Abstract
Scope
High incidence of inflammatory diseases afflicts the increasing aging-population infringing a great health burden. Dietary flavonoids, including the flavone apigenin, are emerging as important anti-inflammatory nutraceuticals due to their health benefits, lack of adverse effects and reduced costs. MicroRNAs (miRs) play a central role in inflammation by regulating gene expression, yet how dietary ingredients affect miRs is poorly understood. The aim of this study was to identify miRs involved in the anti-inflammatory activity of apigenin and apigenin-rich diets and determine their immune regulatory mechanisms in macrophages and in vivo.
Methods and results
A high-throughput quantitative real-time PCR screen of 312 miRs in macrophages revealed that apigenin reduced LPS-induced miR-155 expression. Analyses of miR-155 precursor and primary transcript indicated that apigenin regulated miR-155 transcriptionally. Apigenin-reduced expression of miR-155, led to the increase of anti-inflammatory regulators FOXO3a and SMAD2 in LPS-treated macrophages. In vivo, apigenin or a celery-based apigenin rich diet reduced LPS-induced expression of miR-155 and decreased TNFα in lungs from LPS-treated mice.
Conclusion
These results demonstrate that apigenin and apigenin-rich diets exert effective anti-inflammatory activity in vivo by reducing LPS-induced expression of miR-155, thereby restoring immune balance.
Keywords: flavonoids, sepsis, macrophages, microRNAs, NF-κB, TNFα
1 Introduction
Inflammation is the first line of defense against pathogens and its proper regulation is essential for physiological homeostasis [1]. Dysregulated inflammation, a major contributor to the pathophysiology of sepsis and cardiovascular diseases, is characterized by exacerbated production of inflammatory mediators and uncontrolled immune function [2, 3]. Several non-steroidal anti-inflammatory drugs (NSAIDs) are currently used to ameliorate inflammation, but their long-term consumption is often accompanied by adverse effects [4]. Hence, as the incidence of inflammatory diseases increases worldwide, there is great interest in identifying alternative approaches to restore proper immune balance. Dietary compounds with health beneficial activities (nutraceuticals) are emerging as potential immune modulators, due to their lack of adverse effects, low cost and easy administration [5].
Inflammatory stimuli, including microbial components such as lipopolysaccharide (LPS), trigger a signaling cascade that activates Nuclear Factor Kappa-light-chain-enhancer of activated B cells (NF-κB), a main transcription factor [6]. NF-κB increases the expression of inflammatory regulators including pro-inflammatory cytokines such as tumor necrosis factor α (TNFα) and microRNAs (miRs) [7]. MiRs are single-stranded non-coding RNAs that modulate gene expression by binding to the complementary regions of specific mRNA targets enabling mRNA degradation or inhibiting translation [8]. MiRs are transcribed as long primary transcripts or pri-miRs that are processed into ~60–100 nucleotide hairpins, named precursors or pre-miRs. Pre-miRs are further processed by the endoribonuclease Dicer into mature miRs, averaging 18–22 nucleotides in length [8].
MiRs can act as positive or negative regulators of inflammation [7]. MiR-155 is induced by the NF-κB axis and plays a central role by regulating the duration and intensity of the immune response [9]. Several studies demonstrated that miR-155 regulates TNFα expression levels, a main immune-regulator, by increasing mRNA stability and translation [10–13]. In addition, other targets of miR-155, such as SMAD2 (smooth-muscle-actin and MAD-related 2) [14], a suppressor of the inflammatory molecules TNFα and iNOS (inducible nitric oxide synthase) [15], and FOXO3a (Forkhead Box O3) [16], an inhibitor of NF-κB [17], are also important modulators of inflammation. Yet, despite the central role of miRs in inflammation, how anti-inflammatory dietary compounds affect miRs remains limited studied.
Flavonoids are abundant dietary nutraceuticals [18]. Apigenin (4’,5,7-trihydroxyflavone) is a flavonoid abundant in parsley, celery and chamomile tea [18, 19]. Epidemiological studies have correlated the intake of apigenin with a lower incidence of ovarian cancer and cardiovascular disorders [20, 21]. Apigenin has anti-proliferative and anti-inflammatory activities [22–25]. We showed that apigenin decreases LPS-induced lethality by reducing NF-κB activity and TNFα production [24]. In addition, apigenin reduced LPS-induced endothelial cell death by restoring normal metabolic function [26]. Highlighting the specificity of apigenin, naringenin, a structurally related flavonoid, lacks anti-proliferative and anti-inflammatory activities [22, 27]. Apigenin, similar to other flavonoids, is usually found in plants linked to sugars (glycosylated) [28]. We reported that glycosylated flavonoids showed reduced absorption and anti-inflammatory activity [29]. To overcome these limitations, we developed celery-based apigenin-rich diets with increased aglycone (non-glycosylated) content [29]. Mice fed with these diets showed increased absorption of apigenin, reaching serum concentrations that effectively reduced inflammation in macrophages [29]. Yet, the immuno-regulatory activity of celery-based apigenin-rich diets in vivo has not been studied.
Here, we used a high-throughput quantitative reverse transcription-PCR (qRT-PCR) screening to identify the miRs differentially regulated by apigenin during LPS-induced inflammation in macrophages. MiR-155 was the only miR affected by apigenin during LPS-induced inflammation. We found that apigenin regulates pri-miR-155 at the transcriptional level. Importantly, we showed that consumption of a celery-based apigenin rich diet results in the reduction of LPS-induced miR-155 and inflammatory modulators in vivo at inflammatory organ sites. These studies demonstrate that celery-based diets rich in apigenin confer immune-regulatory activity in vivo, reaching levels of effectiveness similarly found with pure apigenin and suggest a mechanism by which dietary flavones, through miRs regulation, contribute to restore immune balance.
2 Material and Methods
2.1 Chemical, cell lines and cell culture
LPS (E. coli serotypes O111:B4 and 0128:B8), apigenin, naringenin and DMSO were obtained from Sigma (St. Louis, MO). TRIzol®, DNAse-I, TaqMan® Universal PCR Master Mix, SYBR® Green PCR Master Mix, ThermoScript™ RT-PCR system, TaqMan® miR Reverse Transcription kit and the miR assay kits for murine miR-155 (miR-155-5p strand, assay ID 002571), miR-lethal-7a (miR-let-7a, assay ID 000377) and small nucleolar RNA-202 (snoRNA-202, assay ID 001232) were purchased from Life Technologies (Carlsbad, CA). The mouse TNFα DuoSet ELISA kit was obtained from R&D systems (Minneapolis, MN).
RAW 264.7 murine macrophages (referred hereafter as macrophages) were obtained from ATCC and cultured in endotoxin-free RPMI supplemented with 5% FBS and 1% penicillin/streptomycin, as previously described [24, 29]. Cells (5 × 105 cells/ml) were incubated for 16 h prior treatment with 100 ng/ml LPS (Escherichia coli, O111:B4, Sigma) or diluent DMSO (referred as control) added concurrently with different concentrations of apigenin or celery-based apigenin rich extract (ECE). Specific times and concentrations are mentioned in the legends.
