Summary
The ongoing SARS-CoV-2 pandemic has highlighted the importance of the rapid development of vaccines and antivirals. However, the potential for the emergence of antibiotic resistances due to the increased use of antibacterial cleaning products and therapeutics presents an additional, underreported threat. Most antibacterial cleaners contain simple quaternary ammonium compounds (QACs); however, these compounds are steadily becoming less effective as antibacterial agents. QACs are extensively used in SARS-CoV-2-related sanitization in clinical and household settings. Similarly, due to the danger of secondary infections, antibiotic therapeutics are increasingly used as a component of COVID-19 treatment regimens, even in the absence of a bacterial infection diagnosis. The increased use of antibacterial agents as cleaners and therapeutics is anticipated to lead to novel resistances in the coming years.
Subject areas: Multi-Drug Resistant Organisms, Applied Chemistry, Microbiology
Graphical abstract
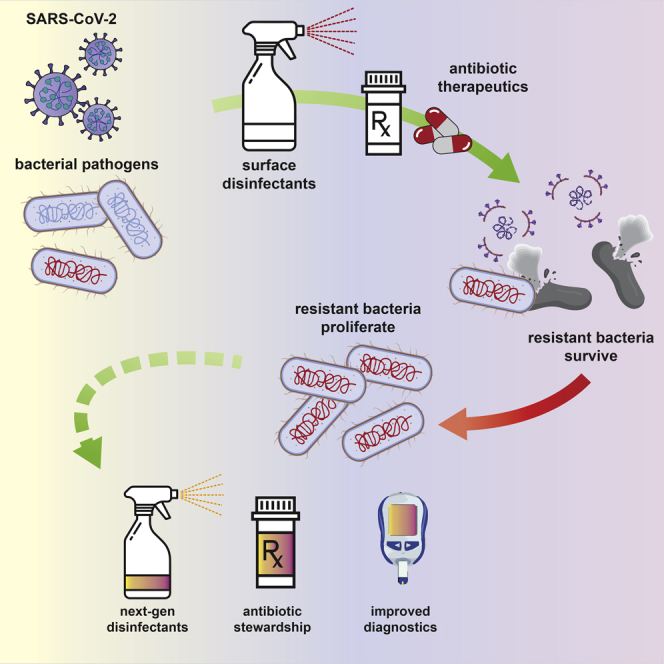
Multi-Drug Resistant Organisms; Applied Chemistry; Microbiology
Introduction
The global coronavirus disease (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has impacted the lives of millions of people worldwide since its initial identification in 2019. As of December 1, 2020, the pathogen has infected more than 97 million people and has caused over 2 million deaths globally (Johns Hopkins University, 2020). In addition to the acute effects of this deadly virus, troubling long-term symptoms that present for months following the initial infection impact an estimated 50%–80% of patients (Greenhalgh et al., 2020). In addition, dangerous bacterial secondary infections are prevalent, especially among those in intensive care due to COVID-19 (Mirzaei et al., 2020). Along with the direct threat of infection to patients, increased use of disinfectants, including hand sanitizers and surface cleaners, is anticipated to cause increased rates of antimicrobial resistance in pathogenic microbes in the coming years (Bengoechea and Bamford, 2020).
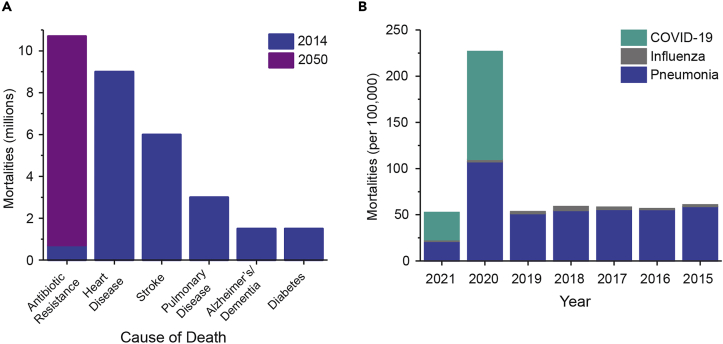
The SARS-CoV-2 pandemic has exacerbated the existing global crisis of antimicrobial resistance (AMR) (Mirzaei et al., 2020). An estimated 700,000 mortalities occur annually due to unsuccessful antibiotic treatments and that number is growing rapidly. In the United States alone, 35,000 deaths are attributed to AMR annually at a cost of $20 billion in healthcare-related expenses (U.S. Department of Health and Human Services, 2019; O'Neill, 2014). Overuse of both therapeutic antibiotic agents and disinfectants continues to cause emergent resistances in pathogens (U.S. Department of Health and Human Services, 2019; Bragg et al., 2014); at the current rate of resistance-related deaths, it is estimated that mortalities will reach 10 million annually by 2050 (Figure 1A) (O'Neill, 2014). This trend is anticipated to be accelerated by the increased use of antibiotics and antimicrobials due to the current pandemic. The impact of the pandemic on the United States, shown as excess mortalities as a result of COVID-19 as compared with other major respiratory illnesses such as influenza and pneumonia, is shown in Figure 1B (CDC, 2020). Here, we summarize two key areas in which COVID-19 is anticipated to yield emergent bacterial resistances: quaternary ammonium compounds used as disinfectants and antibiotic therapeutics often administered unnecessarily with COVID-19 hospitalizations.
Figure 1.
