 0.2 |
| Gm44393 | 138 | Gm11567 | 1043 | − 18.79 | − 0.2 |
| Gm44460 | 129 | Ucp2 | 1116 | − 18.97 | − 0.2 |
| Gm44393 | 138 | Krt35 | 1722 | − 20.46 | − 0.2 |
| Gm44460 | 129 | Marcksl1 | 1605 | − 19.99 | − 0.2 |
| Gm44393 | 138 | Krtap3-2 | 1031 | − 18.26 | − 0.2 |
| Gm44460 | 129 | Gja1 | 3071 | − 17.45 | − 0.2 |
| Gm44393 | 138 | Krtap1-3 | 879 | − 21.63 | − 0.2 |
| Gm44383 | 129 | Tnc | 7100 | − 19.86 | − 0.2 |
| Gm44393 | 138 | Dsg4 | 3478 | − 24.15 | − 0.2 |
| Gm44393 | 138 | Padi1 | 3754 | − 20.22 | − 0.2 |
| Gm44393 | 138 | Atp12a | 3950 | − 25.21 | − 0.2 |
| Gm44393 | 138 | Padi3 | 3072 | − 24.83 | − 0.2 |
| Gm44393 | 138 | Cpm | 5195 | − 19.25 | − 0.2 |
| Gm44393 | 138 | Krt27 | 1550 | − 22.84 | − 0.2 |
| Gm44393 | 138 | Tchh | 5823 | − 29.20 | − 0.2 |
| Gm44393 | 138 | Krt33b | 1581 | − 19.68 | − 0.2 |
| Gm44393 | 138 | S100a3 | 712 | − 19.14 | − 0.2 |
| Gm44393 | 138 | Fhod3 | 4956 | − 30.56 | − 0.2 |
| Gm44460 | 129 | Slc25a37 | 5927 | − 23.27 | − 0.3 |
| Gm44393 | 138 | Krt87 | 2278 | − 57.09 | − 0.4 |
| Gm44393 | 138 | Krt72 | 1953 | − 60.81 | − 0.5 |
| Gm44393 | 138 | Krt83 | 1757 | − 60.84 | − 0.5 |
| Gm44393 | 138 | Krt81 | 1843 | − 61.96 | − 0.5 |
| Gm44393 | 138 | Krt86 | 1963 | − 61.96 | − 0.5 |
| Gm44393 | 138 | Krt71 | 2186 | − 61.62 | − 0.5 |
| Gm13032 | 2001 | Padi1 | 3754 | − 1344.53 | − 0.7 |
| Gm44393 | 138 | Krt73 | 2168 | − 93.22 | − 0.7 |
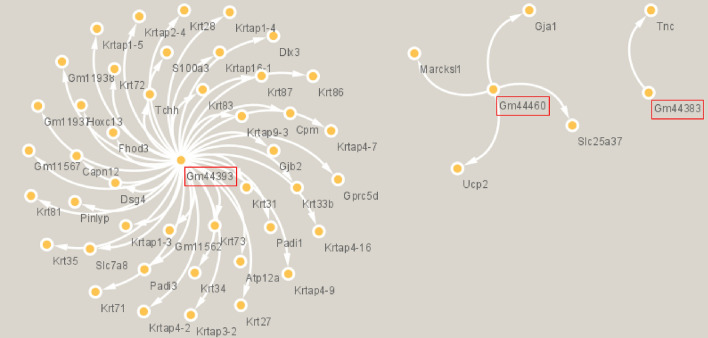
Among the 14 ncRNAs differentially expressed, 3 ncRNAs (Gm44393, Gm44460 and Gm44383) were identified as potential mRNA regulators (Fig. 5). Gm44393 should be implicated in 43 of the 49 potentially regulation identified.
Figure 5.
Representation of potential mRNA regulations by the 3 miRNAs identified after PBS irradiation (CYTOSCAPE analysis). Red rectangles represent the 3 miRNAs potentially involved in mRNA regulation.
Keratinization as a major actor of the late effects observed with PBS?
Table 5 shows that GO-terms enrichment analysis of the 43 genes potentially regulated by Gm44393 miRNA confirmed the impact of PBS irradiation on keratin formation pathway.
Table 5.
