Abstract
The recent pandemic caused by the novel coronavirus resulted in the greatest global health crisis since the Spanish flu pandemic of 1918. There is limited knowledge of whether SARS-CoV-2 is physically associated with human metalloproteins. Recently, high-confidence, experimentally supported protein-protein interactions between SARS-CoV-2 and human proteins were reported. In this work, 58 metalloproteins among these human targets have been identified by a structure-based approach. This study reveals that most human metalloproteins interact with the recently discovered SARS-CoV-2 orf8 protein, whose antibodies are one of the principal markers of SARS-CoV-2 infections. Furthermore, this work provides sufficient evidence to conclude that Zn2+ plays an important role in the interplay between the novel coronavirus and humans. First, the content of Zn-binding proteins in the involved human metalloproteome is significantly higher than that of the other metal ions. Second, a molecular linkage between the identified human Zn-binding proteome with underlying medical conditions, that might increase the risk of severe illness from the SARS-CoV-2 virus, has been found. Likely perturbations of host cellular metal homeostasis by SARS-CoV-2 infection are highlighted.
Keywords: SARS-CoV-2, SARS-CoV-2 orf8, PPI, Metalloproteome, Metal homeostasis
Abbreviations: AD, Alzheimer's Disease; AMD, age macular degeneration; Cb5, cytochrome b5; CNS, central nervous system; COVID-19, corona virus disease 2019; CPS1, carbamoyl-phosphate synthase 1; CSNK2B, casein kinase II subunit beta; DNMT1, DNA methyltransferase 1; HNSCC, head and neck squamous cell carcinoma; HT, hypertension; ITGB1, integrin beta-1; KRCC, kidney renal clear cell carcinoma; PD, Parkinson's Diseases; PPI, protein-protein interaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SIRT5, sirtuin-5; VPS11, vacuolar protein sorting-associated protein 11; ZNF318, zinc finger protein 318
Graphical abstract
1. Introduction
Since the beginning of the 21st century, the world has faced severe crises due to deadly viruses such as Ebola, Zika, avian influenza, AIDS, SARS, and MERS, to name a few. The large-scale explosion of these diseases' viral epidemics resulted from the evolution of pre-existing viruses or the appearance of new viral species, causing enormous damage to society in terms of health, economic and social crisis. Since December 2019, another virus named Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) has spread worldwide. The resulting pandemic is severely affecting global healthcare systems. Around a year after the pandemic began, about 125 million cases and over 2.7 million deaths have been reported worldwide [1]. COVID-19, the disease caused by the SARS-CoV-2, can result in pneumonia and respiratory failure that can be fatal, especially in the presence of other comorbidity factors [2]. The disease spread rapidly, causing a dramatic upheaval in people's everyday lives and in the local and global economies. Despite intense efforts by academic groups and pharmaceutical companies towards developing SARS-CoV-2 vaccines, their approvals by the appropriate organizations are now (and rather delayed) emerging, and vaccination campaigns for mass immunization are just beginning. Therefore, the search for effective antiviral drugs must continue together with the need to contain the viral spread by proper everyday preventive actions following the advice provided by the health authorities (which includes social distancing, wearing a mask, and cleaning the hands), and a rational immunomodulatory approach for host immunity-boosting [[3], [4], [5]].
In COVID-19 infection, like in other coronavirus infections, SARS-CoV-2 needs to interact with the host cells and replicate its genome [6]. Consequently, virus-host protein-protein interaction (PPI) identification is useful for better understanding the virus invasion mechanism and for designing proper therapeutic strategies. Experimental and computational studies have revealed the PPIs between human targets and SARS-CoV-2 [7,8], as well as in for other viruses [[9], [10], [11]], involved in important host biological processes. Specifically, SARS-CoV-2 interacts with i) innate immune pathways (the interferon and the NF-κB pathway and E3 ubiquitin ligases that regulate antiviral innate immune signaling: TRIM59 and MIB1), ii) bromodomain proteins (BRD2 and BRD4 members of the extra-terminal (BET) domain family of epigenetic readers that bind to acetylated histones to regulate gene transcription), and iii) hijacks ubiquitination pathways for replication and pathogenesis (interacts with members of a cullin-2 RING E3 ligase complex) [7].
In this work, metal-binding molecular targets of COVID-19 infection have been identified by applying a systematic structure-based approach used previously by our group to propose putative metalloproteomes in various organisms and viruses [[11], [12], [13]]. Furthermore, possible perturbations in cellular metal homeostasis and molecular linkage between the involved human metalloproteome with several disorders were in silico investigated.
2. Methods
All the human targets of SARS-CoV-2 were retrieved from the supplementary material of the study by Gordon et al. [7]. The metal-binding proteins were predicted using the strategy applied previously to identify putative metalloproteomes in various organisms [[11], [12], [13], [14], [15]]. The approach combines strategies based on structural data and annotation to identify putative metallo-binders by searching for known metal-binding domains in their sequences [16]. The list of known metal-binding domains has been extracted from the Pfam library (http://pfam.xfam.org/) [17], and the 3D structures of known metalloproteins among those available from the Protein Data Bank (PDB; http://www.rcsb.org/pdb/) [18] and MetalPDB databases (https://metalpdb.cerm.unifi.it/) [19]. All the human targets were analyzed for the relevant Pfam metal-binding domains with the search tool HMMER (http://hmmer.org/) [20].
