Abstract
Non-human primates (NHPs) have been proposed as good models for neurodevelopmental disorders due to close similarities to humans in terms of brain structure and cognitive function. The recent development of genome editing technologies has opened new avenues to generate and investigate genetically modified NHPs as models for human disorders. Here, we review the early successes of genetic NHP models for neurodevelopmental disorders and further discuss the technological challenges and opportunities to create next generation NHP models with more sophisticated genetic manipulation and faithful representations of the human genetic mutations. Taken together, the field is now poised to usher in a new era of research using genetically modified NHP models to empower a more rapid translation of basic research and maximize the preclinical potential for biomarker discovery and therapeutic development.
A new era for non-human primate models in neurodevelopmental disorders research
Recent large-scale genomic studies have revealed the significant contribution of genetic factors in the etiology of neurodevelopmental disorders. In the case of autism spectrum disorders (ASD), the largest classification of neurodevelopmental disorders, rare genetic mutations or chromosome abnormalities are thought to be causal for up to 10-20% of cases [1]. Genetically modified mice targeting these identified risk genes and associated chromosomal abnormalities have provided important insights into gene function and the potential pathophysiological mechanisms of neurodevelopmental disorders. Although mouse models have been the dominant model for preclinical research, direct translation of research outcomes from mice into new treatments for human patients has proven difficult, or impossible. 80 million years of evolutionary distance between the human and the mouse is evident across all domains of scientific investigation, including genomic, transcriptome, molecular, cellular, circuit, anatomical, physiological, cognitive, and behavioral studies [2-4]. Thus, additional models with brain structure and function closer to that of human are needed to complement rodent models to advance our understanding of neurobiological mechanisms as well as development of effective treatment. [3,4].
Non-human primates (NHPs), such as cynomolgus (crab-eating) and rhesus macaque monkeys or the common marmoset, are phylogenetically much closer to humans than rodents. NHPs share with humans many brain regions, circuits, and cell types critical for higher cognitive capacity [5]. For example, NHPs have a well-developed prefrontal cortex (PFC), a region dramatically expanded in primates and one of the largest and most-developed regions in the human brain, whereas rodents have a much smaller PFC. The dorsolateral PFC forms neural circuits connected with other cortical areas and subcortical regions, and mediates various cognitive functions such as decision making, working memory, executive function, and attention control, which are often affected in brain disorders. At the cellular level, recent single cell transcriptomic analyses revealed significant differences in molecular signatures and primate-specific cell types [6]. In addition, NHPs have higher cognitive and social communication abilities expressed through complex behavioral repertoires including face recognition, eye gazing, and vocalization that more closely resemble humans and are difficult to study in rodents [2-4]. Thus NHPs are expected to be better animal models for studying the function and dysfunction involving PFC-related circuits and behaviors in brain disorders [2-4]. Despite these potential advantages, the value of NHPs to study the biological mechanisms underlying human disorders has not been realized due to the lack of technologies to precisely manipulate NHP genomes to model the genetic mutations and chromosomal abnormalities found in human patients. However, recent advances in CRISPR-mediated genome editing technologies are changing the field by enabling precise modification of genomes virtually in any cell type in any species, including NHPs.
In the past decade, we have witnessed a second “Big Bang” in scientific advancement via the CRISPR revolution [7]. CRISPR (clustered regularly interspaced short palindromic repeats) / Cas (CRISPR-associated) systems are programmable nucleases that cut target genomic site(s) with extremely high efficiency and specificity. At the target site, a CRISPR-induced DNA double strand break (DSB) is repaired through cellular DNA repair machinery called non-homologous end-joining (NHEJ) with frequent formation of small insertions and deletions (indels) that lead to functional knockout of the target gene. DSBs can also be repaired through homology-directed repair (HDR), which introduces nucleotide substitution or transgene insertion under the presence of repair template leading to precise knockin into the target gene, although HDR efficiency is much lower than that of NHEJ in many cell types. CRISPR has been engineered to be a multi-operational molecular platform that can be harnessed for single base substitution without DSB by base editing, HDR-independent substitution and modification with less indels by prime editing, transposon-based transgene insertion, RNA editing, transcriptional activation and repression, epigenome editing, live imaging of the genome and RNA, manipulation of chromosomal locations, and more [7] (Table 1).
Table 1. Potential genome editing strategies for next generation NHP models.
G: generation, KO: knockout, CNV: copy number variation, LD: large deletion, DSBs: DNA double strand breaks, RNP: ribonucleoprotein, HDR: homology-directed repair.
| Types | Methods | Pros | Cons | New improvements | |
|---|---|---|---|---|---|
| 1st Generation | KO | Indel | Very efficient | Unpredictable outcomes Unexpected truncated protein |
Predictable algorithms [65] Entire gene deletion [66] |
| CNV (LD) | 2-DSBs | Efficient | Unpredictable outcomes | Bridging donor with RNP [67] | |
| 2nd Generation | SNV | HDR | Precise | Variable efficiency Indel formation Needs donor DNA |
RNP [34] Donor tethering [68] HDR enhancers [69] |
| Base Editing | Precise Efficient No DSB |
Deaminase DNA and RNA off-target mutations C>T/G>A, A>G/T>C only Bystander editing Limited target window |
New variants without deaminase off-target [39] PAMless Cas9 [70] |
||
| Prime Editing | Precise Efficient Any SNVs Few DSB |
Not tested in animals yet Needs pegRNA and nicking sgRNA optimization |
None yet | ||
| 3rd Generation | Humanized | HDR | Precise Up to several kb |
Variable efficiency Indel formation Needs donor DNA |
RNP [34] Donor tethering and 2-cell stage injection [71] HDR enhancers [72] |
| Prime Editing | Precise Efficient Few DSB |
Needs optimization Not tested in animals yet Currently, up to ~40bp |
None yet |
CRISPR-mediated genome editing has been rapidly applied to generate mutant NHP models by manipulation of genome directly in zygotes [8]. More than 10 mutant NHP lines, mainly knockout by NHEJ, but also a few knockin lines by HDR, have been reported in cynomolgus, rhesus, and marmoset (also by other tools such as ZFN (zinc-finger nuclease) [9] and TALEN (transcription activator-like effector nuclease) [10]) (Table 2). These recent advances in generating mutant NHPs have demonstrated the feasibility, robustness, and power of CRISPR-mediated genome editing in NHPs, and opened exciting avenues to investigate genetically modified NHPs as models for the biological underpinnings of human disorders.
