Abstract
Coronary artery disease remains the leading cause of death globally and is a major burden to every health system in the world. There have been significant improvements in risk modification, treatments, and mortality; however, our ability to detect asymptomatic disease for early intervention remains limited. Recent discoveries regarding the inflammatory nature of atherosclerosis have prompted investigation into new methods of diagnosis and treatment of coronary artery disease. This article reviews some of the highlights of the important developments in cardioimmunology and summarizes the clinical evidence linking the immune system and atherosclerosis. It provides an overview of the major serological biomarkers that have been associated with atherosclerosis, noting the limitations of these markers attributable to low specificity, and then contrasts these serological markers with the circulating immune cell subtypes that have been found to be altered in coronary artery disease. This review then outlines the technique of mass cytometry and its ability to provide high‐dimensional single‐cell data and explores how this high‐resolution quantification of specific immune cell subpopulations may assist in the diagnosis of early atherosclerosis in combination with other complimentary techniques such as single‐cell RNA sequencing. We propose that this improved specificity has the potential to transform the detection of coronary artery disease in its early phases, facilitating targeted preventative approaches in the precision medicine era.
Keywords: atherosclerosis, coronary artery disease, immune system, inflammation, mass cytometry
Subject Categories: Biomarkers, Inflammation, Diagnostic Testing, Atherosclerosis, Cell Biology/Structural Biology
Nonstandard Abbreviations and Acronyms
- CANTOS
Canakinumab Antiinflammatory Thrombosis Outcome Study
- CARDIoGRAMplusC4D
Coronary Artery Disease Genome Wide Replication and Meta‐Analysis (CARDIoGRAM) Plus the Coronary Artery Disease (C4D) Genetics
- CITE‐seq
cellular indexing of transcriptomes and epitopes by sequencing
- CONCORDANCE
Cooperative National Registry of Acute Coronary Syndrome Care
- CyTOF
mass cytometry time of flight
- INTERHEART
Effect of Potentially Modifiable Risk Factors Associated With Myocardial Infarction
- mTOR
mechanistic target of rapamycin
- MyD88
myeloid differentiation primary response 88
- pVAT
perivascular adipose tissue
- SCOT‐HEART
Scottish Computed Tomography of the Heart
- scRNA‐seq
single‐cell RNA sequencing
- SMuRFs
standard modifiable cardiovascular risk factors
- TET2
tet methylcystosine dioxygenase 2
- TLR4
toll‐like receptor 4
Coronary artery disease (CAD) persists as a major cause of death in Australia 1 despite decades of improvement in treatments and identification of traditional risk factors for management. Globally, the picture is no better, with ischemic heart disease remaining the leading cause of years of life lost for high and middle sociodemographic groups worldwide, 2 and accounting for one third of all deaths annually. 3
To improve this situation, we need to better understand the causes and mechanisms that promote CAD. The foundation of our current understanding of cardiovascular risk profiling dates back to the 1950s when Framingham and other epidemiology studies associated cigarette smoking, diabetes mellitus, hypertension, and hyperlipidemia with cardiac mortality. 4 While these data were invaluable and saved many lives, they also resulted in the development of a sense of complacency that CAD was “solved” and that CAD is now predominantly self‐induced when individuals fail to manage their risk factors. This damaging myth stems from the misinterpretation of population data such as is seen in the INTERHEART (Effect of Potentially Modifiable Risk Factors Associated With Myocardial Infarction) study, which showed that 9 potentially modifiable risk factors account for >90% of the population‐attributable risk for heart attack. 5 However, population‐attributable risk is not expected to sum to 100%, and should only be used for estimating the societal impact of risk factor control policies, not to argue against the importance of discovering new mechanisms of disease. 6 Indeed, at an individual case level, it is not uncommon for a patient to present with extensive atherosclerosis and a life‐threatening heart attack without adequate explanation—prompting the difficult‐to‐answer question: “Why me?”
Despite our best efforts in primary care and with heart health checks, our reliance on algorithms derived from large community cohort studies overestimate risk in some people and underestimate it in others, sometimes by significant amounts. 7 While the calculations based on standard modifiable cardiovascular risk factors (SMuRFs) reflect statistical risk at a community level, they fail to take into consideration the individual phenotypes that mediate the host response. Locally, this can be seen in the proportion of patients without SMuRFs presenting to an Australian center with life‐threatening ST‐segment–elevation myocardial infarction, which increased from 11% in 2006 to 27% by 2014. 8 This increase in patients with SMuRFs who had ST‐segment–elevation myocardial infarction was confirmed as a national phenomenon in the large CONCORDANCE (Cooperative National Registry of Acute Coronary Syndrome Care) cohort, and, furthermore, the in‐hospital mortality of these patients was found to be higher when compared with their counterparts with traditional risk factors. 9
These patients with disease not explained by traditional risk scoring systems are an archetypal population highlighting the need to improve on precision in cardiovascular medicine. They illustrate the biological variability in signaling pathways that impacts an individual's vulnerability or resistance to the factors that drive CAD, confounding the expected results calculated from population medicine studies. Improved markers that reflect either the level of susceptibility of an individual, or the disease activity itself, are urgently needed to improve personalized preventative approaches to CAD.
Recently, there has been a shift in focus towards precision medicine, which aims to understand unique patient characteristics that can impact disease manifestations and response to treatment. There is a well‐established body of evidence demonstrating that inflammation is an integrating common pathway downstream of the major cardiac risk factors. 10 , 11 , 12 , 13 However, high‐sensitivity C‐reactive protein remains the only inflammatory biomarker routinely used in clinical practice, despite it having a low specificity for atherosclerotic disease.
In the quest to detect subclinical atherosclerotic activity, new tools that can identify the inflammatory biosignature of atherosclerotic plaque will be of great importance. With a view to that aim, this article reviews some of the recently described links between immunology and cardiology, examines the existing data on inflammatory biomarkers in CAD, and explores the potential utility of immunophenotyping using mass cytometry in the precision medicine of tomorrow.
Established Links Between Atherosclerosis and Inflammation: Some Highlights
In the 19th century, pathologists such as the renowned Rudolf Virchow identified the inflammatory nature of atherosclerotic plaques, but most research conducted in the 20th century was preoccupied with atherosclerosis as a passive accumulation of excessive amounts of free and esterified cholesterol. 11 There were initial expectations that we could “cure” CAD with aggressive treatment of hypertension and hyperlipidemia. However, despite decades of aggressive risk factor management we have only seen a partial reduction in cardiovascular death and morbidity. While a component of this will relate to health system challenges, and differences in access and uptake of evidence‐based preventative strategies, improving our knowledge of mechanisms of individual susceptibility and resilience will have broad benefits in targeting additional efforts. In search for the factors that contribute to the residual risk variation between individuals, attention has shifted back towards the basic biology of signaling pathways and cellular interactions, with a renewed focus on inflammation.
The modern era has given us significant advantages over scientists from previous generations who studied CAD. We now have an extensive array of assays, better insight into molecular and cellular biology, impressive animal models of diseases, and a solid foundation in the genetics that underlie it all. Advances in immunology have given us antibodies for molecular targeting and single‐cell analysis that can achieve incredible resolution, differentiating specific subpopulations of the immune response at particular phases of disease development. This has helped us make substantial headway in understanding the key molecular relationships between inflammation and atherosclerosis—we have come a long way from the era of Virchow. But despite the expansive body of knowledge now accumulated on biological mechanisms at a tissue level, we are only now on the brink of the ultimate translation of these data to the clinically useful development of new biomarkers for early atherosclerosis. This “phenotyping” of an individual's vascular inflammatory activity may also identify individuals appropriate for targeted anti‐inflammatory therapies, some of which are already in development.
Outlined below are interesting results from selected studies that highlight some of the key findings that have allowed us to progress to where we are today.
The Genome: Genetic Association Studies
Initially, researchers thought that the missing biology causing CAD would be found in the genome, and indeed in recent years there have been large‐scale genome‐wide association studies that have identified connections between CAD risk and the immune system. The CARDIoGRAMplusC4D (Coronary Artery Disease Genome Wide Replication and Meta‐Analysis [CARDIoGRAM] Plus the Coronary Artery Disease [C4D] Genetics) consortium gathered data from multiple genetic studies in a collaborative effort to identify loci of risk for cardiovascular diseases, 14 and in 2012 they were able to identify a novel CAD risk locus in the major histocompatibility complex, which achieved genome‐wide significance, although the odds ratio was <1.3. This region of the genome contains a dense cluster of genes that have roles in inflammation, immunity, and self‐recognition. In 2013, a large study from the same group reported novel risk loci for CAD, and network analysis demonstrated that the 4 most significant pathways, mapping to 85% of candidate genes, were involved in lipid metabolism and inflammatory pathways. 15
These large studies and a 2015 meta‐analysis of over 185 000 cases and controls 16 have provided sound evidence that genetic predisposition to CAD is largely polygenic and related to multiple single nucleotide polymorphisms with small effect size, and that many of these mutations are associated with lipid and inflammatory pathways. Although these population studies have not given us the answer to how inflammation is causally associated with CAD, they provide concrete evidence of the significant associations between them. Additionally, it is worth considering the ability of the immune system to adapt to environmental changes; as such, genetic association studies may struggle to identify the true magnitude of the relationship between the immune system and CAD in the germline DNA.
The Circulating Milieu: Key Recent Findings in Inflammatory Atherobiology
The vascular endothelium exists in conjunction with an ever‐changing circulatory fluid containing a highly diverse array of proteins, cells, and even pathogens. Humans exist in an environment rich in microbes, and the concept that CAD might relate to bacterial inflammation has long been postulated. The association between periodontal disease and CAD has been known for decades, 17 and the detection of diverse bacterial signatures in atherosclerotic lesions obtained via catheter atherectomy 18 prompted theories of how CAD could be caused by chronic bacterial infection of the arterial wall. Chlamydia pneumoniae, a common intracellular human pathogen, was found to be particularly persistent and detectable within smooth muscle cells, fibroblasts, and macrophages of atherosclerotic tissue. 19 However, the results of antimicrobial drug trials were unfavourable, and a 2005 meta‐analysis of antibiotic therapy for CAD in nearly 20 000 patients showed no impact on all‐cause mortality or incidence of myocardial infarction. 20 These results shifted the focus of research towards other avenues of investigation that might reduce CAD risk.