The Centers for Disease Control and Prevention (CDC) provided a list of underlying medical conditions that might increase the risk of severe illness caused by the SARS-CoV-2 virus [2,21]. All established alleles (at-risk genes) associated with humans disorders were retrieved from the Genome-Wide Association Studies (GWAS) online catalog (https://www.ebi.ac.uk/gwas/) and are shown in Table 1 . Then, the retrieved genetic loci employed were used to identify gene networks using the random walk algorithm with restart, previously described [11], and the Gene Prioritization and Evidence Collection (GPEC) plug-in of Cytoscape [22]. Using this algorithm, GPEC finds any neighboring interactants of at-risk genes with a topological distance of 1. The genes in the developed networks were defined as the “neighboring genes”. Then, each neurological disorder's neighboring genes were individually Venn-analyzed compared to the genes that encode the human targets.
Table 1.
Humans disorders or conditions retrieved from the Genome-Wide Association Studies (GWAS) online catalog.
| Diseases/disorders | |
|---|---|
| Neurology | Alzheimer's Disease, Parkinson's Disease, Multiple Sclerosis, Amyotrophic Lateral Sclerosis, Vascular Dementia, Restless Leg Syndrome, Migraine, Creutzfeldt—Jakob Disease, Narcolepsy, Autism-Autism Spectrum |
| Oncology | Urothelial Bladder Carcinoma, Breast Cancer, Colorectal Adenocarcinoma, Glioblastoma, Head and Neck Squamous Cell Carcinoma, Kidney Cancer, Acute Myeloid Leukemia, Lung Adenocarcinoma, Lung Squamous Cell Carcinoma, Ovarian Cancer, Uterine Corpus Endometrial Carcinoma, Multiple Myeloma |
| Ophthalmology | Age Macular Degeneration, Glaucoma |
| Cardiology | Hypertension, Heart Failure |
| Endocrinology | Diabetes, Obesity |
| Pulmonology | Chronic Obstruct Pulmonary, Lung Cystic Fibrosis, Pulmonary Fibrosis, Asthma, Smoking |
| Nephrology | Chronic Kidney |
| Hematology | Thalassemia, Sickle Cell Anemia |
| Obstetrics | Pregnancy |
Functional annotation of human targets was performed by the DAVID tool (http://david.abcc.ncifcrf.gov/) [23]. Subcellular localization was predicted based on the UniProt catalog's information (https://www.uniprot.org/) [24]. The protein abundance information was taken from PaxDb (https://pax-db.org/) [25].
3. Results
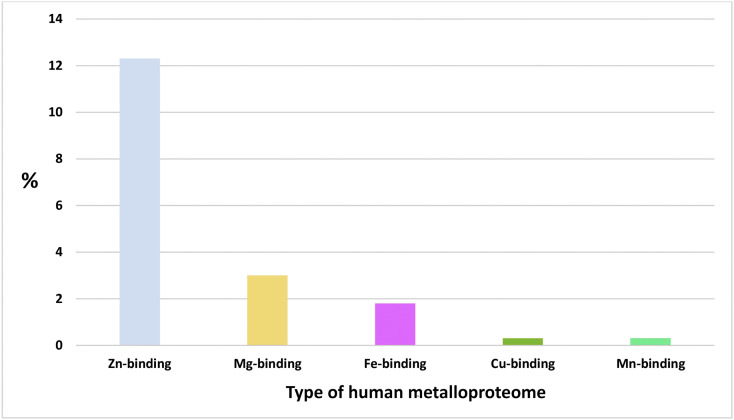
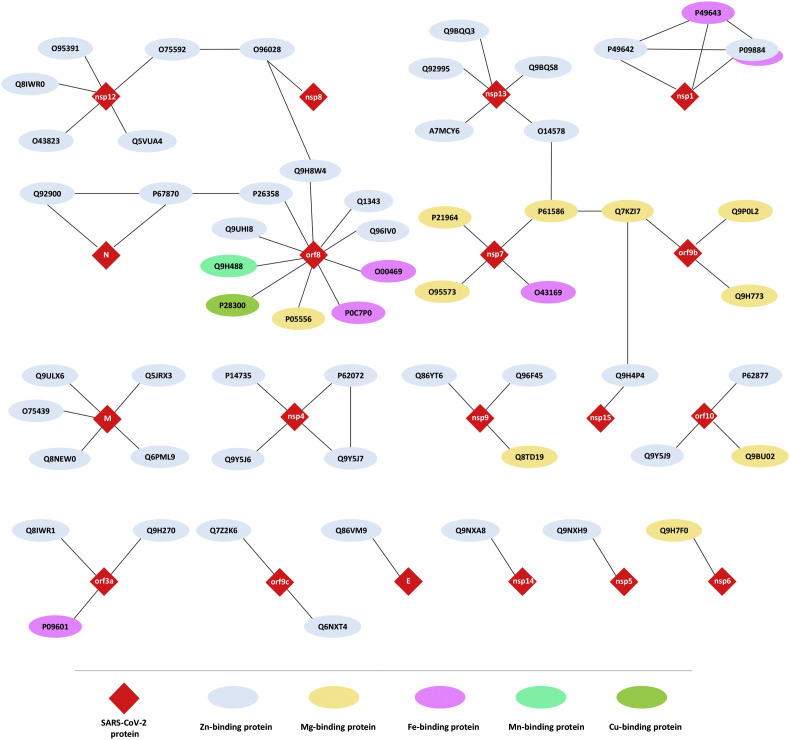
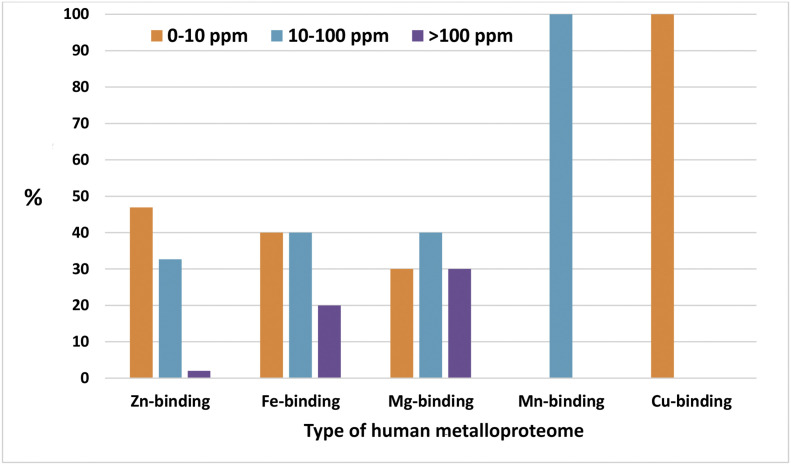
12.3%, 3%, 1.8%, 0.3%, and 0.3% of the studied human proteome interacting with SARS-CoV-2-proteins contains Zn, Mg, Fe, Cu, and Mn metalloproteins, respectively (Fig. 1 ). The complete list of metalloproteins is collected in Supplementary Table S1 and is illustrated in Fig. 2 . The majority of these metalloproteins (17%) interact with accessory protein SARS-CoV-2 orf8, and 8.6% interact with structural membrane protein SARS-CoV-2 M and the non-structural proteins SARS-CoV-2 nsp12 and nsp13 (Table 2 ). The majority of the human metalloproteins that interact with the viral proteins of SARS-CoV-2 localized in membranes represent 58.36%, followed by those in the cytoplasm (23.6%), nucleus (8.7%), and mitochondria (0.6%). According to the protein abundance database (paxdb.org), the human Mg-binding targets are abundantly expressed compared with those of Fe-, Mn- Zn-, and Cu-binders (Fig. 3 ). The most significant molecular function gene ontology terms represented in human metalloproteins refer to binding (85%), catalytic (46%), transporter (7%), and transcription (3%) regulator activity. Venn analysis revealed shared genes between human targeted metalloproteome and certain underlying medical conditions that increase the risk for severe illness