Antimicrobial resistance and COVID-19 mortalities
(A) Projected global annual deaths due to antibiotic resistance by 2050 as compared with the most prevalent causes of mortalities in 2014 (O'Neill, 2014).
(B) Annual mortalities due to influenza, pneumonia, and COVID-19 in the United States per 100,000 people. Data for 2021 is through the beginning of March (CDC, 2020).
Quaternary ammonium compound resistance
The antimicrobial potential of quaternary ammonium compounds (QACs) was first identified over a century ago, when researchers at the Rockefeller Institute described the bactericidal activity of a series of hexamethylenetetraminium salts on Salmonella typhi in 1916 (Jacobs et al., 1916). Since then, QACs have come to constitute one of the largest classes of active ingredients in disinfectant and biocide formulations. Due to their potential for activity against a wide variety of pathogens (bacteria, fungi, enveloped viruses, and amebae), QACs are recommended for general hospital use in the sterilization of noncritical patient care equipment—any surface that has the potential for contact with skin but not mucous membranes (Leas et al., 2015). Outside of hospitals, QACs also see use as surface disinfectants in household and foodservice settings, comprising the active ingredient of many commercially-available cleaning sprays and wipes.
Although the general class of QACs may contain a wide variety of substituents around the ammonium cation, the most commonly employed QACs for sanitization are alkonium chlorides, with benzalkonium (BACs) and dimethyldecyl ammonium chloride (DDAC) being the most common (Figure 2B). The former contains at least one benzyl group in addition to linear or branched alkyl groups ranging from 1 to 18 carbons in length. The most common counterion in QACs by far is chloride, but bromide, saccharide, and acetate are also seen (Hora et al., 2020; Schrank et al., 2020). Commercial QAC formulations are often dilute solutions (generally between 0.01% and 1%) and may contain a combination of multiple QACs. Recently, investigations into the structure-activity relationships of simple QAC scaffolds have revealed that incorporation of multiple ammonium cations and decreasing structural rigidity of QACs may lead to improved efficacy and therapeutic indices (Leitgeb et al., 2020).
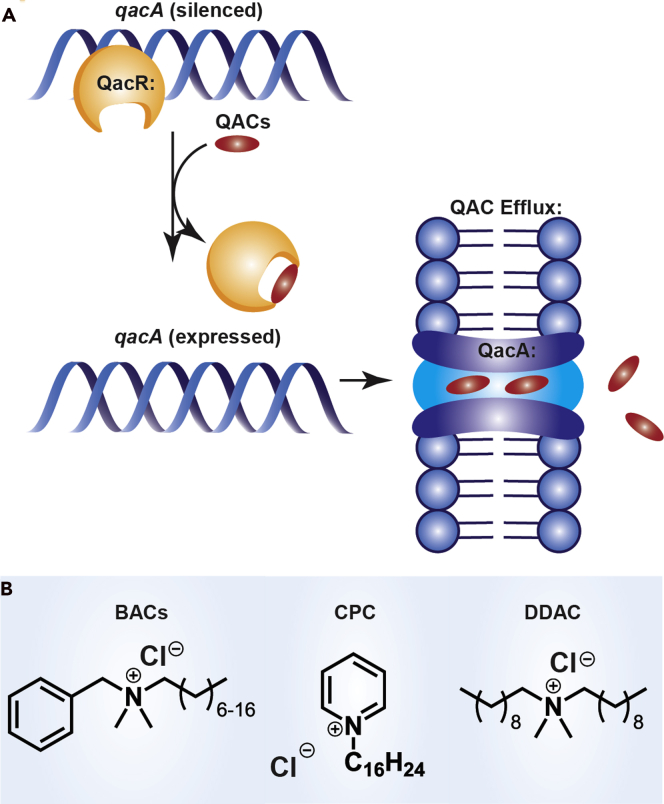
Figure 2.
Quaternary ammonium compound (QAC) resistance and structure
(A) Overview of QAC resistance mechanism in S. aureus. Once a QAC enters the cell it engages with the negative transcriptional regulator QacR. Upon binding qacA is overexpressed, resulting in the production and incorporation of QacA efflux pumps on the cell surface to expel QACs. Homologous mechanisms occur in other organisms.
(B) Structures of common QAC compounds (benzylalkonium chloride (BAC), cetylpyridinium chloride (CPC), didecyldimethylammonium chloride (DDAC)).
Although bacterial resistance to QACs was first identified in the 1980s, the precise mechanisms by which this resistance occurs remain significantly understudied (Russell et al., 1986; Jennings et al., 2016a, 2016b, 2017). As the vast majority of QAC disinfectants function via membrane permeabilization leading to cell lysis, some bacterial species have intrinsic resistance to QACs caused by the composition of their outer membrane. However, it is generally accepted that acquired resistance in Gram-positive bacteria such as Staphylococcus species is the result of the presence of multidrug or QAC-specific efflux pumps (Figure 2A) (Bragg et al., 2014; Sidhu et al., 2002). Studies have shown that methicillin-resistant S. aureus (MRSA) isolates containing qac genes have significantly higher minimum bactericidal concentrations (MBCs) than those without these genes, confirming that qac genes are indeed likely the true mechanism of resistance in these bacteria (Smith et al., 2008). Although QAC resistance in Gram-negative bacteria is less common in the literature, 2- to 8-fold changes in minimum inhibitory concentrations (MICs) in Gram-negative species were observed from species isolated from drains that had repeated QAC exposure (McBain et al., 2004). Additional studies have highlighted the emergence of QAC tolerance in the pathogens Listeria monocytogenes (Xu et al., 2014) and Pseudomonas aeruginosa (Méchin et al., 1999), with the latter noting membrane composition changes that may contribute to this tolerance.