However, our understanding of inflammation and the inflammasome has continued to evolve in the years since some of these bacterial studies were completed in the early and mid‐2000s. We are now starting to identify key links between signaling pathways in inflammation and atherosclerosis. The Chlamydia hypothesis was recently readdressed in a mechanistic fashion, and in an atherosclerosis‐prone mouse model it was shown that both C pneumoniae and metabolic stress secondary to dyslipidemia result in signaling via shared innate immune pathways involving TLR4 (toll‐like receptor 4) and MyD88 (myeloid differentiation primary response 88). 21 This interesting finding has not yet been confirmed in humans, but it suggests potentially valuable avenues of investigation. If the activation of TLR4 by this pathway could be specifically prevented, this theoretically could provide a safe therapeutic option to reduce inflammation without dampening immune responses to other pathogens. Work on vaccines targeting lipid and nonlipid antigens is now in progress, 22 and while the concept of an atherosclerosis vaccine is still preliminary, it holds exciting potential.
In 2017, publication of several influential articles related to circulating cell types helped to solidify the connections between inflammation and CAD in clinical populations. The first set of results involved clonal hematopoiesis of indeterminate potential, a condition that is defined by the presence of an expanded population of bone marrow–derived cells in the peripheral circulation. Clonal hematopoiesis of indeterminate potential was thought to be a precancerous state and was under investigation in the field of hematology, but, somewhat unexpectedly, an almost 2‐fold increase in cardiac mortality was seen in patients with clonal hematopoiesis of indeterminate potential, and this was found to be related to several distinct somatic mutations. 23 Investigation into one of the foremost mutations, TET2 (Tet methylcystosine dioxygenase 2), showed that Tet2‐mutant bone marrow cells underwent clonal expansion, generating a significant increase in inflammatory macrophages and larger atherosclerotic plaque size. Increases in signaling in the NOD‐like receptor pyrin domain containing 3 inflammasome and higher levels of interleukin 1β (IL‐1β) were also identified. 24 Research into these mutations and their clinical implications is ongoing, but the concept that somatic genetic changes within cells in the bone marrow are linked to atherosclerosis in the artery confirms the importance of the systemic circulation in the process of atherogenesis.
Finally, perhaps the most important finding in the inflammatory CAD field came with the first demonstration in humans that blocking an inflammatory signaling pathway could improve cardiovascular outcomes. In this key work achieved by Ridker et al 25 in CANTOS (Canakinumab Antiinflammatory Thrombosis Outcome Study), therapy with canakinumab, an IL‐1β–blocking antibody, reduced rates of recurrent cardiovascular events in patients who had a previous myocardial infarction. The absolute decrease in events was small, and treatment was associated with a higher incidence of fatal infection, but the fact that the decrease was independent of lipid levels provided crucial mechanistic evidence supporting the inflammatory hypothesis of CAD. While this will in no way replace established therapies such as statins, subsequent subgroup analysis showed that response to therapy was related to the magnitude of C‐reactive protein reduction, 26 supporting the notion that suppression of inflammation was causal in reducing cardiovascular events, rather than being the result of off‐target drug effects. While there are practical, economic, and safety issues pertaining to IL‐1β antibody blocking use in chronic disease, this seminal study highlights the value in targeting this pathway and has provided renewed motivation for pharmacological assessment of other components of the inflammasome. IL‐1β may prove to be too broad of a therapeutic target, and other avenues are currently under investigation, 13 , 27 but the causal connection between inflammation and CAD outcomes is now definite and there are many other discrete targets that remain to be investigated, as evident in Figure 1.
Figure 1. Overview of inflammatory associations with atherosclerosis.
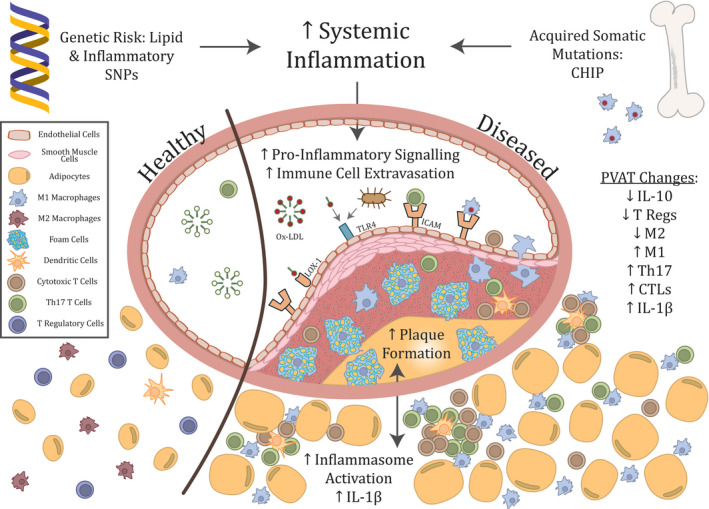
CHIP indicates clonal hematopoiesis of indeterminate potential; CTL, cytotoxic T lymphocyte; ICAM, intercellular adhesion molecule; IL‐1β, interleukin 1β; IL‐10, interleukin 10; LOX‐1, lectin‐type oxidised low‐density‐lipoprotein receptor 1; M1, M1 macrophage; M2, M2 macrophage; ox‐LDL, oxidized low‐density‐lipoprotein; SNPs, single‐nucleotide polymorphisms; T regs, regulatory T cells; Th17, T‐helper 17 cells; and TLR‐4, toll‐like receptor 4.
The Tissue: Atherosclerosis and Perivascular Adipose Tissue
Recent research into perivascular adipose tissue (pVAT) and CAD has identified significant interactions between atheroma and the immune cells contained in the tissue surrounding the vasculature. pVAT was historically thought to be a mechanical support tissue with no other function, but more recently it has been found to be extensively involved in the inflammatory regulation of vascular function. 28 It contains significant numbers of macrophages and effector/memory T cells, as shown in Figure 1, although nearly all immune cell types can be found in the pVAT. 29
Studies of pVAT in mice have shown that endothelial dysfunction is associated with a highly proinflammatory milieu with recruitment of a wide range of immune cells 30 and is present in mouse models of obesity, diabetes mellitus, hypertension, and atherosclerosis. 29 Experiments using human pVAT have been more limited for obvious reasons; however, Antonopoulos et al 31 recently demonstrated that the degree of inflammation in surgically explanted pVAT tissue, as characterized by inflammatory gene expression and cytokine levels, correlated with a computed tomographic coronary angiogram measure known as the fat attenuation index. This imaging measure was significantly associated with atherosclerotic plaque burden in the underlying artery on computed tomographic coronary angiogram. Subsequently, there have been a number of studies assessing the ability of various imaging techniques to quantify the degree of inflammation around a diseased artery, 32 further solidifying the link between the inflammatory changes in the pVAT and the extent of the endothelial dysfunction in the underlying atherosclerotic vessel. Earlier this year, inflammatory features of pVAT were assessed in the SCOT‐HEART (Scottish Computed Tomography of the Heart) 33 computed tomographic coronary angiogram cohort, and a new machine learning algorithm, the fat radiometric profile, was able to significantly improve major adverse cardiac event prediction when compared with tools that incorporated only risk factors and standard disease severity features seen on computed tomographic coronary angiogram. 34
These highly significant studies have provided an imaging correlate of coronary inflammation that may prove effective in risk stratification of patients. However, therapeutic strategies to target atherogenic inflammatory pathways will require a better understanding of the biology that drives it on a cellular level. Ultimately, the addition of serological biomarkers for CAD to imaging measures such as the fat radiometric profile will improve our ability to screen for and detect CAD at early stages, and the drive to search for such markers remains.
Inflammatory Drivers Related to the Western Lifestyle
Ischemic heart disease secondary to CAD is one of many chronic, noncommunicable diseases that have increased in frequency globally over the past decades, along with asthma, inflammatory bowel disease, and rheumatoid arthritis. 35 While CAD has not been traditionally included in the group of conditions that are thought to be caused by inappropriate immune responses to foreign or self antigens, CAD has similarly been associated with a Western lifestyle, which suggests it should be considered as a candidate “immunoinflammatory” disease.
Current theory explaining the increasing incidence of these diseases focuses on changes to our immune system's ability to correctly identify self. Development of self‐tolerance is a complex area in immunology, centrally involving deletion of autoreactive immune cells in the thymus, and peripherally involving networks of regulatory T cells that are capable of dampening immune responses by modulating dendritic cell costimulation. 36 The microbiome also plays a key role in determining the balance between immune activation and immune tolerance. 37 In the healthy individual, these systems work to support a balance between appropriate responses to pathogens and self‐tolerance.
The Western lifestyle is associated with changes in environmental factors that have potential impacts on our immune system. The first factor noted historically was sanitation. Increased hygiene decreases infections, particularly in childhood, and hence reduces the foreign antigens to which our immune system is exposed. The second environmental factor to consider is food, as the type and availability of nutrition has changed significantly with societal development. The diet prevalent in most affluent nations is now associated with high‐calorie, low‐fiber, processed meals that can induce major alterations to the human metabolism. Overnutrition can result in accumulation of adipose tissue, changes to adipokine production, and altered glucose tolerance and insulin resistance, and has been theorized to reduce proliferation of regulatory T cells as a result of overactivity of mTOR (mechanistic target of rapamycin). 38 This type of chronic metabolic change may explain the higher risk of autoimmune disease in obese patients. 39 The diet also has effects on the microbiome, and alterations in microbiota have been shown to have significant impacts on the homeostasis of a variety of T‐cell populations. 37 Indeed, the “hygiene hypothesis,” which is likely to result from failure to establish an optimal microbiome in early life, has long been used to explain the increase in allergic diseases noted since the end of the 19th century. 36 The microbiome of patients with CAD has been shown to be less fermentative and more inflammatory than in healthy patients, and similar changes are seen in obesity and type 2 diabetes mellitus. 40 Research is ongoing to determine whether the microbiotal changes observed represent a primary pathology that could be treated by inducing changes to the flora.