from the virus that causes COVID-19. These comprise three neurological disorders with Multiple Sclerosis (MS) showing the highest number of shared genes (MYCBP2, VPS11, CSNK2B, and ITGB1), followed by Parkinson's Diseases (PD) (CYB5B and RHOA) and Alzheimer's Disease (AD) (RHOA). Also, two types of cancer were highlighted: Head and Neck Squamous Cell Carcinoma (HNSCC) (DNMT1) and Kidney Renal Clear Cell Carcinoma (CCRCC) (KIRC). Between other disorders, we found Glaucoma (SIRT5, RHOA, and LOX), Age Macular Degeneration (AMD) (TGB1 and LOX), Hypertension (HT) (ZNF318), and Smoking (CYB5B), as shown in Table 3 .
Fig. 1.
The content of each type of human metalloproteome that is interacting with SARS-CoV-2-proteins.
Fig. 2.
The PPI interactions between SARS-CoV-2 proteins and human metal-binding proteins (IDs in the nodes correspond to Uniprot protein identifiers).
Table 2.
Distribution of the metallo-binding interactors of SARS-CoV-2 proteins.
| SARS-CoV-2 protein | Number of target-metalloproteins |
|---|---|
| SARS-CoV-2 orf8 | 10 |
| SARS-CoV-2M | 5 |
| SARS-CoV-2 nsp12 | 5 |
| SARS-CoV-2 nsp13 | 5 |
| SARS-CoV-2 nsp4 | 4 |
| SARS-CoV-2 nsp7 | 4 |
| SARS-CoV-2 nsp1 | 3 |
| SARS-CoV-2 nsp9 | 3 |
| SARS-CoV-2 orf10 | 3 |
| SARS-CoV-2 orf3a | 3 |
| SARS-CoV-2 orf9b | 3 |
| SARS-CoV-2N | 2 |
| SARS-CoV-2 orf9c | 2 |
| SARS-CoV-2 E | 1 |
| SARS-CoV-2 nsp14 | 1 |
| SARS-CoV-2 nsp15 | 1 |
| SARS-CoV-2 nsp5 | 1 |
| SARS-CoV-2 nsp6 | 1 |
| SARS-CoV-2 nsp8 | 1 |
Fig. 3.
The protein abundance of target-metalloproteins.
Table 3.
Shared genes between human metalloproteome targeted by SARS-CoV-2 proteins and associated disorders.
| Neurological disorders |
Cancer |
Others |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Proteins | Multiple sclerosis | Alzheimer's | Parkinson's | Head and neck squamous cell carcinoma | Kidney renal clear cell carcinoma | Age macular | Glaucoma | Hypertension | Smoking |
| Zn-binding | MYCBP2,VPS11,CSNK2B | DNMT1 | SIRT5 | ZNF318 | |||||
| Fe-binding | CYB5B | CYB5B | |||||||
| Mg-binding | ITGB1 | RHOA | RHOA | ITGB1 | ITGB1 | RHOA | |||
| Mn-binding | |||||||||
| Cu-binding | LOX | LOX | |||||||
4. Discussion
The human metalloproteome targeted by SARS-CoV-2 proteins has been identified from the experimentally supported, high-confidence, SARS-CoV-2 protein–human protein interactions available in the literature [7]. The results show that the content of Zn-binding proteins (12.3%) in the human proteome is significantly higher than that of the other metal ions, followed by the contents of Mg-, Fe-, Cu- and Mn-binding proteins. Based on the present analysis, the metalloproteome has a notable number of zinc transporters [[26], [27], [28], [29], [30], [31]], indicating that the SARS-CoV-2 infection may cause strong intracellular zinc homeostasis disturbances. Additionally, 27% of the zinc-binding domains of the virus-targeted human metalloproteome are of the zinc finger-type known, by many reports [[32], [33], [34], [35], [36]], to be crucial for the structural stability and catalytic activity of many enzymes [37]. Specifically, four human zinc-binding RING (Really Interesting New Gene) E3 ligases can be identified: E3 ubiquitin-protein ligase MIB, MYCBP2, NRDP1, and RBX1. This result is expected since RING E3s are considered acting as hubs in the immunity-regulating network connecting different signaling pathways or different systems [[38], [39], [40]]. One of the Mg-binding proteins is the cation-transporting ATPase 13A3 [41], which is involved in cation transmembrane transfer through channels, indicating that SARS-CoV-2 infection could interfere with metal uptake and efflux, and sensing pathways.
