Abstract
The third pandemic of coronavirus infection, called COVID-19 disease, began recently in China. The newly discovered coronavirus, entitled SARS-CoV-2, is the seventh member of the human coronaviruses. The main pathogenesis of SARS-CoV-2 infection is severe pneumonia, RNAaemia, accompanied by glass turbidity, and acute cardiac injury. It possesses a single-stranded positive-sense RNA genome which is 60–140 nm in diameter, and has a size of 26–32 kbp. Viral pathogenesis is accomplished with spike glycoprotein through the employment of a membrane-bound aminopeptidase, called the ACE2, as its primary cell receptor. It has been confirmed that various factors such as different national rules for quarantine and various races or genetic backgrounds might influence the mortality and infection rate of COVID-19 in the geographic areas. In addition to various known and unknown factors and host genetic susceptibility, mutations and genetic variabilities of the virus itself have a critical impact on variable clinical features of COVID-19. Although the SARS-CoV-2 genome is more stable than SARS-CoV or MERS-CoV, it has a relatively high dynamic mutation rate with respect to other RNA viruses. It's noteworthy that, some mutations can be founder mutations and show specific geographic patterns. Undoubtedly, these mutations can drive viral genetic variability, and because of genotype-phenotype correlation, resulting in a virus with more/lower/no decrease in natural pathogenic fitness or on the other scenario, facilitating their rapid antigenic shifting to escape the host immunity and also inventing a drug resistance virus, so converting it to a more infectious or deadly virus. Overall, the detection of all mutations in SARS-CoV-2 and their relations with pathological changes is nearly impossible, mostly due to asymptomatic subjects. In this review paper, the reported mutations of the SARS-CoV-2 and related variations in virus structure and pathogenicity in different geographic areas and genotypes are widely investigated. Many studies need to be repeated in other regions/locations for other people to confirm the findings. Such studies could benefit patient-specific therapy, according to genotyping patterns of SARS-CoV-2 distribution.
Keywords: SARS-CoV-2, COVID-19, Mutation, Pathogenicity, Pandemic disease
1. COVID-19 disease
The COVID-19 disease first started spreading in China and then in almost every region of the globe. As more cases were confirmed, many countries worldwide announced restrictions for transportation and implemented quarantine [1]. Findings revealed that not only people with the disease who had a range of symptoms could transmit the virus, but even those who did not develop symptoms were carriers of the virus [2]. In some instances, people infected with the virus while not experiencing any symptoms of COVID-19 or even showing mild symptoms of the disease. On the other hand, signs of the disease may change from mild and moderate to acute (critical) and become severe, necessitating special care for the patient with the development of the acute respiratory distress syndrome (ARDS) syndrome and accumulation of fluids in the lungs [[2], [3], [4], [5], [6], [7]]. According to epidemiological researches, the clinical outcomes (ranging from asymptotic to death) are heavily influenced by factors such as age, demographic characteristics, clinical care quality, and other potential unknown parameters [[8], [9], [10]]. Additionally, a healthy diet is recognized as another substantial factor in preventing the development of the severe form of the disease by boosting the immune system [11]. The presence of some underlying diseases (e.g., diabetes) in a person is another factor affecting the increased probability of COVID-19 development and the incidence of a severe condition of the disease (Fig. 1 ) [12]. At the same time, the virus can cause bacterial pneumonia infections [13]. To ensure the presence of the virus in the body, polymerase chain reaction (PCR) molecular testing is done. The other technique is computed tomography of the chest. Additionally, the number of white blood cells, creatine phosphokinase (CPK), erythrocyte sedimentation rate (ESR), c-reactive protein (CRP), and lactate dehydrogenase D (LDHD) is measured in the blood [2,14].
Fig. 1.
Possible influencing factors on variable symptoms and severity of COVID-19.
2. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
SARS-CoV-2 belongs to a large family of viruses known as coronaviruses. Coronaviruses are the largest group of viruses belonging to the Nidovirales containing Coronaviridae families, Roniviridae, and Arteriviridae. Coronaviridae is a family of positive-sense single-stranded enveloped RNA viruses, with a genome between 26 and 32 kilobases in length. This viral group consists of four different genera, namely Alpha, Beta, Gamma, and Delta, and encompasses various human and animal coronaviruses [15]. Human coronavirus was first identified in the mid-1960s. Seven human coronaviruses have so far been identified that belong to the Alpha and Beta genera; as depicted in Fig. 2 , genus Alpha, including (human coronavirus NL63 (HCoV-NL63) and human coronavirus 229E (HCoV-229E)) and genus Beta, including (human coronavirus OC43 (HCoV-OC43), human coronavirus HKU1 (HCoV-HKU1), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and current SARS-CoV-2) [16]. They bring about a wide range of diseases, including the common cold and respiratory diseases with mild and acute symptoms.
Fig. 2.
Classification and outbreaks of coronaviruses in human society.
To date, five coronaviruses belonging to beta-coronavirus lineage have been detected in association with acute pneumonia in humans (Fig. 2) [17]. SARS-CoV-2 is the third coronavirus after MERS and SARS-CoV [18,19].
Bats have been reported to be an essential host for some coronaviruses. Examples include SARS-CoV, a virus with acute respiratory symptoms that spread through China, killing hundreds of people in 2003–2004. After that, MERS-CoV was reported in the middle east in 2012; both bat-originated viruses caused the infection and death of many people [17,20]. In 2020, a new disease with a growing global prevalence was named COVID-19 by the World Health Organization. The COVID-19 pandemic has recently emerged and then rapidly spread in more than 200 countries worldwide so far. According to the World Health Organization, 39.8 M global cases of COVID-19 were confirmed on October 16th with a mortality rate of 3.4%.