Detailed research into the immune response in atherosclerosis may also give additional insights into the differential risks seen in the sexes. In general, immune responses are higher in women than men, and this has been proposed as one factor contributing to the higher overall incidence of autoimmune disease in women. 41 However, there are some notable exceptions to this trend, including type 1 diabetes mellitus, 42 myocarditis, and idiopathic pulmonary fibrosis, which are all more common in men. Female sex hormones are thought to protect premenopausal women from both CAD and type 2 diabetes mellitus, 43 but why they have different effects on these immunoinflammatory diseases versus autoimmune conditions is not well understood.
As outlined in Figure 2, immune system dysregulation in response to these environmental factors may help to explain how CAD may develop in the absence of traditional risk factors. Considering the evidence linking inflammation and atherosclerosis and the potential for targeted therapies in this domain, the possibility of immunoinflammatory CAD driven by a reduction in self‐tolerance should be strongly considered in future research, with a focus on assessing immunoregulatory function in patients with unexplained atherosclerosis.
Figure 2. Impact of Western lifestyle on immunoregulatory function.
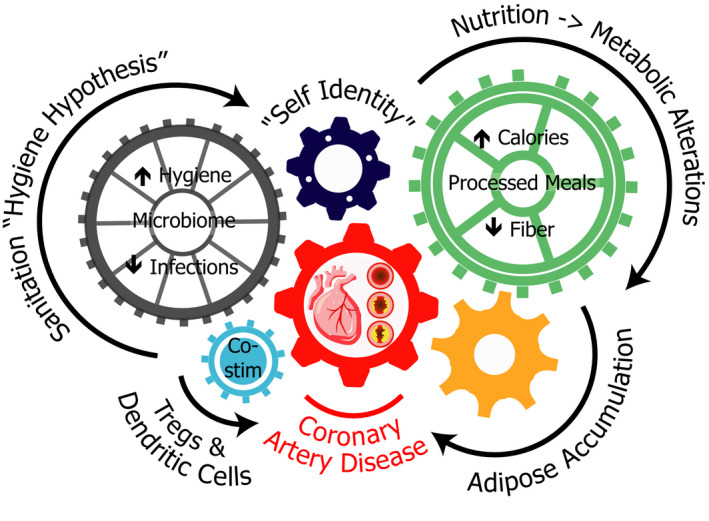
T regs indicates regulatory T cells.
Need for Precision Markers
While remarkable progress has been made in understanding inflammatory atherogenesis and we now have some robust methods of identifying the presence of high‐risk atherosclerosis via imaging, the test sought for by many investigators is a serological, small‐molecule biomarker or an equivalent blood‐based test. We need to be able to diagnose CAD earlier and with increased precision, ideally without putting patients through extensive testing or any form of radiation exposure. A particularly useful biomarker would correlate with the degree of atherosclerosis present in the arterial system, would decrease with sucessful treatment reflecting disease activity, and would be highly sensitive and specific for CAD. To date, a biomarker of this quality has been elusive.
Inflammatory Soluble and Cellular Biomarkers in Predicting Development of CAD
The current repertoire of cardiac biomarkers that are primarily related to immune function includes soluble small molecules detectable in the peripheral circulation, primarily cytokines, acute‐phase proteins, and cleaved cell surface receptors. Over 50 inflammatory biomarkers that relate to CAD in some way have been identified, and these have been reviewed extensively elsewhere. 44 , 45 , 46 , 47 Shown in Table 1 are some of the best‐studied inflammatory biomarkers, all of which have been shown to clinically relate to prediction of CAD or major adverse cardiac events. 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 This is presented alongside an abbreviated list of other conditions with which the relevant biomarker is also associated, highlighting the broad relationships these markers have among disease states, and the difficulty this presents in utilizing them for precision diagnosis of CAD.
Table 1.
Selected Soluble Inflammatory Biomarkers of CAD
| Class | Biomarker | Biology | Usual Sample Type | Most Significant Clinical CAD Evidence | Other Selected Associated Conditions |
|---|---|---|---|---|---|
| Acute‐phase protein | High‐sensitivity C‐reactive protein | Pentameric protein activator of the complement system, produced by the liver 48 | Serum/plasma | Elevated level improves prediction of MACE by C index of 0.0039 in a large (n≈250,000) meta‐analysis 49 | Infections, allergic reactions, inflammatory diseases, necrosis, trauma, and malignancy 48 |
| Acute‐phase protein | Fibrinogen | Glycoprotein coagulation factor 1, precursor of fibrin, produced by the liver 50 | Plasma | Elevated level improves prediction of MACE by C index of 0.0027 in a large (n≈250,000) meta‐analysis 49 | Inflammatory diseases, stroke, brain injury, spinal cord injury, multiple sclerosis, and Alzheimer disease 50 |
| Acute‐phase protein | SAA | Small fibrillar lipophilic proteins, functions incompletely characterized, likely roles in immune and vascular signaling, produced by the liver 51 | Serum | Elevated levels associated with MACE in a large case‐control study of women (n≈28,000)—relative risk 3.0, but not an independent predictor of risk in multivariate analysis 52 | Inflammatory diseases, preterm labor, sarcoidosis, COPD exacerbations, and malignancy 51 |
| Cytokine | IL‐10 | Anti‐inflammatory small soluble protein, acts mainly on other immune cells, produced by immune cells 53 | Serum/plasma | No differences in circulating IL‐10 levels seen in a case‐cohort study (n≈740), 54 some small studies showing higher IL‐10 levels associated with less CAD 55 , 56 | Various rheumatological conditions, 57 multiple metabolic risk factors, 58 and COPD severity 59 |
| Cytokine | IL‐6 | Proinflammatory small soluble protein, acts on immune cells and triggers release of acute‐phase proteins by the liver, produced by immune and epithelial cells 60 | Serum/plasma | Elevated levels associated with CAD in large meta‐analysis (n≈25,000) using the Mendelian randomization approach 61 | Various malignancies and mortality risk, 60 asthma and inflammatory lung diseases, 62 rheumatological conditions, 57 and heart failure 63 |
| Cytokine | TNF‐α | Proinflammatory small soluble protein, acts by binding TNF receptor superfamily receptors, produced by circulating myeloid cells 64 | Serum | Associated with 6‐y incidence of CAD with a risk factor–adjusted hazard ratio of 1.87 54 | Heart failure, 63 periodontal disease, 65 and obstructive sleep anpea 66 |
| Cell adhesion molecule | ICAM‐1 | Transmembrane surface molecule expressed on leukocytes and endothelial cells, which mediates leukocyte adhesion and facilitates extravasation, soluble form results from receptor shedding 67 | Plasma | Elevated levels associated with MACE in a large case‐control study of women (28,000)—a relative risk of 2.6, but not an independent predictor or risk in multivariate analysis. 52 A large cohort study (n≈10,000) showed prediction of CAD development in healthy men with a relative risk of 1.9. 68 | Endometriosis, 69 diabetic retinopathy, 70 frailty, 71 and community‐acquired pneumonia 72 |
| Complement proteins | C3/C5a | Protein components of the innate immune complement system, tagging foreign or damaged cells for destruction, produced by the liver and some circulating immune cells 73 | Serum | Small studies (n≈200) showed C3 associated with severe CAD and increased MACE in women, 74 and C5a associated with cardiac MACE in patients with symptomatic peripheral arterial disease 75 | Various rheumatological conditions, vasculitis, thrombotic microangiopathies, glomerulonephritis, and antibody‐mediated transplant rejection 76 |
C3 indicates complement component 3; C5a, complement component 5a; CAD, coronary artery disease; COPD, chronic obstructive pulmonary disease; ICAM‐1, intercellular adhesion molecule‐1; IL‐6, interleukin 6; IL‐10, interleukin 10; MACE, major adverse cardiac events; SAA, serum amyloid A; and TNF‐α, tumour necrosis factor‐α.
Associations between the quantity and phenotype of circulating immune cells and CAD have also been reported frequently in the past decade, but research in this area has been nowhere near as extensive as with the small molecules. Human peripheral blood contains B cells, basophils, dendritic cells, eosinophils, monocytes, natural killer cells, neutrophils, natural killer T cells, T cells, and stem cells. 77 Studies have identified relationships for most of these cell types with atherosclerosis, although the associations with granulocytes (neutrophils, basophils, and eosinophils) and B cells are mainly seen in tissues rather than the circulation. Listed in Table 2 are some of the significant clinical reports linking circulating immune cell populations and CAD. 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 89 , 90 , 91 , 92 , 93 , 94 , 95 , 96 , 97
Table 2.
