Abstract
The SnRK (Snf1-Related protein Kinase) gene family plays crucial roles in various plant signaling pathways and stress-adaptive responses including biotic and abiotic stresses via activating protein phosphorylation pathways. However, there is no information available on the role of the SnRK gene family in Hedychium coronarium. H. coronarium is an important crop widely cultivated as an ornamental plant, herb, spice, or condiment. In this study, 60 HcSnRK genes were identified from the H. coronarium genomic and transcriptome data. Phylogenetic and gene structure analysis showed that the HcSnRK genes were divided into three groups (HcSnRK1, HcSnRK2 and HcSnRK3) and among them HcSnRK3 subfamily was further subdivided into two clades according to the number of introns. Chromosome localization analysis showed that HcSnRK genes were unevenly mapped onto all chromosomes, and the Ka/Ks ratio of 24 paralogues includes four tandems and 20 segmental duplications indicated that the HcSnRK gene family underwent a purifying selection. Cis-regulatory elements analysis suggested that the HcSnRK genes respond to multiple hormones and other stresses. The responsiveness of HcSnRK genes to several hormones was analyzed by quantitative real-time PCR. Based on the different transcriptome data, two candidates HcSnRK genes (HcSnRK2.2 and HcSnRK2.9) were screened out for further characterization . The subcellular localization experiment revealed that both genes were located in the nucleus and cytoplasm. Moreover, virus-induced gene silencing (VIGS) of HcSnRK2.2 and HcSnRK2.9 significantly reduced the floral volatile contents by suppressing the expression of terpene synthase genes (HcTPS1, HcTPS3, and HcTPS5), indicating that HcSnRK2.2 and HcSnRK2.9 genes play an important role in the regulatory mechanism of floral aroma. These results will provide novel insights into the functional dissection of H. coronarium SnRK gene family.
Keywords: SnRK, Hedychium coronarium, Hormones, Stress responses, Floral scent
Introduction
Floral scent is an important plant secondary metabolite that plays key roles in different developmental processes. Floral scent also plays a crucial role in plant communication both above and below-ground and stress signaling (Dudareva et al., 2006; Das et al., 2013; Muhlemann, Klempien & Dudareva, 2014; Abbas et al., 2017). The main role of floral scent is to protect the plant against external stimuli (biotic and abiotic stresses) and attract pollinators (Dudareva et al., 2006; Das et al., 2013). According to biosynthetic pathway, terpenoids, benzenoids/phenylpropanoid, and derivatives of fatty acids are the main classes of volatile organic compounds (VOCs) emitted from the plant surface. Among them, terpenoids constitute the largest class of plant secondary metabolites playing several functions throughout plant life and the expression profile of terpene synthase genes determines the involvement of terpenoid metabolites in different ecological and physiological functions in response to external stimuli. Protein kinases are considered as central components in defense mechanisms, which activate several protein phosphorylation pathways to regulate the expression of downstream genes related to stress response (Tena, Boudsocq & Sheen, 2011). In recent years, protein kinases such as mitogen-activated protein kinase (MAPK) (Xu & Zhang, 2015), calcium-dependent protein kinases (CDPK) (Baba, Rigó & Ayaydin, 2018), glycogen synthase kinase 3 (GSK3) (Beurel, Grieco & Jope, 2015), and sucrose non-fermenting 1 (SNF1) related protein kinases (SnRK) (Hrabak et al., 2003; Wang et al., 2015; Colina et al., 2019; Wang et al., 2019; Zhang et al., 2020) have been extensively studied. Among them, SnRK proteins play essential roles throughout plant life.
SnRK protein kinases contain a similar Ser/Thr kinase domain, conserved UBA, and the KA1 domains in SnRK1. Meanwhile, the osmotic stress-activated domain I was found in SnRK2, and a unique NAF domain in the SnRK3 subfamily (Coello, Hey & Halford, 2011). Furthermore, some SnRK2 protein kinases have an acidic amino acid–base sequence called domain II that can participate in abscisic acid (ABA)mediated responses to abiotic stresses (Yoshida et al., 2006). The SnRK1 subfamily is evolved in early eukaryotes, before the divergence of fungi, animals, and plants (Halford & Hey, 2009). Thus, SnRK1 protein kinases in plants are highly homologous to SNF1 genes in yeast and AMP-activated protein kinases in mammals, which is mainly involved in carbon and nitrogen response metabolism and energy-sensing (Coello, Hey & Halford, 2011). In Arabidopsis thaliana, AKIN10 and IDD8 constitute a sugar metabolic pathway that mediates flowering time under low-sugar conditions (Jeong et al., 2015). Unlike the SnRK1 subfamily, SnRK2 and SnRK3 are unique in plants and are considered to be evolved from the SnRK1 family via gene duplication during plant evolution, playing a key role in the stress, calcium and ABA signaling pathway with epigenetic and metabolic responses (Halford & Hey, 2009). SnRK2 is the most widely studied subfamily and mainly focused on the participation of SnRK2 protein kinases in ABA-dependent and ABA-independent abiotic stress. In A. thaliana, 8/10 AtSnRK2 (Boudsocq, Barbier-Brygoo & Laurière, 2004; Boudsocq et al., 2007) and in Oryza sativa, all 10 stress/ABA-activated Serine/threonine-protein kinase 1 (OsSAPK1 to OsSAPK10) (Kobayashi et al., 2004) can be activated by hyperosmotic and saline stress. Among them, AtSnRK2.2/2.3/2.6 and OsSAPK8/9/10 were strongly activated by ABA and act as core positive regulators to regulate ABA-dependent stress responses (Fujita et al., 2009). The SnRK2 subfamily also plays a key role in the regulation of gene expression via activating basic region-leucine zipper (bZIP) transcription factors connected to an epigenetic mechanism that controls the activation or repression of a gene (Baena-González & Sheen, 2008; Fujii, Verslues & Zhu, 2011). The SnRK3 subfamily commonly called calcineurin B-like interacting protein kinases (CIPK) with a self-inhibitory NAF domain that can interact with calcineurin B-like (CBL) protein (Cheong et al., 2007). The CBL-CIPK protein complex constitutes a precise calcium signaling system, which plays a vital role in the process of achieving information regarding integration and physiological coordination to resist various stresses in plants (Tang et al., 2020; Tripathi et al., 2009). In Arabidopsis, AtCIPK1 can interact with CBL1 to participate in the ABA-independent signaling pathway and interact with CBL9 in response to ABA-dependent pathways (D’Angelo et al., 2006). Furthermore, numerous shreds of evidence indicate that the SnRK family is widely involved in almost all hormone signaling pathways. For example, SnRK1 phosphorylates FUSCA3 (FUS3) and MYC transcription factors regulate ABA, ethylene, gibberellin synthesis and jasmonic acid signaling (Gazzarrini et al., 2004; Lumba et al., 2012; Im et al., 2014; Chan et al., 2017). SnRK2.8 phosphorylation NONEXPRESSER OF PATHOGENESIS-RELATED GENES1 (NPR1) respond to the systemic immunity in SA-independent systemic signals (Lee et al., 2015). In short, hormone signals are important for the metabolism of floral aromas and SnRK proteins are deeply involved in the hormone signaling pathway (Mai, Wang & Yang, 2011; Ma et al., 2018; Ke et al., 2019). However, either SnRK protein kinases respond to plant hormone signals or participates in the regulation of floral fragrance in H. coronarium is still unknown.