In Staphylococcus, the genes encoding for these efflux pumps are located on mobile genetic elements such as plasmids (Bragg et al., 2014; Sidhu et al., 2002). This allows for efficient horizontal gene transfer, resulting in the potential for the rapid spread of QAC resistance between species (Figure 2A). One study found that of 238 human clinical isolates of Staphylococci from Norway, 50% were phenotypically resistant to BAC. Furthermore, the authors suggested this resistance to be a direct consequence of selective pressure resulting from the use of disinfectants (Sidhu et al., 2002). Additional studies have shown that Staphylococci isolated from surfaces frequently treated with QACs have resistances to BAC (one the most common QACs), whereas Staphylococci isolated from other, nontreated surfaces remained susceptible to this compound (He et al., 2014), providing additional evidence that it is in fact the use of QACs that drive resistance acquisition.
The ongoing SARS-CoV-2 (COVID-19) pandemic has been implicated in the exacerbation of QAC resistance. As a component of the response to this pandemic, the manufacture, sale, and use of QAC disinfectants have all increased dramatically (Hora et al., 2020), Of the 508 products on the Environmental Protection Agency's list of recommended disinfectants against SARS-CoV-2, 238 contain a QAC as the active ingredient (EPA, 2019). Disinfection is also occurring in more environments during the pandemic; once mostly relegated to indoor high-risk settings, QACs are now being used to disinfect public transportation, public benches, and other outdoor spaces. Critically, QACs are non-volatile and are not generally rinsed off of surfaces after application, enabling these compounds to remain on these surfaces or in the environment at potentially sub-inhibitory levels for extended periods of time.
The bactericidal activity of QACs is highly dependent on the context of use. Factors such as surface composition, temperature, method of application, and dwell time have all been shown to significantly impact the efficacy of sanitizers containing these compounds (Leas et al., 2015). It is likely that, due to the overwhelming number of hospital admissions during the COVID-19 pandemic, increased pressure to ensure rapid room turnaround between patients may have led to a relaxation in sanitization protocols; the pandemic has been linked to shortages in PPE, supplies and testing, and logistical support (McMullen et al., 2020). These challenges have further been linked to increases in hospital-acquired infections (McMullen et al., 2020; Richterman et al., 2020) and increased rates of sepsis (Li et al., 2020; Liu et al., 2020). Furthermore, in non-clinical environments, there is much less oversight of proper disinfection guidelines than in hospitals. Thus, QAC resistance has also been detected in non-clinical settings; testing S. aureus strains from the surfaces of ATMs in Hong Kong yielded strains containing the QAC resistance genes qacA and qacB (Zhang et al., 2012).
Reassessment and continuous auditing of disinfection protocols in all workplace and home settings is crucial to minimize the risk of infection and prevent the development of resistances in clinical bacteria. QACs have also been shown to have synergistic antimicrobial effects with phenolic compounds, so multi-component sanitizers may alleviate the immediate threat (Lambert et al., 2003). Finally, future work to investigate the regulation and function of multidrug efflux pumps in more detail is warranted. Previous studies have shown the potential for rational design of QACs to circumvent efflux as a resistance mechanism, and the use of efflux pump inhibitors in combination with QACs such as BAC may prolong the useful lifespan of these now-ubiquitous products (Garrison et al., 2020; Morrison et al., 2019).
Antibiotic therapeutic resistance
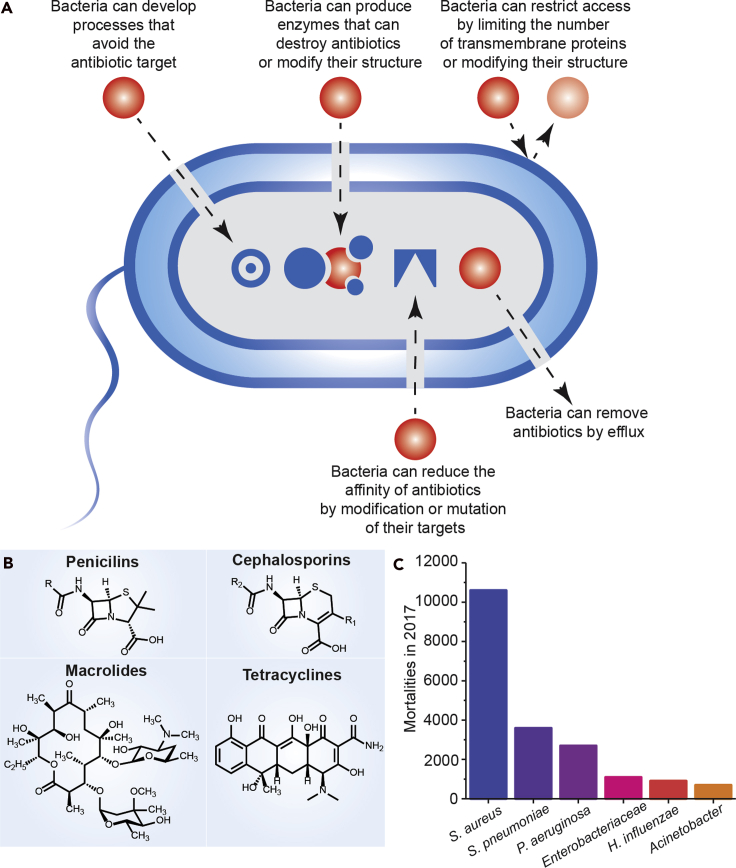
The emergence and spread of bacterial infections that are resistant to antibiotic therapeutics is an increasing global problem. Such antibiotic resistance leads to longer hospitalizations, higher mortality rates, and a significant economic burden (Tyers and Wright, 2019; Unemo and Jensen, 2017; Klein and Hultgren, 2020). Challenges with resistant strains have also limited our ability to treat new and emerging infections (Bush et al., 2011). Although microorganisms naturally acquire resistance mechanisms, the overuse and misuse of available antibiotics has impacted the rate of resistance acquisition (Figure 3A) (Tyers and Wright, 2019). In addition to inappropriate prescription of these therapeutics, improper antibiotic disposal and patient non-compliance have exacerbated the AMR problem. Along with these challenges, novel classes of antibiotics are rare, and resistances have emerged nearly immediately to those that have been introduced. Common classes of antibiotics are shown in Figure 1B. Therefore, to efficiently treat bacterial infections and prevent the continuing spread of the drug resistance, alternative treatment strategies and improved diagnostics must be developed (Tyers and Wright, 2019; Bush et al., 2011). Among various approaches to combat and treat resistant infections, multi-drug therapies and repurposing existing treatments could sufficiently improve efficacy and help avert the emergence of new resistances (Tyers and Wright, 2019).