Circulating Cellular Inflammatory Biomarkers of CAD
| Class | Cell Type | Biology | Summary of Significant Clinical CAD Evidence |
|---|---|---|---|
| Stem cell | EPCs | Bone marrow– and tissue‐derived progenitor cells (CD34+, CD133+, VEGFR2+), which can differentiate into mature endothelial cell types 78 | Reduced EPC levels were an independent predictor of poor cardiovascular prognosis, adjusted for disease and risk factors (HR, 3.9) in small study (n≈120). 79 Increased levels of nontraditional CD34+CD45− EPCs were associated with atheroma burden and predicted future cardiovascular events in a small study (n≈200) 80 |
| Myeloid cell | Intermediate monocytes | Circulating CD14hiCD16+ monocyte subset with both proinflammatory and anti‐inflammatory roles 81 | Multiple clinical studies (n≈100–1000) showing an association between increased intermediate monocyte populations and severity of CAD, 82 , 83 , 84 , 85 vulnerability of plaque, 86 and future MACE 87 |
| Myeloid cell | Nonclassical monocytes | Circulating CD14lowCD16+ monocyte subset with anti‐inflammatory role 81 | A small clinical study (n≈80) of patients with HIV showed that nonclassical monocyte populations correlated with progression of coronary calcium score. 88 A small clinical study (n≈20) showed that an increased Slan+CXCR6+ subset positively correlated with CAD severity 81 |
| Myeloid cell | DCs | Antigen‐presenting cells, which present self and nonself molecules to the adaptive immune system 77 | Small clinical studies (n≈30–100) showing decreased number of myeloid DCs 89 , 90 or plasmacytoid DCs 91 , 92 in patients with CAD compared with healthy controls |
| Lymphoid cell | CD4 T cells | T‐helper cell subset, which acts primarily to regulate the cellular and humoral immune responses 83 | A small clinical study (n≈80) of patients with rheumatoid arthritis showed increased CD4 CD28‐CD56+CD57+ T cells associated with higher coronary artery calcium after correction for risk factors 83 |
| Lymphoid cell | NK cells | Distinct CD16+CD56+ lymphoid lineage cells with cytotoxic activity, but no antigen‐specific receptors 77 | Multiple clinical studies (n≈50–190) showed reduction of NK cells in patients with CAD compared to healthy controls 93 , 94 , 95 |
| Lymphoid cell | NKT cells | T‐cell lineage CD16+CD56+CD3+ cells with both T‐cell and NK‐cell characteristics 77 | Small clinical studies (n≈30–190) showed reduction of NKT cells in patients with CAD compared with healthy controls 93 , 96 |
| Leukocyte ratio | NLR | Ratio of neutrophils to lymphocytes in peripheral circulation, calculated from full blood count 97 | Elevated ratio is associated with CAD (pooled OR, 1.62) in a large meta‐analysis (n≈76,000) 97 |
CAD indicates coronary artery disease; CD, cluster of differentiation; CXCR6, C‐X‐C chemokine receptor 6; DC, dendritic cell; EPC, endothelial progenitor cell; HR, hazard ratio; MACE, major adverse cardiac events; NK, natural killer; NKT, natural killer T; NLR, neutrophil lymphocyte ratio; OR, odds ratio; and VEGFR2, vascular endothelial growth factor receptor 2.
Circulating immune cells include a dynamic population of leukocytes that are actively responding to the status of the endothelium they flow past. We have extensive evidence that atherosclerosis is an inflammatory process, and it is indisputable that this influences the cells nearby. By sampling the cells in peripheral circulation, we may be able to better detect the dynamic changes that are occurring as atherosclerosis develops.
Previous investigations have tried to find associations between CAD and a single small molecule or cellular subtype, but there are advantages to instead using an unbiased approach to analyze all of the populations of circulating immune cells. We know that the immune system responds as a complex integrated network to combat perceived threats, and the likelihood of detecting a single cell that changes specifically in response to CAD is low. It is possible that measurable changes in the immune system may be directly or indirectly responsible for causing CAD, or it may be that the changes are secondary to the development of plaque as a primary pathology. Either way, the ability to measure the system as a whole and identify a change in the immune signature in response to CAD is likely to involve shifts in multiple cell populations, which can be detected using network analyses. Previously, this may have been impractical because of the difficulty in measuring so many peripheral blood cell populations in a single blood sample, but advances in technology have now made this a practical option for research and, soon, in clinical tests.
Immunophenotyping by Mass Cytometry
Immunophenotyping is defined as a process that uses antibodies to identify cells based on the types of surface or intracellular antigens they possess; this allows the identification of the proportions of cell types of interest within a larger heterogeneous population. 98 Previously, this has been accomplished by flow cytometry, a critically important technique in immunology that dates back to the 1950s. Flow cytometry measures the scatter and emission of light energy across a range of wavelengths, as a single stream of cells passes by a light source, usually a laser. Different fluorophores, each with a distinctive excitation and emission profile, are tagged to antibodies that bind specific cellular targets. As the cell passes the light source, each fluorophore is excited and emits a signal at a particular wavelength; this allows simultaneous measurement of multiple characteristics per cell, allowing identification and quantification of cell populations. The technical limitations of flow cytometry relate to the overlapping spectra of the fluorophores and the limits of discrete detection. The maximum number can be as high as 30 markers with the most advanced cytometers and newly developed fluorophores, although in practice the complexity of the overlapping spectra make this difficult to achieve.
More recently, the development of mass cytometry time of flight (CyTOF) 99 has overcome the limitations of fluorescence flow cytometry by using isotopically pure metal ions conjugated to the antibodies, read by a spectrophotometric approach, as detailed in Figure 3. The technique could theoretically allow the use of 100 distinct labels, and presently is being utilized in the 40 to 50 range without excessive artefact. This allows evaluation of complex cellular systems by measurement of intracellular and surface proteins within the subpopulations of analyzed cells.
Figure 3. Overview of typical sample preparation and analysis of single cells by mass cytometry.
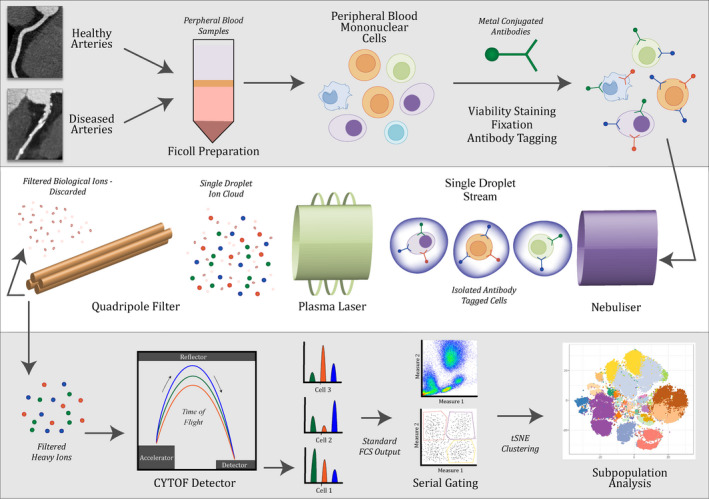
CyTOF indicates mass cytometry time of flight; FCS, flow cytometry standard; and tSNE, t‐distributed stochastic neighbor embedding.
CyTOF generates large data sets, and there are diverse approaches and many tools that have been developed to aid analysis of mass cytometry data. These analysis methods allow for quality assessment of data, cell‐type identification, clustering, and dimensionality reduction, and there are many focused reviews of this rapidly advancing field. 100 , 101 These tools and approaches are readily applicable among multiple high‐dimensional cytometry platforms including spectral flow cytometry, 102 in addition to the mass cytometry strategies of immunophenotyping discussed in the mentioned references.
Another technique that has often been used in conjunction with CyTOF is single‐cell RNA‐sequencing (scRNA‐seq) analysis, which assays RNA transcripts rather than protein expression. There are a wide variety of scRNA‐seq technologies that broadly differ in respect to the following features: (1) transcript coverage; (2) cell isolation and barcoding cells; (3) number of cells captured per experiment (throughput); and (4) inclusion of unique molecular identifiers. Compared with 3′‐end or 5′‐end counting methods, full‐length scRNA‐seq techniques have an advantage in that isoform usage and allelic expression can be determined at the level of a single cell. 103 For immunophenotyping, methods that simultaneously capture 5′ and the V(D)J region have been powerful approaches for immune repertoire profiling. The most widely used methods, particularly for immune cells, employ a droplet‐based microfluidic cell capture, such as the Chromium system from 10x Genomics. 104 These approaches barcode the mRNA from each cell and utilize unique molecular identifiers to quantify the exact number of a given transcript molecule within a cell. The resulting cell‐barcoded library is then sequenced using a next‐generation sequencing platform, 105 and the mRNA data are bioinformatically assigned back to an individual cell. 104 , 106 Sophisticated techniques are able to classify and assign cells to specific immune cell types based on their overall transcriptional profiles, 107 , 108 offering a useful additional framework through which to inform cellular profiling approaches such as mass cytometry.
High‐Dimensional Immunophenotyping in Medicine
CyTOF is an emerging technology being used in different fields, but it has been previously applied to several clinical problems with interesting results. Using CyTOF as a high‐throughput technique has resulted in the identification of cell types associated with colorectal tumors, 109 Hodgkin lymphoma, 110 and postoperative surgical recovery. 111 Immune cell profiling has been used in the field of cancer immunotherapy, identifying which patients will develop certain drug complications. 112 , 113 CyTOF is an appealing tool for clinical situations such as bone marrow transplants, where it has potential for global monitoring of post‐transplant reconstitution of immune cells and for prediction of complications. 114 Although these areas still require further research, some of these early results are encouraging and suggest that mass cytometry will find a permanent role in clinical medicine.
Most of the research in humans has been in smaller studies focused on discovery, but CyTOF also provides the capacity to supply large volumes of data to clinical trials, 115 and has recently been validated for both peripheral blood cells and tissue in comparison to flow cytometry. 116 CyTOF analysis has been utilized in clinical studies in melanoma, 117 , 118 , 119 , 120 hemopoietic stem cell transplants, 121 , 122 rheumatoid arthritis, 123 and diabetes mellitus, 124 and has included examination of tumor or tissue samples in addition to circulating immune cells. One interesting aspect of the inclusion of CyTOF and other unbiased single‐cell data to clinical studies is that data sets for these cohorts are often made available for external review, to confirm findings as well as to answer new scientific questions. This significantly expands the potential of these data sets to address basic and translational inquiries that might otherwise require repetition of expensive studies in clinical medicine. Several data sets from multi‐omics clinical studies, which include CyTOF results, are offering access to their data for external analysis, 117 , 118 , 123 , 125 and while the additional utility that will be derived from each is not yet known, it seems likely to be considerable.