GO-Terms Enrichment of putative Gm44393 linked DEGs.
| Biological process (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0042633 | Hair cycle | 5 of 95 | 0.0017 |
| GO:1905867 | Epididymis development | 2 of 4 | 0.0092 |
| GO:0043588 | Skin development | 5 of 220 | 0.0092 |
| GO:0036414 | Histone citrullination | 2 of 5 | 0.0092 |
| GO:0001942 | Hair follicle development | 4 of 84 | 0.0092 |
| GO:0008544 | Epidermis development | 5 of 237 | 0.0109 |
| Molecular function (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0005198 | Structural molecule activity | 13 of 546 | 1.31e−08 |
| GO:0004668 | Protein-arginine deiminase activity | 2 of 5 | 0.0062 |
| GO:0005243 | Gap junction channel activity | 2 of 14 | 0.0176 |
| Cellular component (GO) | |||
|---|---|---|---|
| GO-term | Description | Count in gene set | False discovery rate |
| GO:0005882 | Intermediate filament | 16 of 120 | 2.49e-22 |
| GO:0045095 | Keratin filament | 9 of 43 | 5.16e-14 |
| GO:0099512 | Supramolecular fiber | 17 of 809 | 1.01e-11 |
| GO:0044430 | Cytoskeletal part | 17 of 1460 | 5.53e-08 |
| GO:0005856 | Cytoskeleton | 17 of 1933 | 2.95e-06 |
| GO:0043232 | Intracellular non-membrane-bounded organelle | 18 of 3809 | 0.0059 |
| GO:0005922 | Connexin complex | 2 of 20 | 0.0095 |
Discussion
This study aims to assess possible differences in the transcriptomic response of skin in C57BL/6 mice after TBI irradiation in the plateau phase of the Bragg peak by active or passive proton beams at the sublethal dose of 6 Gy compared to unirradiated mice. Indeed, few studies have been performed on transcription after these 2 delivery techniques and even less on normal tissues. At the clinical level, Matsubara et al. have shown that beam scanning was not always favorable in patients treated for breast cancer by carbon ions but carbon ions and protons present completely different physical and biological properties45.
Concerning irradiation modalities, the energy (190.6 MeV) was chosen as it was the maximum energy possible with the DS system allowing large field dimensions to be obtained. Indeed, there are limitations in the combination of range, modulation and field size in the clinical DS mode. Moreover, this high energy allowed to have a dose that is as homogeneous as possible in the plateau region, at the entrance of the Bragg peak.
All irradiated mice (active and passive scanning) did not present significant change in their weight during the 3 months of breeding, which confirms that the chosen TBI sub-lethal dose of 6 Gy is adequate to see the appearance of late effects. Indeed, weight loss is a side effect widely described for a very long time in irradiated mice as TBI causes digestive system failure46. After a trend to a decrease after irradiation, mouse weight returned to basal level for both delivery modes. When comparing active and passive delivery, there was a non-significant trend to a lower weight gain from 3 weeks up to 3 months after the passive irradiation compared to the active one. This could be related to the difference in dose rate and delivery between PBS and DS. Indeed, PBS irradiation occurred by pulses and, as described in a previous work on Balb/c mice exposed to X-ray pulsed dose rate versus conventional radiotherapy, pulsed irradiation was shown to limit weight loss compared to conventional radiotherapy47. Further investigations are needed with a higher number of animals and with later times of observations as it could indicate that scattered beams are less well tolerated.
Concerning differential expression procedure and sample size, we followed recommendations as described by Schurch et al.48. Sequencing depth being also a critical point, 9 animals were included per condition, 50 M reads were generated per samples and gene differential expression calculation with DEseq2 (|FC|> 2, padj < 0.05) were performed. The study of DEGs 3 months after irradiation made it possible to identify impairs in gene expression. The total number of DEGs was relatively close: 140 and 167 genes were differentially expressed after active and passive scanning compared to unirradiated, respectively. Concerning the identified ncRNAs, no transcripts could be identified after passive scanning compared to unirradiated. This absence of regulators testifies to the proximity of the expression profiles between unirradiated mice and mice irradiated with passive scattering. We should think that major part of the DEGs would be common to the 2 types of irradiation. However, a single gene is commonly differentially expressed, i.e. RIKEN cDNA 9930021J03. In the literature, a study on human lung epithelial cells exposed to active or proton beams before the Bragg peak has also shown that transcription profiles were completely different between both techniques15.