H. coronarium is a perennial herb, commonly known as “White Butterfly Flower” or “Butterfly Ginger”. H. coronarium is popular due to its elegant shape and refreshing fragrance of flower and widely cultivated in tropical and subtropical regions (Chen et al., 2013; Yue, Yu & Fan, 2014). The blooming of flower results in a strong refreshing scent which is mainly composed of monoterpenes, sesquiterpenes, and some benzenoids (Lan et al., 2013; Yue, Yu & Fan, 2015; Chen et al., 2019; Ke et al., 2019). Some studies also reported that hormone signaling plays an important role in floral scent formation (Schmelz et al., 2003; Dudareva et al., 2013; Cna’ani et al., 2015). SnRK gene family plays an essential role in plants, however, its function in floral scent formation is completely missing. Our previous research showed that auxin and auxin signaling components can alter the amount of floral volatile compounds (Ke et al., 2019). In the present study, a total of 60 Hc SnRK genes were identified and analyzed in H. coronarium genome. The expression patterns of HcSnRK gene family in response to several hormones (ABA, auxin, jasmonic acid and ethylene) and their corresponding hormone inhibitors were measured by qRT-PCR. Furthermore, based on the expression profile, two HcSnRK genes (HcSnRK2.2 and HcSnRK2.9) were screened out and their involvement in the metabolism of floral fragrance was demonstrated by virus-induced gene silencing (VIGS). Also, the subcellular localization of these genes was performed. These results will provide the theoretical basis for better understanding the function of SnRK genes in hormone signaling and the regulatory metabolism of floral scent formation in H. coronarium.
Materials and Methods
Plant growth environment and hormone treatments
The plant materials were grown in a greenhouse under natural light (South China Agricultural University, Guangzhou, China). Plant material was immediately frozen in liquid nitrogen and stored at −80 °C for RNA isolation and further experimentation.
Plants used for hormone treatment were cut into about 40 cm with a wedge-shape and placed in a 500 mL beaker filled with ultrapure water. The concentration of hormones used for treatments was as followed: 400 µM for ABA, 100 µM for nordihydroguaiaretic acid (NDGA), auxin (IAA), methyl jasmonate (MeJA) and acetylsalicylic acid (ASA), 1.5 mM for 2-(4-chlorophenoxy)-isobutyric acid (PCIB), 10 µL/L for ethylene (ET), and 4 µL/L for 1-methylcyclopropene (1-MCP).
RNA extraction, cDNA synthesis and qRT-PCR analysis
Total RNA was extracted using the HiPure plant RNA mini kit (Magen, China) according to the manufacturer’s suggestions. One microgram of total RNA was reverse transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Japan) according to the manufacturer’s instructions. The qRT-PCR experiment was executed in an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) by using iTaq™ Universal SYBR Green Supermix (BIO-RAD, Hercules, CA, USA) with a 20 µL sample volume according to the manufacturer’s protocols. The reaction system was as followed: 95 °C for 1 min, then 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. The relative expression level of each gene was calculated by the standard 2−ΔΔCt method (Livak & Schmittgen, 2001).
Sequence retrieval and genome-wide identification of HcSnRK genes
Arabidopsis protein sequences were obtained from the Phytozome database (http://www.phytozome.net/) (Lamesch et al., 2012), genome data of rice was downloaded from the Rice Annotation Project (RAP) (https://rapdb.dna.affrc.go.jp/) (Sakai et al., 2013), and genomic data (Supplementary raw data) of H. coronarium was obtained from the Beijing Novogene Bioinformatics Technology Corporation (China).
The local BLASTP search (E-value-5) was performed using the 39 and 48 SnRK protein sequences of Arabidopsis and rice according to the Hidden Markov Models profile from the Pfam database (http://pfam.xfam.org/) (Finn et al., 2016). The candidate HcSnRK protein sequences were sent to the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al., 2011), SMART database (http://smart.embl-heidelberg.de/) (Letunic, Doerks & Bork, 2012) and Pfam database (Finn et al., 2016) for domain search. Based on the information from the above three databases, we manually select HcSnRK genes with conserved functional domains. Furthermore, molecular weight (MW) and isoelectric point (pI) of confirmed 60 HcSnRK protein sequences was calculated by ExPASy online software (http://www.expasy.ch/tools/pi_tool.html).
Multiple sequence alignment and phylogenetic analysis of HcSnRKs
The ClustalX software (Thompson et al., 1997) was used for multiple sequence alignment of 60 HcSnRK protein and MEGA 7 software (Kumar et al., 2018) was used to construct a phylogenetic tree using the neighbor-joining method (Saitou & Nei, 1987) with 1,000 replicates of bootstrap values. The DNAMAN software was used to show multiple sequence alignment of 60 HcSnRK genes.
Conserved motifs identification and gene structure analysis of HcSnRK
The exon-intron structure of the HcSnRK genes was performed using the Gene Structure Display Server (http://gsds.gao-lab.org/) (Hu et al., 2015) online program. The conserved motifs of HcSnRK protein sequences were identified by Multiple Expectation Maximization for Motif Elicitation (MEME) online software (http://meme-suite.org/tools/meme) (Bailey et al., 2009) with the following parameters: zero or one occurrence per sequence, 20 motifs should MEME find.
Ka and Ks calculation and selection mode analysis
The ratio of non-synonymous substitutions (Ka) and synonymous substitutions (Ks) were used to analyze the selection modes of HcSnRK genes. Protein sequences without stop codon of HcSnRK were aligned by MEGA 7. The Ka and Ks values were calculated by DnaSP v5 software (Librado & Rozas, 2009) with following parameters: assign coding regions, from start to end; assign genetic code, nuclear universal.
Cis-elements analysis HcSnRK genes
The upstream sequences 2000 bp of each HcSnRK gene were submitted to PlantCARE Database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) to predict the function of HcSnRK genes. Six hormone-related cis-elements including ABA-responsive, auxin-responsive, jasmonic acid-responsive, ethylene-responsive, salicylic acid-responsive, and gibberellin-responsive were identified. Four cis-acting elements involved in plant stress responses, such as defense and stress responses, low-temperature responses, drought responses, and wound responses were analyzed.
Subcellular localization of HcSnRK2.2 and HcSnRK2.9
HcSnRK2.2 and HcSnRK2.9 were fused into the vector pEAQ-HT-GFP using the Age I enzyme followed by transformation into Agrobacterium tumefaciens (strain EHA105). The infection solution (OD600 = 0.6) activated by MES solution (10 mM MgCl2, 10 mM MES and 100 µM acetosyringone, pH = 5.6) was injected into the Nicotiana benthamiana leaves as described previously (Ke et al., 2019). Two to three days later the leaves were visualized using Leica DM RXA2 upright fluorescent microscope with 40 × 0.75 numerical aperture objective as explained previously study (Yue, Yu & Fan, 2014; Abbas et al., 2019). The primers are listed in Table S5.
Headspace floral volatiles analysis
The whole flower was placed in a closed 250 mL glass bottle supplemented with an internal standard. Polydimethylsiloxane (PDMS) fiber was inserted into the bottle for 15 min to adsorb volatiles for 15 min followed by insertion into the gas chromatography-mass spectrometry (GC-MS) system as explained previously (Ke et al., 2019; Yue, Yu & Fan, 2014).