CyTOF in Atherosclerosis
The use of CyTOF in atherosclerosis is in its infancy, but, even at these early stages, the usefulness of the technique can be clearly seen. In murine models of atherosclerosis such as apolipoprotein E and low‐density lipoprotein receptor knockouts, detailed studies have primarily focused on characterization of immune cells within the plaque, comparing the mild atherosclerosis induced by a chow diet with the severe atherosclerosis that develops with a Western diet. Winkels and colleagues 126 used CyTOF in conjunction with scRNA‐seq to generate an atlas of aortic atherosclerotic immune cells, which also looked at gene pathway enrichment, and underscored the need to perform such analyses stratified by immune cell subpopulation. Other murine studies have used similar models to investigate aortic myeloid cell distributions 127 and to do deep phenotyping of monocyte subsets. 128
In humans, the major clinical study to date has been performed by Fernandez and colleagues, 125 who assessed carotid atherosclerotic plaque by scRNA‐seq, cellular indexing of transcriptomes and epitopes by sequencing (CITE‐seq), and CyTOF in patients undergoing carotid endarterectomy. A comparison of the immune cells found in the plaque and the circulating peripheral blood was made, which showed an increase in the heterogenicity of T cells within the atheroma. When symptomatic plaques were compared with asymptomatic plaques, differences in CD4 T‐cell subsets, macrophage activation, and T‐cell exhaustion measures were identified. This study highlights the feasibility of detecting significant differences in immune cell profiles in human atherosclerosis, and provides excellent groundwork towards identifying the immune signatures expected in disease. The next steps to translate this as a diagnostic tool will be to compare diseased populations with healthy controls, and peripheral blood is a far more clinically accessible target than atheroma or the blood vessel wall.
At the time of writing, circulating immune cell profiles in humans with coronary disease had not been extensively assessed. One early CyTOF study used the technique to better characterize monocyte subsets in patients with CAD, 129 and a more recent study looking into monocyte heterogeneity used clustering analysis of CyTOF data from healthy individuals to identify markers that distinguished meta‐clusters within nonclassical monocytes, and then showed that equivalent monocyte subpopulations defined as Slan+ were associated with disease severity in patients with CAD. 81 However, no study to date has assessed the peripheral blood profile as a whole, and this opportunity to assess the immune cell populations of patients by simple venesection should not be missed. Considering the long‐term nature of atherogenesis, the involvement of adaptive immune cells conveying antigenic memory is likely to be significant and may be a peripherally detectable signature of early atherosclerosis.
The potential implications of improved immunophenotyping to inform decisions about therapy are also fascinating. We know from the CANTOS trial 25 that anti‐inflammatory therapies have potential morbidity and mortality benefit, and that the subset of patients whose C‐reactive protein decreased after canakinumab use had more benefit from this therapy. 26 Patient selection seems to be a critical issue here; however, it appears that high‐sensitivity C‐reactive protein is not specific enough to identify those patients who will benefit from immune therapies. Precision immunophenotyping has the potential to identify the immune activity that is directly related to CAD, letting us precisely treat the patients who will benefit from these highly targeted therapies.
In view of the growing body of evidence that links the immune system to CAD, CyTOF should be comprehensively assessed as a potential tool in precision medicine. To determine the utility in a clinical setting, examination of immune signatures in circulating cells that could be collected as part of a routine heart health check is essential. It is highly feasible, with the rapidly improving technology of CyTOF and the ability for samples to be studied after freezing and transport, that such a tool could help us in identifying patients with active subclinical atherosclerosis, leading to earlier diagnosis and prevention of cardiac events in patients who had SMuRFs without traditional risk factors.
Insights From scRNA‐Seq
scRNA‐seq has become a key technique in the era of single‐cell unbiased discovery research and is particularly important in diseases such as atherosclerosis that involve tissues with heterogenous cell populations. Using next‐generation sequencing and modern microfluidics techniques, high‐throughput scRNA‐seq has been commercially available since 2017, and has recently been comprehensively reviewed from the atherosclerosis perspective. 130 scRNA‐seq studies of atherosclerosis have primarily been performed in mouse models and have focused on plaque composition, often in conjunction with other single‐cell techniques such as CyTOF, as outlined in the previous section. 125 , 126 Additional murine studies have led to the discovery of a novel subset of triggering receptor expressed on myeloid cells 2hi macrophages involved in aortic atherosclerosis, 131 helped to characterize the proinflammatory characteristics of foamy versus nonfoamy macrophages, 132 assessed the functional features of subsets of migratory and dancing macrophages, 133 and defined a spectrum of macrophage activation states that also resulted in the discovery of an unexpected subset of proliferating, stem cell–like resident monocytes in aortic plaque. 134 scRNA‐seq has been shown to be an especially powerful discovery tool in combination with CyTOF, with the transcriptomic data used to inform the CyTOF panel design and provide synergistic information about gene expression versus surface marker expression. One exciting link in profiling cells using scRNA‐seq and CyTOF is the simultaneous epitope and transcriptome measurement in individual cells (CITE‐seq). 135 CITE‐seq uses oligonucleotide‐labeled antibodies in conjunction with existing single‐cell RNA capture techniques to profile a combination of cell surface proteins and cellular RNA. Compared with CyTOF, CITE‐seq has a number of distinctions in type and quantification of protein capture. While both approaches provide a quantitative estimate of protein molecules per cell with similar dynamic ranges (4 logs), 135 , 136 CITE‐seq is currently unable to assay intracellular proteins. However, one of the key differences is the number of cells in a given experiment from which data can be generated. As CITE‐seq uses microfluidic systems, the upper bounds of cell capture is bounded by an increasing doublet rate, with the best commercial platforms limited to capturing ≈15 000 cells per experiment. CyTOF, on the other hand, is able to assay millions of cells in a single experiment, providing a deeper resolution of cellular heterogeneity as defined by protein measurement. However, as far as we are aware, there has not yet been a direct comparison of CyTOF, CITE‐seq, or associated techniques such as proximal ligation assay for RNA 137 and RNA expression and protein sequencing assay. 138 Such work would be of considerable value in helping inform under which scenarios respective approaches are best suited. However, when both technologies are used on cells from the same sample, it provides a framework to study the relationship between post‐translational gene expression and protein abundance. The resulting transformation of our knowledge on inflammatory signaling in atherosclerosis has potential to provide a plethora of novel therapeutic targets and potential biomarkers.
Conclusions
CyTOF is an exciting new development in modern science that should be effectively utilized in the search for new links between atherosclerosis and the immune system. Moreover, CyTOF is not just an unbiased approach for discovery; the information provided, particularly in combination with scRNA‐seq data, offers unrivaled capacity for further enhancing our understanding of the complex immune responses that occur in the setting of atherosclerosis. The ability to sample the blood circulating directly past atherosclerotic plaques has potential to quantify the presence of disease and elucidate the inflammatory properties of an individual's vascular endothelium. A broader understanding of immune system dynamics in individual patients may also provide evidence of immune factors involved in causing CAD, particularly in patients with SMuRFs. Thus, these immune signatures could have a variety of uses in the diagnosis, monitoring, and targeting of therapy for CAD.
There is now extensive evidence linking inflammation and CAD, and identification of an immune cell profile that could be used in clinical assessment of atherosclerosis would be an ideal outcome of research in this area. Additionally, other findings may prompt exciting lines of scientific inquiry that could improve our understanding of the biology of inflammatory atherogenesis, suggesting novel therapeutic targets and methods of monitoring therapies. CyTOF has potentially broad applications in the field of cardiology and brings inspiring immunological cross‐disciplinary insights to the field of vascular biology.
Sources of Funding
The authors report the following financial support for the research, authorship, and/or publication of this article: Kott is supported by an Australian Commonwealth Government Research Training Program Stipend Scholarship; Vernon is supported by a University of Sydney Postgraduate Research Scholarship funded by Heart Research Australia; Powell is supported by a National Health and Medical Research Council Investigator Fellowship (grant number AP1175781); McGuire is supported by the International Society for the Advancement of Cytometry Marylou Ingram Scholars Program; and Figtree is supported by a National Health and Medical Research Council Practitioner Fellowship (grant number APP11359290), Heart Research Australia, and the New South Wales Office of Health and Medical Research.
Disclosures
None.
(J Am Heart Assoc. 2020;9:e017759. DOI: 10.1161/JAHA.120.017759.)
For Sources of Funding and Disclosures, see page 13.
References
- 1. Australian Bureau of Statistics . Australia's leading causes of death, 2018. 2018; catergory no. 3303.0, viewed 30 April 2020. Available at: https://www.abs.gov.au/ausstats/abs@.nsf/mf/3303.0. Accessed April 30, 2020.