Interestingly, the present study reveals that most metalloproteins identified (17%) interact with the accessory protein SARS-CoV-2 orf8. The orf8 region is poorly conserved among related coronaviruses and is prone to mutations or deletions during interspecies transmission. Although the function of orf8 remains to be elucidated, its structural plasticity and high diversity suggest an important role in SARS-CoV-2 pathogenicity and the ability of the virus to spread [42].
This study shows a significant molecular association between certain underlying medical conditions that might increase the risk for severe illness from the virus that causes COVID-19 and the involved human metalloproteome. The zinc-binding proteins show the highest number of shared genes with underlying medical conditions. Specifically, six Zn-binding proteins are encoded by shared genes with multiple sclerosis, glaucoma, HNSCC, and KIRC cancer types and hypertension:
-
i)
E3 ubiquitin-protein ligase encoded by the MYCBP2 gene plays a key role in neural development. It is involved in different processes such as regulation of neuronal growth, synaptogenesis, and synaptic plasticity by modulating several signaling pathways, including the p38 MAPK signaling cascade [42].
-
ii)
Vacuolar protein sorting-associated protein 11 homolog (encoded by the VPS11 gene) plays a role in vesicle-mediated protein trafficking to lysosomal compartments, including the endocytic membrane transport and autophagic pathways [43,44]. C846G mutation in its RING-H2 domain causes aberrant ubiquitination and accelerates the turnover of VPS11 protein. Zebrafish harboring a vps11 mutation with truncated RING-H2 domain, demonstrates a significant reduction in central nervous system (CNS) myelination following extensive neuronal death in the hindbrain and midbrain [45].
-
iii)
Casein kinase II subunit beta (encoded by the CSNK2B gene) is a ubiquitous protein kinase that regulates metabolic pathways, signal transduction, transcription, translation, and replication. The enzyme is localized in the endoplasmic reticulum and in the Golgi apparatus [46]. CK2 is present in high levels in the brain, and mutations encoding the α-subunit of CK2 were previously identified in patients with neurodevelopmental disorders and dysmorphic features [47].
-
iv)
NAD-dependent protein deacetylase sirtuin-5, mitochondrial (encoded by the SIRT5 gene) activates CPS1 and contributes to the regulation of blood ammonia levels during prolonged fasting: it acts by mediating desuccinylation and deglutarylation of CPS1, thereby increasing CPS1 activity in response to elevated NAD levels during fasting [48,49]. SIRT5 plays a significant role in inhibiting mitochondrial ROS levels. SIRT5 alleviates 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigrostriatal dopaminergic degeneration by suppressing mitochondrial-derived ROS levels. Given the crucial roles of mitochondria in neuronal viability, SIRT5 seems well-positioned to suppress the onset and/or pace of neurodegenerative disease [50].
-
v)
DNA methyltransferase 1 (encoded by the DNMT1 gene) is an enzyme that transfers a methyl group to cytosine nucleotides of genomic DNA. This protein is the major enzyme responsible for maintaining methylation patterns following DNA replication and shows a preference for hemimethylated DNA. DNMTs may be a potential target for enhancing HNSCC chemotherapy by using inhibitors of DNMTs and reversal of gene methylation [51,52]. It has been reported that DNMT1 enhances the radiosensitivity of neck squamous cell carcinomas via downregulating SMG1 [53].
-
vi)
The zinc finger protein 318 (encoded by the ZNF318 gene) is already recognized as a blood pressure regulator [54].
The Fe-binding Cytochrome b5 (Cb5) is encoded by the CYB5B gene shared with Parkinson's disease and smoking behavior-related traits. This protein is a membrane-bound hemoprotein functioning as an electron carrier for several membrane-bound oxygenases [55]. Cb5 has a high potential as a biomarker of health and disease in the brain because it regulates metabolic pathways essential to maintain normal neuronal function, like lipid biosynthesis, steroid and xenobiotics metabolism, neuronal bioenergetics, and production of reactive oxygen species. Cb5 is highly expressed in pyramidal neurons of the primary and secondary motor areas of the frontoparietal cerebral cortex, hippocampus, vestibular, reticular, and motor nuclei of the cerebellum and brain stem, and also in Purkinje and granule neurons of the cerebellum cortex [56]. Two Mg-binding proteins are encoded by shared genes with Multiple Sclerosis, Alzheimer's Disease, Parkinson's Disease, Glaucoma, and KIRC:
-
i)
integrin beta-1 (encoded by the ITGB1 gene) is a cell surface receptor associated with integrin alpha 1 and integrin alpha 2 to form integrin complexes that function as collagen receptors [57]. β1 integrin subunit has been implicated in neuronal migration and in the laminar organization of the brain. Formation, maturation, and function of synaptic connections require the engagement of several integrins. Presynaptic β1 integrins serve as counter-receptors for postsynaptic intercellular adhesion molecule-5 (ICAM-5; aka telencephalin) [58]. Integrin beta-1 is mainly expressed in normal cells and in tumor-associated cells, where they control various developing processes, including angiogenesis, tumor progression, apoptosis, and metastasis [59]. Recently, a flow cytometry study showed that ITGB1 is expressed at high levels in clear cell renal cell carcinoma [60].