3. The biology of SARS-CoV-2
Structural and non-structural proteins in the virus genome are encoded in the open reading frame (ORF) sequences (Fig. 3 ). Almost 14 ORF have been sequenced in the SARS-CoV-2 genome so far [21]. Molecular analysis of the new coronavirus indicated that it possesses a single-stranded positive-sense RNA genome, which its size varies between 29.8 kb and 29.9 kb, being 60–140 nm in diameter. Its genome is almost 80% homologous with SARS-CoV, and nearly 96% of its genes are shared with the bat coronavirus (Bat-CoV) [22]. Each of these ORFs is responsible for coding 17 proteins, both structural and non-structural, that drive various processes during the entire life of the virus from its survival to virulence power [23,24]. The genome starts with 5′ UTR, which consists of a variable number (between 6 and 11) of ORFs, including ORF1ab, ORF3, ORF6, ORF 7a, ORF 8, and ORF10. SARS-COV 2 has eight accessory proteins (3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14), encoded by ORF3a, ORF6, ORF7a, ORF7b, and ORF8 genes, respectively. These genes contain vital information for virus replication, that has been preserved through successive generations [15].
Fig. 3.
The genomic organization of SARS-CoV-2. ORF1ab, as the largest gene, encodes the pp1ab protein that contains 15 Nsps (nsp1-nsp10 and nsp12-nsp16). It is located at the 5′ end of the genome. The 3′ terminus of the SARS-CoV-2 genome contains four structural genes, which encode four essential structural proteins: envelope (E), spike protein (S), nucleocapsid protein (N), membrane (M) and a gene for RNA-dependent RNA polymerase.
3.1. Non-structural proteins
During the virus transformation phase, the first event is the expression of orf1ab and orf1a gene segments to produce two large polyproteins named pp1ab and pp1a. These two polyproteins are then cleaved into 16 Nsps, including Nsp1 to Nsp10 and Nsp12 to Nsp16 by the action of two protease enzymes as part of each of those polypeptides named papain-like proteases (PLpro) [25]. These Nsps participate in many viral processes; for example, the Nsp3 is the largest component among other constituents that forms the replication and transcription complex of coronaviruses, which its cleavage by the PL proteinase would result in the promotion of cytokine expression and suppression of the host's innate immune response. 3CLPro protease as another Nsp has an essential role in RNA replication. Many of the Nsps encoded in ORF1ab, including Nsp5, Nsp3, and the RNA-dependent RNA polymerase (RdRp, Nap12), are responsible for the assembly of the replicas-transcriptase complex in double-membrane vesicles (DMVs) [16]. Other replicas consist of proteins that facilitate viral replication and block the intrinsic host immune system by controlling the cellular machinery. Nsp8 and Nsp12 are two monomeric RdRps that are involved in the process of SARS-CoV-2 replication. Nsp8 replicates the virus genome through interacting with ORF6 accessory protein and Nsp9 replicas protein, by binding to RNA. In contrast to Nsp12, Nsp8 has primase capacity that makes it needless of any primers to initiate the viral replication process. In addition to these polymerases, the SARS-CoV-2 has a unique multimeric RNA polymerase composed of Nsp7 and Nsp8, which is responsible for both the initiation and elongation of the newly synthesized fragment of the viral genome [16,23].
3.2. Structural proteins
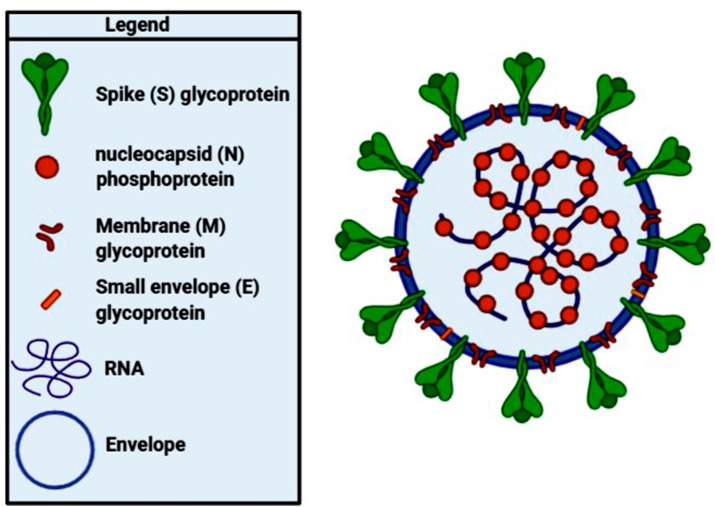
The 3′ end of the SARS-CoV-2 genome consists of four structural protein-encoding genes, which include spike (S), membrane (M), envelope (E), and nucleocapsid (N). From entering the host cells to virus particle formation, various viral processes are dependent on these proteins (Fig. 4 ) [21,25,26].
Fig. 4.
Schematic structure of SARS-CoV-2 with the minimal set of structural proteins.
3.2.1. S protein
The spike or S glycoprotein is a transmembrane protein that resides on the outer surface of the virus, with nearly 150 kDa molecular weight that determines the host range and pathogenicity of the virus by its contribution to viral entry through mediating the attachment of the virus particle to the plasma membrane of the host cell. The S protein of SARS-CoV-1 and SARS-CoV-2 forms homo-trimers protruding in the viral surface that facilitates viral entry into the host cells via interacting with angiotensin-converting enzyme 2 (ACE2) that is their main receptor expressed in lower respiratory tract cells. However, in some circumstances, they might use CD209L as an alternative receptor. In contrast, MERS-CoV uses dipeptidyl peptidase 4 (DPP4) as the primary receptor. These viruses spread between different cell types with the help of this mechanism that suppresses virus-neutralizing antibody responses [16,21,25].