Figure 3.
Antibiotic therapeutic resistance and bacterial co-infections
(A) Overview of therapeutic resistance.
(B) Structures of common classes of broad-spectrum antibiotics.
(C) Mortalities in 2017 due to common bacterial infections (U.S. Department of Health and Human Services, 2019). These infections are prevalent as co-infections and secondary infections with SARS-CoV-2 (Zhu et al., 2020).
Despite ongoing efforts to decrease the overuse of antibiotic therapeutics, prescribing antibiotics remains inevitable, especially in severe cases such as bacterial co-infections or secondary infections with another illness to ensure that the pathogen load is eliminated (Mirzaei et al., 2020; Lai et al., 2020b). Lower- and upper-respiratory tract viral infections substantially predispose the patients to bacterial infections by providing more sites for bacterial adhesion, impairing immune responses, and causing cell and tissue damage that facilitate the spread of bacteria and development of an invasive infection (Mirzaei et al., 2020; Lai et al., 2020a, 2020b; Morris et al., 2017; Vincent et al., 2020). As a result, the severity of the illness, as well as morbidity and mortality rates, are often significantly increased (Mirzaei et al., 2020). Viral diseases can be complicated by secondary bacterial infections caused by numerous strains, including Streptococcus pneumoniae, Streptococcus pyogenes, S. aureus and Haemophilus influenzae (Morris et al., 2017; Rawson et al., 2020). It is important to note that bacterial co-infections are less prevalent than secondary infections. In a study of nearly 1,400 COVID-19 patients, only 2.7% were diagnosed with a bacterial infection that co-occurred with the viral infection (Wang et al., 2021). In another study, 5.1% of the cases (5 out of 99) with COVID-19 were reported to have co-infections including Acinetobacter baumannii and Klebsiella pneumoniae (Chen et al., 2020). In a larger study by Zheng et al., of 221 patients with COVID-19, 25.8% of the patients (57 out of 221) were diagnosed with co-infections and 29.8% of these co-infected cases were reported to have bacterial co-infections (Blasco et al., 2020). Moreover, a review of nine reports published on hospitalized COVID-19 patients showed that 8% of the cases (62 out of 806) had bacterial/fungal co-infections (Rawson et al., 2020).
In contrast, the increasing number of the COVID-19 cases diagnosed with secondary bacterial infections following the viral infection has become a substantial concern, as it can lead to poorer patient outcomes and longer hospitalizations (Mirzaei et al., 2020; Rawson et al., 2020; Hendaus and Jomha, 2020; Chen et al., 2020). In a study by Huang et al., 9.8% of cases (4 out of 41) with COVID-19 had secondary bacterial infections (Huang et al., 2020). A second study of COVID-19 secondary infections found that 9.3% had a secondary infection (Ripa et al., 2021). In addition, increased rates of hospital-acquired infections (McMullen et al., 2020; Richterman et al., 2020) and sepsis (Li et al., 2020; Liu et al., 2020) have occurred during the COVID-19 pandemic, emphasizing the importance of accessible testing and diagnostics for both co-infections and secondary infections; such testing has been limited during the pandemic (McMullen et al., 2020; Ripa et al., 2021). One key challenge, as can be seen with the highly variable rate of diagnosed co-infections and secondary infections, is that effective diagnostics for these illnesses remain lacking, especially those that can identify bacteria and resistances rapidly, even when hospital resources are stretched or testing is needed in resource-limited settings (McMullen et al., 2020).
In the majority of cases, antibiotic administration and combination therapies have been reported as the most frequently practiced treatment strategies for co-infections or secondary infections (Lai et al., 2020b; Rawson et al., 2020). There is also evidence indicating that 70% of hospitalized COVID-19 patients received at least one antibiotic whether or not they were diagnosed with a bacterial infection (Rawson et al., 2020; Nori et al., 2021). In another study on 138 patients with COVID-19, 64.4%, 24.6%, and 18.1% of the patients received moxifloxacin, ceftriaxone, and azithromycin, respectively (Wang et al., 2020). Taken together, the danger of bacterial secondary infections along with the preemptive administration of antibiotic therapeutics clearly shows an environment ripe for the emergence of new resistances.