Virus-induced gene silencing (VIGS)
The barley stripe mosaic virus (BSMV) system was successfully applied in H. coronarium. The BSMV-VIGS system which consists of pCaBS-α, pCaBS-β, and pCaBS-γ vectors was kindly provided by Professor Dawei Li (China Agricultural University). Linearization of pCaBSγ vector using Apa I endonuclease and about 300 bp specific base sequence of HcSnRK2.2 and HcSnRK2.9 were fused to the vector according to the protocol system (Yuan et al., 2011) and optimized by extending the connection time to 2 min. The solution contains a mixture of an equal proportion of pCaBS-α, pCaBS-β, and pCaBS-γ/HcSnRK2.2/HcSnRK2.9 with a final OD600 of 0.5 to 0.6. The flowers at Stage S1 were dipped into the solution followed by vacuum infiltration at -0.8 atmosphere standard for 10 min. After vacuum infiltration, the flowers were immediately washed with sterilized water and were placed in an incubator with following conditions: 12 h day/night period at 16 °C for 5 days (Ke et al., 2019). The floral volatile contents were measured as described above, and the experiment was repeated three to four times.
Results
Identification of HcSnRK gene family in H. coronarium
Base on the BLAST and Hidden Markov Model search, a total of 60 candidate genes were identified in H. coronarium genome. According to subfamily and chromosomal localization of genes, HcSnRK genes were named HcSnRK1.1 ∼HcSnRK1.4, HcSnRK2.1 ∼HcSnRK2.13, and HcSnRK3.1 ∼HcSnRK3.43, respectively. The physical parameters of these genes are summarized in Table 1. The amino acid (aa) length ranged from 326 aa (HcSnRK2.8) to 526 aa (HcSnRK3.14), and the length of HcSnRK2 subfamily is shorter than the other two subfamilies. The average length of HcSnRK1, HcSnRK2, and HcSnRK3 subfamily were 500, 352, and 451 aa, respectively. Meanwhile, protein molecular weight varies greatly from 36.87 kDa (HcSnRK2.8) to 58.80 kDa (HcSnRK3.14), and the isoelectric point from 4.81 (HcSnRK2.11) to 9.48 (HcSnRK3.12).
Table 1. The characteristics of the HcSnRK gene family in H. coronarium.
| Gene Name | Gene ID | Position | CDS (bp) | Amino Acids | Exons | pI | MW (kDa) |
|---|---|---|---|---|---|---|---|
| HcSnRK1.1 | Hc42.68 | Chr4:60322670-60328745(−) | 1,497 | 498 | 11 | 8.43 | 56.70 |
| HcSnRK1.2 | Hc154.85 | Chr14:13692396-13697801(−) | 1,533 | 510 | 12 | 8.71 | 58.24 |
| HcSnRK1.3 | Hc25.126 | Chr0:1124760-1129850(−) | 1,497 | 498 | 11 | 8.63 | 56.16 |
| HcSnRK1.4 | Hc1223.4 | Chr0:18406-24187(+) | 1,482 | 493 | 12 | 7.08 | 40.51 |
| HcSnRK2.1 | Hc479.26 | Chr5:3118456-3123025(+) | 1,092 | 363 | 9 | 4.86 | 41.10 |
| HcSnRK2.2 | Hc163.43 | Chr5:35295876-35298554(+) | 1,098 | 365 | 9 | 5.48 | 41.76 |
| HcSnRK2.3 | Hc39.21 | Chr5:45454971-45460547(+) | 1,098 | 365 | 9 | 4.95 | 41.27 |
| HcSnRK2.4 | Hc782.12 | Chr8:7883604-7886707(−) | 1,125 | 374 | 10 | 6.00 | 43.00 |
| HcSnRK2.5 | Hc54.19 | Chr8:40661379-40668342(−) | 1,002 | 333 | 9 | 5.35 | 38.48 |
| HcSnRK2.6 | Hc843.18 | Chr11:43521897-43525336(+) | 1,020 | 339 | 9 | 5.67 | 38.47 |
| HcSnRK2.7 | Hc275.28 | Chr13:2181732-2184478(+) | 1,032 | 343 | 10 | 5.35 | 38.73 |
| HcSnRK2.8 | Hc326.23 | Chr14:223114-225278(−) | 981 | 326 | 9 | 5.26 | 36.87 |
| HcSnRK2.9 | Hc326.60 | Chr14:473891-477474(+) | 1,020 | 339 | 9 | 5.43 | 38.51 |
| HcSnRK2.10 | Hc57.125 | Chr14:8431879-8435942(+) | 1,092 | 363 | 9 | 4.95 | 41.08 |
| HcSnRK2.11 | Hc25.205 | Chr0:1763461-1767704(+) | 1,059 | 352 | 9 | 4.81 | 39.83 |
| HcSnRK2.12 | Hc191.34 | Chr0:484266-493850(−) | 1,077 | 358 | 9 | 5.80 | 41.17 |
| HcSnRK2.13 | Hc407.47 | Chr0:377348-381541(+) | 1,080 | 359 | 9 | 6.00 | 41.53 |
| HcSnRK3.1 | Hc430.44 | Chr1:6617049-6620752(−) | 1,452 | 483 | 2 | 6.56 | 54.12 |
| HcSnRK3.2 | Hc438.56 | Chr1:11474898-11476860(+) | 1,323 | 440 | 2 | 8.37 | 49.78 |
| HcSnRK3.3 | Hc438.54 | Chr1:11603538-11605298(+) | 1,314 | 437 | 1 | 8.96 | 48.32 |
| HcSnRK3.4 | Hc108.24 | Chr1:28981635-28984540(+) | 1,317 | 438 | 2 | 9.32 | 50.06 |
| HcSnRK3.5 | Hc253.154 | Chr1:53838578-53842095(+) | 1,377 | 458 | 12 | 8.58 | 51.54 |
| HcSnRK3.6 | Hc304.12 | Chr2:35890951-35893118(+) | 1,545 | 514 | 3 | 8.64 | 57.18 |
| HcSnRK3.7 | Hc219.55 | Chr2:61929342-61931157(+) | 1,338 | 445 | 1 | 7.31 | 49.19 |
| HcSnRK3.8 | Hc115.24 | Chr3:6979416-6980852(−) | 1,437 | 478 | 1 | 9.06 | 53.69 |
| HcSnRK3.9 | Hc171.23 | Chr3:11928832-11930468(+) | 1,236 | 411 | 1 | 9.31 | 44.60 |
| HcSnRK3.10 | Hc685.16 | Chr3:24071137-24074279(+) | 1,317 | 438 | 2 | 9.17 | 49.75 |
| HcSnRK3.11 | Hc55.127 | Chr3:38306358-38309493(−) | 1,341 | 446 | 14 | 8.94 | 50.68 |