3.2.2. M protein
Membrane or M protein determines the shape of the virus envelope. It can function as a small transmembrane protein through binding to all other structural proteins. The M protein also contributes to the packaging of the viral RNA genome into a helical ribonucleocapsid during virion formation [16].
3.2.3. N protein
The Nucleocapsid or N protein is the structural constituent of SARS-CoV-2 that is bound to the RNA genome of the virus. This protein takes part in various processes related to the viral genome, including the replication cycle of viruses, viral genome signaling, and the host cells' response to viral infections [22,25].
3.2.4. E protein
The structural evidence shows that the N-amino terminal of the envelope or E protein is a short hydrophilic sequence consisting of 7–12 amino acids. It is followed by a large hydrophobic middle segment with 25 amino acids known as the transmembrane domain and possesses a long hydrophilic C-terminus which constitutes the most of the protein. E protein is involved in the production and maturation processes of the virus through interacting with the host cell membrane protein [22].
4. The mechanism of SARS-CoV-2 infection
The presence of S protein in the process of pathogenesis of SARS-CoV-2 is critical. Spike protein uses a membrane-bound aminopeptidase called ACE2 as the primary cell receptor that binds to dipeptidyl peptidase-4 on the virus [27]. ACE2 is widely expressed in the various organs, including the heart, lungs, gastrointestinal tract, and kidneys [27]. Signal transmission through ACE2 is disrupted when severe myocardial damage and dysfunction in the lungs and other organs such as the kidneys and heart are induced when the virus attaches to the ACE2 receptor [28]. Besides, the determination of target cell specificity is influenced by the S glycoprotein interaction with cellular receptors. The location of the receptor binding domain (RBD) in S1 domain of S protein is determined by the structure of each virus, which can be variable. The RBD regions are located at N Terminal or C1 Terminal of S1. SARS-CoV-2 uses some peptidases as their cellular receptors, although their entry occurs even in the absence of secondary enzymes [25,27].
After fastening to the receptor, the virus enters the host cell cytosol by transmembrane protease serine 2 (TMPRSS2) or other proteases such as furin, and fuses the viral and cell membranes. SARS-CoV-2 spike glycoprotein contains a cleft-like site. The furin detection location is crucial for detection by pyrolysis and, thus, in viral infection of the virus. This glycoprotein is broken down by furin proteinase into a subunit S1 and S2 like a host cell. Infection begins with the interaction of S1, containing RBD, with the host receptor ACE2, into the cell membrane. The S2 domain is a viral fusion protein and is involved in a mixture of virion glands. Immediately after the virus entrance to the cell, the S glycoprotein is cleaved by the CTSL cathepsin to kill the S2 fusion peptide, and then forms endocytosis and low-pH endosomes, hence activating the fusion of membranes within the endosomes. At the junction between the virus membrane and the cell membrane, a bundle is formed that is likely to release the virus genome into the cytoplasm. The following step in the infection cycle is the translation of replica genes from the RNA virus. The two polymer proteins pp1a and pp1 ab are encoded by two large proteins, ORF rep1a and rep1b. A sliding sequence (5′-UUUAAAC-3′) and a pseudoknot RNA help express these proteins, leading to a deformation of the rep1a reading form in the ORF rep1b. Usually, the ribosome opens the pseudoknot structure and continues the translation process until it encounters a pause or reach the end of 11 repetitions in the codon. But sometimes, the ribosome is stopped by the pseudoknot, which prevents stretching and staying in the slippery sequence, and stops the nucleotide reading frame with a backward shift that results in the translation of pp1ab. After the translation process, viral RNA is synthesized, and viral replication complexes are collected. The set of viral replications by the transcription process results in cytopathic effects such as abnormalities in the vascular complexes. In the downstream region of the ridiculous proteins, subgenomic RNAs act as mRNAs for the structural genes of the virus [29].
The next step is the binding and translating the of structural glycoproteins of virus, including S, M and E to ER. These proteins are secreted into the ER-Golgi intermediate chamber (ERGIC). Here, the viral genomes that encode structural proteins are encapsulated by the N-germ protein in the ERGIC membrane and become mature viruses. Protein-protein interaction is mainly mediated by M glycoprotein, which is responsible for collecting CoVs. M protein, expressed with E protein, is involved in the formation of virus-like particles (VLPs) as well as in the formation of coatings for CoVs. While N glycoprotein causing the formation of VLPs, a combination of genomes encoded in ERGIC enhances virus growth. In the ERGIC chamber, S glycoprotein interacts with M glycoprotein due to being essential for its synthesis. Because M protein is abundant compared to E protein, M protein interaction can be a major source of motivation to mask puberty. Glycoprotein changes the host secretion pathway and releases the accumulation of the virus in the host. The M protein attaches to the nucleocapsid located at the C-terminus of endo domain M and signals viral assembly completion. After assembly, the viruses are transmitted by vesicles to the host cell membrane and are released by exocytosis. By being transported to the cell surface, the S protein combines cell fusion between infected and healthy cells, resulting in a large multinucleated cell that spreads the virus in the host and provides the conditions for the detection of specific antibodies against the virus (Fig. 5 ) [29].