2. 2 High-throughput miRs screening and validation
Total RNA was isolated from 1 × 106 macrophages using 500 µl TRIzol® following manufacture’s recommendations (Life technologies). After lyses, RNA was extracted with 100 µl chloroform followed by precipitation with 200 µl isopropanol and two washes of 75% Ethanol. RNA pellets were air dried and resuspended in 50 µl RNAse-free water. RNA (1 µg) was treated with 1 U of DNAse I for 10 min at room temperature following manufacture’s suggestions (Life technologies). For high-throughput screening of miRs, DNAse-treated RNA (1 µg) was converted to cDNA by priming with a 10 µM mix of 450 looped primers specific to mature mouse miRs (Mega Plex kit, Life Technologies) using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies), following manufacture’s instructions and as previously reported [30]. Quantitative Real-Time PCR (qRT-PCR) was performed using the TaqMan® Universal PCR Master Mix and 312 TaqMan® miR probes in an Applied Biosystems 7900HT qRT-PCR equipped with a 384 well reaction plate and the Sequence Detection System (SDS) 2.2 software (Life Technologies). Liquid-handling and the Zymak Twister robots were used to increase throughput and reduce error, as previously reported [30]. MiRs expression was normalized against 5S rRNA [ΔCt (miR – 5S rRNA)], a common standard when different conditions are compared. Fold change was calculated as: 2−Δ;Ct(treatment)/2−ΔCt(control). MiRs expression levels were fit to a general linear model and statistical significance between treatments was determined by pair-wise comparisons [31]. Results were display by Volcano plots, p-value vs. Log2[Fold Change]), using p-value less than 0.05 and fold change greater than 2 as threshold [32]. Data were analyzed using SAS 9.3 (SAS, Inc; Cary, NC).
For miR validation, 200 ng DNAse-treated RNA was reversed transcribed using TaqMan® miR assay kits for mouse miR-155 and miR-let-7a following manufacturer’s conditions and expression determined by qRT-PCR in an Applied Biosystems 7900HT qRT-PCR system using a 96 well reaction plate and following conditions: 40 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min. Validated miR-155 and miR-let-7a expression levels were normalized to the expression of the internal control snoRNA-202, [ΔCt (miR – snoRNA202)]. Fold change was calculated as: 2−ΔCt(treatment)/2−ΔCt(control).
2.3 Analyses of miR-155 immature forms and targets
DNAse-treated RNA (200 ng) was reverse transcribed to cDNA using the ThermoScript RT-PCR system according to manufacture’s instructions. cDNA template (20 ng), 0.25 µM primers and 1× SYBR Green Master Mix was run in an ABI 7900HT RT-PCR (SDS 2.2 software) with the following conditions: 95°C 10 min, 40 cycles of 95°C 1 min, 60°C 1 min, and 72°C 1 min. Pri-miR-155 and pre-miR-155 were evaluated using primers: PAO-1143 (pri-miR-155 forward) 5'-CTGTTAATGCTAATTGTGATAGG-3'; PAO-1144 (pre-miR-155 forward) 5'-TGCATATCCCTTATCCTCTGG-3'; and the same reverse primer PAO-1145: 5'-GCTAACAGGTAGGAGTCAGTCAG-3'. Since primers PAO-1144/1145 recognize both pri-miR-155 and pre-miR-155, to obtain the pre-miR-155 expression values, we applied the following formula: 2−ΔCt (1144/1145) - 2−ΔCt (1143/1145) [30]. MiR-155 immature forms were normalized against 18S: PAO-296 (forward) 5'-GTAACCCGTTGAACCCCATT-3', PAO-297 (reverse) 5'-CCATCCAATCGGTAGTAGCG-3' and transformed to fold change.
SMAD2 and FOXO3a were evaluated using primers: SMAD2, PAO-1101 (Forward) 5'-CAAACTCGGAGAGGTTCTGC-3', PAO-1102 (Reverse) 5'-GCCAGCCGTATCTCTGGTTA-3'; FOXO3a: PAO-1107 (Forward) 5'-GCTGACAGGCGGTTCCTC-3', PAO-1108 (Reverse) 5'-CAGCTACCTCGGCTCCTTC-3' and normalized against the house keeping genes GAPDH and CAP, amplified using primers: GAPDH, PAO-292 (Forward) 5'-TTCACCACCATGGAGAAGGC-3', PAO-293 (Reverse) 5'-GGCATGGACTGTGGTCATGA-3'; CAP, PAO-330 (Forward) 5'-GAAGGCGGTGATTTTAACGA-3'; PAO-331 (Reverse) 5'-TCCAGCGATTTCTGTCACTG-3'.
2.4 Preparation of celery-based apigenin-rich extracts and diets
Celery-based apigenin rich extracts with increased aglycone content (referred hereafter as ECE) and AIN-93G control diet supplemented with 10% w/w ECE (referred hereafter as ECP) were prepared as previously described [29]. In serum from mice consuming ECP for 7 days apigenin reached ~ 1 µM concentration, as demonstrated by HPLC analyses and previously reported [29].
2.5 In vivo experiments
Male C57BL/6J mice, 6–8 weeks of age (Jackson Laboratories, Bar Harbor, ME), were taken care according to OSU-Institutional Animal Care and Use Committee regulations approved protocol A0208 and used after 7 days acclimation. Mice received 50 mg/kg apigenin or diluent DMSO intraperitoneally (i.p.) 3 h prior administration of 37.5 mg/kg LPS (E. coli 0128:B8) or PBS by i.p. for additional 3 h. Alternatively, mice were fed ad libitum for 7 days with either control diet or ECP prior receiving 37.5 mg/kg LPS or PBS by i.p. for 3 h. Mice were sacrificed, lungs, broncho-alveolar lavage fluids (BALFs) and serum were collected and stored at −80°C. Total RNA was isolated from 50 mg lung tissue using TRIzol® and miR-155 determined using the TaqMan® assay kit according to manufacturer’s instructions (Life Technologies). TNFα in lung tissues was determined by qRT-PCR using primers: PAO-393 (forward) 5'-CCCCAAAGGGATGAGAAGTT-3' and PAO-394 (reverse) 5'-CACTTGGTGGTTTGCTACGA-3' and normalized to GAPDH and CAP expression as described for miR-155 targets. TNFα protein was measured in BALFs and serum by ELISA using the mouse TNFα DuoSet kit and following manufacture’s instructions (R&D Systems).
2.6 Statistical analyses
Data are expressed as mean ± SEM and analyzed by one-way analysis of variance (ANOVA) followed by Bonferroni's post hoc comparisons. For analyses of TNFα levels in BALF and serum, two-tailed student’s t-test was used. Data were analyzed with GraphPad Prism software (GraphPad Software, San Diego, CA). Number of mice used and p values are noted in the figure legends.