Despite the demonstrated efficacy of antibiotic treatments, the increased use of antibiotics in COVID-19 hospitalizations (Rawson et al., 2020; Nori et al., 2021) during this pandemic is anticipated to result in a strong selective pressure on bacterial pathogens to evolve resistances, leading to an increased rate of drug-resistant bacterial infections (Karbelkar and Furst, 2020). Reported bacterial infections in COVID-19 patients include Streptococcus pneumoniae, Klebsiella pneumoniae, Haemophilus influenzae, Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Acinetobacter baumannii (Rawson et al., 2020). These are also among the most prevalent deadly antibiotic-resistant infections, as reported by the CDC (Figure 3C) (U.S. Department of Health and Human Services, 2019; Zhu et al., 2020). As AMR will likely become an even larger challenge in the post-pandemic era, developing novel therapeutic strategies as well as strictly implementing antimicrobial stewardship programs are crucial to combat this emergence. Although treatments are continuously developed for evolving resistances, therapeutic alternatives such as combination therapies, targeted delivery systems, and alternative small molecules are crucial to combat the fast-paced development of AMR (Brooks and Brooks, 2014).
Summary and outlook
As highlighted earlier, increasing levels of antibacterial resistant organisms are inevitable, given the extreme circumstances facing us. This increase has been directly shown in studies of COVID-19 patients; in one study from a New York City hospital, it was found that 98% of patients received antibiotics during their hospitalization for COVID-19, and multiple species (K. pneumoniae, P. aeruginosa, and Enterobacter cloacae) had a significant (>10%) decline in susceptibility to at least one antibiotic (Nori et al., 2021). As a result, a holistic, all-encompassing approach will be needed to ward off a future wave of pan-resistant organisms. Effective communication and breakthroughs in scientific research, namely in new therapeutics and diagnostics, are of paramount importance to mitigate the impact of such a wave.
First and foremost (as evidenced by this perspective), there will need to be greater scientific communication of the upcoming threats in order to increase awareness not only to the general public but also to governments as well as non-profit organizations and foundations. The latter is essential to provide the necessary funding levels to conduct the tremendous amount of work facing us in the future. CARB-X (Carb-X, 2016), a global non-profit partnership, is one example of an efficient and well-organized effort focused on supporting innovative and necessary antibiotic development. In addition, media campaigns by both the WHO and CDC have increased awareness to the public about best practices for hand-washing and sanitation. This work will need to be extended more broadly to continue educating the community about the perils of antibacterial drug resistance, as in many cases, the severity and speed of this challenge remain poorly understood by the public (Ancillotti et al., 2018; Röing et al., 2020; Wernli et al., 2017).
On the research front, antibiotic stewardship will be essential. To prolong the effective lives of the available antibiotics, three clinical questions should always be answered prior to antibiotic prescription: first, whether the patient is infected with a microbial pathogen; second, whether they need an antimicrobial treatment; and third, what antimicrobial treatment would have the highest efficacy (Burnham et al., 2017). Developing rapid, low-cost, point-of-care diagnostic technologies is urgently needed to answer these questions in a timely manner. Such rapid diagnostic tests must be able to identify bacteria, determine their antimicrobial susceptibility, and facilitate the prescription of the proper narrow-spectrum antibiotics at the point of care. Such easy-to-use sensors will also facilitate diagnosis of asymptomatic patients infected with the resistant pathogens, particularly in nosocomial settings.
In addition to the development of diagnostics for microbial pathogens, engineering assays to rapidly distinguish between viral and bacterial infections would significantly decrease the rate of unnecessary antibiotic therapeutic administration by preventing their preemptive prescription (Burnham et al., 2017). Conventional diagnostics for bacterial infections are either genotypic methods that detect resistance genes or immunological assays such as ELISA (enzyme-linked immunosorbent assay) that are based on antigen-antibody interactions. The major drawback of the traditional techniques is that they are time-consuming and require centralized laboratory facilities (Burnham et al., 2017; Karbelkar and Furst, 2020). Further, the genotypic methods are not capable of detecting antimicrobial susceptibility and emerging resistance patterns for which a gene has not been defined (Burnham et al., 2017). In contrast, biosensing technologies have demonstrated significant potential for rapid point-of-care testing with low-cost instruments (Karbelkar and Furst, 2020). Electrochemical biosensors, particularly, are favorable for their low-cost assembly, rapid detection, quantitative readout, and extremely low limits of detection (Furst and Francis, 2019). We believe electrochemical biosensors incorporated into low-cost, high-throughput platforms will become the gold standard for point-of-care diagnostics to differentiate between viral and bacterial infections, identify bacterial pathogens present in an infection, and determine the susceptibility of bacterial pathogens to multiple antibiotics to prevent a surge of antibiotic resistances during the COVID-19 pandemic and in the post-pandemic era (Karbelkar and Furst, 2020).