-
ii)
Transforming protein RhoA (encoded by RHOA gene) is a small GTPase protein primarily associated with cytoskeleton regulation, mostly actin stress fibers formation and actomyosin contractility [61].
Rho GTPases dysregulated in various neurological diseases/disorders show synaptic irregularity, such as Huntington's Disease, Parkinson's Disease, Amyotrophic Lateral Sclerosis, and Schizophrenia. They are also characterized as key regulators in Alzheimer's disease (AD)-related signals and studied as AD targets [62]. The Cu-binding Protein-lysine 6-oxidase is an enzyme encoded by the LOX gene linked to multiple sclerosis and glaucoma. It catalyzes the conversion of lysine molecules into highly reactive aldehydes that form cross-links in extracellular matrix proteins [63]. Localization of LOX at the CNS has been reported previously. In these studies, LOX is localized at the vascular wall and intracellularly in the brain matrix's cortical and subcortical neurons. Also, LOX is one of the genes known to be up-regulated in Amyotrophic Lateral Sclerosis (ALS) patients [64].
5. Conclusions
In this work, the human metalloproteome, which is physically associated with SARS-CoV-2 proteins, has been mined from available experimental data and highlighted for the first time. This work strongly indicates the important role of Zn2+ in the interplay between SARS-CoV-2 and humans for two main reasons:
-
i)
The content of Zn-binding proteins (12.3%) is significantly higher than that of the other metal ions.
-
ii)
The Zn-binding proteins have shown the highest number of shared genes with underlying medical conditions that might increase the risk of severe illness from the SARS-CoV-2 virus.
Additionally, most virus-targeted metalloproteins (17%) interact with the accessory protein SARS-CoV-2 orf8, a region with high variability to structural changes, closely related to the virus's ability to spread. Furthermore, PPIs that could cause possible significant perturbation of human zinc homeostasis from viral infection have been identified. Integrated gene network analysis reveals gene circuits shared among the identified metalloproteome and three specific neurological disorders like Multiple Sclerosis, Alzheimer's Disease, Parkinson's Disease, and other specific conditions are known to be implicated in COVID-19 severity such as hypertension and smoking. Bioinorganic chemistry can play an important role in the current pandemic. Recently, it was reported that Cys-rich proteins expressed in SARS-CoV-2 by genes ORF7a and ORF8 are likely involved in zinc binding and in the interactions with bone marrow stromal antigen 2 (BST2) activated by extensive disulfide bonds. The stability of Zn-S bonds, with S belonging to reduced sulfide groups, can, in turn, favor the breaking of some of the disulfide bridges of the folded BST2 tetherin, allowing the inhibition of the BST2 antiviral activity [65]. As pointed out in a recent remarkable report by Kozak, Gray, and Garza-Lopez [66], metal ions might affect the function of structural stability of the SARS-CoV-2 main protease (Mpro). Based on a detailed analysis of Mpro stability, the authors identified the regions where inorganic therapeutic agents could compromise the coronavirus, by targeting histidines and/or cysteines. In one scenario, the main protease could be inhibited by Co(III) attachment to His 41, and in another scenario, Co(III) binding to the active-site Cys 145 thiolate would be lethal to enzyme function. Finally, we hope that this study opens the doors for developing metallodrugs as alternatives antiviral strategy in the fight against SARS-CoV-2. This is a topic of current intense scientific interest [67,68], and the well-established abilities of coordination complexes to inhibit different stages of the replicative cycles of several viruses may be a good starting point. Furthermore, we believe that this type of study could be useful for developing more efficient techniques for the reliable detection of SARS-CoV-2, which is also an area of intense research efforts [[69], [70], [71]], in order to control, prevent and therefore reduce its pathogenic spread.
Declaration of Competing Interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jinorgbio.2021.111423.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasmi A., Peana M., Pivina L., Srinath S., Benahmed A.G., Semenova Y., Menzel A., Dadar M., Bjørklund G. Interrelations between COVID-19 and other disorders. Clin. Immunol. 2020:108651. doi: 10.1016/j.clim.2020.108651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Tian H., Zhang L., Zhang M., Guo D., Wu W., Zhang X., Kan G.L., Jia L., Huo D., Liu B., Wang X., Sun Y., Wang Q., Yang P., MacIntyre C.R. Reduction of secondary transmission of SARS-CoV-2 in households by face mask use, disinfection and social distancing: a cohort study in Beijing, China. BMJ Glob. Health. 2020;5 doi: 10.1136/bmjgh-2020-002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasmi A., Tippairote T., Mujawdiya P.K., Peana M., Menzel A., Dadar M., Gasmi Benahmed A., Bjørklund G. Micronutrients as immunomodulatory tools for COVID-19 management. Clin. Immunol. 2020;220:108545. doi: 10.1016/j.clim.2020.108545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasmi A., Chirumbolo S., Peana M., Noor S., Menzel A., Dadar M., Bjørklund G. The role of diet and supplementation of natural products in COVID-19 prevention. Biol. Trace Element Res. 2021 doi: 10.1007/s12011-021-02623-3. vol. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.V'kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khorsand B., Savadi A., Naghibzadeh M. SARS-CoV-2-human protein-protein interaction network. Inform. Med. Unlocked. 2020;20:100413. doi: 10.1016/j.imu.2020.100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh A. Human–virus interactome atlas. Nat. Methods. 2019;16:1081. doi: 10.1038/s41592-019-0635-0. [DOI] [PubMed] [Google Scholar]