3 Results
3.1 Apigenin regulates inflammatory miR expression in LPS-stimulated macrophages
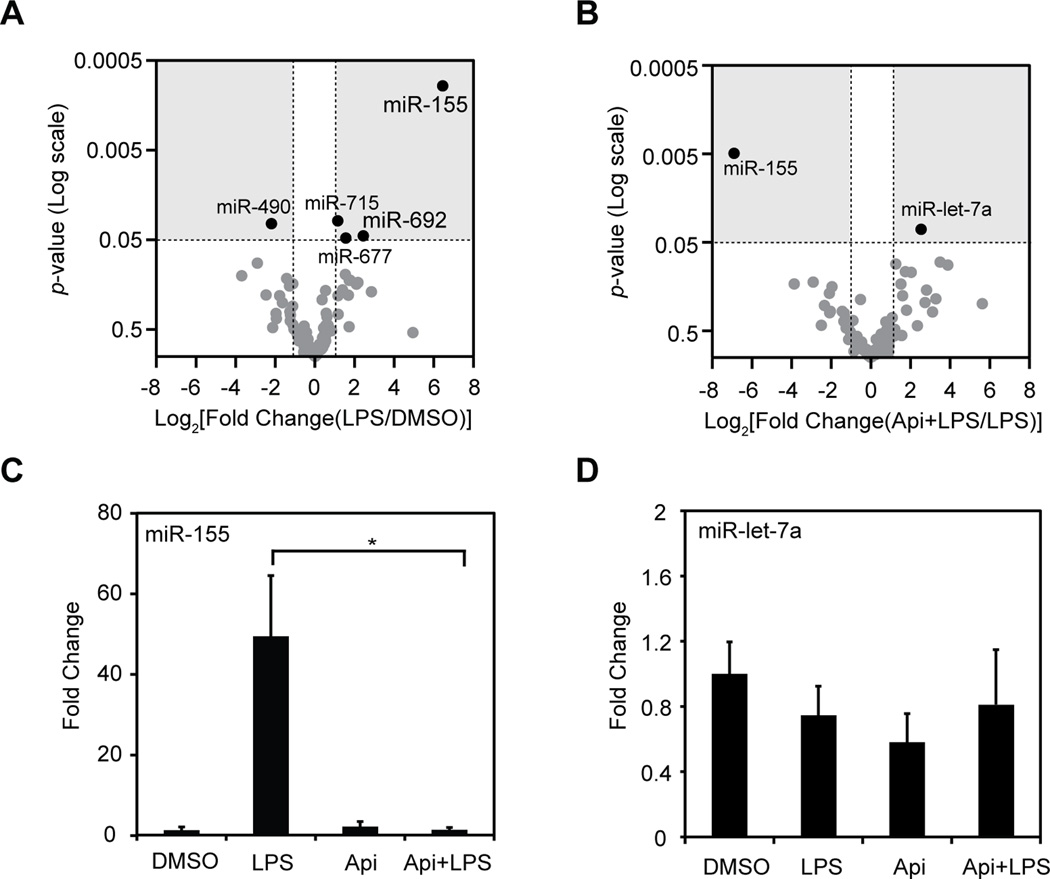
To identify miRs responsible for the immune regulatory activity of apigenin, a high-throughput screening of miRs was conducted, as previously described [30]. Total RNA was isolated from macrophages treated concurrently with 100 ng/ml LPS and 50 µM apigenin (Api+LPS) or diluent DMSO (LPS), or treated with PBS and apigenin (Api) or vehicle PBS and DMSO (DMSO) for 8 h. Changes in miR expression between LPS and DMSO (Fig. 1A) or LPS and Api+LPS (Fig. 1B) were represented as Volcano plots and considered significant when p values were less than 0.05 and fold changes were greater than 2 fold (Fig. 1A and B, gray areas). Out of the 312 miRs included in the array, 154 were expressed in macrophages. LPS significantly changed the expression of five miRs as compared with macrophages treated with DMSO (Fig. 1A, black dots). LPS increased subtly the expression of miR-715 (2.2 fold, p = 0.031), miR-677 (2.9 fold, p = 0.048), miR-692 (5.4 fold, p = 0.045) and greatly increased miR-155 (82.3 fold, p = 0.00096), whereas miR-490 was moderately decreased by LPS (−4.6 fold, p = 0.033, Fig. 1A). Our results indicated that miR-155 was the only miR highly affected by LPS in macrophages, in agreement with previous reports [9, 11, 33], while very few other miRs were only slightly changed by LPS. To evaluate the effect of apigenin on miRs involved in inflammation, the high-throughput screening was used to identify miRs affected by apigenin in LPS-treated macrophages. Apigenin dramatically decreased the LPS-induced expression of miR-155 by ~120.5 fold (p = 0.0049, Fig. 1B), whereas a subtle increased of miR-let-7a by ~5.8 fold was observed (p = 0.035, Fig. 1B).
Figure 1. Identification of apigenin-regulated miRs in LPS-induced inflammation.
Macrophages were treated with 100 ng/ml LPS in the presence of 50 µM apigenin (Api+LPS) or DMSO vehicle (LPS) or with PBS in the presence of apigenin (Api) or DMSO (DMSO) for 8 h. A. MiRs high-throughput qRT-PCR-based array from macrophages treated with LPS or DMSO. Expression values were converted to Log2[Fold change(LPS/DMSO)]. B. MiRs high-throughput qRT-PCR-based array from macrophages treated with LPS or Api+LPS. Expression levels were converted to Log2[Fold Change(Api+LPS/LPS]. For A and B, Log2(Fold change) was compared to p-values and represented as Volcano plots. Dots represent mean of three independent biological replicates. C. Individual qRT-PCR of miR-155 or, D. miR-let-7a. MiR expression is represented as fold change relative to DMSO control. Mean ± SEM, n=5, *p < 0.05.
We next performed validation of miR-155 and miR-let-7a, the only two miRs affected by apigenin during LPS-induced inflammation as identified by the high-throughput screening. Consistent with high-throughput screening results, apigenin reduced the LPS-induced expression of miR-155 by ~45-fold reaching levels found in controls (Fig. 1C). In contrast, miR-let-7a expression was unaffected in all conditions tested (Fig. 1D), and therefore was not further studied. Altogether, these results identified miR-155 as the only miR regulated by apigenin during LPS-induced inflammation in macrophages.
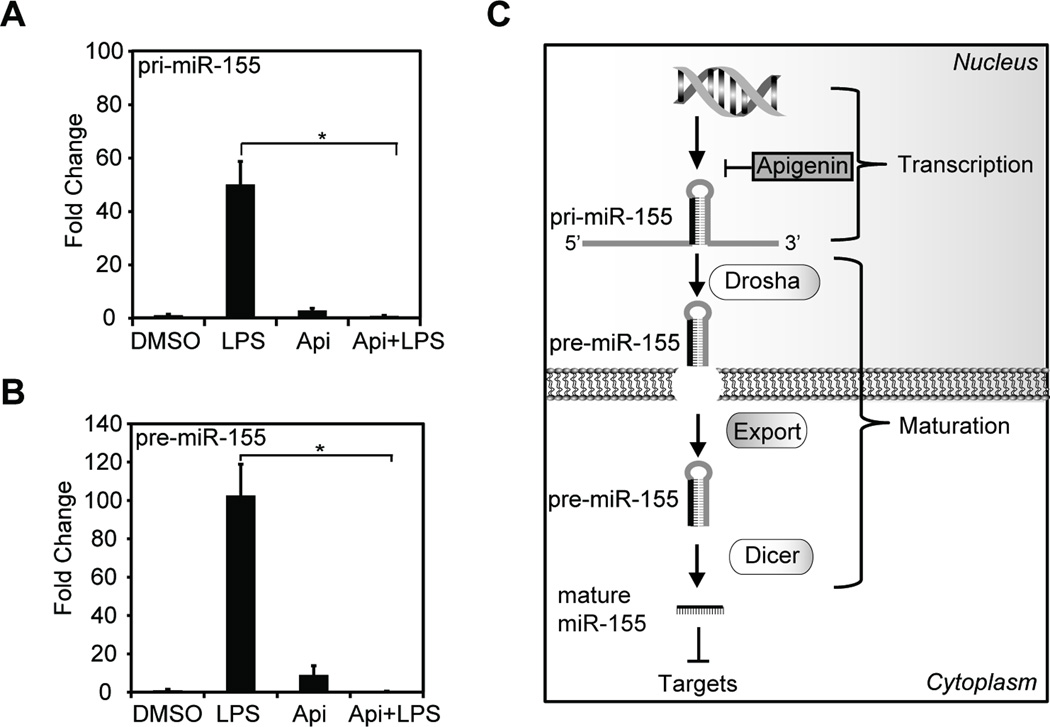
3.2 Apigenin reduces the LPS-induced expression of miR-155 primary transcript
To investigate the molecular mechanism involved in the regulation of miR-155 by apigenin, the expression of pri-miR-155 and pre-miR-155 were evaluated in macrophages treated with 100 ng/ml LPS in the presence of 50 µM apigenin (Api+LPS) or diluent DMSO (LPS) or treated with PBS in the presence of apigenin (Api) or DMSO for 8 h. LPS increased pri-miR-155 expression by ~50 fold, as compared with DMSO or apigenin controls (Fig. 2A). Apigenin significantly decreased LPS-induced pri-miR-155 to levels found in macrophages treated with DMSO control (Fig. 2A, Api+LPS vs. LPS). The precursor pre-miR-155 was induced ~100 fold in the presence of LPS as compared with control (Fig. 2B, LPS vs. DMSO). Apigenin reduced pre-miR-155 in LPS-treated macrophages as compared with LPS-treated macrophages (Fig. 2B, Api+LPS vs. LPS). These findings suggest that apigenin regulates miR expression at the level of transcription, thereby resulting in lower levels of mature miR-155 (Fig. 2C).