Finally, continued research efforts toward antibacterial agents with novel mechanisms of action are necessary. Significant progress has been made over the past years, aided substantially by the CARB-X initiatives and academic endeavors. However, continued discoveries will be necessary. In a similar fashion, refocused studies toward novel antiseptics are also needed. As alluded to earlier, disinfectant efficacy is being challenged as never before, and resistance to QACs is rising. New cocktails or formulations will be necessary to maintain activity. Recent work in one of our labs (W.M.W.) has demonstrated that QACs containing two or more ammonium moieties (multiQACs) are not as susceptible to the same resistance mechanisms as BAC and might serve as new additives to broaden the efficacy scope of this heralded class of disinfectants (Forman et al., 2016; Mitchell et al., 2015; Jennings et al., 2016a). Work is ongoing to test this hypothesis against a number of multi-drug resistant organisms.
In summary, the current COVID-19 pandemic, caused by the SARS-CoV-2 virus, has brought society to a standstill and caused hundreds of thousands of deaths. The need for disinfection strategies and treatments for deadly infections cannot be understated (Hardie, 2020; Hsu, 2020; Strathdee et al., 2020). Importantly, though, both the disinfection and therapeutic strategies used to combat the spread and severity of COVID-19 are anticipated to exacerbate another global crisis: antimicrobial resistance. The increased use of antibiotic therapeutics and QAC disinfectants during this pandemic is anticipated to result in the rapid emergence of novel, deadly, AMR pathogens. This secondary crisis will only be mitigated with active efforts to improve communication to the public about the risk of these infections, increase funding to research new and alternative disinfectants and therapies to combat the spread of these microbes, and develop precision point-of-care diagnostics to reduce unnecessary antibiotic prescriptions.
Limitations of the study
This work was written with information available at the time of publication. Data on the COVID-19 pandemic are constantly being updated.
Resource availability
Lead contact
Ariel L. Furst (afurst@mit.edu).
Materials availability
There are no materials that were generated.
Data and code availability
There is no data or code generated unique to this work.
Acknowledgments
The Furst Lab acknowledges the MIT Research Support Committee for funding. The Wuest lab acknowledges funds from the NIH (GM119426). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Writing, Reviewing, & Editing, A.R.M. and M.M.S.; Funding Acquisition, W.M.W. and A.L.F.; Supervision, W.M.W. and A.L.F.
Declaration of interests
W.M.W. has intellectual property covering novel quaternary ammonium compounds. A.L.F. is a member of the Editorial Advisory Board of iScience.
References
- Ancillotti M., Eriksson S., Veldwijk J., Nihlén Fahlquist J., Andersson D.I., Godskesen T. Public awareness and individual responsibility needed for judicious use of antibiotics: a qualitative study of public beliefs and perceptions. BMC Public Health. 2018;18:1153. doi: 10.1186/s12889-018-6047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . CDC; 2020. Coronavirus Disease 2019 (COVID-19) [Google Scholar]
- Bengoechea J.A., Bamford C.G. SARS -CoV-2, bacterial co-infections, and AMR : the deadly trio in COVID -19? EMBO Mol. Med. 2020:10–13. doi: 10.15252/emmm.202012560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.L., Buesa J., Colomina J., Forner M.J., Galindo M.J., Navarro J., Noceda J., Redón J., Signes-Costa J., Navarro D. Co-detection of respiratory pathogens in patients hospitalized with Coronavirus viral disease-2019 pneumonia. J. Med. Virol. 2020;92:1799–1801. doi: 10.1002/jmv.25922. [DOI] [PubMed] [Google Scholar]
- Bragg R., Jansen A., Coetzee M., van der Westhuizen W., Boucher C. Bacterial resistance to quaternary ammonium compounds (QAC) disinfectants. Adv. Exp. Med. Biol. 2014;808:1–13. doi: 10.1007/978-81-322-1774-9_1. [DOI] [PubMed] [Google Scholar]
- Brooks B.D., Brooks A.E. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 2014;78:14–27. doi: 10.1016/j.addr.2014.10.027. [DOI] [PubMed] [Google Scholar]
- Burnham C.A.D., Leeds J., Nordmann P., O’Grady J., Patel J. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 2017;15:697–703. doi: 10.1038/nrmicro.2017.103. [DOI] [PubMed] [Google Scholar]
- Bush K., Courvalin P., Dantas G., Davies J., Eisenstein B., Huovinen P., Jacoby G.A., Kishony R., Kreiswirth B.N., Kutter E. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carb-X . 2016. Home - Carb-X. https://carb-x.org/ [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA . 2019. List N Tool: COVID-19 Disinfectants. https://cfpub.epa.gov/giwiz/disinfectants/index.cfm. [Google Scholar]
- Forman M.E., Fletcher M.H., Jennings M.C., Duggan S.M., Minbiole K.P.C., Wuest W.M. Structure-resistance relationships: interrogating antiseptic resistance in bacteria with multicationic quaternary ammonium dyes. ChemMedChem. 2016;11:958–962. doi: 10.1002/cmdc.201600095. [DOI] [PubMed] [Google Scholar]
- Furst A.L., Francis M.B. Impedance-based detection of bacteria. Chem. Rev. 2019;119:700–726. doi: 10.1021/acs.chemrev.8b00381. [DOI] [PubMed] [Google Scholar]