| HcSnRK3.12 | Hc256.98 | Chr4:8831260-8833000(−) | 1,305 | 434 | 1 | 9.48 | 47.86 |
| HcSnRK3.13 | Hc280.50 | Chr4:44679903-44683582(+) | 1,332 | 443 | 3 | 8.75 | 50.59 |
| HcSnRK3.14 | Hc484.68 | Chr5:184915-186806(+) | 1,581 | 526 | 1 | 9.03 | 58.80 |
| HcSnRK3.15 | Hc484.67 | Chr5:189058-191912(−) | 1,401 | 466 | 3 | 8.45 | 52.34 |
| HcSnRK3.16 | Hc316.81 | Chr5:44515665-44521645(−) | 1,305 | 434 | 13 | 6.41 | 48.87 |
| HcSnRK3.17 | Hc32.21 | Chr6:8904624-8906421(+) | 1,356 | 451 | 1 | 8.80 | 50.82 |
| HcSnRK3.18 | Hc769.7 | Chr6:51928707-51942543(+) | 1,326 | 441 | 14 | 8.67 | 50.28 |
| HcSnRK3.19 | Hc3.374 | Chr7:3703792-3708273(−) | 1,365 | 454 | 14 | 5.66 | 51.08 |
| HcSnRK3.20 | Hc971.16 | Chr7:7639489-7645951(+) | 1,446 | 481 | 13 | 7.87 | 55.94 |
| HcSnRK3.21 | Hc33.46 | Chr7:8729879-8733633(+) | 1,311 | 436 | 2 | 9.06 | 49.17 |
| HcSnRK3.22 | Hc247.6 | Chr8:47314082-47316537(−) | 1,353 | 450 | 3 | 8.68 | 49.82 |
| HcSnRK3.23 | Hc102.99 | Chr9:1148877-1153212(+) | 1,323 | 440 | 16 | 6.82 | 49.96 |
| HcSnRK3.24 | Hc369.81 | Chr9:2688438-2692404(+) | 1,389 | 462 | 13 | 5.95 | 51.98 |
| HcSnRK3.25 | Hc71.72 | Chr10:906032-912303(−) | 1,134 | 377 | 14 | 5.78 | 42.95 |
| HcSnRK3.26 | Hc48.119 | Chr11:9190841-9192641(−) | 1,317 | 438 | 10 | 6.65 | 49.63 |
| HcSnRK3.27 | Hc439.14 | Chr11:16527358-16532095(−) | 1,386 | 461 | 15 | 8.70 | 52.46 |
| HcSnRK3.28 | Hc286.62 | Chr11:46635686-46645148(−) | 1,470 | 489 | 15 | 9.31 | 55.72 |
| HcSnRK3.29 | Hc158.15 | Chr12:1277125-1278952(+) | 1,449 | 482 | 2 | 6.15 | 53.97 |
| HcSnRK3.30 | Hc5.4 | Chr12:34636145-34637497(+) | 1,353 | 450 | 1 | 9.11 | 50.94 |
| HcSnRK3.31 | Hc79.41 | Chr12:43896038-43897396(+) | 1,359 | 452 | 1 | 6.74 | 50.06 |
| HcSnRK3.32 | Hc114.20 | Chr14:2279142-2284718(−) | 1,323 | 440 | 2 | 8.72 | 49.65 |
| HcSnRK3.33 | Hc279.1 | Chr14:16032098-16047415(−) | 1,332 | 443 | 8 | 8.75 | 50.19 |
| HcSnRK3.34 | Hc132.10 | Chr15:33199307-33201621(+) | 1,476 | 491 | 10 | 8.11 | 54.23 |
| HcSnRK3.35 | Hc74.133 | Chr16:2385971-2387422(−) | 1,452 | 483 | 1 | 6.11 | 54.51 |
| HcSnRK3.36 | Hc747.7 | Chr16:36843545-36847614(+) | 1,335 | 444 | 2 | 9.01 | 50.38 |
| HcSnRK3.37 | Hc26.164 | Chr17:1572756-1575773(−) | 1,308 | 435 | 2 | 9.16 | 49.33 |
| HcSnRK3.38 | Hc259.23 | Chr17:6843287-6866867(−) | 1,335 | 444 | 16 | 8.70 | 49.91 |
| HcSnRK3.39 | Hc259.98 | Chr17:7354852-7357698(+) | 1,392 | 463 | 13 | 7.22 | 51.51 |
| HcSnRK3.40 | Hc15.312 | Chr0:2384884-2386204(−) | 1,215 | 404 | 2 | 9.37 | 44.25 |
| HcSnRK3.41 | Hc15.375 | Chr0:2948868-2950520(+) | 1,446 | 481 | 1 | 8.88 | 53.95 |
| HcSnRK3.42 | Hc414.48 | Chr0:563909-565482(+) | 1,308 | 435 | 1 | 8.81 | 47.59 |
| HcSnRK3.43 | Hc444.50 | Chr0:650222-651310(+) | 1,089 | 362 | 1 | 7.01 | 40.66 |
Phylogeny and multiple sequence alignment of HcSnRK gene family
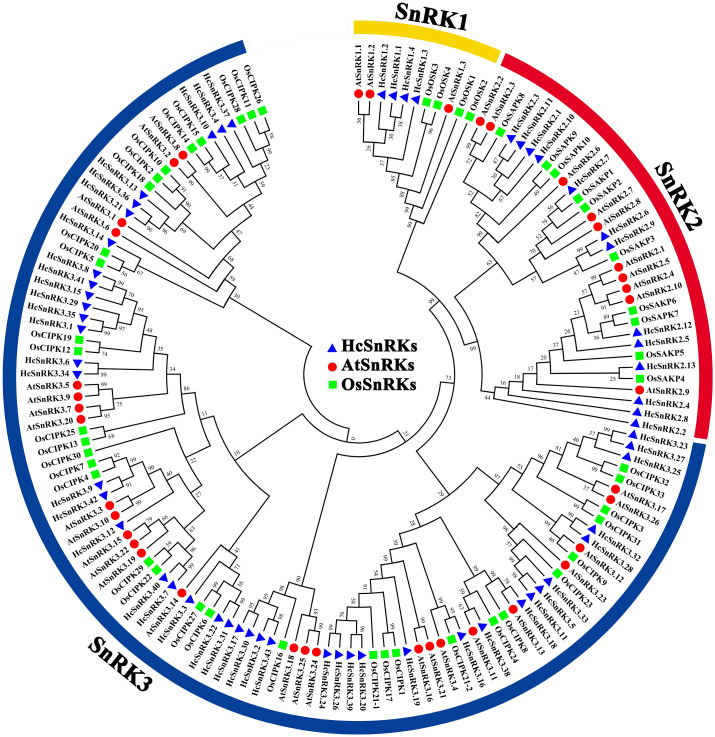
The evolutionary relationships of the SnRK genes in H. coronarium, A. thaliana, and O. sativa was revealed by constructing the phylogenetic tree based on multiple sequence alignment of amino acids. The full-length protein sequence of 39 AtSnRK, 48 OsSnRK, and 60 HcSnRK genes were used to construct a phylogenetic tree using MEGA 7 and by choosing the neighbor-joining method (Fig. 1). The results showed that 60 HcSnRK genes were divided into three groups as expected. Alike Arabidopsis and rice, the member of HcSnRK3 family were the highest (43) followed by HcSnRK2 (13) and HcSnRK1 (4), respectively.
Figure 1. Phylogenetic tree of SnRK genes from H. coronarium, Arabidopsis, and rice.
Sixty HcSnRK genes, 39 AtSnRK genes, and 48 OsSnRK genes are clustered as SnRK1, SnRK2, and SnRK3 subgroups. The detailed information of these genes from different plants are provided in Table S4. Clustal X software was used for multiple sequence alignment and MEGA 7 software used to construct a phylogenetic tree by the neighbor-joining method.
To explore the gene structure of the HcSnRK gene family, multiple sequence alignment was performed using DNAMAN 8.0 software. As shown in Fig. 2, ATP binding site and protein kinase active-site were found at N-terminal, and domain I was identified at C- terminal of HcSnRK2 subfamily. Meanwhile, the amino acid sequence of the complete protein kinase domain and NAF domain were recognized in HcSnRK3 subfamily. In summary, conserved domain analysis and multiple sequence alignment validated that all 60 HcSnRK genes have complete functional domains.
Figure 2. MSA of SnRK genes in H. coronarium.