Fig. 5.
The mechanism of coronavirus infection and spread. Coronaviruses bind to ACE-2 receptors on the surface of the target cell via their spike proteins. Next, the ACE2 receptor will be cleaved by the TMPRSS2 that results in the activation of the spike protein through which the virus enters the cell.
5. The SARS-CoV-2 mutations and related alterations in the virus
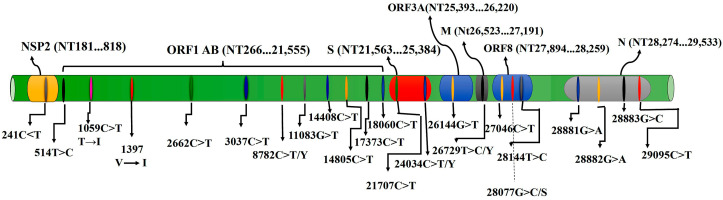
The pace of mutation in SARS-CoV-2 as an RNA virus is possibly replacement of bp for every year. Besides, variations may appear in the course of each genome replication cycle [30]. Single-nucleotide polymorphism (SNPs) is applied for the comparison of DNA sequences. It can be used for evolutionary investigations, being useful in identifying mutations in the coronavirus genome, where many mutations may occur by cause of an RdRp in the process of genome replication [31,32]. Table 1 and Fig. 6 summarize the reported mutations in various parts of SARS-CoV-2 over the last months.
Table 1.
Some reported mutations in various parts of SARS-CoV-2.
| Virus Genome Regions | Mutation Site | Amino Acid Change | Mutation Type | Ref. |
|---|---|---|---|---|
| Nsp2 (Nt181...818) | 241C>T | NA* | NA | [16, 34] |
| Orf1ab (Nt266...21,555) | 1397 | V→I | Missense | [35] |
| 2662C>T | NA | NA | [24] | |
| 2891 | NA | NA | [35] | |
| 3036 | NA | Synonymous | [35] | |
| 3037C>T | NA | Synonymous | [16, 34] | |
| 8782C>T/Y | NA | Synonymous | [24, 35, 36] | |
| 11083G>T | NA | NA | [24, 35] | |
| 14408C>T | P→L | Missense | [16, [34], [35]] | |
| 17373C>T | NA | NA | [24] | |
| 17746 | P→L | Missense | [35] | |
| 17857 | C→Y | Missense | [35] | |
| 18060C>T | NA | Synonymous | [24, 35] | |
| S (Nt21,563...25,384) | 21707C>T | NA | NA | [24] |
| 23403A>G | D→G | Missense | [16, 35] | |
| 24034C>T/Y | NA | NA | [24] | |
| Orf3a(Nt25,393...26,220) | 26143 | G→V | Missense | [35] |
| 26144G>T | G→V | Missense | [24] | |
| M (Nt26,523...27,191) | 26729T>C/Y | NA | NA | [24] |
| Orf8 (Nt27,894...28,259) | 28077G>C/S | NA | NA | [24] |
| 28144T>C | L→S and G→R | Missense | [16, 24, 35, 36] | |
| N (Nt28,274...29,533) | 28881G>A | NA | NA | [35] |
| 28882G>A | NA | NA | [16] | |
| 28883G>C | NA | NA | [16] | |
| 29095C>T | NA | Synonymous | [36] |
Fig. 6.
Linear positions of the reported mutations in various regions of the SARS-CoV-2 genome.
In a study done by Yin, 558 COVID-19 strains were investigated to calculate the frequencies of mutations. The first regular SNP mutation in the virus genome was in nsp2 (241 C > T), which is a critical genomic location. It has been discovered that the nsp2 mutation (241 C > T) has emerged with three other critical mutations including, (3037 C > T), (14408 C > T), and (23403 A > G), that bring about mutations at the amino acid level of RNA primase, and S protein. The mutations as mentioned earlier (241 C > T, 14408 C > T, and 23403 A > G) are located in proteins which are essential for the replication of RNA (241C > T, 14408 C > T), and the spike protein (23403A > G) since it attaches to receptors. This paper recognized that these four mentioned-mutations are common in the SARS-CoV-2 European isolates genomes, where the severity of the infection is mostly more intense than in the other geographical regions. The second amplest SNP mutation in the isolates was (28144 T > C), which has appeared at nsp8, where the amino acid is changed (Leu to Ser), and the third one (26144 G > T) was in nsp3. The nsp3 collaborates with two other proteins called nonstructural protein-4 and 6, to bring about DMVs, which are complexes of the membrane that assist in replicating and assembling RNA. The most common mutation (23403 A > G) was positioned in the S protein sequence. This mutation in D614G (23403 A > G) was placed in the supposed S1–S2 junction part, which splits the spike protein. Analysis of the results demonstrated that nsp8, which is an RNA primase, had the greatest number of mutations (28881 G > A, 28144 T > C, 28882 G > A and 28883 G > C) in comparison to the remaining proteins. Mutations in RNA polymerase or RNA primase might increase fidelity and present obstruction to the usage of nucleotide analogs that are mutagenic. In strains from the US, three mutations in the sequences of nsp8 (28881 G > A, 28882 G > A, and 28883 G > C) have been discovered [16].
ORF1ab gene, which has the most length in the ORFs in the virus genome and is split into viral non-structural proteins (nsp1 to nsp16), was reported as a gene with a high number of missense mutations [23,37].