- 10.Lasso G., Mayer S.V., Winkelmann E.R., Chu T., Elliot O., Patino-Galindo J.A., Park K., Rabadan R., Honig B., Shapira S.D. A structure-informed atlas of human-virus interactions. Cell. 2019;vol. 178:1526–1541. doi: 10.1016/j.cell.2019.08.005. e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chasapis C.T. Interactions between metal binding viral proteins and human targets as revealed by network-based bioinformatics. J. Inorg. Biochem. 2018;186:157–161. doi: 10.1016/j.jinorgbio.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Chasapis C.T., Andreini C., Georgiopolou A.K., Stefanidou M.E., Vlamis-Gardikas A. Identification of the zinc, copper and cadmium metalloproteome of the protozoon Tetrahymena thermophila by systematic bioinformatics. Arch. Microbiol. 2017;199:1141–1149. doi: 10.1007/s00203-017-1385-y. [DOI] [PubMed] [Google Scholar]
- 13.Peana M., Chasapis C.T., Simula G., Medici S., Zoroddu M.A. A model for manganese interaction with Deinococcus radiodurans proteome network involved in ROS response and defense. J. Trace Elem. Med. Biol. 2018;50:465–473. doi: 10.1016/j.jtemb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Andreini C., Bertini I., Cavallaro G., Decaria L., Rosato A. A simple protocol for the comparative analysis of the structure and occurrence of biochemical pathways across superkingdoms. J. Chem. Inf. Model. 2011;51:730–738. doi: 10.1021/ci100392q. [DOI] [PubMed] [Google Scholar]
- 15.Chasapis C.T. Shared gene-network signatures between the human heavy metal proteome and neurological disorders and cancer types. Metallomics. 2018;10:1678–1686. doi: 10.1039/c8mt00271a. [DOI] [PubMed] [Google Scholar]
- 16.Chasapis C.T. Building bridges between structural and network-based systems biology. Mol. Biotechnol. 2019;61:221–229. doi: 10.1007/s12033-018-0146-8. [DOI] [PubMed] [Google Scholar]
- 17.Mistry J., Chuguransky S., Williams L., Qureshi M., Gustavo A., Salazar E.L.L., Sonnhammer S.C.E., Tosatto L., Paladin S., Raj L.J., Richardson R.D., Finn A. Bateman, Pfam: The protein families database in 2021. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman H.M. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putignano V., Rosato A., Banci L., Andreini C. MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018;46:D459–D464. doi: 10.1093/nar/gkx989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler T.J., Eddy S.R. Nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention People with Certain Medical Conditions. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html Availabe online. (accessed on Jan. 5, 2020)
- 22.Le D.-H., Kwon Y.-K. GPEC: a Cytoscape plug-in for random walk-based gene prioritization and biomedical evidence collection. Comput. Biol. Chem. 2012;37:17–23. doi: 10.1016/j.compbiolchem.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The UniProt Consortium Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M., Herrmann C.J., Simonovic M., Szklarczyk D., Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bafaro E., Liu Y., Xu Y., Dempski R.E. The emerging role of zinc transporters in cellular homeostasis and cancer. Signal Transduct. Targeted Therapy. 2017:2. doi: 10.1038/sigtrans.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kambe T., Hashimoto A., Fujimoto S. Current understanding of ZIP and ZnT zinc transporters in human health and diseases. Cell. Mol. Life Sci. 2014;71:3281–3295. doi: 10.1007/s00018-014-1617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreini C., Banci L., Bertini I., Rosato A. Zinc through the three domains of life. J. Proteome Res. 2006;5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 29.Colomar-Carando N., Meseguer A., Company-Garrido I., Jutz S., Herrera-Fernández V., Olvera A., Kiefer K., Brander C., Steinberger P., Vicente R. Zip6 transporter is an essential component of the lymphocyte activation machinery. J. Immunol. 2019;202:441–450. doi: 10.4049/jimmunol.1800689. [DOI] [PubMed] [Google Scholar]
- 30.Anzilotti C., Swan D.J., Boisson B., Deobagkar-Lele M., Oliveira C., Chabosseau P., Engelhardt K.R., Xu X., Chen R., Alvarez L., Berlinguer-Palmini R., Bull K.R., Cawthorne E., Cribbs A.P., Crockford T.L., Dang T.S., Fearn A., Fenech E.J., de Jong S.J., Lagerholm B.C., Ma C.S., Sims D., van den Berg B., Xu Y., Cant A.J., Kleiner G., Leahy T.R., de la Morena M.T., Puck J.M., Shapiro R.S., van der Burg M., Chapman J.R., Christianson J.C., Davies B., McGrath J.A., Przyborski S., Santibanez Koref M., Tangye S.G., Werner A., Rutter G.A., Padilla-Parra S., Casanova J.-L., Cornall R.J., Conley M.E., Hambleton S. An essential role for the Zn2+ transporter ZIP7 in B cell development. Nat. Immunol. 2019;20:350–361. doi: 10.1038/s41590-018-0295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas P., Converse A., Berg H.A. ZIP9, a novel membrane androgen receptor and zinc transporter protein. Gen. Comp. Endocrinol. 2018;257:130–136. doi: 10.1016/j.ygcen.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 32.Birkou M., Chasapis C.T., Marousis K.D., Loutsidou A.K., Bentrop D., Lelli M., Herrmann T., Carthy J.M., Episkopou V., Spyroulias G.A. A residue specific insight into the Arkadia E3 ubiquitin ligase activity and conformational plasticity. J. Mol. Biol. 2017;429:2373–2386. doi: 10.1016/j.jmb.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Chasapis C.T., Kandias N.G., Episkopou V., Bentrop D., Spyroulias G.A. NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase. Proteins. 2012;80:1484–1489. doi: 10.1002/prot.24048. [DOI] [PubMed] [Google Scholar]