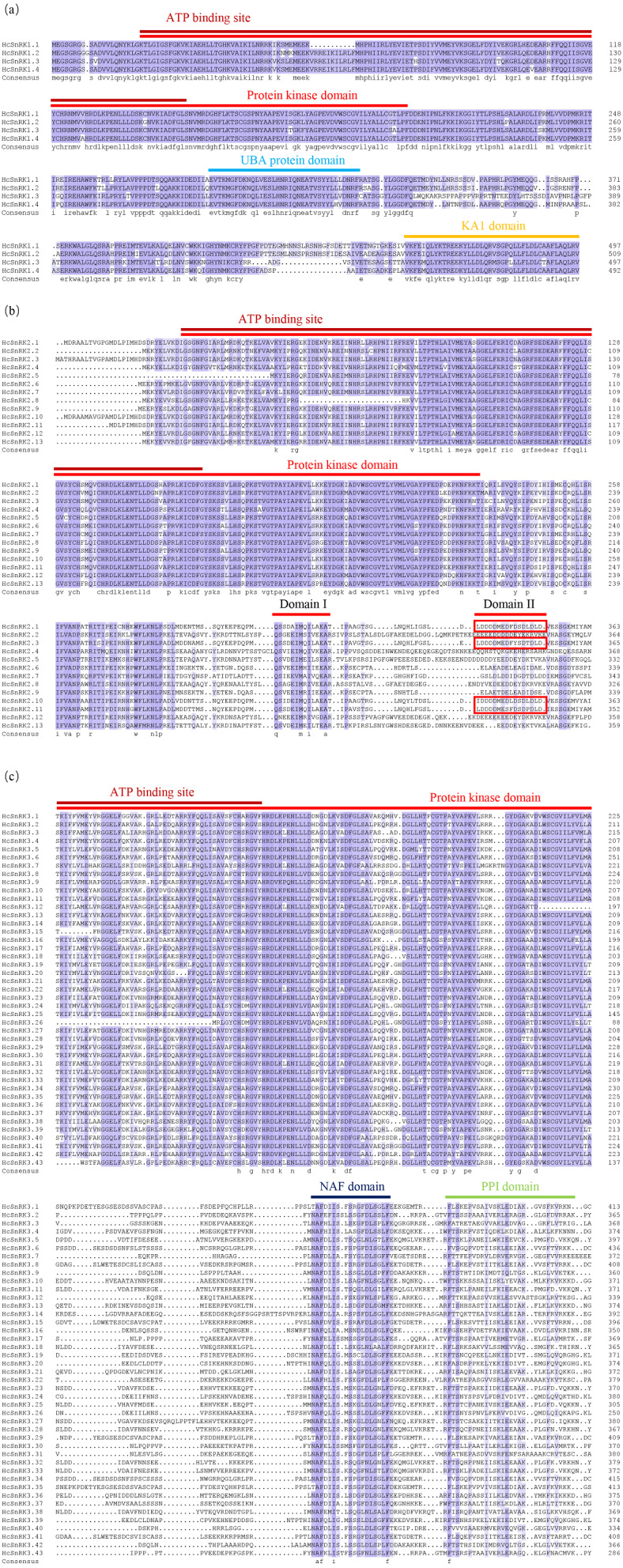
Sequences were aligned using Clustal W software and edited in Photoshop.
Gene structural analysis of HcSnRK gene family
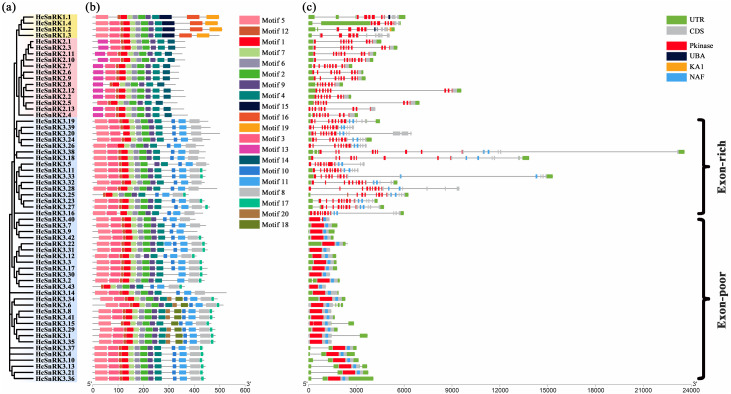
The sequence structure of 60 HcSnRK proteins was analyzed using the MEME program by choosing the 20 motifs with default parameters (Table S1). The results showed that motif 1 encoded a protein kinase domain and was present in all HcSnRK genes, while different subfamilies retained an obvious difference in motif composition (Figs. 3A and 3B). Furthermore, motifs 1, 2, 6, 7, and 9 were present in all HcSnRK genes. Meanwhile, motif 19 was only found in the HcSnRK1 subfamily which encoded the KA1 domain, whereas, motif 10 and 11 only appeared in the HcSnRK3 subfamily, which encoded the NAF domain. Moreover, HcSnRK genes in the same subfamily have a similar motif, indicating that those genes have the same gene structure and functional domain. As for other motifs, the Pfam database did not find any functional annotations.
Figure 3. Phylogenetic relationships, conserved motifs, and gene structures of HcSnRK genes.
(A) The phylogenetic tree of 60 HcSnRK genes was built via choosing the NJ method. Yellow, red, and blue background color represents SnRK1, SnRK2, and SnRK3 subgroups. (B) The conserved motifs were analyzed by MEME software. (C) Gene structure and conserved domains of SnRK genes.
The combined phylogenetic tree and web server Gene Structure Display Server (GSDS) analysis was performed to determine the intron/exon structure of HcSnRK genes (Fig. 3C). The data showed that the members of the same subfamily share similar features. The HcSnRK1 subfamily genes have 11 to 12 introns, while the HcSnRK2 subfamily contains 9 to 10 introns. However, the number of introns in the HcSnRK3 subfamily varies. The 27 HcSnRK3 genes contained less than 3 introns, and the 16 HcSnRK3 genes contained 8 to 13 introns. Therefore, HcSnRK3 genes can be divided into two subgroups, intron-rich and intron-poor subgroups, respectively, based on the number of introns. Previously, similar intron numbers and classification pattern was observed in both monocots and dicots species, such as Arabidopsis, rice, maize, poplar, etc. The characteristic of intron number indicates that the evolution of SnRK genes was conserved in plants.