After analyzing around 48,000 SARS-CoV-2 genome sequences and comparing them with the reference sequence, researchers demonstrated that there reside at least three clades according to geological and genomic relevance. The clade common in Europe obtains the p.614(Asp to Gly) mutation associated with the S protein along with (14408 C > T), (241 C > T), and (3037 C > T). The other significant mutation was in the N protein (p.RG203KR) [34].
Based on a study that analyzed 220 genome sequences from COVID-19 patients, the scientists appraised the disposition of the virus mutations within four geographic regions by standardizing the number of genomes containing a likely mutation in each region. The appearance of mutations in nucleotide locations 3036, 8782, 11083, 28144, and 26143 has been confirmed. Besides, mutations were found in positions 1397, 2891, 14408, 17746, 17857, 18060, 23403, 28881, existing in sequences of ORF-1ab (1397, 2891, 14408, 17746, 17857, and 18060), S (23403) and N (28881). three silent mutations (locations 3036, 8782, and 18060) were observed among the twelve most common ones, considering that one mutation (location 11083) was not in the sequence of ORF. Also, mutations in these locations cause amino acids to change in this order: 1397 (Val to Ile), 14408 (Pro to Leu), 17746 (Pro to Leu), 17857 (Cys to Tyr), 23403 (Asp to Gly), 26143 (Gly to Val), 28144 (L to Ser). One mutation (location 28881) causes the substitution of 2 amino acids, particularly 28881 (Arg to Lys) and (Gly to Arg) [38].
According to a paper that has analyzed the SARS-CoV-2 genomes, 10 out of the 48 sequences merely had lost their base pairs at the start and the end. However, 80 alternatives contained mutations including, 43 missenses, 21 synonymous, three deletions, and 13 noncoding deletion types. The most frequent alternatives (13 specimens) were in ORF-1ab (8782 C > T) and ORF-8 (28144 T > C), and after that, in N (29095 C > T) (5 specimens). Mutations in locations (8782 C > T) and (28144T > C) were observed at the same time. Also, (29095 C > T) is detected in both of them. The two mutations (8782 C > T) and (29095 C > T) were synonymous ones; nonetheless, (28144 T > C) has altered amino acid in the ORF8 region (p. L84S). Out of the 43 missense alternatives, 30 ones were observed in ORF-1ab. All three deletion mutations found in this study, are in-frame ones and were observed in nsp1, which is in ORF1ab. All of the noncoding deletion mutations were detected in 3′-UTR or 5′-UTR positions, and researchers suggest that they do not significantly influence the functions of the virus. Through various alignments with other SARS-CoV-2 ORF8 regions, they have found out that L84, which is engaged in (28144 T > C) (p.L84S), is not conserved [36]. Khailany et al. have confirmed these results in their separate research [23].
Wang et al. also analyzed 95 SARS-CoV-2 genome sequences. They compared the isolates with the reference sequence and depicted that the total homology was 99.99%, whereas 15 isolates were identical with the reference one. Also, the homology of the ORF-1a in the genome was 99.99%, whereas the homology in connection with the bulk of other parts being 100% while no alteration in regions of E, ORF-6, and ORF-7b, was found. The results show that by comparing the amino acids of the sequences, the overall homology was 99.99%, while the homology between the bulk of the isolates in individual parts was 100%. Mutations which occurred in more than three variants were observed in particular nucleotide positions including, ORF-1ab (2662, 8782, 11083, 17373, 18060), S (21707, 24034), ORF3A (26144), M (26729), ORF8 (28077, 28144) and N (28854, 29095). Notably, the highest mutation rate was detected in ORF8 (28144) which was about 30% (29 out of 95), where C took the place of T. Likewise, the mutation rate of position ORF-1ab (8782) was about 29% (28 out of 95), where mostly T took the place of C. Mutations in amino acid sequences appearing in more than three strains were p.3606(Leu to Phe), p.49(His to Tyr), p.860(Val to Gln), p.251(Gly to Val), p.62(Val to Leu), p.84(Leu to Ser), p.194(Ser to Leu). Besides, six deletion mutations were observed in 5 of the sequences. These mutations led to 4 various shortened variations in the sequence of amino acids. Moreover, two more deletion-type mutations were observed in 3′-UTR or 5′-UTR of the genome, respectively [24].
Yu et al. stated that 120 substitution locations were observed in 8 coding regions of the viral genomes, encompassing 79 transitions (66%) and 41 transversions (34%). The 120 substitution locations were engaged in 119 codons, containing 79 nonsynonymous (66%) and 40 synonymous (34%) mutations. Forty nonsynonymous mutations (51%) altered the biochemical characteristics of the original amino acids, and thus are probably engaged in the adjustment of the virus [39].
Multiple sequence alignment of 86 patient-isolated strains of the SARS-CoV-2 disclosed 93 mutations including 42 missense mutations in viral proteins except for the E protein, 29 missense mutations in the ORF1ab protein, and 8 of them in the S protein [40].
In another paper, ten viral genomes derived out of the NCBI database were analyzed through sequence alignment. They did not see any divergence in the amino acid arrangement of N and M proteins. Two alternatives in terms of amino acid level, in spike protein, were detected. According to the reference sequence, the authors of this study suggested two SNP subtypes named “L" and “S". Two likely “L" and “S" mutations within the sequences of ORF1ab and ORF8 were discovered. One mutation was found through evaluating the sequence alignment of E protein, in which the mutation “His” was detected from a South Korean genome in contrast with “Leu” from alternative nine ones in the alignment. In the course of studying the spike protein, one mutation of “Trp” was found in a South Korean genome in contrast with “Ser” from the alternative nine ones. Also, one mutation of “Arg” from an Australian sequence was observed considering “Ser” from alternative nine ones [22].