This first clear difference in DEGs (Supplementary Tables S1 and S2) could have its origin in the physicochemical properties of the 2 types of irradiations. Indeed, if the protons are identical and the doses equivalent from a physical point of view, their mode of delivery is different. Passive scattering and active scanning lead to strong differences in dose delivery7,9. Secondarily, the DEGs after passive scattering did not seem to react to a particular stimulus as evidenced by the absence of GO-terms enrichment in the biological process category. These results highlight again the relatively close profiles between irradiated mice and unirradiated controls. In opposition, it is very clear that skin cells of mice irradiated with a proton beam actively scanned continue to respond to particular stimuli inducing an activation of the synthesis of 17 cytoskeletal and cuticular keratins of type I and II (Supplementary Tables S1 and S2) 3 months after the irradiation. These results should be waited as irradiation is known to induce acute keratin gene expression impair49. But we could expect 3 months after irradiation a return to basal state of keratinocyte with an expression of Keratin 5. Instead of that, Kr6 and Kr16 were overexpressed highlighting that keratinocyte activation was occurring and due to cutaneous late damage still under wounding50.
After analysis of potential interactions between DEGs transcripts and differentially expressed ncRNAs, 3 miRNAs were highlighted: Gm44393, Gm44460 and Gm44383 (Supplementary Table S3). Gm44393 seems to be of particular interest as it appears to interact with 43 of the 49 identified DEGs transcripts. The miRNA Gm44393 appears to target genes of keratinization pathway. Strategies based on miRNA expression modulation are promising tools to new treatment protocols. Jiang et al.51 have discovered a specific miRNA pattern expression in psoriatic epidermis. Indeed, miR-486-3p is not expressed allowing Keratin 17 protein overexpression and leading to the pathogenesis of psoriasis. miRNA Gm44393 activation specifically after active scanning proton beam irradiation is a step forward in fundamental knowledge of healthy tissue recovery and bring new perspectives on the modulation of PT cutaneous side effects.
To conclude, this study highlighted at the transcriptomic level great differences in skin response 3 months after irradiation. Profiles of DEGs were very distinct. On one hand, after passive proton beam, mRNAs expression was slightly different from control and no differences could be observed in ncRNAs. On the other hand, active scanning led to an overexpression of mRNAs related to keratin and putative miRNAs keratin regulators. Future investigations are strongly needed to explain the differences in biological responses observed between PBS and DS. Indeed, differences were also recently shown in terms of genotoxicity, oxidative stress and inflammation in various organs in total body irradiated C57BL/6 mice exposed to scattered versus scanned proton beams52. These findings pointed out the absolute need to adapt the PT protocols according to the type of scanning.
Supplementary Information
Acknowledgements
The authors would like to thank Pr Marc-André Mahé, head of the Cancer Centre François Baclesse.
Author contributions
Conceptualization, C.L., F.P., L.D.M., F.M.C., F.S., D.S. and J.-L.H.; methodology, C.L., F.P., L.D.M. and F.M.C.; formal analysis, A.L., E.C., S.C., and C.L.; investigation, A.L., S.C., L.D.M., F.P. and C.L.; writing—original draft preparation, A.L. and C.L.; writing—review and editing, E.C., F.S., J.-L.H., F.P., F.M.C., L.D.M. and C.L.; supervision, C.L.; funding acquisition, C.L., F.P., L.D.M., F.M.C., F.S., D.S. and J.-L.H. All authors have read and agreed to the published version of the manuscript.
Funding
The European project CPIER ESPRITS is co-funded by the Normandy County Council and the European Union in the framework of the ERDF-ESF operation programme 2014–2020. This research was also funded by the Cancéropole Nord-Ouest, EDF (RB2018-07) and the National Research Agency under the Programme Investissements d'Avenir—Institut Carnot (ANR-11CARN-008-01).
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained a repeated error, where the unit of measurement “MeV” was incorrectly given as “meV”.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/30/2021
A Correction to this paper has been published: 10.1038/s41598-021-97156-z
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85394-0.