Chromosomal location and gene duplication analysis
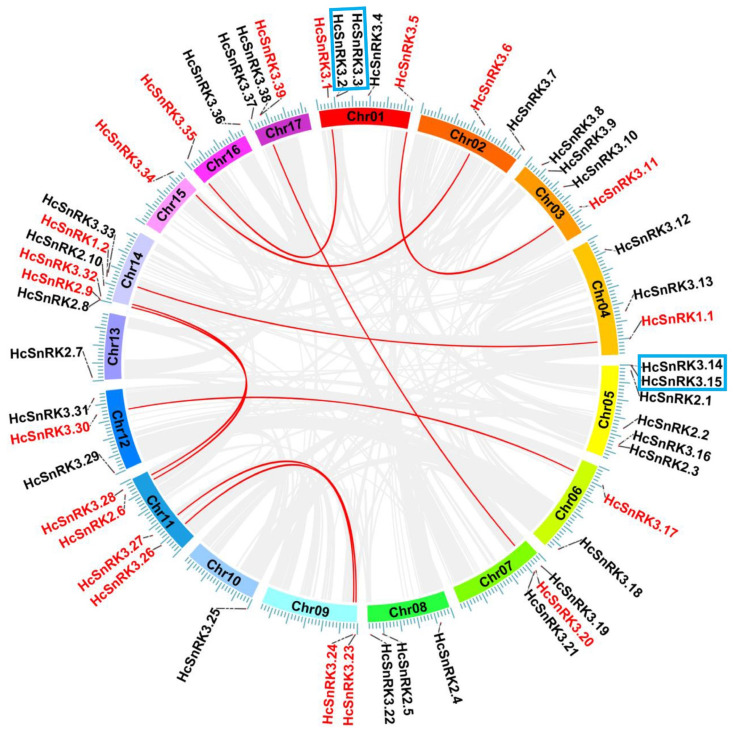
The chromosomal localization analysis showed that 60 HcSnRK genes were unevenly mapped onto all 17 chromosomes, including four HcSnRK1 genes, 13 HcSnRK2 genes and 43 HcSnRK3 (Fig. 4), whilst nine HcSnRK genes were localized on an unknown chromosome which will, later on, be assigned on anyone among 17 via refining whole-genome sequencing. Six HcSnRK genes were located on chromosome 5, and 14, while chromosome 1 has five HcSnRK genes distribution. However, chromosomes 10, 13, and 15 only contain one HcSnRK gene. In short, all the HcSnRK genes were randomly distributed on all chromosomes in H. coronarium genome.
Figure 4. Chromosomal location and synteny analysis of HcSnRK in H. coronarium genome.
HcSnRK genes are mapped onto all 17 chromosomes, gray background lines represent synteny blocks in whole H. coronarium genome and red lines showed the gene duplication pairs of HcSnRK genes.
To identify the segmental duplication of HcSnRK genes, BLAST and MCScanX methods were used. Among 60 HcSnRK genes, 10 duplicated pairs derived from segmental duplication and 2 from tandem duplication were observed (Fig. 4). The majority of the duplicated gene pairs were found on chromosome 11 and 14. To demonstrate and reveal the effect of selection pressure on the evolution of HcSnRK genes, synonymous (Ks), non-synonymous substitutions (Ka), and Ka/Ks ratios per site between every duplicated pair were calculated by DnaSP 5.0 software. More importantly, Ka/Ks = 1 indicates neutral selection, Ka/Ks<1 represents purifying selection, and Ka/Ks> 1 represents positive selection and accelerated evolution. The Ka/Ks ratio of 12 duplicated gene pairs ranged from 0.095 to 0.465 (Table S2), suggesting that all duplicated gene pairs of HcSnRK had undergone purifying selection.
Cis-regulatory elements analysis
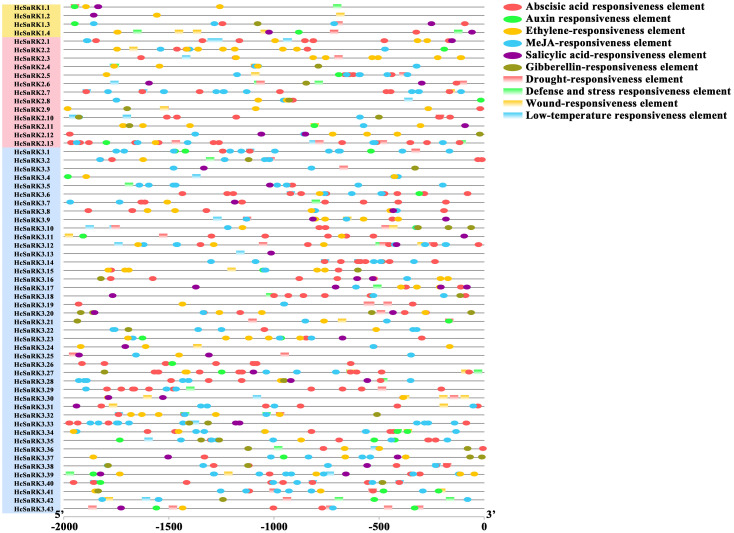
The upstream 2 kb of promoter sequences of 60 HcSnRK genes were submitted to the PlantCARE database, to analyze the function and regulatory mechanism of HcSnRK genes. The results showed that almost all HcSnRK genes contained hormone-responsive cis-elements; however, less than half of HcSnRK genes contained abiotic stresses cis-elements. Interestingly, 53 out of 60 HcSnRK promoters contained ABRE cis-elements, while 50 and 44 out of 60 HcSnRK promoters contained MeJA-responsive and ethylene responsive cis-acting regulatory elements, respectively. Meanwhile, about half of the promoters of the HcSnRK gene contained auxin, salicylic acid, and gibberellin responsive cis-elements. On the other hand, less than half of the promoters of HcSnRK genes contained low-temperature and drought-inducible cis-elements, and about one-third of promoters contained defense, stresses, and wound-responsive element (Fig. 5; Table S3). Cis-regulatory element analysis data suggested that HcSnRK genes might respond to multiple hormones and abiotic stresses.
Figure 5. Cis-acting elements analysis of the HcSnRK genes promoter region.
The 2,000 bp sequence before the start codon was used to analyze cis-acting elements. Different color boxes indicate different cis-acting elements.
Expression pattern of HcSnRK genes in different varieties, tissues and flower developmental stages
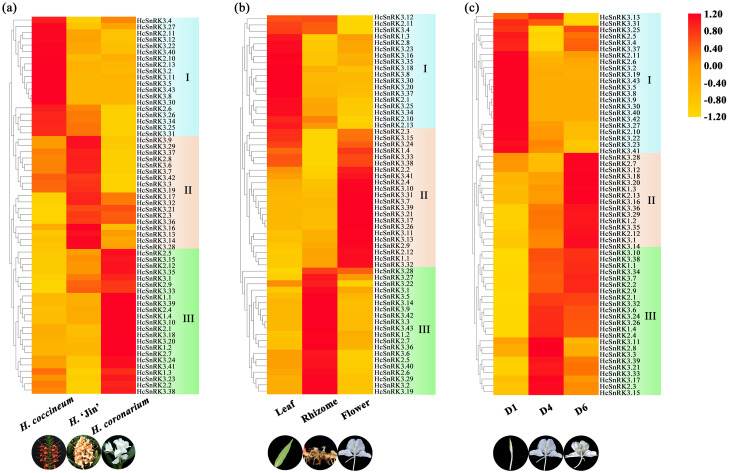
The analysis of differential expression pattern of HcSnRK genes was performed by using the transcriptome data of three different tissue (leaf, rhizome, and flower) and three flower development stages (D1; bud stage, D4; full-bloom and D6; flower senescence stage) of H. coronarium and three different varieties of Hedychium (Yue, Yu & Fan, 2015). The volatile compounds among different varieties vary significantly. The GC-MS analysis showed that the emission of volatile compounds from H. coronarium were higher as compared to H. ‘Jin’, while volatile compounds in H. coccineum are very low (Fan et al., 2007). The expression level of HcSnRK genes is presented in a heat map and HcSnRK genes with similar expression patterns were grouped into distinct groups. Cluster I represent the group of HcSnRK genes which had the highest expression in H. coccineum, Cluster II in H. ‘Jin’ and cluster III had preferential expression H. coronarium, respectively (Fig. 6A). Previous studies showed that the number of volatile contents were higher in the flowers compared to leaf and rhizome. Moreover, the amount of floral volatiles was low at the D1 stage and peak at the full-bloom stage (D4) with flower development and declined at the D6 stage (Ke et al., 2019). Similarly, expression of HcSnRK genes in different tissues and different flower development stages of H. coronarium were also grouped in three clusters. The tissue-specific expression is important for gene functioning. Cluster I showed preferential expression in leaf, Cluster II represents the group of genes that had the highest expression in flower, and cluster III represents specific expression in the rhizome, respectively (Fig. 6B). Based on different transcriptome data and preferential expression of cluster III in H. coronarium, flower-specific expression and highest expression at full-bloom stage of flower indicate its potential role in the flower development and in the regulation of floral scent formation processes (Fig. 6C). Based on transcriptome data, HcSnRK2.2 and HcSnRK2.9 were screened out for further experimental analysis.