- 34.Kandias N.G., Chasapis C.T., Bentrop D., Episkopou V., Spyroulias G.A. High yield expression and NMR characterization of Arkadia E3 ubiquitin ligase RING-H2 finger domain. Biochem. Biophys. Res. Commun. 2009;378:498–502. doi: 10.1016/j.bbrc.2008.11.055. [DOI] [PubMed] [Google Scholar]
- 35.Chasapis C.T., Loutsidou A.K., Orkoula M.G., Spyroulias G.A. Zinc binding properties of engineered RING finger domain of Arkadia E3 ubiquitin ligase. Bioinorg. Chem. Appl. 2010;2010:1–7. doi: 10.1155/2010/323152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chasapis C.T. Hierarchical core decomposition of RING structure as a method to capture novel functional residues within RING-type E3 ligases: a structural systems biology approach. Comput. Biol. Med. 2018;100:86–91. doi: 10.1016/j.compbiomed.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 37.Chasapis C.T., Ntoupa P.-S.A., Spiliopoulou C.A., Stefanidou M.E. Recent aspects of the effects of zinc on human health. Arch. Toxicol. 2020;94:1443–1460. doi: 10.1007/s00204-020-02702-9. [DOI] [PubMed] [Google Scholar]
- 38.Mintis D.G., Chasapi A., Poulas K., Lagoumintzis G., Chasapis C.T. Assessing the direct binding of ark-like E3 RING ligases to ubiquitin and its implication on their protein interaction network. Molecules. 2020;25:4787. doi: 10.3390/molecules25204787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medvar B., Raghuram V., Pisitkun T., Sarkar A., Knepper M.A. Comprehensive database of human E3 ubiquitin ligases: application to aquaporin-2 regulation. Physiol. Genomics. 2016;48:502–512. doi: 10.1152/physiolgenomics.00031.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Xie P., Lu L., Wang J., Diao L., Liu Z., Guo F., He Y., Liu Y., Huang Q., Liang H., Li D., He F. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat. Commun. 2017:8. doi: 10.1038/s41467-017-00299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seddigh S. Comprehensive comparison of two protein family of P-ATPases (13A1 and 13A3) in insects. Comput. Biol. Chem. 2017;68:266–281. doi: 10.1016/j.compbiolchem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect. Genet. Evol. 2020;85:104525. doi: 10.1016/j.meegid.2020.104525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wartosch L., Günesdogan U., Graham S.C., Luzio J.P. Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic. 2015;16:727–742. doi: 10.1111/tra.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chirivino D., Del Maestro L., Formstecher E., Hupé P., Raposo G., Louvard D., Arpin M., Gruenberg J.E. The ERM proteins interact with the HOPS complex to regulate the maturation of endosomes. Mol. Biol. Cell. 2011;22:375–385. doi: 10.1091/mbc.e10-09-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akey J.M., Zhang J., Lachance V., Schaffner A., Li X., Fedick A., Kaye L.E., Liao J., Rosenfeld J., Yachelevich N., Chu M.-L., Mitchell W.G., Boles R.G., Moran E., Tokita M., Gorman E., Bagley K., Zhang W., Xia F., Leduc M., Yang Y., Eng C., Wong L.-J., Schiffmann R., Diaz G.A., Kornreich R., Thummel R., Wasserstein M., Yue Z., Edelmann L. A founder mutation in VPS11 causes an autosomal recessive leukoencephalopathy linked to autophagic defects. PLoS Genet. 2016;vol. 12:e1005848. doi: 10.1371/journal.pgen.1005848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheusova T., Khan M.A., Schubert S.W., Gavin A.-C., Buchou T., Jacob G., Sticht H., Allende J., Boldyreff B., Brenner H.R., Hashemolhosseini S. Casein kinase 2-dependent serine phosphorylation of MuSK regulates acetylcholine receptor aggregation at the neuromuscular junction. Genes Dev. 2006;20:1800–1816. doi: 10.1101/gad.375206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poirier K., Hubert L., Viot G., Rio M., Billuart P., Besmond C., Bienvenu T. CSNK2B splice site mutations in patients cause intellectual disability with or without myoclonic epilepsy. Hum. Mutat. 2017;38:932–941. doi: 10.1002/humu.23270. [DOI] [PubMed] [Google Scholar]
- 48.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H., Kim J., Woo J., Kim J.H., Choi B.H., He B., Chen W., Zhang S., Cerione R.A., Auwerx J., Hao Q., Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan M., Peng C., Kristin A., Anderson P., Chhoy Z., Xie L., Dai J., Park Y., Chen H., Huang Y., Zhang J.Ro., Wagner G.R., Green M.F., Madsen A.S., Schmiesing J., Peterson B.S., Xu G., Ilkayeva O.R., Muehlbauer M.J., Braulke T., Mühlhausen C., Backos D.S., Olsen C.A., McGuire P.J., Pletcher S.D., Lombard D.B., Hirschey M.D., Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S., Lombard D.B. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit. Rev. Biochem. Mol. Biol. 2018;53:311–334. doi: 10.1080/10409238.2018.1458071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki M., Shinohara F., Endo M., Sugazaki M., Echigo S., Rikiishi H. Zebularine suppresses the apoptotic potential of 5-fluorouracil via cAMP/PKA/CREB pathway against human oral squamous cell carcinoma cells. Cancer Chemother. Pharmacol. 2008;64:223–232. doi: 10.1007/s00280-008-0833-4. [DOI] [PubMed] [Google Scholar]