Table 2. Genome edited NHPs.
Only mutant NHPs with relevant-phenotypes are shown. RTT: Rett Syndrome, PMS: Phelan-McDermid Syndrome, AHC-HH: Adrenal hypoplasia congenital and hypogonadotropic hypogonadism, ADPKD: Autosomal dominant polycystic kidney disease, PD: Parkinson’s disease, DMD: Duchenne muscular dystrophy, SCID: Severe combined immunodeficiency.
| Diseases | Species | Genes | Mutations | Phenotypes | Refs |
|---|---|---|---|---|---|
| RTT | Cynomolgus | MECP2 | KO TALEN | RTT-like symptoms in het females | [12] |
| PMS | Cynomolgus | SHANK3 | KO CRISPR | PMS-like symptoms in homo and het monkeys | [24,26] |
| Longevity | Cynomolgus | SIRT6 | KO CRISPR | Prenatal death, developmental retardation | [52] |
| Microcephaly | Cynomolgus | MCPH1 | KO TALEN | Microcephaly | [73] |
| Circadian rhythm | Cynomolgus | BMAL1 | KO CRISPR | Reduced sleep | [74] |
| AHC-HH | Cynomolgus | DAX1 | KO CRISPR | AHC-HH-like symptoms | [75] |
| ADPKD | Cynomolgus | PKD1 | KO CRISPR | Cysts formation in homo and het monkeys | [63] |
| PD | Rhesus | PINK1 | KO CRISPR | Neurodegeneration | [76] |
| DMD | Rhesus | DMD | KO CRISPR | Muscular degeneration | [77] |
| Hypercholesterolemia | Rhesus | PCSK9 | in vivo KO Meganuclease | Reduced serum cholesterol | [78] |
| SCID | Marmoset | IL2RG | KO ZFN/TALEN | Immunodeficiency | [9] |
Early success of genetically modified NHPs for neurodevelopmental disorder research
Rett syndrome
Rett syndrome (RTT) is a severe monogenic neurodevelopmental disorder caused by heterozygous mutation in the X-linked MECP2 gene. Hemizygous MECP2 mutation in males is largely embryonic lethal, therefore RTT patients are predominantly female. Females exhibit normal development in the first 6-18 months of life, but subsequently lose previously acquired motor function and social communication, eventually developing symptoms shared with other neurodevelopmental disorders, such as stereotypical hand movements, seizures, and intellectual disability. Although mutant mice have been used in multiple studies exploring the mechanisms of Rett syndrome, neurological abnormalities are less severe than what is seen in human RTT patients. Additionally, Mecp2 hemizygous mutant male mice are viable while heterozygous mutant female mice exhibit only mild symptoms in adulthood [11].
MECP2 knockout cynomolgus monkeys are one of the earliest successful cases of mutant NHP production and founder (F0) generation mutant monkeys have been deeply characterized [10,12-15]. Mutant MECP2 monkeys demonstrate remarkable similarities to human RTT syndrome patients in several aspects [12]: 1) Male mutant monkeys are embryonic lethal, consistent with human male RTT patients. 2) Female mutant monkeys (with mosaicism) are viable and MECP2 protein levels are decreased to roughly half of control wild-type monkeys, resembling heterozygous MECP2 mutation in human female RTT patients. 3) Behavioral analyses have revealed a wide variety of abnormalities in female mutant monkeys that are consistent with human RTT patients, including fragmented sleep, reduced sensitivity to sensory stimuli, such as pain and noise, reduced social interaction, and increased stereotyped repetitive behaviors. 4) Eye-tracking tests have shown that mutant monkeys exhibit poor recognition of emotional facial expressions such as aggressive and submissive monkey faces, consistent with human RTT patients. 5) Structural MRI (Magnetic Resonance Imaging) has revealed decreases in subregional gray matter and white matter volumes, cortical surface area, and cortical thickness in mutant monkeys, consistent with human RTT patients. 6) Mutant monkeys showed reduced heart rate and extended QT interval, consistent with cardiac-related changes in human RTT patients. 7) Finally, blood transcriptome analyses have revealed downregulated immune pathways and upregulated RNA processing and protein translation pathways in both mutant monkeys and human RTT patients. These close similarities in genetics, transcriptome, anatomy, physiology, and behavior of MECP2 mutant monkeys to human RTT patients demonstrate the potential value of genetically modified NHPs for RTT research. Although several other important RTT symptoms such as regression of previously acquired skills, seizures, and cognitive deficits have not yet been addressed in F0 mutant monkeys, next generation mutant monkeys with pure heterozygous mutations may provide further insights of pathogenesis of RTT as well as developmental trajectories of the symptoms, both of which are critical for understanding disease mechanisms and testing potential therapeutics.