Holland et al. discovered that one of the viral genomes had an 81 nucleotide in-frame deletion in the ORF-7a sequence, leading to a loss of 27 amino acids [41]. Comparable deletion mutations in the virus genomes has appeared, especially in the ORF-8 sequence, which may likely lessen the fitness of the SARS-CoV-2 [42,43].
By computational tools, established a reference genome sequence of SARS-CoV-2, and revealed 80 mutated bases at 47 locations. Besides, the most significant SNP mutation in the S protein was found to be A > G.
6. The SARS-CoV-2 mutations and its related variabilities in clinical manifestations
It is confirmed that the different mortality and infection rate of COVID-19 in geographic areas might be influenced by various factors such as different national rules for quarantine and movement restrictions, and also different races or the genetic backgrounds of people [38]; in particular, SNP array data analyzing revealed that a specific haplotype of TMPRSS2 gene in a European population which results in upregulation of TMPRSS2 gene expression (as a crucial enzyme for SARS-CoV-2 entry into the host cell) in an androgen-dependent manner, showed association with disease severity in Italian population [45]. Moreover, viral mutations over time are regarded as the contributing factors in transmission and severity of the COVID-19 infection [38]. Whereas the SARS-CoV-2 genome is more stable than SARS-CoV or MERS-CoV, it has a relatively high dynamic mutation rate comparable to other RNA viruses [46]. It's noteworthy that, some mutations can be as founder mutations and show specific geographic patterns. For example, the D614G mutation in S protein is a founder mutation in Europe (appearance from February 16, 2020) [47]. Undoubtedly, these mutations can drive viral genetic variability, and because of genotype-phenotype correlation, resulting in a virus with more/lower/no decrease in natural pathogenic fitness or on the other hand, facilitating their rapid antigenic shifting in order to escape the host immunity and also creating a drug resistance virus, hence converting it to a more infectious or deadly virus (Table 2 ) [[47], [48], [49]]. Issa et al. discovered 51 distinct non-synonymous amino-acid substitutions in the ORF3a proteins by analyzing 2782 SARS-CoV-2 sequenced isolates. The functional domains of the ORF3a protein are related to pathogenicity and virulence [50].
Table 2.
Correlation of SARS-CoV-2 mutations with pathogenicity.
| Location/Source | SARS-CoV-2 Genome Variation | Associated Phenotypic Changes | Ref. |
|---|---|---|---|
| Various Genome Sequences Database (Such As Gisaid Database) Analyzing | p.614(Asp to Gly) in S (Missense) | Higher fatality rate | [51] |
| p.367(Val to Phe) in RBD of S (Missense) p.436(Trp to Arg) in RBD of S (Missense) p.364(Asp to Tyr) in RBD of S (Missense) |
Higher infectivity (higher affinity to human ACE2 receptors) | [52] | |
| p.614(Asp to Gly) in S (Missense) | Higher infectivity | [52] | |
| p.475(Ala to Val) in RBD of S (Missense) p.452(Leu to Arg) in RBD of S (Missense) p.483(Val to Ala) in RBD of S (Missense) p.490(Phe to Leu) in RBD of S (Missense) |
Resistant to multiple neutralizing antibodies | [52] | |
| p.408(Arg to Ile) in RBD of S (Missense) p.455(Leu to Tyr) in RBD of S (Missense) p.486(Phe to Leu) in RBD of S (Missense) p.493(Gln to Asn) in RBD of S (Missense) p.498(Gln to Tyr) in RBD of S (Missense) p.501(Asn to Thr) in RBD of S (Missense) p.930(Ala to Val) in heptad repeat 1 (HR1) domain of S (Missense) p.936(Asp to Tyr) in heptad repeat 1 (HR1) domain of S (Missense) |
Reducing stability of spike proteins | [53] | |
| p.3691 in NSP6 p.9659 in ORF-10 |
Reducing the stability of the proteins structures | [54] | |
| 8782C > T in ORF-1ab (Synonymous Mutation) 28144T > C in ORF-8 (Missense) 29095C > T in N (Synonymous Mutation) |
Potential Effects On Transmission and Severity of COVID-19 | [23] | |
| Singapore | Elimination Of ORF8 (Deletion Of 382 nt) | Attenuated Phenotype | [43] |
| During Passaging Virus in Vero-E6 Cells | 15-30 nt Deletion in S1/S2 Junction Region | Attenuated Phenotype | [49] |
| China | 23403A > G in S (Missense) 23902A > C in S (Missense) 23607A > A in S (Missense) 22303T > G and 22301A > C in S (Missense) 27775T > C, 27776T > G, And 27777G > A in S (Missense) 22205G > C in S (Missense) |
Strengthen Phenotype | [47] |
Eleven SARS-CoV-2 Chinese patient-isolated viral genome sequences were analyzed through ultra-deep sequencing regarding the mutations in the genome. Then researchers evaluated their infectivity in the Vero-E6 cells by Cycle threshold values (Ct) to quantify the viral load through in-vitro assays that resulted in identification of a total of 33 mutations. They recognized that these viral isolates demonstrate up to 270-fold differences in viral load and cytopathogenic effects (CPE) during spreading among Vero-E6 cells. After researching 12,343 SARS-CoV-2 genome patient-isolated sequences worldwide, Toyoshima et al. found 131 mutations with frequencies of more than 10%. After multiple-regression analysis, they discovered that the frequency of p.614(Asp to Gly) in spike protein, and P4715L in ORF1ab had depicted positive correlations with the fatality rate of the virus (r = 0.45, P = 0.070; r = 0.49, P = 0.047, respectively). They also found out that several epitopes which include locations of the two mentioned-mutations are likely to attach to HLA molecules [51].