- 52.Gaździcka J., Gołąbek K., Strzelczyk J.K., Ostrowska Z. Epigenetic modifications in head and neck cancer. Biochem. Genet. 2019;58:213–244. doi: 10.1007/s10528-019-09941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang C., Mi J., Deng Y., Deng Z., Long D., Liu Z. DNMT1 enhances the radiosensitivity of HPV-positive head and neck squamous cell carcinomas via downregulating SMG1. OncoTargets Ther. 2020;13:4201–4211. doi: 10.2147/ott.s227395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewin A., Marques F.Z., Campain A.E., Davern P.J., Yang Y.H.J., Head G.A., Morris B.J. PLoS One. 2011;6:e19203. doi: 10.1371/journal.pone.0019203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schenkman J.B., Jansson I. The many roles of cytochrome b5. Pharmacol. Ther. 2003;97:139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- 56.Samhan-Arias A. Biochemical and anatomical basis of brain dysfunctions caused by cytochrome b5 reductase deficiency or dysregulation. J. Neurol. Neuromed. 2016;1:61–65. doi: 10.29245/2572.942x/2016/6.1066. [DOI] [Google Scholar]
- 57.Jokinen J., Dadu E., Nykvist P., Käpylä J., White D.J., Ivaska J., Vehviläinen P., Reunanen H., Larjava H., Häkkinen L., Heino J. Integrin-mediated cell adhesion to type I collagen fibrils. J. Biol. Chem. 2004;279:31956–31963. doi: 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
- 58.Jaudon F., Thalhammer A., Cingolani L.A. Integrin adhesion in brain assembly: from molecular structure to neuropsychiatric disorders. Eur. J. Neurosci. 2020 doi: 10.1111/ejn.14859. [DOI] [PubMed] [Google Scholar]
- 59.Song J.I.A., Zhang J., Wang J., Cao Z., Wang J.U.N., Guo X., Dong W. β1 integrin modulates tumor growth and apoptosis of human colorectal cancer. Oncol. Rep. 2014;32:302–308. doi: 10.3892/or.2014.3168. [DOI] [PubMed] [Google Scholar]
- 60.Yoon J.-Y., Gedye C., Paterson J., Ailles L. Stem/progenitor cell marker expression in clear cell renal cell carcinoma: a potential relationship with the immune microenvironment to be explored. BMC Cancer. 2020:20. doi: 10.1186/s12885-020-06733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vabres P., Sorlin A., Kholmanskikh S.S., Demeer B., St-Onge J., Duffourd Y., Kuentz P., Courcet J.-B., Carmignac V., Garret P., Bessis D., Boute O., Bron A., Captier G., Carmi E., Devauchelle B., Geneviève D., Gondry-Jouet C., Guibaud L., Lafon A., Mathieu-Dramard M., Thevenon J., Dobyns W.B., Bernard G., Polubothu S., Faravelli F., Kinsler V.A., Thauvin C., Faivre L., Ross M.E., Rivière J.-B. Postzygotic inactivating mutations of RHOA cause a mosaic neuroectodermal syndrome. Nat. Genet. 2019;51:1438–1441. doi: 10.1038/s41588-019-0498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aguilar B.J., Zhu Y., Lu Q. Rho GTPases as therapeutic targets in Alzheimer’s disease. Alzheimer’s Res. Ther. 2017:9. doi: 10.1186/s13195-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo D.C., Regalado E.S., Gong L., Duan X., Santos-Cortez R.L., Arnaud P., Ren Z., Cai B., Hostetler E.M., Moran R., Liang D., Estrera A., Safi H.J., G. University of Washington Center for Mendelian, Leal S.M., Bamshad M.J., Shendure J., Nickerson D.A., Jondeau G., Boileau C., Milewicz D.M. LOX mutations predispose to thoracic aortic aneurysms and dissections. Circ. Res. 2016;118:928–934. doi: 10.1161/CIRCRESAHA.115.307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li P.-A., He Q., Cao T., Yong G., Szauter K.M., Fong K.S.K., Karlsson J., Keep M.F., Csiszar K. Up-regulation and altered distribution of lysyl oxidase in the central nervous system of mutant SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Mol. Brain Res. 2004;120:115–122. doi: 10.1016/j.molbrainres.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Morante S., La Penna G., Rossi G., Stellato F. SARS-CoV-2 Virion Stabilization by Zn Binding. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozak J.J., Gray H.B., Garza-López R.A. Structural stability of the SARS-CoV-2 main protease: can metal ions affect function? J. Inorg. Biochem. 2020;211:111179. doi: 10.1016/j.jinorgbio.2020.111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Paiva R.E.F., Marçal Neto A., Santos I.A., Jardim A.C.G., Corbi P.P., Bergamini F.R.G. What is holding back the development of antiviral metallodrugs? A literature overview and implications for SARS-CoV-2 therapeutics and future viral outbreaks. Dalton Trans. 2020;49:16004–16033. doi: 10.1039/d0dt02478c. [DOI] [PubMed] [Google Scholar]
- 68.Augustin T.L., Hajbabaie R., Harper M.T., Rahman T. Novel small-molecule scaffolds as candidates against the SARS coronavirus 2 main protease: a fragment-guided in silico approach. Molecules. 2020;25:5501. doi: 10.3390/molecules25235501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokhrel P., Hu C., Mao H. Detecting the coronavirus (COVID-19) ACS Sens. 2020;5:2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhalla N., Pan Y., Yang Z., Payam A.F. Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iravani S. Nano- and biosensors for the detection of SARS-CoV-2: challenges and opportunities. Mater. Adv. 2020;1:3092–3103. doi: 10.1039/d0ma00702a. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.