- 2. Wang H, Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd‐Allah F, Abera SF, Abraha HN, Abu‐Raddad LJ, Abu‐Rmeileh NM, et al. Global, regional, and national under‐5 mortality, adult mortality, age‐specific mortality, and life expectancy, 1970–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1084–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roth GA, Johnson C, Abajobir A, Abd‐Allah F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K, et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017;70:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham heart study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yusuf S, Hawken S, Ôunpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case‐control study. Lancet. 2004;364:937–952. [DOI] [PubMed] [Google Scholar]
- 6. Rowe AK, Powell KE, Flanders WD. Why population attributable fractions can sum to more than one. Am J Prev Med. 2004;26:243–249. [DOI] [PubMed] [Google Scholar]
- 7. DeFilippis AP, Young R, Carrubba CJ, McEvoy JW, Budoff MJ, Blumenthal RS, Kronmal RA, McClelland RL, Nasir K, Blaha MJ. An analysis of calibration and discrimination among multiple cardiovascular risk scores in a modern multiethnic cohort. Ann Intern Med. 2015;162:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vernon ST, Coffey S, Bhindi R, Soo Hoo SY, Nelson GI, Ward MR, Hansen PS, Asrress KN, Chow CK, Celermajer DS, et al. Increasing proportion of ST elevation myocardial infarction patients with coronary atherosclerosis poorly explained by standard modifiable risk factors. Eur J Prev Cardiol. 2017;24:1824–1830. [DOI] [PubMed] [Google Scholar]
- 9. Vernon ST, Coffey S, D'Souza M, Chow CK, Kilian J, Hyun K, Shaw JA, Adams M, Roberts‐Thomson P, Brieger D, et al. ST‐segment‐elevation myocardial infarction (STEMI) patients without standard modifiable cardiovascular risk factors‐how common are they, and what are their outcomes? J Am Heart Assoc. 2019;8:e013296. DOI: 10.1161/JAHA.119.013296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu MY, Li CJ, Hou MF, Chu PY. New insights into the role of inflammation in the pathogenesis of atherosclerosis. Int J Mol Sci. 2017;18:2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Libby P. Inflammation in atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32:2045–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rocha VZ, Libby P. Obesity, inflammation, and atherosclerosis. Nat Rev Cardiol. 2009;6:399. [DOI] [PubMed] [Google Scholar]
- 13. Nguyen MT, Fernando S, Schwarz N, Tan JT, Bursill CA, Psaltis PJ. Inflammation as a therapeutic target in atherosclerosis. J Clin Med. 2019;8:1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies RW, Wells GA, Stewart AF, Erdmann J, Shah SH, Ferguson JF, Hall AS, Anand SS, Burnett MS, Epstein SE, et al. A genome‐wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ Cardiovasc Genet. 2012;5:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, et al. Large‐scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nikpay M, Goel A, Won H‐H, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 genomes‐based genome‐wide association meta‐analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carrizales‐Sepúlveda EF, Ordaz‐Farías A, Vera‐Pineda R, Flores‐Ramírez R. Periodontal disease, systemic inflammation and the risk of cardiovascular disease. Heart Lung Circ. 2018;11:1327–1334. [DOI] [PubMed] [Google Scholar]
- 18. Ott SJ, El Mokhtari NE, Musfeldt M, Hellmig S, Freitag S, Rehman A, Kühbacher T, Nikolaus S, Namsolleck P, Blaut M, et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation. 2006;113:929–937. [DOI] [PubMed] [Google Scholar]
- 19. Borel N, Summersgill JT, Mukhopadhyay S, Miller RD, Ramirez JA, Pospischil A. Evidence for persistent chlamydia pneumoniae infection of human coronary atheromas. Atherosclerosis. 2008;199:154–161. [DOI] [PubMed] [Google Scholar]
- 20. Andraws R, Berger JS, Brown DL. Effects of antibiotic therapy on outcomes of patients with coronary artery disease: a meta‐analysis of randomized controlled trials. JAMA. 2005;293:2641–2647. [DOI] [PubMed] [Google Scholar]
- 21. Chen S, Shimada K, Crother TR, Erbay E, Shah PK, Arditi M. Chlamydia and lipids engage a common signaling pathway that promotes atherogenesis. J Am Coll Cardiol. 2018;71:1553–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chyu KY, Shah PK. In pursuit of an atherosclerosis vaccine. Circ Res. 2018;123:1121–1123. [DOI] [PubMed] [Google Scholar]
- 23. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu CL, Sano S, Muralidharan S, Rius C, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 26. Ridker PM, MacFadyen JG, Everett BM, Libby P, Thuren T, Glynn RJ; Group CT . Relationship of C‐reactive protein reduction to cardiovascular event reduction following treatment with canakinumab: a secondary analysis from the CANTOS randomised controlled trial. Lancet. 2018;391:319–328. [DOI] [PubMed] [Google Scholar]
- 27. Martinez GJ, Celermajer DS, Patel S. The NLRP3 inflammasome and the emerging role of colchicine to inhibit atherosclerosis‐associated inflammation. Atherosclerosis. 2018;269:262–271. [DOI] [PubMed] [Google Scholar]
- 28. Antoniades C. 'Dysfunctional' adipose tissue in cardiovascular disease: a reprogrammable target or an innocent bystander? Cardiovasc Res. 2017;113:997–998. [DOI] [PubMed] [Google Scholar]
- 29. Guzik TJ, Skiba DS, Touyz RM, Harrison DG. The role of infiltrating immune cells in dysfunctional adipose tissue. Cardiovasc Res. 2017;113:1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nosalski R, Guzik TJ. Perivascular adipose tissue inflammation in vascular disease. Br J Pharmacol. 2017;174:3496–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antonopoulos AS, Sanna F, Sabharwal N, Thomas S, Oikonomou EK, Herdman L, Margaritis M, Shirodaria C, Kampoli AM, Akoumianakis I, et al. Detecting human coronary inflammation by imaging perivascular fat. Sci Transl Med. 2017;9:eaal2658. [DOI] [PubMed] [Google Scholar]
- 32. Ohyama K, Matsumoto Y, Shimokawa H. Coronary artery spasm and perivascular adipose tissue inflammation: insights from translational imaging research. Eur Cardiol. 2019;14:6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. The SCOT‐HEART Investigators . CT coronary angiography in patients with suspected angina due to coronary heart disease (SCOT‐HEART): an open‐label, parallel‐group, multicentre trial. Lancet. 2015;385:2383–2391. [DOI] [PubMed] [Google Scholar]
- 34. Oikonomou EK, Williams MC, Kotanidis CP, Desai MY, Marwan M, Antonopoulos AS, Thomas KE, Thomas S, Akoumianakis I, Fan LM, et al. A novel machine learning‐derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J. 2019;40:3529–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd‐Allah F, Abdulkader RS, Abdulle AM, Abebo TA, Abera SF, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de St Groth BF. Regulatory T‐cell abnormalities and the global epidemic of immuno‐inflammatory disease. Immunol Cell Biol. 2012;90:256–259. [DOI] [PubMed] [Google Scholar]
- 37. Lee N, Kim WU. Microbiota in T‐cell homeostasis and inflammatory diseases. Exp Mol Med. 2017;49:e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Rosa V, La Cava A, Matarese G. Metabolic pressure and the breach of immunological self‐tolerance. Nat Immunol. 2017;18:1190–1196. [DOI] [PubMed] [Google Scholar]
- 39. Harpsøe MC, Basit S, Andersson M, Nielsen NM, Frisch M, Wohlfahrt J, Nohr EA, Linneberg A, Jess T. Body mass index and risk of autoimmune diseases: a study within the Danish National Birth Cohort. Int J Epidemiol. 2014;43:843–855. [DOI] [PubMed] [Google Scholar]
- 40. Jie Z, Xia H, Zhong S‐L, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–638. [DOI] [PubMed] [Google Scholar]
- 42. Blohmé G, Nyström L, Arnqvist HJ, Lithner F, Littorin B, Olsson PO, Scherstén B, Wibell L, Östman J. Male predominance of type 1 (insulin‐dependent) diabetes mellitus in young adults: results from a 5‐year prospective nationwide study of the 15–34‐year age group in Sweden. Diabetologia. 1992;35:56–62. [DOI] [PubMed] [Google Scholar]
- 43. Gubbels Bupp MR. Sex, the aging immune system, and chronic disease. Cell Immunol. 2015;294:102–110. [DOI] [PubMed] [Google Scholar]
- 44. Yayan J. Emerging families of biomarkers for coronary artery disease: inflammatory mediators. Vasc Health Risk Manag. 2013;9:435–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Christodoulidis G, Vittorio TJ, Fudim M, Lerakis S, Kosmas CE. Inflammation in coronary artery disease. Cardiol Rev. 2014;22:279–288. [DOI] [PubMed] [Google Scholar]
- 46. Soeki T, Sata M. Inflammatory biomarkers and atherosclerosis. Int Heart J. 2016;57:134–139. [DOI] [PubMed] [Google Scholar]
- 47. Mirzaei H, Ferns GA, Avan A, Mobarhan MG. Cytokines and microRNA in coronary artery disease. Adv Clin Chem. 2017;82:47–70. [DOI] [PubMed] [Google Scholar]
- 48. Pepys MB, Hirschfield GM. C‐reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaptoge S, Di Angelantonio E, Pennells L, Wood AM, White IR, Gao P, Walker M, Thompson A, Sarwar N, Caslake M, et al. C‐reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med. 2012;367:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Davalos D, Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin Immunopathol. 2012;34:43–62. [DOI] [PubMed] [Google Scholar]
- 51. Sack GH Jr. Serum amyloid A—a review. Mol Med. 2018;24:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ridker PM, Hennekens CH, Buring JE, Rifai N. C‐reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med. 2000;342:836–843. [DOI] [PubMed] [Google Scholar]
- 53. Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL‐10 in immunity and cancer. Cancer Lett. 2015;367:103–107. [DOI] [PubMed] [Google Scholar]
- 54. Subirana I, Fito M, Diaz O, Vila J, Frances A, Delpon E, Sanchis J, Elosua R, Munoz‐Aguayo D, Degano IR, et al. Prediction of coronary disease incidence by biomarkers of inflammation, oxidation, and metabolism. Sci Rep. 2018;8:3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liang K, Dong SR, Peng H. Serum levels and clinical significance of IFN‐γ and IL‐10 in patients with coronary heart disease. Eur Rev Med Pharmacol Sci. 2016;20:1339–1343. [PubMed] [Google Scholar]