Phelan–McDermid syndrome and autism spectrum disorders
Phelan–McDermid syndrome (PMS) is a rare neurodevelopmental disorder characterized by hypotonia, generalized developmental delay, intellectual disability, ASD-like behaviors, and delayed or absent speech [16]. About 75% of PMS patients are diagnosed with ASD, and PMS is one of the most frequent causes of ASD. The core symptoms of PMS are caused by the heterozygous deletions in 22q13.3 encompassing SHANK3 or the haploinsufficiency of SHANK3. Mutations in SHANK3 also lead to neurodevelopmental and neurobehavioral deficits and account for up to 1% of ASD [17,18]. SHANK3 encodes a postsynaptic scaffolding protein critical for the development and function of glutamatergic synapses. We and many other groups have generated various Shank3 mutant mouse lines [19]. Homozygous Shank3 knockout mice demonstrate increased compulsive/repetitive behaviors, impaired social behaviors, motor defects and increased anxiety (and seizure in some cases) [20-22]. These knockout models have greatly facilitated the dissection of Shank3 function at synapses. However, similar to RTT mouse models, heterozygous Shank3 knockout mice demonstrate only subtle [22] or no [23] phenotypes. Here again, data suggest a difference in dosage effect of the Shank3 gene mutation between mice and humans.
To address this question, we used CRISPR/Cas9 method to generate SHANK3 mutant cynomolgus monkeys mimicking a frameshift mutation found in ASD patients and characterized juvenile F0 mutant monkeys [24]. Among five SHANK3 mutant monkeys, 2 do not have wild-type allele at the genomic level, and thus are virtually full knockouts (1 homozygous and 1 compound heterozygous which consist of 3 different indel alleles), and 3 carried 50% wild-type alleles, and thus are heterozygous (2 are mosaic with more than one indel alleles). Post-behavior brain biopsy analysis showed that SHANK3 protein in the brain are mostly gone in the full knockout monkeys and reduced approximately to 50% of wildtype levels in heterozygous mutants. Both full knockout and heterozygous mutant SHANK3 monkeys exhibited multiple behavioral abnormalities including disrupted sleep, increased stereotyped/repetitive behaviors, reciprocal social interaction deficits, reduced vocalization, cognitive impairment, and muscular hypotonia, all of which are hallmarks of PMS and ASD. Consistent with human ASD patients, mutant monkeys showed alterations in pupillary reflex and gaze fixation. Furthermore, resting-state fMRI studies revealed altered global and local functional connectivity, consistent with a dysregulated resting-state connectivity reported in human ASD patients as a potential biomarker. Taken together, these results as well as a case study from another group [25,26] suggest that the SHANK3 mutant monkey is a good model for studying higher brain function and related behaviors disrupted in ASD and PMS. This model may also facilitate the discovery of translatable biomarker and preclinical evaluation of therapeutics.
Towards next generation NHP models
Although initial studies of MECP2 and SHANK3 mutant NHPs have shown promises for modeling and studying neurodevelopmental disorders, several technological challenges and opportunities warrant discussion in preparation for “next generation” genetically modified NHP models that could maximize translational value of the approach.
Target selection
The use of genetically engineered NHP models must be scientifically and ethically well justified for each research project. It is required to have a clear rationale to justify the use of genetically engineered NHP models as the best way to address the critical issues with respect to NHP-specific brain functions, such as PFC-related function and dysfunction as well as primate-specific genes [27], cell types [6], and epigenetic regulations [28].
Faithful genomic modification
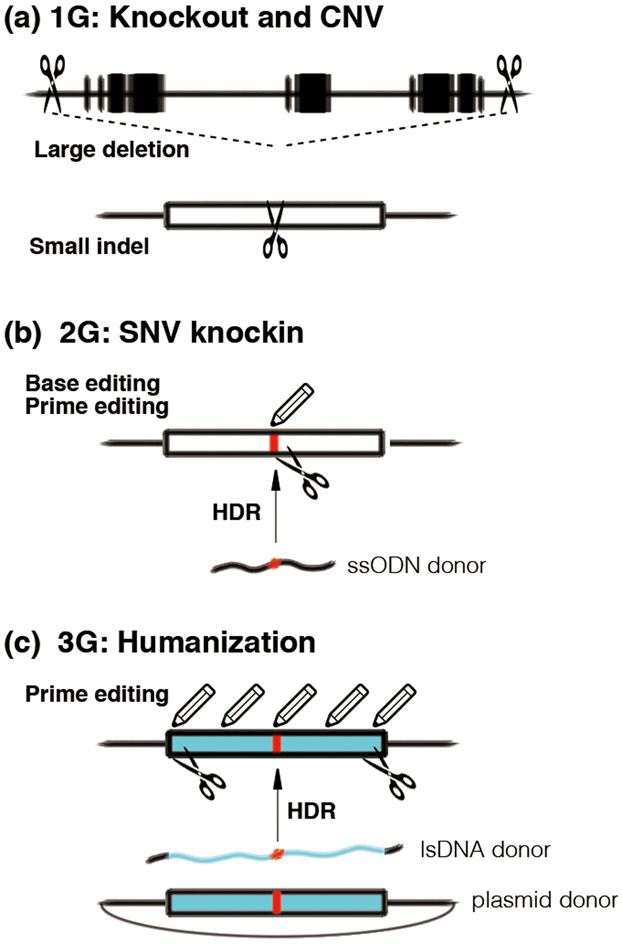
InDel/knockout: MECP2 and SHANK3 mutants were generated by NHEJ-mediated indel induction using multiple guideRNAs with CRISPR or multiple pairs of TALENs, both make multiple DSBs (Figure 1a). This approach induces efficient functional gene knockout through random indel formation, or defined large deletion between two DSBs. Chromosomal abnormalities, such as megabase-sized chromosomal deletions and duplications and inversions, are one of the frequent causes of neurodevelopmental disorders [29]. The two DSBs approach has shown to induce these chromosomal abnormalities between two DSBs in mouse embryos [30], with inversion as a primary outcome followed by deletion, and much less duplication [30]. Employing this approach will be useful to generate NHPs harboring pathogenic chromosomal abnormalities, yet further improvement of efficiency is essential for NHP application. The outcome of NHEJ-mediated DSB repair is generally unpredictable and often induces in-frame deletion, which can result in truncated, but still functional protein products. Thus, more predictable editing methods are needed for 2nd generation genetically modified NHP models if we are to precisely engineer disease-relevant mutation into NHPs.