Figure 6. Expression Patterns of HcSnRK genes from different transcriptome data.
(A) Heat map of HcSnRK genes expression pattern in three different varieties. (B) Expression patterns of HcSnRK genes in different tissues. (C) Expression profiles of HcSnRK genes in different flower developmental stages. Levels of upregulated expression (red) and downregulated expression (yellow) are shown on a log2 scale from the highest to lowest expression.
Expression patterns of HcSnRK genes in response to hormone treatments
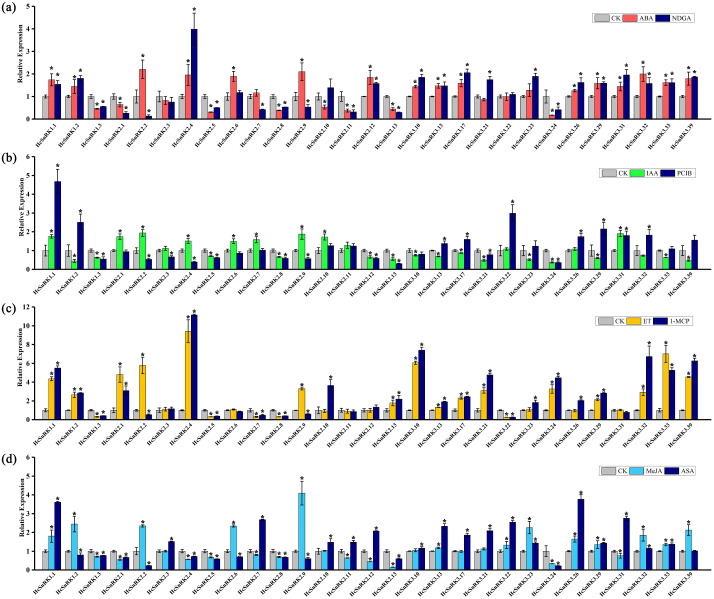
Auxin, ethylene, and ABA are the major hormones involved in the development of the flower. H. coronarium flowers were subjected to various hormone treatments. The results revealed that the volatile compounds of H. coronarium flowers were increased by 16%, 21%, 20%, and 22% under ABA, IAA, ethylene, and methyl jasmonate treatment, respectively. Meanwhile, the emission of volatile compounds decreased by 30%, 35%, 52%, and 34% under their corresponding hormone inhibitors nordihydroguaiaretic acid (NDGA), 2-(4-chlorophenoxy)-isobutyric acid (PCIB), 1-methyl cyclopropane (1-MCP) and acetylsalicylic acid (ASA), respectively (Ke et al., 2019). The expression level of selected HcSnRK genes was measured by qRT-PCR under hormone treatments (Fig. 7). The data showed that the expression level of HcSnRK2.2 and HcSnRK2.9 significantly up-regulated by ABA treatment, while down-regulated by NGDA (Fig. 7A). Overall, 24 HcSnRK genes were significantly up or downregulated under ABA treatment and 25 genes significantly changed under NDGA treatment. Under IAA and PCIB treatments, 24 HcSnRK genes showed significant difference in their expression pattern (Fig. 7B). Notably, HcSnRK2.2, 2.4, and 2.9 genes significantly increased in IAA treatment and decreased in PCIB treatment. Furthermore, 21 HcSnRK genes significantly up or down-regulated after ethylene treatment, and 24 genes significantly changed after 1-MCP treatment (Fig. 7C). Moreover, HcSnRK2.2 and 2.9 were highly up-regulated under ethylene treatment and down-regulated after 1-MCP treatment. In addition, 24 HcSnRK genes significantly increase or decrease in methyl jasmonate treatment, and 28 genes significantly changed in acetylsalicylic acid treatment (Fig. 7D). As expected HcSnRK2.2, 2.6, and 2.9 genes significantly increased in methyl jasmonate treatment and decreased in acetylsalicylic acid treatment. In particular, the expression level of the HcSnRK2.9 gene increased by 4 times under MeJA treatment. In short, plant hormones, such as ABA, IAA, ethylene, and jasmonic acid have a crucial effect on the regulation of floral aroma and the response pattern of HcSnRK2.2 and 2.9 genes towards hormones were consistent with the changes of floral aroma contents. The results indicate that HcSnRK2.2 and 2.9 genes play an important role in regulating the metabolism of floral aroma substances via crosstalk in hormone signaling.
Figure 7. HcSnRK genes respond to several hormone treatments.
The relative expression levels of HcSnRK genes in response to ABA (A), IAA (B), ET (C), MeJA (D), and corresponding inhibitor NDGA, PCIB, 1-MCP, AS was analyzed by qRT-PCR. The expression level of the control group was set to 1, error bars represent standard deviation from three to four biological replicates. Significant differences between the control group and hormone treatment samples are indicated by an asterisk (p < 0.05).
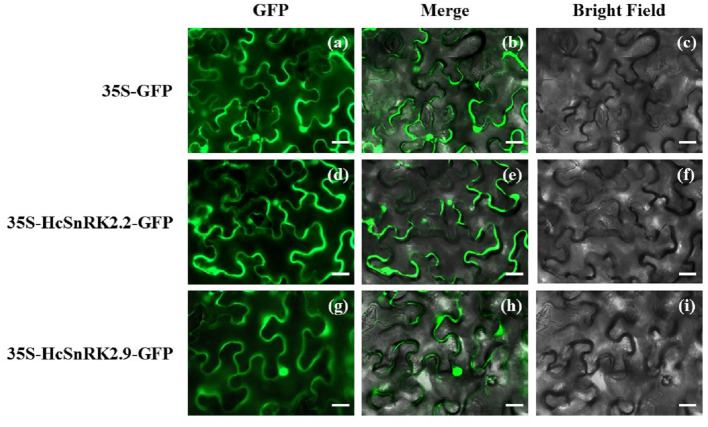
Subcellular localization of HcSnRK2.2 and HcSnRK2.9
The amino acid sequence of HcSnRK2.2/2.9 was submitted to WoLP PSORT (https://wolfpsort.hgc.jp/) to predict subcellular localization. The predicted results showed that all HcSnRK2.2/2.9 proteins were expressed in the nucleus and cytoplasm. To experimentally verify the subcellular localization, full-length sequences of candidate HcSnRK2.2 and HcSnRK2.9 were fused to a GFP reporter gene and transferred to N. benthamiana leaves (Fig. 8). Subcellular localization experiments results revealed that both HcSnRK2.2 and HcSnRK2.9 were localized in the cell nucleus and cytoplasm as predicted. Similarly, multiple SnRK proteins have been reported previously from Arabidopsis and rice, which were in the nucleus and cytoplasm and were involved in the regulation of several ABA responses.
Figure 8. Subcellular localization of HcSnRK2.2 and HcSnRK2.9 proteins.