To clarify the effect of a mutation on virus fitness, Yao et al. infected Vero-E6 cells (high similarity of its ACE2 or virus gate protein to human one) by employing 11 patient-derived SARS-CoV-2 isolates. They showed that mutations in RBD of S protein, as one of the most variable segments of SARS-CoV-2, has influenced the pathogenicity of SARS-CoV-2. According to the mentioned study, S-D614G has a significantly positive effect on viral replication [47]. Following Yao et al. study, another research showed that S protein is highly mutated, and some of its mutations such as D614G can potentially increase virus infection activity and also the severity of disease [48]. One study showed that a 15-30 nt deletion in the S1/S2 junction region which happened during passaging virus in Vero-E6 cells, reduced the infection rate of theSARS-CoV-2. S1/S2 junction region is cleaved during virus entry to the cell by TMPRSS2 and therefore, deletion mutations in this region can attenuate the infection rate of the virus. So far, the pre-mentioned deletion in the S1/S2 junction region has not been detected in isolated sequences from COVID-19 patients [49].
Interestingly, it was suggested that mutations of the S1/S2 junction region could potentially increase vaccine development [49]. In another study, it was reported that 8782 C > T in ORF1-ab (synonymous mutation), 28144 T > C in ORF-8 (missense), 29095 C > T in the N gene (synonymous mutation) are the most common mutations till April 5, 2020 and may have some effects on transmission and severity of COVID-19 in most area. Notably, genomic sequences analysis showed that the transition of C to T is considered the most common nucleotide variation, leading to significantly higher frequencies of synonymous mutation and missense mutations rather than deletion and insertion ones [23].
After bio-informatic analysis of 351 genome sequences of the SARS-CoV-2 patients, researchers detected two mutations in the recent isolates, involving nsp6 and adjacent regions of the ORF10 (at AA locations: 3691 and 9659). Analytics applying a Maximum Likelihood tree implies that both of the mentioned-mutations reduce the stability of the structures of the proteins [54].
Causative agents of the virus genome mutations are various, including fidelity of RdRp (as a main replication/transcription enzyme of the virus), host enzymes, effects of physical and chemical mutagens, and genetic recombination [38]. By analyzing genomic sequences from the GISAID database derived from COVID-19 patients, it was reported that viruses with a missense (P to L) mutation in position 14408 of RdRp have a higher number of mutations compared to viruses without the mutation in RdRp [38]. Also, after RdRp 14408 nt mutation occurrence (from February 16, 2020), the pattern of other variations has been changed, and new versions have been introduced to the virus genome in Europe. However, similar mutation pattern in North America was not observed in the SARS-CoV-2 genome sequences isolated from infected people, which had RdRp 14408 nt mutation. As though, the simultaneous occurrence of RdRp 14408 nt mutation with nsp3 3036 nt, S protein 23403 nt, and nucleocapsid phosphoprotein 28881 nt in Europe and with nsp13 (helicase) 17746 nt and 17857 nt, and nsp14 (ExoN) 18060 nt in North America had happened [38].
Therefore, the co-appearance of the other mutations with RdRp 14408 nt mutation is also geographically distinct. Despite the possible effect of RdRp 14408 nt mutation in the occurrence of the aforementioned mutations, other unknown factors have also influenced them. The frequency of RdRp 14408 nt in Europe is significantly higher than in North American and Asia. Detected virus genome variation from European and North American patients (mainly Europe) showed a higher number than Asia during the viral spread out of China, and as a result, the infection has a higher mortality rate out of the China [38].
Despite the reported effect of some virus genome mutations on the severity of COVID-19, so far, the researchers found less evidence of the imposed modification by the identified mutations (such as D614G in virus S protein) on the virus's transmissibility.
Through in-silico methods, researchers identified the most damaging mutations in two of the functional domains of the viral S protein, including RBD (p.408 (Arg to Ile), p.455 (Leu to Tyr), p.486 (Phe to Leu), p.493 (Gln to Asn), p.498 (Gln to Tyr), and p.501 (Asn to Thr)) and heptad repeat 1 (HR1) domain (p.930 (Ala to Val), p.936 (Asp to Tyr)) [53]. Another study investigated the S protein mutations and their biological importance by analyzing 80 SARS-CoV-2 variants and 26 mutant ones. They discovered that variants with p.614 (Asp to Gly) mutation were quite more infectious and most of the variants with a mutation in the RBD domain were less infectious. Sequences containing p.475 (Ala to Val), p.452 (Leu to Arg), p.483 (Val to Ala), and p.490 (Phe to Leu) showed resistance to several neutralizing antibodies [52].
7. Conclusion
In addition to various known and unknown factors and host genetic susceptibility, mutations and genetic variabilities of the virus itself have a critical impact on variable clinical features of COVID-19 infection. Overall, at this time, detection of all the appeared mutations in SARS-CoV-2 and their relation with pathological changes is nearly impossible, mainly because of asymptomatic subjects. Notably, the asymptomatic situation in some affected persons with the mentioned disease could be due to attenuated occurred mutations in SARS-CoV-2, which needs to be clarified in future studies. Furthermore, to confirm the results of the findings, many studies need to be repeated in other regions/locations for other people. Such studies could favor patient-specific therapy, by revealing genotyping patterns of SARS-CoV-2 distribution.