Figure 1. Genome editing technologies for next generation NHP models.
(a) 1st generation (1G) genetically modified NHP models for knockout and genomic deletion by NHEJ and large deletion. (top) large deletion of multiple genes (black boxes) by simultaneous DSBs (scissors), mimicking CNV (loss of genomic region). (bottom) Small indel of target exon by NHEJ, resulting functional gene knockout. (b) 2nd generation (2G) genetically modified NHP models for precise genome engineering and SNV knockin. SNV (red) can be precisely installed by base editing and prime editing (pencil) without DSB, or HDR with ssODN donor after DSB induction. (c) 3rd generation (3G) genetically humanized NHP models for clinical translation. Target region or exon can be humanized (blue) with SNV by prime editing without DSB or HDR with lsDNA or plasmid donor after DSB induction. NHP: non-human primate, NHEJ: non-homologous end-joining, CNV: copy number variation, SNV: single nucleotide variation, ssODN: single strand oligodeoxynucleotide, HDR: homology-directed repair, lsDNA: long single strand DNA.
Precise genome engineering/knockin: Since the majority of de novo mutations found in ASD patients are single nucleotide variations (SNVs) [31], precise installation of patient’s SNVs into NHP genomes is essential (Figure 1b). For this purpose, HDR approaches using ssODN (single strand oligodeoxynucleotide) donors have been explored in NHP embryos, however, initial attempts in cynomolgus monkey embryos resulted in very little HDR-mediated knockin with low allele frequency [32], or complete failure [33]. We have developed highly efficient RNP (ribonucleoprotein)-based HDR approaches in mice [34,35] and demonstrate robust knockin efficiency in marmoset embryos [36,37], yet further improvement and reduction of undesired mutation such as indel are essential. DSB-independent, direct substitution of target nucleotide by base editing and prime editing [7] are promising approaches to generate “indel-free” knockin NHPs. Base editing has shown efficient installation of precise C-to-T (or G-to-A) and A-to-G (T-to-C) substitutions into NHP [38] embryos. Although, guideRNA-independent, genome-wide random DNA and RNA off-target mutations have been major concerns associated with base editing, the latest engineered base editors have been reported to overcome off-target concerns [39], and are now ready for application in NHP embryos.
An additional limitation of base editing is that only transitions such as C-to-T (or G-to-A) and A-to-G (T-to-C) located within specific editing windows within guideRNAs can be installed, thus only a fraction of human pathogenic mutations can be targeted. Alternatively, the recently published prime editing approach provides the potential for a more flexible method to install any type of substitution around guideRNA target sites with few indel formation. Given the relatively recent development of these methods, it is strongly recommended that in vivo testing occur in mouse embryos first to validate the approach for animal model production and eventual translation to NHP models. Utilizing these new methods, 2nd generation models could drastically expand the range of human mutations that can be investigated in NHPs.
Translational considerations: The correction of causal mutations by genome editing in monogenic neurodevelopmental disorders is getting attention as a potential promising gene therapy. Therefore, genomic humanization of target regions will be critical for the 3rd generation mutant NHP models (Figure 1c), enabling direct transfer of therapeutic CRISPR reagents to clinical trials, and thereby maximizing the value of NHP models. Although genomic sequences between humans and NHPs are highly conserved (~98.77%), in addition to target mutations, multiple SNVs can be found within or flanking CRISPR target sites. These differences in genomic sequences may impact the behavior and function of human mutations on NHP genomic background. Humanization of target exons, in addition to target mutations, has shown to improve the recapitulation of human patient’s pathology in genetically modified mice [40]. HDR-mediated precise replacement of wild-type NHP genomic target sites with humanized small mutant gene fragments is one important avenue for further study. The precise knockin of long transgenes such as fluorescent reporter proteins into target sites have been successfully archived in NHPs by our group (Aida T et al., unpublished) and others [41,42]. Since DSB-stimulated HDR frequently induces indel, trans nicking [43] and CRISPR-transposon [44], both are indel-free, will be explored. Employing a combination of these approaches will allow 3rd generation NHP models to have an even greater value for rapid translation of preclinical research.