Figure 2. Apigenin reduces the expression of LPS-induced primary miR-155 transcript.
A. Macrophages were treated with 100 ng/ml LPS in the presence of 50 µM apigenin (Api+LPS) or diluent DMSO (LPS), apigenin with PBS (Api) or both vehicles PBS and DMSO (DMSO) for 8 h. Pri-miR-155 expression was analyzed by qRT-PCR. B. qRT-PCR of pre-miR in the same samples described in (A). For B and C, miR expression is represented as fold change relative to DMSO control. Mean ± SEM, n=4, *p < 0.05. C. Schematic representation of miR processing. Primary transcript, pri-miR-155, is transcribed and cleaved into a precursor or pre-miR-155 and subsequently exported to the cytoplasm and further processed into mature miR-155 by a Dicer-containing complex.
3.3 Celery-based apigenin-rich diets reduce miR-155 in LPS-induced inflammation
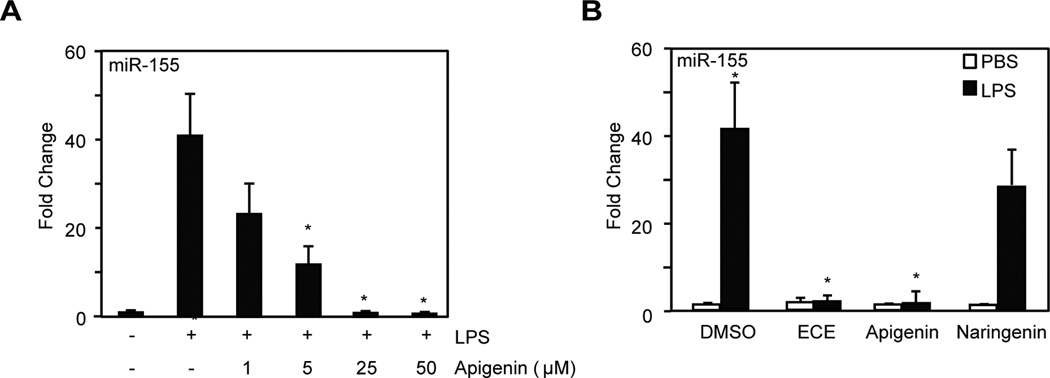
We previously prepared celery-based aglycone apigenin-rich extracts (ECE) and showed their effectiveness in reducing LPS-induced NF-κB activity in macrophages [29]. To study the effect of ECE in inflammation, we first performed a dose response with pure apigenin added concurrently with 100 ng/ml LPS for 8 h in macrophages and determine its effect on the expression of miR-155. Apigenin reduced LPS-induced miR-155 expression in a dose dependent manner reaching statistical significance at 5 µM and at 25 µM apigenin the level found in controls (Fig. 3A).
Figure 3. Celery-based apigenin rich diets reduce LPS-induced miR-155 expression in macrophages.
A. Macrophages were treated with 100 ng/ml LPS in the presence of different concentrations of apigenin or diluent DMSO (indicated as +/−) or with diluents PBS and DMSO (−/−) for 8 h. MiR-155 expression was analyzed by qRT-PCR. B. Macrophages were treated with 100 ng/ml LPS (black bars) or PBS (white bars) concurrently with ECE (25 µM apigenin equivalents), 25 µM pure apigenin, 50 µM naringenin or DMSO for 8 h. MiR-155 expression was determined by qRT-PCR. Data represent fold change relative to cells treated with DMSO. Mean ± SEM, n=5, *p < 0.05.
Next, macrophages were treated with 100 ng/ml LPS or diluent PBS control (Fig. 3B, black and white bars respectively) in the presence of ECE (25 µM apigenin-equivalent, as determined by HPLC analyses [29]), 25 µM pure apigenin, diluent DMSO or 50 µM naringenin, a structurally related flavonoid with no anti-inflammatory activity [27]. ECE, apigenin and naringenin had no effect on basal miR-155 expression as compared with non-stimulated macrophages (Fig. 3B, white bars). ECE significantly reduced LPS-induced miR-155 expression (Fig. 3B, ECE vs. DMSO, black bars) to levels observed in non-stimulated macrophages (Fig. 3B, DMSO, white bar). Similar results were obtained in cells treated with LPS in the presence of pure apigenin (Fig. 3B, Apigenin vs. DMSO, black bars). In contrast, naringenin had no significant effect on LPS-induced miR-155 expression (Fig. 3B, Naringenin vs. DMSO, black bars). Together, these results show that the celery-based apigenin-rich extracts reduce macrophage miR-155 expression in inflammation to levels achieved with pure apigenin.