Funding
The authors received no specific funding for this work.
Declaration of competing interest
The authors declare no conflicts of interest.
References
- 1.Pourghasemi H.R., Pouyan S., Heidari B., Farajzadeh Z., Shamsi S.R.F., Babaei S., et al. Spatial modelling, risk mapping, change detection, and outbreak trend analysis of coronavirus (COVID-19) in Iran (days between 19 February to 14 June 2020) Int. J. Infect. Dis. 2020;98:90–108. doi: 10.1016/j.ijid.2020.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Cai H., Hu J., Lian J., Gu J., Zhang S., et al. Epidemiological, clinical characteristics of cases of SARS-CoV-2 infection with abnormal imaging findings. Int. J. Infect. Dis. 2020;94:81–87. doi: 10.1016/j.ijid.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microb. Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect. Dis. 2020;20(6):689–696. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K., Chen Y., Lin R., Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J. Infect. 2020;80(6):e14–e18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakhshandeh B., Sorboni S.G., Javanmard A.R., Mottaghi S.S., Mehrabi M.R., Sorouri F., et al. Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect. Genet. Evol. 2021 doi: 10.1016/j.meegid.2021.104773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant W.B., Lahore H., McDonnell S.L., Baggerly C.A., French C.B., Aliano J.L., et al. Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients. 2020;12:988. doi: 10.3390/nu12040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W., Li M., Dong Y., Zhou H., Zhang Z., Tian C., et al. Diabetes is a risk factor for the progression and prognosis of COVID ‐19. Diabetes Metab Res Rev. 2020:e3319. doi: 10.1002/dmrr.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghandi N.A., Allameh S.F., Saffarpour R. All about COVID-19 in brief. New Microbes New Infect. 2020;35 doi: 10.1016/j.nmni.2020.100678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112(5):3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020:1–28. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wille M., Holmes E.C. Wild birds as reservoirs for diverse and abundant gamma-and deltacoronaviruses. FEMS Microbiol. Rev. 2020;44:631–644. doi: 10.1093/femsre/fuaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dashraath P., Jeslyn W.J.L., Karen L.M.X., Min L.L., Sarah L., Biswas A., et al. Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am. J. Obstet. Gynecol. 2020;222(6):521–531. doi: 10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China, Cell Host & Microbe Commentary. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang T.J., Yang D.M., Wang M.L., Liang K.H., Tsai P.H., Chiou S.H., et al. Genomic analysis and comparative multiple sequences of SARS-CoV2. J. Chin. Med. Assoc. 2020;83:537–543. doi: 10.1097/JCMA.0000000000000335. [DOI] [PubMed] [Google Scholar]
- 23.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Astuti I., Srafil Y. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaul D. An overview of coronaviruses including the SARS-2 coronavirus e Molecular biology, epidemiology and clinical implications. Current Medicine Research and Practice. 2020;10:54e64. doi: 10.1016/j.cmrp.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratre Y.K., Kahar N., Bhaskar L.V.K.S., Bhattacharya A., Verma H.K. Molecular mechanism, diagnosis, and potential treatment for novel coronavirus (COVID-19): a current literature review and perspective. 3 Biotech. 2021;11:94. doi: 10.1007/s13205-021-02657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bansal M. Cardiovascular disease and COVID-19. Diabetes & metabolic syndrome. Clin. Res. Rev. 2020;14(3):247–250. doi: 10.1016/j.dsx.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj S., Chandel V., Rathi B., Kumar D. 2020. Understanding the Molecular Mechanism(s) of SARS-CoV2 Infection and Propagation in Human to Discover Potential Preventive and Therapeutic Approach. 1020944/preprints2020040285v1. [Google Scholar]
- 30.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 32.May R.M., McLean A.R., Pattison J., Weiss R.A., Holmes E.C., Rambaut A. Viral evolution and the emergence of SARS coronavirus. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2004;359:1059–1065. doi: 10.1098/rstb.2004.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020:11. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyama T., Platt D., Parida L. 2020. Variant Analysis of COVID-19 Genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu W.B., Tang G.D., Zhang L., Corlett R.T. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2/HCoV-19) using whole genomic data. Zool. Res. 2020;41:247–257. doi: 10.24272/j.issn.2095-8137.2020.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., et al. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (january to march 2020) J. Virol. 2020;94(14):e00711–e00720. doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Y.C., Anderson D.E., Young B.E., Zhu F., Linster M., Kalimuddin S., et al. bioRxiv; 2020. Discovery of a 382-nt Deletion during the Early Evolution of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asselta R., Paraboschi E.M., Mantovani A., Duga S. medRxiv; 2020. ACE2 and TMPRSS2 Variants and Expression as Candidates to Sex and Country Differences in COVID-19 Severity in Italy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao H., Lu X., Chen Q., Xu K., Chen Y., Cheng L., et al. medRxiv; 2020. Patient-derived Mutations Impact Pathogenicity of SARS-CoV-2. [Google Scholar]
- 48.Korber B., Fischer W., Gnanakaran S.G., Yoon H., Theiler J., Abfalterer W., et al. 2020. Spike Mutation Pipeline Reveals the Emergence of a More Transmissible Form of SARS-CoV-2. [Google Scholar]
- 49.Lau S.Y., Wang P., Mok B.W.Y., Zhang A.J., Chu H., Lee A.C.Y., et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microb. Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020;65(12):1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahamad S., Kanipakam H., Gupta D. Insights into the structural and dynamical changes of spike glycoprotein mutations associated with SARS-CoV-2 host receptor binding. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1811774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]