- 56. Barcelos AL, de Oliveira EA, Haute GV, Costa BP, Pedrazza L, Donadio MV, de Oliveira JR, Bodanese LC. Association of IL‐10 to coronary disease severity in patients with metabolic syndrome. Clin Chim Acta. 2019;495:394–398. [DOI] [PubMed] [Google Scholar]
- 57. Chun HY, Chung JW, Kim HA, Yun JM, Jeon JY, Ye YM, Kim SH, Park HS, Suh CH. Cytokine IL‐6 and IL‐10 as biomarkers in systemic lupus erythematosus. J Clin Immunol. 2007;27:461–466. [DOI] [PubMed] [Google Scholar]
- 58. Kulshrestha H, Gupta V, Mishra S, Mahdi AA, Awasthi S, Kumar S. Interleukin‐10 as a novel biomarker of metabolic risk factors. Diabetes Metab Syndr. 2018;12:543–547. [DOI] [PubMed] [Google Scholar]
- 59. Silva BS, Lira FS, Ramos D, Uzeloto JS, Rossi FE, Freire AP, Silva RN, Trevisan IB, Gobbo LA, Ramos EM. Severity of COPD and its relationship with IL‐10. Cytokine. 2018;106:95–100. [DOI] [PubMed] [Google Scholar]
- 60. Unver N, McAllister F. IL‐6 family cytokines: key inflammatory mediators as biomarkers and potential therapeutic targets. Cytokine Growth Factor Rev. 2018;41:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Niu W, Liu Y, Qi Y, Wu Z, Zhu D, Jin W. Association of interleukin‐6 circulating levels with coronary artery disease: a meta‐analysis implementing Mendelian randomization approach. Int J Cardiol. 2012;157:243–252. [DOI] [PubMed] [Google Scholar]
- 62. Poynter ME, Irvin CG. Interleukin‐6 as a biomarker for asthma: hype or is there something else? Eur Respir J. 2016;48:979–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bozkurt B, Mann DL, Deswal A. Biomarkers of inflammation in heart failure. Heart Fail Rev. 2010;15:331–341. [DOI] [PubMed] [Google Scholar]
- 64. Idriss HT, Naismith JH. TNFα and the TNF receptor superfamily: structure‐function relationship(s). Microsc Res Tech. 2000;50:184–195. [DOI] [PubMed] [Google Scholar]
- 65. Gomes FI, Aragão MG, Barbosa FC, Bezerra MM, de Paulo Teixeira Pinto V, Chaves HV. Inflammatory cytokines interleukin‐1β and tumour necrosis factor‐α—novel biomarkers for the detection of periodontal diseases: a literature review. J Oral Maxillofac Res. 2016;7:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Q, Zheng X. Tumor necrosis factor alpha is a promising circulating biomarker for the development of obstructive sleep apnea syndrome: a meta‐analysis. Oncotarget. 2017;8:27616–27626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Blankenberg S, Barbaux S, Tiret L. Adhesion molecules and atherosclerosis. Atherosclerosis. 2003;170:191–203. [DOI] [PubMed] [Google Scholar]
- 68. Luc G, Arveiler D, Evans A, Amouyel P, Ferrieres J, Bard JM, Elkhalil L, Fruchart JC, Ducimetiere P. Circulating soluble adhesion molecules ICAM‐1 and VCAM‐1 and incident coronary heart disease: the PRIME study. Atherosclerosis. 2003;170:169–176. [DOI] [PubMed] [Google Scholar]
- 69. Li R, Qiu Y. Diagnostic value of serum ICAM‐1 for endometriosis: a meta‐analysis. Medicine. 2018;97:e11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yao Y, Du J, Li R, Zhao L, Luo N, Zhai JY, Long L. Association between ICAM‐1 level and diabetic retinopathy: a review and meta‐analysis. Postgrad Med J. 2019;95:162. [DOI] [PubMed] [Google Scholar]
- 71. Lee WJ, Chen LK, Liang CK, Peng LN, Chiou ST, Chou P. Soluble ICAM‐1, independent of IL‐6, is associated with prevalent frailty in community‐dwelling elderly taiwanese people. PLoS One. 2016;11:e0157877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang PY, Tsao SM, Chang JH, Chien MH, Hung WY, Huang YW, Yang SF. Plasma levels of soluble intercellular adhesion molecule‐1 as a biomarker for disease severity of patients with community‐acquired pneumonia. Clin Chim Acta. 2016;463:174–180. [DOI] [PubMed] [Google Scholar]
- 73. Li HSK, Zhao R, Hu J, Hao Z, Wang F, Lu Y, Liu F, Zhang Y. Inflammatory biomarkers of coronary heart disease. Front Biosci (Landmark Ed). 2017;1:504–515. [DOI] [PubMed] [Google Scholar]
- 74. Széplaki G, Prohászka Z, Duba J, Rugonfalvi‐Kiss S, Karádi I, Kókai M, Kramer J, Füst G, Kleiber M, Romics L, et al. Association of high serum concentration of the third component of complement (C3) with pre‐existing severe coronary artery disease and new vascular events in women. Atherosclerosis. 2004;177:383–389. [DOI] [PubMed] [Google Scholar]
- 75. Speidl WS, Exner M, Amighi J, Kastl SP, Zorn G, Maurer G, Wagner O, Huber K, Minar E, Wojta J, et al. Complement component C5a predicts future cardiovascular events in patients with advanced atherosclerosis. Eur Heart J. 2005;26:2294–2299. [DOI] [PubMed] [Google Scholar]
- 76. Ekdahl KN, Persson B, Mohlin C, Sandholm K, Skattum L, Nilsson B. Interpretation of serological complement biomarkers in disease. Front Immunol. 2018;9:2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chong Mark Seow K, Ng WK, Chan Jerry Kok Y. Concise review: Endothelial progenitor cells in regenerative medicine: applications and challenges. Stem Cells Transl Med. 2016;5:530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schmidt‐Lucke C, Rössig L, Fichtlscherer S, Vasa M, Britten M, Kämper U, Dimmeler S, Zeiher AM. Reduced number of circulating endothelial progenitor cells predicts future cardiovascular events. Circulation. 2005;111:2981–2987. [DOI] [PubMed] [Google Scholar]
- 80. Padfield GJ, Tura‐Ceide O, Freyer E, Barclay GR, Turner M, Newby DE, Mills NL. Endothelial progenitor cells, atheroma burden and clinical outcome in patients with coronary artery disease. Heart. 2013;99:791. [DOI] [PubMed] [Google Scholar]
- 81. Hamers AA, Dinh HQ, Thomas GD, Marcovecchio P, Blatchley A, Nakao CS, Kim C, McSkimming C, Taylor AM, Nguyen AT, et al. Human monocyte heterogeneity as revealed by high‐dimensional mass cytometry. Arterioscler Thromb Vasc Biol. 2019;39:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lo SC, Lee WJ, Chen CY, Lee BC. Intermediate CD14(++)CD16(+) monocyte predicts severe coronary stenosis and extensive plaque involvement in asymptomatic individuals. Int J Cardiovasc Imaging. 2017;33:1223–1236. [DOI] [PubMed] [Google Scholar]
- 83. Winchester R, Giles JT, Nativ S, Downer K, Zhang HZ, Bag‐Ozbek A, Zartoshti A, Bokhari S, Bathon JM. Association of elevations of specific T cell and monocyte subpopulations in rheumatoid arthritis with subclinical coronary artery atherosclerosis. Arthritis Rheumatol. 2016;68:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ozaki Y, Imanishi T, Taruya A, Aoki H, Masuno T, Shiono Y, Komukai K, Tanimoto T, Kitabata H, Akasaka T. Circulating CD14+CD16+ monocyte subsets as biomarkers of the severity of coronary artery disease in patients with stable angina pectoris. Circ J. 2012;76:2412–2418. [DOI] [PubMed] [Google Scholar]
- 85. Höpfner F, Jacob M, Ulrich C, Russ M, Simm A, Silber RE, Girndt M, Noutsias M, Werdan K, Schlitt A. Subgroups of monocytes predict cardiovascular events in patients with coronary heart disease. The PHAMOS trial (Prospective Halle Monocytes Study). Hellenic J Cardiol. 2019;60:311–321. [DOI] [PubMed] [Google Scholar]
- 86. Yamamoto H, Yoshida N, Shinke T, Otake H, Kuroda M, Sakaguchi K, Hirota Y, Toba T, Takahashi H, Terashita D, et al. Impact of CD14++CD16+ monocytes on coronary plaque vulnerability assessed by optical coherence tomography in coronary artery disease patients. Atherosclerosis. 2018;269:245–251. [DOI] [PubMed] [Google Scholar]
- 87. Kashiwagi M, Imanishi T, Ozaki Y, Taruya A, Nishiguchi T, Katayama Y, Tanimoto T, Kuroi A, Kubo T, Tanaka A, et al. Prognostic value of human peripheral monocyte subsets for future coronary events in patients without significant coronary artery stenosis. Circ J. 2019;83:2250–2256. [DOI] [PubMed] [Google Scholar]
- 88. Zungsontiporn N, Tello RR, Zhang G, Mitchell BI, Budoff M, Kallianpur KJ, Nakamoto BK, Keating SM, Norris PJ, Ndhlovu LC, et al. Non‐classical monocytes and monocyte chemoattractant protein‐1 (MCP‐1) correlate with coronary artery calcium progression in chronically HIV‐1 infected adults on stable antiretroviral therapy. PLoS One. 2016;11:e0149143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wen J, Wen Y, Zhiliang L, Lingling C, Longxing C, Ming W, Qiang F. A decrease in the percentage of circulating mDC precursors in patients with coronary heart disease: a relation to the severity and extent of coronary artery lesions? Heart Vessels. 2013;28:135–142. [DOI] [PubMed] [Google Scholar]
- 90. Yilmaz A, Weber J, Cicha I, Stumpf C, Klein M, Raithel D, Daniel WG, Garlichs CD. Decrease in circulating myeloid dendritic cell precursors in coronary artery disease. J Am Coll Cardiol. 2006;48:70–80. [DOI] [PubMed] [Google Scholar]
- 91. Van Vré EA, Hoymans VY, Bult H, Lenjou M, Van Bockstaele DR, Vrints CJ, Bosmans JM. Decreased number of circulating plasmacytoid dendritic cells in patients with atherosclerotic coronary artery disease. Coron Artery Dis. 2006;17:243–248. [DOI] [PubMed] [Google Scholar]
- 92. Van Brussel I, Van Vré EA, De Meyer Guido RY, Vrints Christiaan J, Bosmans Johan M, Bult H. Decreased numbers of peripheral blood dendritic cells in patients with coronary artery disease are associated with diminished plasma FLT3 ligand levels and impaired plasmacytoid dendritic cell function. Clin Sci. 2011;120:415–426. [DOI] [PubMed] [Google Scholar]
- 93. Jabir NR, Firoz CK, Ahmed F, Kamal MA, Hindawi S, Damanhouri GA, Almehdar HA, Tabrez S. Reduction in CD16/CD56 and CD16/CD3/CD56 natural killer cells in coronary artery disease. Immunol Invest. 2017;46:526–535. [DOI] [PubMed] [Google Scholar]
- 94. Backteman K, Ernerudh J, Jonasson L. Natural killer (NK) cell deficit in coronary artery disease: no aberrations in phenotype but sustained reduction of NK cells is associated with low‐grade inflammation. Clin Exp Immunol. 2014;175:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jonasson L, Backteman K, Ernerudh J. Loss of natural killer cell activity in patients with coronary artery disease. Atherosclerosis. 2005;183:316–321. [DOI] [PubMed] [Google Scholar]