3.4 Celery-based apigenin-rich diets reduce LPS-induced miR-155 expression modulating inflammatory regulators
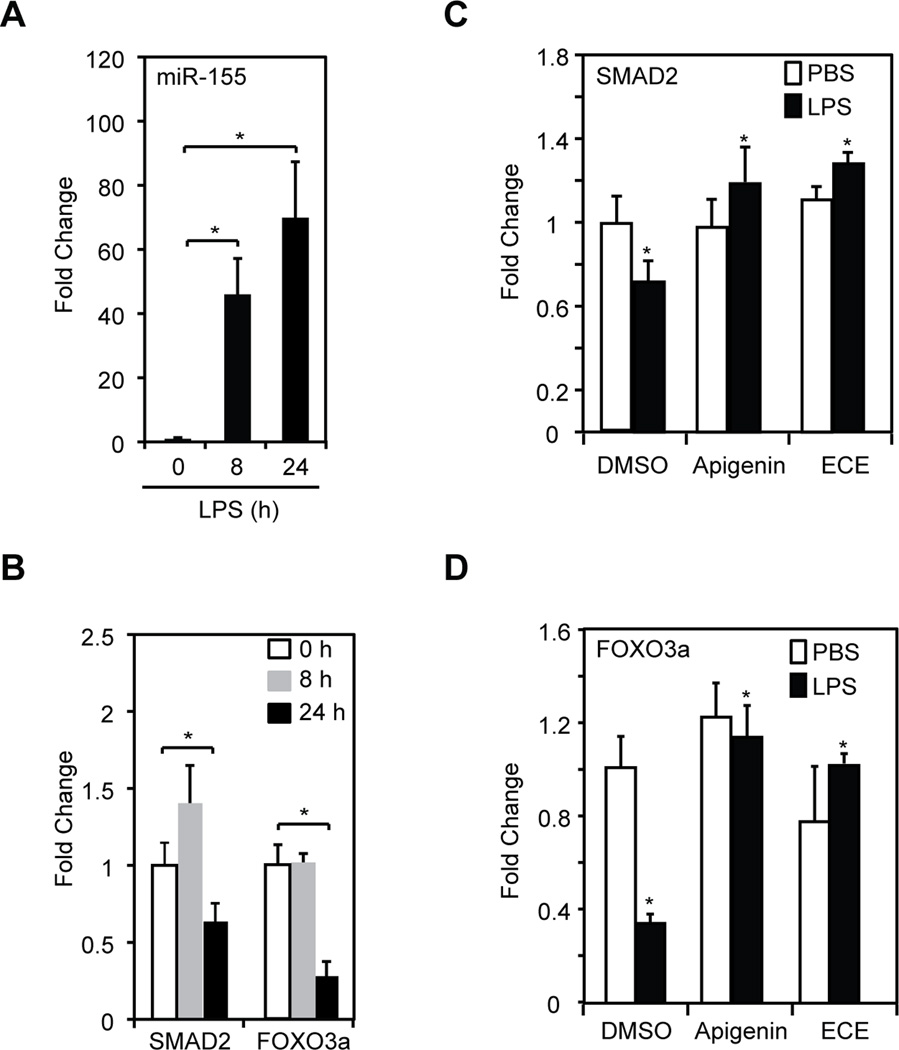
Inflammatory regulators act with distinct kinetics that is in part dependent on the stimulus and the length of stimulation [34, 35]. Hence, we determined the kinetics of miR-155 expression in macrophages treated with 100 ng/ml LPS. We found that miR-155 expression significantly increased at 8 h after LPS stimulation and continued similarly high at 24 h, in agreement with previous studies (Fig. 4A and [33]). Next, we studied the expression of SMAD2 and FOXO3a, targets of miR-155 that regulate inflammation [14, 16]. We found that the expression of SMAD2 and FOXO3a was not significantly decreased in macrophages stimulated with LPS for 8 h compared with non-stimulated macrophages treated with PBS diluent control (Fig. 4B, gray vs. white bars). Yet, at 24 h, SMAD2 and FOXO3a expression were significantly decreased in LPS-treated macrophages (Fig. 4B, black vs. white bars).
Figure 4. Celery-based apigenin rich foods and pure apigenin restored the expression of miR-155 targets.
A. Mir-155 expression in macrophages treated with 100 ng/ml LPS for different periods of time as determined by qRT-PCR. B. SMAD2 and FOXO3a were determined by qRT-PCR in same samples described in (A). For A and B, data represent fold change relative to time 0 h. Mean ± SEM, n=3, *p < 0.05. C. Macrophages were treated with 100 ng/ml LPS (black bars) or PBS (white bars) in the presence of ECE (25 µM apigenin equivalents), 25 µM apigenin or DMSO for 24 h. SMAD2 expression was evaluated by qRT-PCR. D. FOXO3a expression was examined by qRT-PCR in same samples described in (C). For C and D, data represent fold change compared to cells treated with PBS and DMSO (DMSO, white bars). Mean ± SEM, n=5, *p < 0.05.
We next examined the effect of apigenin and celery-based apigenin-rich extracts in the LPS-induced expression of SMAD2 and FOXO3a. Macrophages were stimulated with 100 ng/ml LPS (Fig. 4C–D, back bars) or diluent PBS (Fig. 4C–D, white bars) in the presence of 25 µM apigenin, ECE (at 25 µM apigenin-equivalent) or diluent DMSO for 24 h, a time corresponding to a high level of miR-155 and low levels of both SMAD2 and FOXO3. ECE or apigenin, when administered alone, had no effect on the basal expression of SMAD2 or FOXO3a (Fig. 4C–D, white bars). The addition of ECE to LPS-stimulated macrophages significantly increased the expression of SMAD2 and FOXO3a (Fig. 4C–D, ECE vs. DMSO, black bars). Similar effects were observed with apigenin (Fig. 4C–D, Apigenin vs. DMSO, black bars). These results indicate that both the celery-based apigenin-rich diet and pure apigenin effectively modulate miR-155 biological targets during inflammation.
3.5 Celery-based apigenin-rich diets decrease LPS-induced expression of miR-155 and TNFα in vivo during inflammation
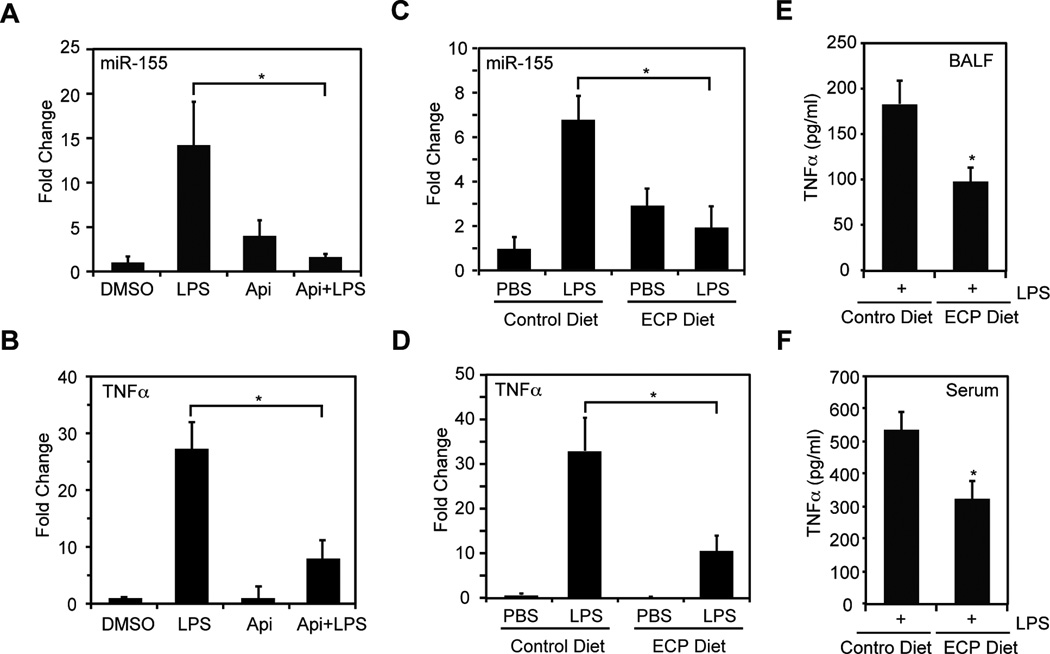
We previously reported that apigenin reduces LPS-induced mortality and decreases the expression of TNFα in vivo [24]. In this model, LPS increased miR-155 expression in mouse lungs [36]. Hence, we examined the effects of apigenin in LPS-induced miR-155 expression in lungs procured from mice treated with 50 mg/kg apigenin or DMSO 3 h prior stimulation with 37.5 mg/kg LPS or diluent PBS for 3 h. We found that LPS increased miR-155 expression in lungs by ~14 fold as compared with lungs from mice treated with vehicle DMSO control or apigenin alone (Fig. 5A, LPS vs. DMSO or Api). Administration of apigenin significantly decreased LPS-induced miR-155 expression to levels found in control mice (Fig. 5A, Api+LPS vs. LPS). In addition, qRT-PCR analyses showed that apigenin significantly reduced LPS-induced TNFα in the lungs (Fig. 5B, Api+LPS vs. LPS).