Reducing mosaicism
Genetic mosaicism, in which two or more populations of cells with different genomes are present in an individual monkey, is now recognized as a major concern in genetically modified NHP production. In mosaicism, a single cell stage embryo has two copies of genomes, and after DNA replication, four copies, thus genome editing after DNA replication often results in mosaic embryos due to multiple CRISPR events. In addition, after cell division, delayed CRISPR events may only happen in some cells. “Hit-and-Away” editing at earlier timing is one of the major courses taken to reduce mosaicism in embryos and successful reduction of mosaicism has been reported in human embryos [45] by delivery of CRISPR components as RNPs [34,35], which act immediately at target sites and degrade quickly, into MII-stage human embryos. Cas9 messenger RNA (mRNA), which acts slowly in later stage due to translation, has been commonly used for production of NHP models and is thought to be one of the causes of mosaicism due to its delayed and prolonged action potentially at the 2-cell or later stage. In line with this idea, destabilized Cas9 mRNA can reduce mosaicism formation in NHP embryos [46]. Saturated genome editing activity can also reduce mosaicism in human [47] and NHP [38] embryos. Emerging reproductive technologies such as somatic cell cloning by somatic cell nuclear transfer (SCNT) are also likely to have a significant impact on generation of NHP models. The successful productions of cloned live cynomolgus monkeys by SCNT[48], combined with CRISPR-mediated knockout [49], has opened a completely new avenue to produce genetically modified mutant monkeys. SCNT may allow the field to potentially overcome almost all the current challenges in mutant monkey production including precise modification of target genome (by selecting correctly targeted somatic cells), mosaicism, allele dosage, multiplex knockin, and CRISPR off-target. Although current SCNT success rate in NHPs is low (based on recent reports of only ~1-5% live births from transferred embryos [48,49]), this approach still represents a significant methodological advancement and may become a standard approach for genome editing, similar to approaches for other livestock animals. In vitro induction of primordial germ cells (PGCs) following differentiation into functional sperm and oocytes from pluripotent stem cells (PSCs) such as embryonic stem cells (ESCs) and iPSCs (induced PSC) is another promising reproductive technology [50]. Taken together, the improvement of genome editing and emerging reproductive technologies will likely eliminate the mosaicism from genetically modified NHP production.
Critical need for robust ON- and OFF-target analyses
Unintended, off-target mutation is a major concern in CRISPR-mediated genome editing [7]. Genome-wide off-target analyses in genetically modified monkeys have reported few guideRNA-dependent off-target mutations [42,51-54]. The use of engineered high-fidelity Cas proteins, RNP, and sophisticated guideRNA design algorithms further eliminate the possibility of guideRNA-dependent CRISPR off-target mutation [7]. Importantly, due to highly divergent NHP genomes across colonies compared to public databases, unique SNVs in each colony can potentially create new off-target sites (similar to genetic divergence reported in human [55]). Thus, it is important to carefully design off-target analyses to include appropriate controls such as parents [53], siblings [51], and unedited endogenous cells [47,56]. Other unexpected on-target editings such as large deletions [57] and tandem integration of multiple copies of exogenous DNA donors [58] are frequently missed by standard PCR genotyping, thus careful design of on-target site analysis is also important. Circular DNA or end modification of linear DNA donors [59,60] may prevent the tandem integration of DNA donor and future studies are needed to further integrate this approach into existing frameworks. Although guideRNA-independent mutations such as deaminase-induced genome-wide DNA [56] and RNA [61] off-target mutations by base editing as well as ADAR-mediated genome-wide RNA off-target mutations [62] have also been reported, guideRNA-independent off-target mutations have been almost completely eliminated with the latest engineered base editors [39] and the possibility of off-target mutation is now much lower. In addition to sequencing based analyses, the reporter NHP lines for on- and off-target mutations are critical to evaluate the efficiency and safety of genome editing especially for therapeutics, and are under development in NIH Somatic Cell Genome Editing program.
Allele dosage
Haploinsufficiency is a cause of many neurodevelopmental disorders, therefore, the precise control of allele dosage in genetically modified NHPs, in other words, heterozygous or homozygous animals, is critical for generating more faithful models of human diseases. Several approaches have been proposed to achieve this goal: 1) If a SNV is located on or surrounding a target site, allele-specific editing should be considered [63]. This approach may be flexible for knockout purposes by using two DSBs targeting specific alleles in introns where more SNVs are available, and thus eliminate the target exon. 2) Allele-specific targeting often induces homozygous conversion of heterozygous alleles by interhomolog repair (recombination between two homologous chromosomes) [35,45]. This process can be promoted by exogenous supplementation of RAD51 [35], enabling one-step production of homozygous NHPs or self-repair of the heterozygous mutations. 3) Alternatively, asymmetric editing capacity between paternal and maternal alleles during development of one-cell stage embryo provides a unique time window to specifically edit paternal or maternal alleles [64] and may also help to generate heterozygous mutant NHPs. 4) Finally, as stated above, we believe the best approach to control genotype is to employ new reproductive technologies, such as SCNT and PGCs.
Conclusions
In this review, we have discussed early examples of successful modeling of neurodevelopmental disorders RTT, ASD, and PMS with genome edited mutant NHPs, including MECP2 and SHANK3 knockouts. These “new models” have shown the potentials of genetically modified NHPs for disease-relevant behavioral analysis, circuit dissection and translatable biomarker discovery that are beyond rodent models. Based on these early successes, we propose a framework for creating the next generation NHP models that may increase their translational value. We have also identified important technological challenges and potential solutions to accelerate and disseminate genetically modified NHPs for research and therapeutic development of neurodevelopmental disorders. Fortunately, the advancement of genome editing technologies is incredibly fast and thus we expect most technological challenges to be overcome in the near future. However, the most important question, whether genetically modified NHPs can help biomarker discovery and therapeutic development for human neurodevelopmental disorders, has not yet been answered and it will take time. The field urgently needs a proof-of-concept pre-clinical study (and subsequent successful translation to human trials) to truly assess the value of genetically modified NHP models.
Highlights.