References
- 1.Yock TI, et al. Long-term toxic effects of proton radiotherapy for paediatric medulloblastoma: a phase 2 single-arm study. Lancet Oncol. 2016;17:287–298. doi: 10.1016/S1470-2045(15)00167-9. [DOI] [PubMed] [Google Scholar]
- 2.Gunther JR, et al. Imaging changes in pediatric intracranial ependymoma patients treated with proton beam radiation therapy compared to intensity modulated radiation therapy. Int. J. Radiat. Oncol. 2015;93:54–63. doi: 10.1016/j.ijrobp.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Bölling T, et al. Acute and late side effects to salivary glands and oral mucosa after head and neck radiotherapy in children and adolescents. Results of the “Registry for the evaluation of side effects after radiotherapy in childhood and adolescence”: Side effects after head and neck radiotherapy in children. Head Neck. 2015;37:1137–1141. doi: 10.1002/hed.23715. [DOI] [PubMed] [Google Scholar]
- 4.Green DM, et al. Pulmonary function after treatment for embryonal brain tumors on SJMB03 that included craniospinal irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2015;93:47–53. doi: 10.1016/j.ijrobp.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lomax AJ. Intensity modulated proton therapy and its sensitivity to treatment uncertainties 2: the potential effects of inter-fraction and inter-field motions. Phys. Med. Biol. 2008;53:1043–1056. doi: 10.1088/0031-9155/53/4/015. [DOI] [PubMed] [Google Scholar]
- 6.Laurent C. Oxidative Stress in Hadrontherapy. Nov. Prospects Oxidative Nitrosative Stress. 2018 doi: 10.5772/intechopen.73238. [DOI] [Google Scholar]
- 7.Wang D. A critical appraisal of the clinical utility of proton therapy in oncology. Med. Devices Auckl. NZ. 2015;8:439–446. doi: 10.2147/MDER.S65594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDonald SM, et al. Proton radiotherapy for chest wall and regional lymphatic radiation; dose comparisons and treatment delivery. Radiat. Oncol. 2013;8:71. doi: 10.1186/1748-717X-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Marzi, L. Effets physiques et biologiques des faisceaux de protons balayés : mesures et modélisation pour des balayages séquentiels à haut débit. (Université Paris Saclay (COmUE), 2016).
- 10.Gridley DS, et al. Comparison of proton and electron radiation effects on biological responses in liver, spleen and blood. Int. J. Radiat. Biol. 2011;87:1173–1181. doi: 10.3109/09553002.2011.624393. [DOI] [PubMed] [Google Scholar]
- 11.Chang J, et al. Whole-body proton irradiation causes long-term damage to hematopoietic stem cells in mice. Radiat. Res. 2015;183:240–248. doi: 10.1667/RR13887.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao XW, Green LM, Mekonnen T, Lindsey N, Gridley DS. Gene expression analysis of oxidative stress and apoptosis in proton-irradiated rat retina. Vivo. 2010;24:425–430. [PubMed] [Google Scholar]
- 13.Baluchamy S, et al. Differential oxidative stress gene expression profile in mouse brain after proton exposure. Vitro Cell. Dev. Biol. Anim. 2010;46:718–725. doi: 10.1007/s11626-010-9330-2. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen S, et al. Comparison of coding transcriptomes in fibroblasts irradiated with low and high LET proton beams and cobalt-60 photons. Int. J. Radiat. Oncol. 2019;103:1203–1211. doi: 10.1016/j.ijrobp.2018.11.065. [DOI] [PubMed] [Google Scholar]
- 15.Gridley DS, Pecaut MJ, Mao XW, Wroe AJ, Luo-Owen X. Biological effects of passive versus active scanning proton beams on human lung epithelial cells. Technol. Cancer Res. Treat. 2015;14:81–98. doi: 10.7785/tcrt.2012.500392. [DOI] [PubMed] [Google Scholar]
- 16.Goodhead DT. New radiobiological, radiation risk and radiation protection paradigms. Mutat. Res. Mol. Mech. Mutagen. 2010;687:13–16. doi: 10.1016/j.mrfmmm.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Belli M, Tabocchini MA. Ionizing radiation-induced epigenetic modifications and their relevance to radiation protection. Int. J. Mol. Sci. 2020;21:5993. doi: 10.3390/ijms21175993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 20.Li XH, Ha CT, Fu D, Xiao M. Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS ONE. 2012;7:e48700. doi: 10.1371/journal.pone.0048700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat. Res. Mol. Mech. Mutagen. 2011;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat. Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhry MA, Kreger B, Omaruddin RA. Transcriptional modulation of micro-RNA in human cells differing in radiation sensitivity. Int. J. Radiat. Biol. 2010;86:569–583. doi: 10.3109/09553001003734568. [DOI] [PubMed] [Google Scholar]