Figure 5. Celery-based apigenin rich foods reduce LPS-induced expression of miR-155 and TNFα in vivo.
A. Lungs were procured from mice treated with 50 mg/kg apigenin or DMSO for 3 h prior administration of 37.5 mg/kg LPS or PBS for additional 3 h. MiR-155 was determined by qRT-PCR. B. TNFα was determined by qRT-PCR in same lung samples used in (A). For A and B, data represent fold change expression relative to DMSO. Mean ± SEM, n = 8. * p < 0.05. C. Lungs were obtained from mice fed with control or ECP diets for seven days prior administration of 37.5 mg/kg LPS or PBS by i.p. for 3 h. MiR-155 expression was determined by qRT-PCR. D. TNFα was determined by qRT-PCR in same lung samples used in (C). E. TNFα was determined in broncho-alveolar lavage fluids (BALFs) by ELISA. F. TNFα was determined in serum by ELISA. For C–F, data represent fold change compared to control diet. Mean ± SEM, n = 6. * p < 0.05.
We next evaluated the effect of ECP in LPS-induced miR-155 expression in vivo, mice were fed ad libitum with ECP or control diet for 7 days prior stimulation with 37.5 mg/kg LPS or diluent PBS for 3 h. Remarkably, ECP significantly decreased the LPS-induced miR-155 expression in the lungs by ~3.5 fold as compared with LPS-treated mice fed with control diet (Fig. 5C, ECP+LPS vs. Control Diet+LPS), reaching similar levels found in non-stimulated mice (Fig. 5C, ECP+LPS vs. ECP+PBS). Consistently, we found that lungs from LPS-treated mice fed with ECP had a significant ~3.5 fold reduction of TNFα as compared with LPS-treated mice fed with control diet (Fig. 5D, ECP+LPS vs. Control Diet+LPS). Underscoring the physiological activity of the celery-based apigenin rich diet, we observed that ECP decreased TNFα protein levels in broncho-alveolar lavage fluids (BALF) and serum of LPS-treated mice (Figs. 5E and 5F). Together, these results demonstrate the immune-regulatory effectiveness of apigenin-rich diets in vivo, highlighting the potential benefits of dietary interventions in the restoration of proper immune function.
4 Discussion
Increased incidence of inflammatory diseases and health care costs has ignited the interest in the use of nutraceuticals for the prevention and treatment of these diseases. Yet the underlying immune-regulatory mechanisms associated to dietary compounds remain unclear. Even less understood is how foods rich in nutraceuticals exert their health beneficial effects. Here we showed that the flavonoid apigenin and a celery-based apigenin-rich diet regulate the expression of miR-155 during LPS-induced inflammation in macrophages and in vivo, thereby helping to restore immune balance.
Using a high-throughput miRs screening in macrophages, we found miR-155 expression dramatically increased (~80 fold) by LPS, in agreement with other reports [9, 11, 33]. Previous studies showed a subtle transient increase of miR-132 (~1.5 fold) in bone marrow derived macrophages stimulated with LPS [33] and a small decrease of miR-125b (−1.3 fold) in RAW264.7 macrophages stimulated with LPS for 6 h [11]. The differences on the LPS-affected miRs identified may rely on the length of stimulation and the cell type used. Two miRs, miR-155 and miR-let-7a, were modulated by apigenin in LPS-induced macrophages. However, only miR-155 was validated and therefore further studied. Differences between high-throughput screenings and qRT-PCR have been reported [33, 37], and may reflect noise inherent to high-throughput platforms, suggesting the need to use more stringent threshold for screening analyses.
Few reports have investigated the biological relationship of flavonoids and miRs [38, 39]. Apigenin improved glucose tolerance by decreasing maturation of miR-103 in epithelial cells [40], indicating that apigenin may modulate different miRs depending on the cell type and stimulus used. In addition, quercetin decreased LPS-induced expression of miR-155 in macrophages [41]. We showed that apigenin and ECP, but not naringenin, reduced LPS-induced miR-155 expression in macrophages. Similarly, apigenin, but not naringenin, induces apoptosis of various cancer cell lines [22] and lacked anti-inflammatory activity [27]. Naringenin also failed to interact with the direct targets of apigenin [42], suggesting that despite structural similarities, naringenin and apigenin elicit significantly different biological activities.
Regulation of miR levels by polyphenols has been associated to indirect effects on the transcription of miRs or to direct interaction with mature miRs [41, 43]. Apigenin binds yeast RNA in vitro [44], yet whether apigenin binds miRs has not been shown. Nevertheless, treatment with apigenin reduced LPS-induced pri-miR-155, pre-miR-155 and matured miR-155 expression, indicating that apigenin decreases miR-155 at the transcriptional level. Up-regulation of miR-155 during inflammation is controlled in part by NF-κB [9, 45–47]. We previously showed that apigenin inhibits the transcriptional activity of NF-κB in mouse macrophages and human monocytes [24], suggesting that apigenin may regulate the transcription of pri-miR-155 in an NF-κB mediated pathway. Post-transcriptional regulation of miR-155 by KHSRP (KH-type splicing regulatory protein) has been reported in LPS-treated macrophages [33], highlighting the role of RNA binding proteins in the control of miR-155 maturation during LPS-stimulation. Recent studies observed that apigenin inhibits maturation of miRs such as miR-103 by inhibiting the activation of TRBP (Tar RNA-binding protein) [40]. The lack of effect of apigenin on the LPS-induced miR-155 maturation compared to the effect of apigenin reported for miR-103 maturation might reflect the ability of apigenin to modulate different sets of RNA-binding proteins implicated in miR maturation. We have actually identified several RNA-binding proteins as direct targets of apigenin [42]. Together, these results suggest that in macrophages, apigenin reduces pri-miR-155 transcriptionally during inflammation, likely through the inhibition of NF-κB, leading to the decrease of mature miR-155.