Genome editing technologies have made it feasible to generate genetically modified NHP models
Genetically modified NHP models have certain advantages over rodent models for studying higher brain function and dysfunction in neurodevelopmental disorders
We identify challenges for pushing genetic NHP models forward identified and propose opportunities for next generation genetic NHP models
Acknowledgements
The work in the laboratory of Guoping Feng is supported by the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT, the James and Patricia Poitras Center for Psychiatric Disorders Research at MIT, the Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard, the McGovern Institute for Brain Research at MIT, the Rett Syndrome Research Trust, Annie and Alex Phillips, NIH-BRAIN Initiative (U01 MH114819), NIH/NIMH Conte Center Grant (P50 MH094271), NIH Office of the Director (U24 OD026638). We would like to thank Tyler Huff (MIT) and Tyler Brown (Broad Institute) for their editing and critical reading, the members of Feng lab at MIT and Broad Institute, especially Yang Zhou (McGill University), Tobias Kaiser, Steven M. Colvin, Xian Gao, Jonathan Wilde, Martin Wienisch, and Qiangge Zhang for helpful discussions, Shihua Yang (South China Agriculture University) and his lab members, especially Wenhui Zhang, and Zhonghua Lu (Shenzhen Institutes of Advanced Technology) and his lab members for collaborations.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Geschwind DH, State MW: Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol 2015, 14:1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser T, Feng G: Modeling psychiatric disorders for developing effective treatments. Nat Med 2015, 21:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izpisua Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, Lee K-F, Leopold DA, Miller CT, Mitchell JF, et al. : Brains, genes, and primates. Neuron 2015, 86:617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jennings CG, Landman R, Zhou Y, Sharma J, Hyman J, Movshon JA, Qiu Z, Roberts AC, Roe AW, Wang X, et al. : Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci 2016, 19:1123–1130. [DOI] [PubMed] [Google Scholar]
- 5.Wise SP: Forward frontal fields: phylogeny and fundamental function. Trends Neurosci 2008, 31:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krienen FM, Goldman M, Zhang Q, del Rosario R, Florio M, Machold R, Saunders A, Levandowski K, Zaniewski H, Schuman B, et al. : Innovations in Primate Interneuron Repertoire. bioRxiv 2019, doi: 10.1101/709501.• This work identified primate-specific interneuron subtypes by single cell RNA-seq.
- 7.Doudna JA: The promise and challenge of therapeutic genome editing. Nature 2020, 578:229–236.•This is an excellent review for the latest genome editing technologies.
- 8.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. : Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156:836–843. [DOI] [PubMed] [Google Scholar]
- 9.Sato K, Oiwa R, Kumita W, Henry R, Sakuma T, Ito R, Nozu R, Inoue T, Katano I, Sato K, et al. : Generation of a Nonhuman Primate Model of Severe Combined Immunodeficiency Using Highly Efficient Genome Editing. Cell Stem Cell 2016, 19:127–138. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, et al. : TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 2014, 14:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoghbi HY: Rett Syndrome and the Ongoing Legacy of Close Clinical Observation. Cell 2016, 167:293–297. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G, Hu Y, Wang J, Lu Y, Kang Y, et al. : Modeling Rett Syndrome Using TALEN-Edited MECP2 Mutant Cynomolgus Monkeys. Cell 2017, 169:945–955.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Zhou X, Zhu Y, Chen Z-F, Yu B, Wang Y, Zhang C-C, Nie Y-H, Sang X, Cai Y-J, et al. : Generation of a monkey with MECP2 mutations by TALEN-based gene targeting. Neurosci Bull 2014, 30:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Zhou Z-G, Zhou Y, Chen Y-C: Increased attention to snake images in cynomolgus monkeys: an eye-tracking study. Zool Res 2020, 41:32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Zhou Z, Zhou Y, Zhang T, Ma Y, Niu Y, Ji W, Chen Y: Social-valence-related increased attention in rett syndrome cynomolgus monkeys: An eye-tracking study. Autism Res 2019, 12:1585–1597. [DOI] [PubMed] [Google Scholar]
- 16.Harony-Nicolas H, De Rubeis S, Kolevzon A, Buxbaum JD: Phelan McDermid Syndrome: From Genetic Discoveries to Animal Models and Treatment. J Child Neurol 2015, 30:1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, et al. : Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet 2013, 21:310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betancur C, Buxbaum JD: SHANK3 haploinsufficiency: a “common” but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Mol Autism 2013, 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monteiro P, Feng G: SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci 2017, 18:147–157. [DOI] [PubMed] [Google Scholar]
- 20.Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G: Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 2011, 472:437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mei Y, Monteiro P, Zhou Y, Kim J-A, Gao X, Fu Z, Feng G: Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 2016, 530:481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Kaiser T, Monteiro P, Zhang X, Van der Goes MS, Wang D, Barak B, Zeng M, Li C, Lu C, et al. : Mice with Shank3 Mutations Associated with ASD and Schizophrenia Display Both Shared and Distinct Defects. Neuron 2016, 89:147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Bey AL, Katz BM, Badea A, Kim N, David LK, Duffney LJ, Kumar S, Mague SD, Hulbert SW, et al. : Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat Commun 2016, 7:11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Sharma J, Ke Q, Landman R, Yuan J, Chen H, Hayden DS, Fisher JW, Jiang M, Menegas W, et al. : Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature 2019, 570:326–331.•• This work revealed behavioral and connectivity defects in SHANK3-mutant macaques, resembling features in human ASD and PMS patients.