- Garrison M.A., Mahoney A.R., Wuest W.M. Tricepyridinium-inspired QACs yield potent antimicrobials and provide insight into QAC resistance. ChemMedChem. 2020 doi: 10.1002/cmdc.202000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T., Knight M., A’Court C., Buxton M., Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370:2020–2021. doi: 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- Hardie K.R. Antimicrobial resistance: the good, the bad, and the ugly. Emerg. Top. Life Sci. 2020;4:129–136. doi: 10.1042/ETLS20190194. [DOI] [PubMed] [Google Scholar]
- He G.-X., Landry M., Chen H., Thorpe C., Walsh D., Varela M.F., Pan H. Detection of benzalkonium chloride resistance in community environmental isolates of staphylococci. J. Med. Microbiol. 2014;63:735–741. doi: 10.1099/jmm.0.073072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus M.A., Jomha F.A. Covid-19 induced superimposed bacterial infection. J. Biomol. Struct. Dyn. 2020;0:1–7. doi: 10.1080/07391102.2020.1772110. [DOI] [PubMed] [Google Scholar]
- Hora P.I., Pati S.G., McNamara P.J., Arnold W.A. Increased use of quaternary ammonium compounds during the SARS-CoV-2 pandemic and beyond: consideration of environmental implications. Environ. Sci. Technol. Lett. 2020;7:622–631. doi: 10.1021/acs.estlett.0c00437. [DOI] [PubMed] [Google Scholar]
- Hsu J. How covid-19 is accelerating the threat of antimicrobial resistance. BMJ. 2020;369:m1983. doi: 10.1136/bmj.m1983. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs W.A., Heidelberger M., Amoss H.L. The bactericidal properties of the quaternary salts of hexamethylenetetramine: II. The relation between constitution and bactericidal action in the substituted benzylhexamethylenetetraminium salts. J. Exp. Med. 1916;23:569–576. doi: 10.1084/jem.23.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M.C., Buttaro B.A., Minbiole K.P.C., Wuest W.M. Bioorganic investigation of multicationic antimicrobials to combat QAC-resistant Staphylococcus aureus. ACS Infect. Dis. 2016;1:304–309. doi: 10.1021/acsinfecdis.5b00032. [DOI] [PubMed] [Google Scholar]
- Jennings M.C., Minbiole K.P.C., Wuest W.M. Quaternary ammonium compounds: an antimicrobial mainstay and platform for innovation to address bacterial resistance. ACS Infect. Dis. 2016;1:288–303. doi: 10.1021/acsinfecdis.5b00047. [DOI] [PubMed] [Google Scholar]
- Jennings M.C., Forman M.E., Duggan S.M., Minbiole K.P.C., Wuest W.M. Efflux pumps might not Be the major drivers of QAC resistance in methicillin-resistant Staphylococcus aureus. ChemBioChem. 2017;18:1573–1577. doi: 10.1002/cbic.201700233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University . Johns Hopkins Coronavirus Resource Center; 2020. COVID-19 Map - Johns Hopkins Coronavirus Resource Center. [Google Scholar]
- Karbelkar A.A., Furst A.L. Electrochemical diagnostics for bacterial infectious diseases. ACS Infect. Dis. 2020;6:1567–1571. doi: 10.1021/acsinfecdis.0c00342. [DOI] [PubMed] [Google Scholar]
- Klein R.D., Hultgren S.J. Urinary tract infections: microbial pathogenesis, host–pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 2020;18:211–226. doi: 10.1038/s41579-020-0324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Wang C.-Y., Hsueh P.-R. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020;53:505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R.J.W., Johnston M.D., Hanlon G.W., Denyer S.P. Theory of antimicrobial combinations: biocide mixtures - synergy or addition? J. Appl. Microbiol. 2003;94:747–759. doi: 10.1046/j.1365-2672.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Leitgeb A.J., Feliciano J.A., Sanchez H.A., Allen R.A., Morrison K.R., Sommers K.J., Carden R.G., Wuest W.M., Minbiole K.P.C. Further investigations into rigidity-activity relationships in BisQAC amphiphilic antiseptics. ChemMedChem. 2020;15:667–670. doi: 10.1002/cmdc.201900662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., Su X., Cao B. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Wang Q., Zhang H., Cui L., Shen F., Chen Y., Sun Jiali, Gan L., Sun Jianhui, Wang J. Viral sepsis is a complication in patients with novel corona virus disease (COVID-19) Med. Drug Discov. 2020;8:100057. doi: 10.1016/j.medidd.2020.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain A.J., Ledder R.G., Moore L.E., Catrenich C.E., Gilbert P. Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 2004;70:3449–3456. doi: 10.1128/AEM.70.6.3449-3456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen K.M., Smith B.A., Rebmann T. Impact of SARS-CoV-2 on hospital acquired infection rates in the United States: predictions and early results. Am. J. Infect. Control. 2020;48:1409–1411. doi: 10.1016/j.ajic.2020.06.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchin L., Dubois-Brissonnet F., Heyd B., Leveau J.Y. Adaptation of Pseudomonas aeruginosa ATCC 15442 to didecyldimethylammonium bromide induces changes in membrane fatty acid composition and in resistance of cells. J. Appl. Microbiol. 1999;86:859–866. doi: 10.1046/j.1365-2672.1999.00770.x. [DOI] [PubMed] [Google Scholar]
- Mirzaei R., Goodarzi P., Asadi M., Soltani A., Aljanabi H.A.A., Jeda A.S., Dashtbin S., Jalalifar S., Mohammadzadeh R., Teimoori A. Bacterial co-infections with SARS-CoV-2. IUBMB Life. 2020;72:2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M.A., Iannetta A.A., Jennings M.C., Fletcher M.H., Wuest W.M., Minbiole K.P.C. Scaffold-hopping of multicationic amphiphiles yields three new classes of antimicrobials. ChemBioChem. 2015;16:2299–2303. doi: 10.1002/cbic.201500381. [DOI] [PubMed] [Google Scholar]