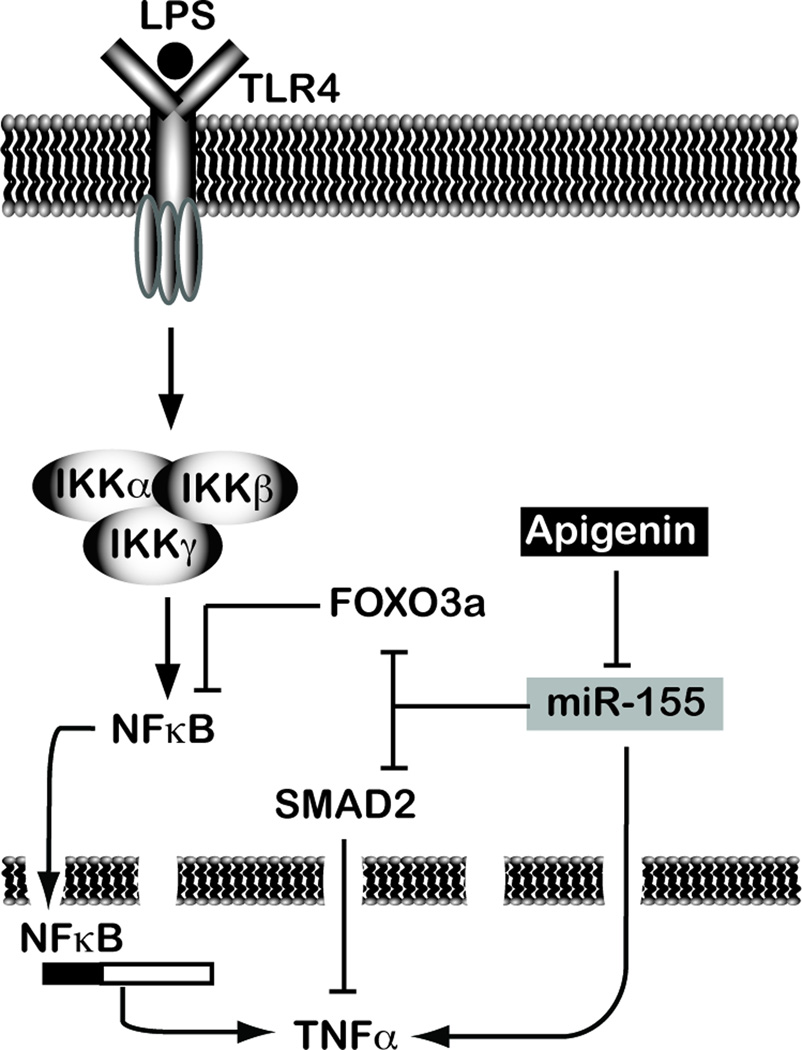
MiR-155 has been suggested as an immune modulatory checkpoint [45], by targeting several molecules involved in the regulation of the immune response including SMAD2 and FOXO3a [14, 16]. SMAD2 have anti-inflammatory activity by suppressing, among others, TNFα production, a main marker of inflammation [15], while FOXO3a is an inhibitor of NF-κB activity, a master regulator of TNFα expression [17]. Additionally, miR-155 increases TNFα levels by stabilizing its mRNA and potentiating TNFα translation [10–13]. Hence, miR-155 is a main regulator of the key inflammatory molecule TNFα. In LPS-treated macrophages SMAD2 and FOXO3a expression was significantly decreased at 24 h. Differences in the kinetics of expression between miR-155 and some of its targets have been previously reported, showing delays of 8 to 12 h [48, 49]. These differences have been attributed to a combination of the rate of transcription, the rate of miR loading into the RISC complex and the rate of mRNA decay [50]. Hence, our observation on the lag of time between miR-155 induction and SMAD2/FOXO3a silencing are aligned with reports on other miR-155 targets. We observed that apigenin and ECE restored the expression of SMAD2 and FOXO3a to levels found in non-stimulated cells. Consistently, we previously reported that apigenin and ECE decreased LPS-induced TNFα in macrophages [29]. Highlighting the physiological activity of dietary apigenin, we found that the celery-based apigenin-rich diet decreased miR-155 and TNFα levels in LPS-treated mice. Thus, apigenin relieves the inhibition of SMAD2 and FOXO3a and decreases TNFα levels by reducing miR-155, thereby restoring immune-balance (Fig. 6).
Figure 6.
Model of the immune regulatory activity of apigenin.
LPS induces dysregulated inflammation leading to metabolic dysfunction, sepsis and severe lung injury [26, 51]. Intraperitoneal apigenin decreased LPS-induced lethality and reduced TNFα levels in serum in animal models. Yet, while promising effects have been shown with apigenin, limited solubility makes this route of administration unfeasible for clinical applications. Overcoming this common limitation of flavonoids, we demonstrated that a celery diet rich in apigenin reduced LPS-induced miR-155 and effectively restored TNFα expression in vivo. Effective concentrations of flavonoids normally range ~5–100 µM in cellular models [24, 26, 29, 41, 52]. Importantly, our studies showed that in vivo, concentrations of apigenin of ~ 1 µM, found in serum of mice fed with the celery-based apigenin rich diets [28] effectively confer immune-regulatory activity. Future experiments are guarantee to evaluate the therapeutic as well as the preventive potential of this diet.
Together, these findings identify miR-155 as a central apigenin-regulated miR in inflammation and provide evidence of the underlying mechanism by which apigenin and diets rich in apigenin contribute to restore homeostasis, highlighting the benefits of dietary interventions as a strategy to restore proper immune function in vivo.
Acknowledgements
This work was supported by Grant R01 HL075040–01 and CAFRE research grant to AID and the FIC Doctoral Research Grant to D.A. D.A. was supported by the PHPID and the CCC-Pelotonia fellowships. We thank the Nutrient and Phytochemical Analytical Shared Resource (NPASR) for flavone’s identification studies.
Abbreviations
- MicroRNAs
miRs
- SMAD2
smooth-muscle-actin and MAD-related proteins 2
- FOXO3a
forkhead Box O3a
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- ECE
celery-based apigenin rich food extracts
- ECP
control diet supplemented with 10% ECE
- LPS
lipopolysaccharide
- TNFα
tumor necrosis factor
Footnotes
Author contributions: A.I.D. and D.A. designed research; D.A., M.D., and L.S.R. performed experiments; S.S., J.L.C. and T.D.S. contributed new analytic tools; J.J. and T.D.S contributed the high-throughput qRT-PCR screening; X.M. helped with statistical analyses. A.I.D. and D.A. interpreted data and wrote the manuscript.
The authors have declared no conflict of interest.