- 25.Zhao H, Tu Z, Xu H, Yan S, Yan H, Zheng Y, Yang W, Zheng J, Li Z, Tian R, et al. : Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res 2017, 27:1293–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu Z, Zhao H, Li B, Yan S, Wang L, Tang Y, Li Z, Bai D, Li C, Lin Y, et al. : CRISPR/Cas9-mediated disruption of SHANK3 in monkey leads to drug-treatable autism-like symptoms. Hum Mol Genet 2019, 28:561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ataman B, Boulting GL, Harmin DA, Yang MG, Baker-Salisbury M, Yap E-L, Malik AN, Mei K, Rubin AA, Spiegel I, et al. : Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 2016, 539:242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doan RN, Bae B-I, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, et al. : Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 2016, 167:341–354.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takumi T, Tamada K: CNV biology in neurodevelopmental disorders. Curr Opin Neurobiol 2018, 48:183–192. [DOI] [PubMed] [Google Scholar]
- 30.Boroviak K, Doe B, Banerjee R, Yang F, Bradley A: Chromosome engineering in zygotes with CRISPR/Cas9. Genesis 2016, 54:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, Peng M, Collins R, Grove J, Klei L, et al. : Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180:568–584.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan H, Feng C, Teng F, Yang S, Hu B, Niu Y, Xiang AP, Fang W, Ji W, Li W, et al. : One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res 2015, 25:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Midic U, Hung P-H, Vincent KA, Goheen B, Schupp PG, Chen DD, Bauer DE, VandeVoort CA, Latham KE: Quantitative assessment of timing, efficiency, specificity and genetic mosaicism of CRISPR/Cas9-mediated gene editing of hemoglobin beta gene in rhesus monkey embryos. Hum Mol Genet 2017, 26:2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K: Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 2015, 16:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilde JJ, Aida T, Wienisch M, Zhang Q, Qi P, Feng G: Efficient Zygotic Genome Editing via RAD51-Enhanced Interhomolog Repair. bioRxiv 2018, doi: 10.1101/263699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumita W, Sato K, Suzuki Y, Kurotaki Y, Harada T, Zhou Y, Kishi N, Sato K, Aiba A, Sakakibara Y, et al. : Efficient generation of Knock-in/Knock-out marmoset embryo via CRISPR/Cas9 gene editing. Sci Rep 2019, 9:12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimatsu S, Okahara J, Sone T, Takeda Y, Nakamura M, Sasaki E, Kishi N, Shiozawa S, Okano H: Robust and efficient knock-in in embryonic stem cells and early-stage embryos of the common marmoset using the CRISPR-Cas9 system. Sci Rep 2019, 9:1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Aida T, del Rosario RCH, Wilde JJ, Ding C, Zhang X, Baloch Z, Huang Y, Tang Y, Li D, et al. : Multiplex precise base editing in cynomolgus monkeys. Nat Commun (in press).•• This work first demonstrated precise, highly efficient, and multiplex base editing in monkey embryos.
- 39.Zuo E, Sun Y, Yuan T, He B, Zhou C, Ying W, Liu J, Wei W, Zeng R, Li Y, et al. : High-fidelity base editor with no detectable genome-wide off-target effects. bioRxiv 2020, doi: 10.1101/2020.02.07.939074.•• This work developed engineered base editors without deaminase-dependent DNA and RNA off-target mutations, making base editing ready for NHP model production as well as gene therapy.
- 40.Saito T, Matsuba Y, Mihira N, Takano J, Nilsson P, Itohara S, Iwata N, Saido TC: Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci 2014, 17:661–663. [DOI] [PubMed] [Google Scholar]
- 41.Yao X, Liu Z, Wang X, Wang Y, Nie Y-H, Lai L, Sun R, Shi L, Sun Q, Yang H: Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing. Cell Res 2018, 28:379–382.•• This work reported the first transgene knockin monkey with high efficiency.
- 42.Cui Y, Niu Y, Zhou J, Chen Y, Cheng Y, Li S, Ai Z, Chu C, Wang H, Zheng B, et al. : Generation of a precise Oct4-hrGFP knockin cynomolgus monkey model via CRISPR/Cas9-assisted homologous recombination. Cell Res 2018, 28:383–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Janssen JM, Liu J, Maggio I, ’t Jong AEJ, Mikkers HMM, Gonçalves MAFV: In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat Commun 2017, 8:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, Zhang F: RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019, 365:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma H, Marti-Gutierrez N, Park S-W, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, et al. : Correction of a pathogenic gene mutation in human embryos. Nature 2017, 548:413–419. [DOI] [PubMed] [Google Scholar]
- 46.Tu Z, Yang W, Yan S, Yin A, Gao J, Liu X, Zheng Y, Zheng J, Li Z, Yang S, et al. : Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci Rep 2017, 7:42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M, Zhou C, Wei Y, Xu C, Pan H, Ying W, Sun Y, Sun Y, Xiao Q, Yao N, et al. : Human cleaving embryos enable robust homozygotic nucleotide substitutions by base editors. Genome Biol 2019, 20:101.• This work revealed non-mosaic base editing in human embryos.
- 48.Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M, et al. : Cloning of Macaque Monkeys by Somatic Cell Nuclear Transfer. Cell 2018, 172:881–887.e7.•• This work reported the first landmark success of SCNT in NHP, opened a new avenue for production of genetic NHP models.
- 49.Liu Z, Cai Y, Liao Z, Xu Y, Wang Y, Wang Z, Jiang X, Li Y, Lu Y, Nie Y, et al. : Cloning of a gene-edited macaque monkey by somatic cell nuclear transfer. Natl Sci Rev 2019, 6:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurimoto K, Saitou M: Germ cell reprogramming. Curr Top Dev Biol 2019, 135:91–125.• This is an excellent review on in vitro induction of PGCs followed by differentiation into functional sperm and oocytes.