- Morris D.E., Cleary D.W., Clarke S.C. Secondary bacterial infections associated with influenza pandemics. Front. Microbiol. 2017;8:1041. doi: 10.3389/fmicb.2017.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison K.R., Allen R.A., Minbiole K.P.C., Wuest W.M. More QACs, more questions: recent advances in structure activity relationships and hurdles in understanding resistance mechanisms. Tetrahedron Lett. 2019;60:150935. doi: 10.1016/j.tetlet.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nori P., Cowman K., Chen V., Bartash R., Szymczak W., Madaline T., Punjabi Katiyar C., Jain R., Aldrich M., Weston G. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect. Control Hosp. Epidemiol. 2021;42:84–88. doi: 10.1017/ice.2020.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. 2014. Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. The Review on Antimicrobial Resistance chaired by Jim O'Neill. Available from: https://wellcomecollection.org/works/rdpck35v. [Google Scholar]
- Rawson T.M., Moore L.S.P., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Cooke G., Holmes A. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richterman A., Meyerowitz E.A., Cevik M. Hospital-acquired SARS-CoV-2 infection: lessons for public Health. JAMA. 2020;324:2155–2156. doi: 10.1001/jama.2020.21399. [DOI] [PubMed] [Google Scholar]
- Ripa M., Galli L., Poli A., Oltolini C., Spagnuolo V., Mastrangelo A., Muccini C., Monti G., De Luca G., Landoni G. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röing M., Björkman I., Eriksen J., Stålsby Lundborg C. The challenges of implementing national policies to contain antibiotic resistance in Swedish healthcare-A qualitative study of perceptions among healthcare professionals. PLoS One. 2020;15:e0233236. doi: 10.1371/journal.pone.0233236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.D., Hammond S.A., Morgan J.R. Bacterial resistance to antiseptics and disinfectants. J. Hosp. Infect. 1986;7:213–225. doi: 10.1016/0195-6701(86)90071-X. [DOI] [PubMed] [Google Scholar]
- Schrank C.L., Minbiole K.P.C., Wuest W.M. Are quaternary ammonium compounds, the workhorse disinfectants, effective against severe acute respiratory syndrome-coronavirus-2? ACS Infect. Dis. 2020;6:1553–1557. doi: 10.1021/acsinfecdis.0c00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu M.S., Heir E., Leegaard T., Wiger K., Holck A. Frequency of disinfectant resistance genes and genetic linkage with beta-lactamase transposon Tn552 among clinical staphylococci. Antimicrob. Agents Chemother. 2002;46:2797–2803. doi: 10.1128/aac.46.9.2797-2803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., Gemmell C.G., Hunter I.S. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J. Antimicrob. Chemother. 2008;61:78–84. doi: 10.1093/jac/dkm395. [DOI] [PubMed] [Google Scholar]
- Strathdee S.A., Davies S.C., Marcelin J.R. Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet. 2020;396:1050–1053. doi: 10.1016/S0140-6736(20)32063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Wright G.D. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019;17:141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- Unemo M., Jensen J.S. Antimicrobial-resistant sexually transmitted infections: gonorrhoea and Mycoplasma genitalium. Nat. Rev. Urol. 2017;14:139–152. doi: 10.1038/nrurol.2016.268. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services . Centers for Disease Control and Prevention; 2019. Antibiotic Resistance Threats in the United States; pp. 1–113. [Google Scholar]
- Vincent J.L., Sakr Y., Singer M., Martin-Loeches I., MacHado F.R., Marshall J.C., Finfer S., Pelosi P., Brazzi L., Aditianingsih D. Prevalence and outcomes of infection among patients in intensive care units in 2017. JAMA. 2020;323:1478–1487. doi: 10.1001/jama.2020.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Amin A.K., Khanna P., Aali A., McGregor A., Bassett P., Gopal Rao G. An observational cohort study of bacterial co-infection and implications for empirical antibiotic therapy in patients presenting with COVID-19 to hospitals in North West London. J. Antimicrob. Chemother. 2021;76:796–803. doi: 10.1093/jac/dkaa475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernli D., Jørgensen P.S., Harbarth S., Carroll S.P., Laxminarayan R., Levrat N., Røttingen J.-A., Pittet D. Antimicrobial resistance: the complex challenge of measurement to inform policy and the public. PLoS Med. 2017;14:e1002378. doi: 10.1371/journal.pmed.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Li Y., Zahid M.S.H., Yamasaki S., Shi L., Li J., Yan H. Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int. J. Food Microbiol. 2014;190:24–30. doi: 10.1016/j.ijfoodmicro.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Zhang M., O’Dononghue M., Boost M.V. Characterization of staphylococci contaminating automated teller machines in Hong Kong. Epidemiol. Infect. 2012;140:1366–1371. doi: 10.1017/S095026881100207X. [DOI] [PubMed] [Google Scholar]
- Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., Zhu F., Zhu B., Cui L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leas, B.F., Sullivan, N., Han, J.H., Pegues, D.A., Kaczmarek, J.L., Umscheid, C.A.. Environmental Cleaning for the Prevention of Healthcare-Associated Infections. Rockville (MD): Agency for Healthcare Research and Quality (US); 2015. (Technical Briefs, No. 22.) Available from: https://www.ncbi.nlm.nih.gov/books/NBK311016/. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no data or code generated unique to this work.