References
- 1.Henson PM. Dampening inflammation. Nat Immunol. 2005;6:1179–1181. doi: 10.1038/ni1205-1179. [DOI] [PubMed] [Google Scholar]
- 2.Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8:776–787. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha VZ, Libby P. Obesity, inflammation, and atherosclerosis. Nat Rev Cardiol. 2009;6:399–409. doi: 10.1038/nrcardio.2009.55. [DOI] [PubMed] [Google Scholar]
- 4.Harirforoosh S, Asghar W, Jamali F. Adverse effects of nonsteroidal antiinflammatory drugs: an update of gastrointestinal, cardiovascular and renal complications. J Pharm Pharm Sci. 2013;16:821–847. doi: 10.18433/j3vw2f. [DOI] [PubMed] [Google Scholar]
- 5.Galland L. Diet and inflammation. Nutr Clin Pract. 2010;25:634–640. doi: 10.1177/0884533610385703. [DOI] [PubMed] [Google Scholar]
- 6.Pasparakis M. Regulation of tissue homeostasis by NF-κB signalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 7.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 8.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 9.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bala S, Marcos M, Kodys K, Csak T, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNF α) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tili E, Michaille JJ, Cimino A, Costinean S, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Tian F, Wang F. Rheumatoid arthritis-associated microRNA-155 targets SOCS1 and upregulates TNF-α and IL-1β in PBMCs. Int J Mol Sci. 2013;14:23910–23921. doi: 10.3390/ijms141223910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajaram MV, Ni B, Morris JD, Brooks MN, et al. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A. 2011;108:17408–17413. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-β. J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiyama Y, Kakoi K, Kimura A, Takada I, et al. Smad2 and Smad3 are redundantly essential for the suppression of iNOS synthesis in macrophages by regulating IRF3 and STAT1 pathways. Int Immunol. 2012;24:253–265. doi: 10.1093/intimm/dxr126. [DOI] [PubMed] [Google Scholar]
- 16.Min M, Peng L, Yang Y, Guo M, et al. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm Bowel Dis. 2014;20:652–659. doi: 10.1097/MIB.0000000000000009. [DOI] [PubMed] [Google Scholar]
- 17.Lin L, Hron JD, Peng SL. Regulation of NF-κB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Polyphenols: food sources and bioavailability. Am J Clin Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 19.McKay DL, Blumberg JB. A review of the bioactivity and potential health benefits of chamomile tea (Matricaria recutita L.) Phytother Res. 2006;20:519–530. doi: 10.1002/ptr.1900. [DOI] [PubMed] [Google Scholar]
- 20.Estruch R, Ros E, Salas-Salvado J, Covas MI, et al. Primary prevention of cardiovascular disease with a Mediterranean diet. N Engl J Med. 2013;368:1279–1290. doi: 10.1056/NEJMc1806491. [DOI] [PubMed] [Google Scholar]
- 21.Gates MA, Vitonis AF, Tworoger SS, Rosner B, et al. Flavonoid intake and ovarian cancer risk in a population-based case-control study. Int J Cancer. 2009;124:1918–1925. doi: 10.1002/ijc.24151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargo MA, Voss OH, Poustka F, Cardounel AJ, et al. Apigenin-induced-apoptosis is mediated by the activation of PKCδ and caspases in leukemia cells. Biochem Pharmacol. 2006;72:681–692. doi: 10.1016/j.bcp.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Arango D, Parihar A, Villamena FA, Wang L, et al. Apigenin induces DNA damage through the PKCδ-dependent activation of ATM and H2AX causing down-regulation of genes involved in cell cycle control and DNA repair. Biochem Pharmacol. 2012;84:1571–1580. doi: 10.1016/j.bcp.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholas C, Batra S, Vargo MA, Voss OH, et al. Apigenin blocks lipopolysaccharide-Induced lethality in vivo and proinflammatory cytokines expression by inactivating NF-κB through the suppression of p65 phosphorylation. J Immunol. 2007;179:7121–7127. doi: 10.4049/jimmunol.179.10.7121. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Mejia ME, Voss OH, Murnan EJ, Doseff AI. Apigenin-induced apoptosis of leukemia cells is mediated by a bimodal and differentially regulated residue-specific phosphorylation of heat-shock protein-27. Cell Death Dis. 2010;1:e64. doi: 10.1038/cddis.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duarte S, Arango D, Parihar A, Hamel P, et al. Apigenin protects endothelial cells from lipopolysaccharide (LPS)-induced inflammation by decreasing caspase-3 activation and modulating mitochondrial function. Int J Mol Sci. 2013;14:17664–17679. doi: 10.3390/ijms140917664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha SK, Lee P, Park JA, Oh HR, et al. Apigenin inhibits the production of NO and PGE2 in microglia and inhibits neuronal cell death in a middle cerebral artery occlusion-induced focal ischemia mice model. Neurochem Int. 2008;52:878–886. doi: 10.1016/j.neuint.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Hostetler GL, Riedl KM, Schwartz SJ. Endogenous enzymes, heat, and pH affect flavone profiles in parsley (Petroselinum crispum var. neapolitanum) and celery (Apium graveolens) during juice processing. J Agric Food Chem. 2012;60:202–208. doi: 10.1021/jf2033736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hostetler G, Riedl K, Cardenas H, Diosa-Toro M, et al. Flavone deglycosylation increases their anti-inflammatory activity and absorption. Mol Nutr Food Res. 2012;56:558–569. doi: 10.1002/mnfr.201100596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slinker BK. The statistics of synergism. J Mol Cell Cardiol. 1998;30:723–731. doi: 10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
- 32.Loscher CJ, Hokamp K, Kenna PF, Ivens AC, et al. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruggiero T, Trabucchi M, De Santa F, Zupo S, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 34.Shih VF, Kearns JD, Basak S, Savinova OV, et al. Kinetic control of negative feedback regulators of NF-κB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci U S A. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh CH, Rau CS, Jeng JC, Chen YC, et al. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J Biomed Sci. 2012;19:69. doi: 10.1186/1423-0127-19-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Gelfond JA, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blade C, Baselga-Escudero L, Salvado MJ, Arola-Arnal A. miRNAs, polyphenols, and chronic disease. Mol Nutr Food Res. 2013;57:58–70. doi: 10.1002/mnfr.201200454. [DOI] [PubMed] [Google Scholar]
- 39.Milenkovic D, Jude B, Morand C. miRNA as molecular target of polyphenols underlying their biological effects. Free Radic Biol Med. 2013;64:40–51. doi: 10.1016/j.freeradbiomed.2013.05.046. [DOI] [PubMed] [Google Scholar]
- 40.Ohno M, Shibata C, Kishikawa T, Yoshikawa T, et al. The flavonoid apigenin improves glucose tolerance through inhibition of microRNA maturation in miRNA103 transgenic mice. Sci Rep. 2013;3:2553. doi: 10.1038/srep02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boesch-Saadatmandi C, Loboda A, Wagner AE, Stachurska A, et al. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J Nutr Biochem. 2011;22:293–299. doi: 10.1016/j.jnutbio.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Arango D, Morohashi K, Yilmaz A, Kuramochi K, et al. Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc Natl Acad Sci U S A. 2013;110:E2153–E2162. doi: 10.1073/pnas.1303726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baselga-Escudero L, Blade C, Ribas-Latre A, Casanova E, et al. Resveratrol and EGCG bind directly and distinctively to miR-33a and miR-122 and modulate divergently their levels in hepatic cells. Nucleic Acids Res. 2014;42:882–892. doi: 10.1093/nar/gkt1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nafisi S, Shadaloi A, Feizbakhsh A, Tajmir-Riahi HA. RNA binding to antioxidant flavonoids. J Photochem Photobiol B. 2009;94:1–7. doi: 10.1016/j.jphotobiol.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 2013;41:542–553. doi: 10.1093/nar/gks1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao B, Liu Z, Li BS, Tang B, et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 47.Imaizumi T, Tanaka H, Tajima A, Yokono Y, et al. IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am J Nephrol. 2010;32:462–468. doi: 10.1159/000321365. [DOI] [PubMed] [Google Scholar]
- 48.Thounaojam MC, Kundu K, Kaushik DK, Swaroop S, et al. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J Virol. 2014;88:4798–4810. doi: 10.1128/JVI.02979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H, Huang X, Cui H, Luo X, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 50.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 51.Wang G, Huang X, Li Y, Guo K, et al. PARP-1 inhibitor DPQ attenuates LPS-induced acute lung injury through inhibiting NF-κB-mediated inflammatory response. PLoS One. 2013;8:e79757. doi: 10.1371/journal.pone.0079757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhaskaran N, Shukla S, Srivastava JK, Gupta S. Chamomile: an anti-inflammatory agent inhibits inducible nitric oxide synthase expression by blocking RelA/p65 activity. Int J Mol Med. 2010;26:935–940. doi: 10.3892/ijmm_00000545. [DOI] [PMC free article] [PubMed] [Google Scholar]