- 51.Wang S, Ren S, Bai R, Xiao P, Zhou Q, Zhou Y, Zhou Z, Niu Y, Ji W, Chen Y: No off-target mutations in functional genome regions of a CRISPR/Cas9-generated monkey model of muscular dystrophy. J Biol Chem 2018, 293:11654–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Wan H, Feng G, Qu J, Wang J, Jing Y, Ren R, Liu Z, Zhang L, Chen Z, et al. : SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 2018, 560:661–665. [DOI] [PubMed] [Google Scholar]
- 53.Luo X, He Y, Zhang C, He X, Yan L, Li M, Hu T, Hu Y, Jiang J, Meng X, et al. : Trio deep-sequencing does not reveal unexpected off-target and on-target mutations in Cas9-edited rhesus monkeys. Nat Commun 2019, 10:5525.•• This work provided solid evidence that no guideRNA-dependent, Cas9-mediated genomic off-target mutations by well-designed trio-based whole genome sequencing.
- 54.Zuo E, Cai Y-J, Li K, Wei Y, Wang B-A, Sun Y, Liu Z, Liu J, Hu X, Wei W, et al. : One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res 2017, 27:933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott DA, Zhang F: Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat Med 2017, 23:1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H: Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019, 364:289–292.•• This work reports highly-sensitive and well-controlled genome-wide in vivo off-target analysis scheme, and discovered guideRNA-independent, deaminase-dependent random off-target mutations throughout genome in mouse embryos.
- 57.Kosicki M, Tomberg K, Bradley A: Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018, 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skryabin BV, Kummerfeld D-M, Gubar L, Seeger B, Kaiser H, Stegemann A, Roth J, Meuth SG, Pavenstädt H, Sherwood J, et al. : Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9–mediated genome editing events. Science Advances 2020, 6:eaax2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Canaj H, Hussmann JA, Li H, Beckman KA, Goodrich L, Cho NH, Li YJ, Santos DA, McGeever A, Stewart EM, et al. : Deep profiling reveals substantial heterogeneity of integration outcomes in CRISPR knock-in experiments. bioRxiv 2019, doi: 10.1101/841098. [DOI] [Google Scholar]
- 60.Iyer S, Mir A, Vega-Badillo J, Roscoe BP, Ibraheim R, Zhu LJ, Lee J, Liu P, Luk K, Mintzer E, et al. : Efficient Homology-directed Repair with Circular ssDNA Donors. bioRxiv 2019, doi: 10.1101/864199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK: Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019, 569:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F: RNA editing with CRISPR-Cas13. Science 2017, 358:1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukiyama T, Kobayashi K, Nakaya M, Iwatani C, Seita Y, Tsuchiya H, Matsushita J, Kitajima K, Kawamoto I, Nakagawa T, et al. : Monkeys mutant for PKD1 recapitulate human autosomal dominant polycystic kidney disease. Nat Commun 2019, 10:5517.•• This work reports a strategy for targeted installation of heterozygous mutations, and successful recapitulation of ADPKD phenotypes in happloinsufficiency condition in cynomolgus monkey.
- 64.Suzuki T, Asami M, Perry ACF: Asymmetric parental genome engineering by Cas9 during mouse meiotic exit. Sci Rep 2014, 4:7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, Sherwood RI: Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 2018, 563:646–651.• This work revealed stereotyped patterns of DSB repair by large scale systematic screening and provided a prediction tool, enabling template-free precise and predictable genome editing outcomes.
- 66.El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai S-L, et al. : Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568:193–197.• This work revealed complex mechanisms of genetic compensation, and suggested a strategy for complete functional gene knockout.
- 67.Kato T, Hara S, Goto Y, Ogawa Y, Okayasu H, Kubota S, Tamano M, Terao M, Takada S: Creation of mutant mice with megabase-sized deletions containing custom-designed breakpoints by means of the CRISPR/Cas9 system. Sci Rep 2017, 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma M, Zhuang F, Hu X, Wang B, Wen X-Z, Ji J-F, Xi JJ: Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res 2017, 27:578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riesenberg S, Chintalapati M, Macak D, Kanis P, Maricic T, Pääbo S: Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res 2019, 47:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP: Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368:290–296.•• A landmark paper eliminated PAM constraint in CRISPR-mediated genome editing.
- 71.Gu B, Posfai E, Rossant J: Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat Biotechnol 2018, 36:632–637.•• This work provided a new method for highly efficient transgene knockin in mouse zygotes.
- 72.Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J: RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun 2016, 7:10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ke Q, Li W, Lai X, Chen H, Huang L, Kang Z, Li K, Ren J, Lin X, Zheng H, et al. : TALEN-based generation of a cynomolgus monkey disease model for human microcephaly. Cell Res 2016, 26:1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiu P, Jiang J, Liu Z, Cai Y, Huang T, Wang Y, Liu Q, Nie Y, Liu F, Cheng J, et al. : BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl Sci Rev 2019, 6:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang Y, Zheng B, Shen B, Chen Y, Wang L, Wang J, Niu Y, Cui Y, Zhou J, Wang H, et al. : CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. Hum Mol Genet 2015, 24:7255–7264. [DOI] [PubMed] [Google Scholar]
- 76.Yang W, Liu Y, Tu Z, Xiao C, Yan S, Ma X, Guo X, Chen X, Yin P, Yang Z, et al. : CRISPR/Cas9-mediated PINK1 deletion leads to neurodegeneration in rhesus monkeys. Cell Res 2019, 29:334–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, Tu Z, Si C, Wang H, Xing R, et al. : Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet 2015, 24:3764–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Smith J, Breton C, Clark P, Zhang J, Ying L, Che Y, Lape J, Bell P, Calcedo R, et al. : Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat Biotechnol 2018, 36:717–725. [DOI] [PubMed] [Google Scholar]