- 96. Andoh Y, Fujii S, Iwabuchi K, Yokota T, Inoue N, Nakai Y, Mishima T, Yamashita T, Nakagawa T, Kitabatake A, et al. Lower prevalence of circulating natural killer T cells in patients with angina: a potential novel marker for coronary artery disease. Coron Artery Dis. 2006;17:523–528. [DOI] [PubMed] [Google Scholar]
- 97. Angkananard T, Anothaisintawee T, McEvoy M, Attia J, Thakkinstian A. Neutrophil lymphocyte ratio and cardiovascular disease risk: a systematic review and meta‐analysis. Biomed Res Int. 2018;2018:2703518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the human immunology project. Nat Rev Immunol. 2012;12:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kimball AK, Oko LM, Bullock BL, Nemenoff RA, van Dyk LF, Clambey ET. A beginner's guide to analyzing and visualizing mass cytometry data. J Immunol. 2018;200:3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Keyes TJ, Domizi P, Lo YC, Nolan GP, Davis KL. A cancer biologist's primer on machine learning applications in high‐dimensional cytometry. Cytometry A. 2020;97:782–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ferrer‐Font L, Pellefigues C, Mayer JU, Small SJ, Jaimes MC, Price KM. Panel design and optimization for high‐dimensional immunophenotyping assays using spectral flow cytometry. Curr Protoc Cytom. 2020;92:e70. [DOI] [PubMed] [Google Scholar]
- 103. Hagemann‐Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks GJ, Larsson AJ, Faridani OR, Sandberg R. Single‐cell RNA counting at allele and isoform resolution using Smart‐seq3. Nat Biotechnol. 2020;38:708–714. [DOI] [PubMed] [Google Scholar]
- 104. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Senabouth A, Andersen S, Shi Q, Shi L, Jiang F, Zhang W, Wing K, Daniszewski M, Lukowski SW, Hung SS, et al. Comparative performance of the BGI and Illumina sequencing technology for single‐cell RNA‐sequencing. NAR Genom Bioinform. 2020;2:lqaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Senabouth A, Lukowski SW, Hernandez JA, Andersen SB, Mei X, Nguyen QH, Powell JE. Ascend: R package for analysis of single‐cell RNA‐seq data. Gigascience. 2019;8:gizo87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Alquicira‐Hernandez J, Sathe A, Ji HP, Nguyen Q, Powell JE. scPred: accurate supervised method for cell‐type classification from single‐cell RNA‐seq data. Genome Biol. 2019;20:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Abdelaal T, Michielsen L, Cats D, Hoogduin D, Mei H, Reinders MJT, Mahfouz A. A comparison of automatic cell identification methods for single‐cell RNA sequencing data. Genome Biol. 2019;20:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Norton SE, Ward‐Hartstonge KA, McCall JL, Leman JK, Taylor ES, Munro F, Black MA, de St Groth BF, McGuire HM, et al. High‐dimensional mass cytometric analysis reveals an increase in effector regulatory T cells as a distinguishing feature of colorectal tumors. J Immunol. 2019;202:1871. [DOI] [PubMed] [Google Scholar]
- 110. Cader FZ, Schackmann RC, Hu X, Wienand K, Redd R, Chapuy B, Ouyang J, Paul N, Gjini E, Lipschitz M, et al. Mass cytometry of Hodgkin lymphoma reveals a CD4(+) regulatoryT‐cell-rich and exhausted T‐effector microenvironment. Blood. 2018;132:825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gaudillière B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, Silva J, Ganio EA, Yeh CG, Maloney WJ, et al. Clinical recovery from surgery correlates with single‐cell immune signatures. Sci Transl Med. 2014;6:255ra131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lomax AJ, McGuire HM, McNeil C, Choi CJ, Hersey P, Karikios D, Shannon K, van Hal S, Carr U, Crotty A, et al. Immunotherapy‐induced sarcoidosis in patients with melanoma treated with PD‐1 checkpoint inhibitors: case series and immunophenotypic analysis. Int J Rheum Dis. 2017;20:1277–1285. [DOI] [PubMed] [Google Scholar]
- 113. McGuire HM, Shklovskaya E, Edwards J, Trevillian PR, McCaughan GW, Bertolino P, McKenzie C, Gourlay R, Gallagher SJ, de St Groth BF, et al. Anti‐PD-1-induced high‐grade hepatitis associated with corticosteroid‐resistant T cells: a case report. Cancer Immunol Immunother. 2018;67:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Stern L, McGuire H, Avdic S, Rizzetto S, de St Groth BF, Luciani F, Slobedman B, Blyth E. Mass cytometry for the assessment of immune reconstitution after hematopoietic stem cell transplantation. Front Immunol. 2018;9:1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hartmann FJ, Babdor J, Gherardini PF, Amir ED, Jones K, Sahaf B, Marquez DM, Krutzik P, O'Donnell E, Sigal N, et al. Comprehensive immune monitoring of clinical trials to advance human immunotherapy. Cell Rep. 2019;28:819–831.e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gadalla R, Noamani B, MacLeod BL, Dickson RJ, Guo M, Xu W, Lukhele S, Elsaesser HJ, Razak ARA, Hirano N, et al. Validation of CyTOF against flow cytometry for immunological studies and monitoring of human cancer clinical trials. Front Oncol. 2019;9:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. T‐cell invigoration to tumour burden ratio associated with anti‐PD-1 response. Nature. 2017;545:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, Dummer R, Robinson MD, Levesque MP, Becher B. High‐dimensional single‐cell analysis predicts response to anti‐PD-1 immunotherapy. Nat Med. 2018;24:144–153. [DOI] [PubMed] [Google Scholar]
- 119. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, Madore J, Lim SY, Velickovic R, Wongchenko M, et al. Distinct immune cell populations define response to anti‐PD-1 monotherapy and anti‐PD-1/anti‐CTLA-4 combined therapy. Cancer Cell. 2019;35:238–255.e236. [DOI] [PubMed] [Google Scholar]
- 120. Pirozyan MR, McGuire HM, Emran AA, Tseng HY, Tiffen JC, Lee JH, Carlino MS, Menzies AM, Long GV, Scolyer RA, et al. Pretreatment innate cell populations and CD4 T cells in blood are associated with response to immune checkpoint blockade in melanoma patients. Front Immunol. 2020;11:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lakshmikanth T, Olin A, Chen Y, Mikes J, Fredlund E, Remberger M, Omazic B, Brodin P. Mass cytometry and topological data analysis reveal immune parameters associated with complications after allogeneic stem cell transplantation. Cell Rep. 2017;20:2238–2250. [DOI] [PubMed] [Google Scholar]
- 122. McGuire HM, Rizzetto S, Withers BP, Clancy LE, Avdic S, Stern L, Patrick E, de St Groth BF, Slobedman B, et al. Mass cytometry reveals immune signatures associated with cytomegalovirus (CMV) control in recipients of allogeneic haemopoietic stem cell transplant and CMV‐specific T cells. Clin Transl Immunology. 2020;9:e1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon‐Escoto K, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single‐cell transcriptomics and mass cytometry. Nat Immunol. 2019;20:928–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Eichmann M, Baptista R, Ellis RJ, Heck S, Peakman M, Beam CA. Costimulation blockade disrupts CD4+ T cell memory pathways and uncouples their link to decline in β‐cell function in type 1 diabetes. J Immunol. 2020;204:3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, et al. Single‐cell immune landscape of human atherosclerotic plaques. Nat Med. 2019;25:1576–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AAJ, Cochain C, Vafadarnejad E, Saliba A‐E, et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single‐cell RNA‐sequencing and mass cytometry. Circ Res. 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cole JE, Park I, Ahern DJ, Kassiteridi C, Danso Abeam D, Goddard ME, Green P, Maffia P, Monaco C. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc Res. 2018;114:1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Roberts ME, Barvalia M, Silva J, Cederberg RA, Chu W, Wong A, Tai DC, Chen S, Matos I, Priatel JJ, et al. Deep phenotyping by mass cytometry and single‐cell RNA‐sequencing reveals lyn‐regulated signaling profiles underlying monocyte subset heterogeneity and lifespan. Circ Res. 2020;126:e61–e79. [DOI] [PubMed] [Google Scholar]
- 129. Thomas GD, Hamers AAJ, Nakao C, Marcovecchio P, Taylor AM, McSkimming C, Nguyen AT, McNamara CA, Hedrick CC. Human blood monocyte subsets: a new gating strategy defined using cell surface markers identified by mass cytometry. Arterioscler Thromb Vasc Biol. 2017;37:1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Williams JW, Winkels H, Durant CP, Zaitsev K, Ghosheh Y, Ley K. Single cell RNA sequencing in atherosclerosis research. Circ Res. 2020;126:1112–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cochain C, Vafadarnejad E, Arampatzi P, Jaroslav P, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single‐cell RNA‐seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 132. Kim K, Shim D, Lee JS, Zaitsev K, Williams JW, Kim KW, Jang MY, Seok Jang H, Yun TJ, Lee SH, et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ Res. 2018;123:1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. McArdle S, Buscher K, Ghosheh Y, Pramod AB, Miller J, Winkels H, Wolf D, Ley K. Migratory and dancing macrophage subsets in atherosclerotic lesions. Circ Res. 2019;125:1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lin JD, Nishi H, Poles J, Niu X, McCauley C, Rahman K, Brown EJ, Yeung ST, Vozhilla N, Weinstock A, et al. Single‐cell analysis of fate‐mapped macrophages reveals heterogeneity, including stem‐like properties, during atherosclerosis progression and regression. JCI Insight. 2019;4:e124574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Stoeckius M, Hafemeister C, Stephenson W, Houck‐Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yang YS, Atukorale PU, Moynihan KD, Bekdemir A, Rakhra K, Tang L, Stellacci F, Irvine DJ. High‐throughput quantitation of inorganic nanoparticle biodistribution at the single‐cell level using mass cytometry. Nat Commun. 2017;8:14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. [DOI] [PubMed] [Google Scholar]
- 138. Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP, Gherardini PF. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]