- 24.Acharya SS, et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci. Transl. Med. 2015;7:287ra69–287ra69. doi: 10.1126/scitranslmed.aaa6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F, et al. Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci. Rep. 2017;7:44132. doi: 10.1038/srep44132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh SP, et al. Gamma-tocotrienol modulates radiation-induced MicroRNA expression in mouse spleen. Radiat. Res. 2016;185:485. doi: 10.1667/RR14248.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halimi M, Asghari SM, Sariri R, Moslemi D, Parsian H. Cellular response to ionizing radiation: a MicroRNA story. Int. J. Mol. Cell. Med. 2012;1:178–184. [PMC free article] [PubMed] [Google Scholar]
- 28.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr. Opin. Genet. Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong P, Zhang T, He D, Hsieh J-T. MicroRNA-145 modulates tumor sensitivity to radiation in prostate cancer. Radiat. Res. 2015;184:630. doi: 10.1667/RR14185.1. [DOI] [PubMed] [Google Scholar]
- 30.El Bezawy R, et al. miR-205 enhances radiation sensitivity of prostate cancer cells by impairing DNA damage repair through PKCε and ZEB1 inhibition. J. Exp. Clin. Cancer Res. 2019;38:51. doi: 10.1186/s13046-019-1060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhry MA. Radiation-induced microRNA: discovery, functional analysis, and cancer radiotherapy: RADIATION-INDUCED miRNA. J. Cell. Biochem. 2014;115:436–449. doi: 10.1002/jcb.24694. [DOI] [PubMed] [Google Scholar]
- 32.Cellini F, Morganti A, Genovesi D, Silvestris N, Valentini V. Role of MicroRNA in response to ionizing radiations: evidences and potential impact on clinical practice for radiotherapy. Molecules. 2014;19:5379–5401. doi: 10.3390/molecules19045379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan SY, et al. Distinctive microRNA expression signatures in proton-irradiated mice. Mol. Cell. Biochem. 2013;382:225–235. doi: 10.1007/s11010-013-1738-z. [DOI] [PubMed] [Google Scholar]
- 34.Roberts CJ, Morgan GR, Danford N. Effect of hormones on the variation of radiosensitivity in females as measured by induction of chromosomal aberrations. Environ. Health Perspect. 1997;105:1467–1471. doi: 10.1289/ehp.97105s61467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorriaux J, et al. Consistency in quality correction factors for ionization chamber dosimetry in scanned proton beam therapy. Med Phys. 2017;44:4919–4927. doi: 10.1002/mp.12434. [DOI] [PubMed] [Google Scholar]
- 36.Michaelidesová A, et al. In vitro comparison of passive and active clinical proton beams. Int J Mol Sci. 2020;6:5650. doi: 10.3390/ijms21165650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.IAEA. Absorbed dose determination in external beam radiotherapy: an international code of practice for dosimetry based on standards of absorbed dose to water. Tech. Rep. Ser. 398 (2000).
- 38.Newhauser W, Zhang R. The physics of proton therapy. Phys Med Biol. 2015;60:R155–R209. doi: 10.1088/0031-9155/60/8/R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mering C. v. STRING: a database of predicted functional associations between proteins. Nucl. Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, et al. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015;16:806–812. doi: 10.1093/bib/bbu048. [DOI] [PubMed] [Google Scholar]
- 45.Matsubara H, et al. Comparison of passive and scanning irradiation methods for carbon-ion radiotherapy for breast cancer. J. Radiat. Res. (Tokyo) 2018;59:625–631. doi: 10.1093/jrr/rry052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bacq, Z. M. & Alexander, P. Fundamentals of radiobiology (1966).
- 47.Zhang P, et al. Local tumor control and normal tissue toxicity of pulsed low-dose rate radiotherapy for recurrent lung cancer: an in vivo animal study. Dose Response. 2015;13:1559325815588507. doi: 10.1177/1559325815588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schurch NJ, et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA. 2016;22:839–851. doi: 10.1261/rna.053959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nanashima N, Ito K, Ishikawa T, Nakano M, Nakamura T. Damage of hair follicle stem cells and alteration of keratin expression in external radiation-induced acute alopecia. Int. J. Mol. Med. 2012;30:579–584. doi: 10.3892/ijmm.2012.1018. [DOI] [PubMed] [Google Scholar]
- 50.Freedberg IM, Tomic-Canic M, Komine M, Blumenberg M. Keratins and the keratinocyte activation cycle. J. Invest. Dermatol. 2001;116:633–640. doi: 10.1046/j.1523-1747.2001.01327.x. [DOI] [PubMed] [Google Scholar]
- 51.Jiang M, et al. TGFβ/SMAD/MicroRNA-486-3p signaling axis mediates keratin 17 expression and keratinocyte hyperproliferation in psoriasis. J. Invest. Dermatol. 2017;137:2177–2186. doi: 10.1016/j.jid.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Chaouni S, et al. Biological effects of scattered versus scanned proton beams on normal tissues in total body irradiated mice: survival, genotoxicity, Oxidative Stress and Inflammation. Antioxidants. 2020;9:1170. doi: 10.3390/antiox9121170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.