HcSnRK2.2-GFP and HcSnRK2.9-GFP fusion vectors were transformed into N. benthamiana leaves and subcellular localization was carried out 48 h after infiltration using Leica DM RXA2 upright fluorescent microscope. The bar indicates 20 µm.
Silencing of HcSnRK2.2 and HcSnRK2.9 genes
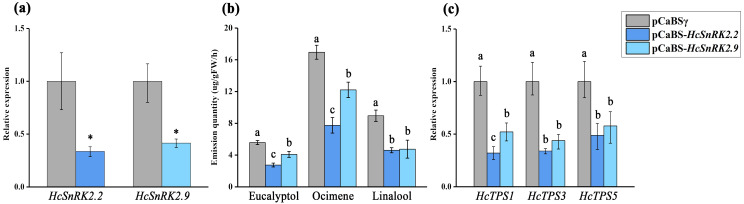
To verify the function of HcSnRK2.2 and 2.9 in the regulation of floral volatile synthesis, the virus-induced gene silencing (VIGS) system was used to suppress gene expression in H. coronarium. As shown in Fig. 9, the expression of HcSnRK2.2 and HcSnRK2.9 genes were significantly reduced by 66% and 58% compared to control, after silencing HcSnRK2.2 and 2.9, respectively. Moreover, the content of the main floral volatile substance, such as eucalyptol, ocimene, and linalool decreased significantly by 51%, 54%, and 48% after silencing HcSnRK2.2 gene, and decreased by 27%, 28%, and 47% after silencing HcSnRK2.9, respectively. Furthermore, the expression levels of main volatile synthesis genes significantly decreased, such as HcTPS1 which is responsible for eucalyptol synthesis, HcTPS3 for ocimene, and HcTPS5 for linalool synthesis. HcTPS1, HcTPS3 and HcTPS5 were down-regulated 68%, 66% and 51% after the suppression of HcSnRK2.2, and down-regulated 48%, 56% and 42% after silencing HcSnRK2.9. These findings indicate that HcSnRK2.2 and HcSnRK2.9 play an important key role in the regulation of floral aroma synthesis.
Figure 9. The silencing of HcSnRK2.2 and HcSnRK2.9 genes reduces the amount of the volatile compounds in H. coronarium flowers.
(A) The relative expression level of HcSnRK2.2 and HcSnRK2.9 genes after BSMV-HcSnRK2.2/2.9 silencing were analyzed by qRT-PCR. (B) The main volatile compounds measured by GC-MS after silencing of HcSnRK2.2 and HcSnRK2.9 genes. (C) The relative expression level of key volatile biosynthesis genes. Error bars represent standard deviations from three to four biological replicates and asterisks indicate statistically significant differences (p < 0.05).
Discussion
The SnRK gene family plays a crucial role in different physiological processes and is conserved in all eukaryotes. The SnRK1 is involved in the functioning of cell energy sensing, while SnRK2 and SnRK3 play fundamental roles in the signaling pathway and the regulation of gene expression (Halford et al., 2004; Li et al., 2010; Colina et al., 2019). The SnRK2 and SnRK3 subfamily are unique in plants and originate from the duplication of the SnRK1 subfamily (Colina et al., 2019; Halford & Hey, 2009). The expansion of the SnRK family may be partly since plants are sessile organisms and are forced to face more biotic and abiotic stresses than animals (Colina et al., 2019). In plants, the SnRK family represents an interface between stress signaling and metabolic pathway and is widely involved in ABA-dependent and ABA-independent abiotic stress. The SnRK family has been reported from a wide range of plant species, including Arabidopsis, maize (Chen et al., 2011), rice (Kanwar et al., 2014), and cotton (Cui et al., 2020), however, the SnRK family from H. coronarium has not been studied.
In the current study, 60 HcSnRK genes including 4 HcSnRK1, 13 HcSnRK2, and 43 HcSnRK3 in H. coronarium were identified. Previously, 34, 48, 39, 44, and 52 SnRK genes have been identified form Eucalyptus grandis (Wang et al., 2019), Oryza sativa (Kobayashi et al., 2004), A. thaliana (Hrabak et al., 2003), Brachypodium distachyon (Wang et al., 2015) and Glycine max (Zhu et al., 2016), respectively. Moreover, different SnRK gene subfamily encompasses various conserved domains, however, all genes included a protein kinase domain present at the N-terminal. Phylogenetic analyses showed that like Arabidopsis and rice, H. coronarium also contain a similar number of SnRK1 subfamily members (3 to 4) and SnRK2 subfamily (10 or 13). However, the number of SnRK3 subfamily genes varies from species to species, such as 26 in A. thaliana, 34 in O. sativa, 24 in E. grandis, 52 in G. max, and 43 in H. coronarium.
The different number of exon-intron also plays an important role in the evolution and function of a different gene family (Jo & Choi, 2015). The SnRK3 subfamily not only varies on the number of genes, but previous findings also showed that the SnRK3 subfamily can be subdivided into two clades according to the number of introns (Tang et al., 2016; Zhu et al., 2016). Likewise, the HcSnRK3 subfamily can be subdivided into an intron-rich and intron-poor clade. The 16 HcSnRK3 genes were grouped into an intron-rich clade (more than 8 introns) and 27 HcSnRK3 genes in the intron-poor clade (less than 3 introns). Similarly, in Arabidopsis and rice, the SnRK3 subfamily was subdivided according to the number of introns indicating that an increase or decrease in the number of introns can promote the structural evolution of the SnRK3 gene family before eudicot–monocot divergence (Zhu et al., 2016). Recent findings suggest that the SnRK3 subfamily originated in green algae, and the intron-poor group first appeared in the seed plants (Colina et al., 2019). It has been assumed that when seed plants will face great environmental pressure during evolution, intron-rich groups will lose intron and become intron-poor groups (Colina et al., 2019). Also, HcSnRK1 has 11 to 12 introns, HcSnRK2 subfamily has 9 or 10 introns. These results indicated that the number of introns in HcSnRK genes is similar to other plants. The conserved motif analysis revealed that gene structure and conserved motifs were similar in the same subfamily, indicating the close evolutionary relationship within the same subfamily, but different subfamilies involved in different stress response pathways.
A large number of cis-elements related to hormone response were found in the promoter sequences of HcSnRK genes, suggesting that HcSnRK genes respond to multiple hormone signals and interact with other metabolic pathways. Plants have developed unique strategies to cope with the external environment. Numerous evidences indicated that the SnRK family is widely involved in the response to various biotic and abiotic stresses, including salt, high or low temperature, and drought (Tang et al., 2016; Zhu et al., 2016; Wang et al., 2019). Many cis-elements related to stresses, wounding and defense response were identified in the promoter sequences of HcSnRK genes. Previous findings indicate that hormones play essential roles in the flower development and regulation of aroma (Chandler, 2011; Iqbal et al., 2017; Ke et al., 2019). The ethylene, Auxin, ABA, and MeJA responsive cis-elements were found in the majority of the promoters of HcSnRK genes suggesting their significant functions by crosstalk with HcSnRK genes in H. coronarium flower. The above results are in line with the previous findings from tomato and H. coronarium (Audran-Delalande et al., 2012; Ke et al., 2019). In our previous research, we describe that Auxin/IAA genes are involved in the regulation of floral scent and the volatile contents of H. coronarium flower were altered under different hormone treatments (Ke et al., 2019). Moreover, ethylene and ABA are also involved in floral scent regulation and flower senescence. To verify the response of the HcSnRK genes to several hormones, the expression levels of 29 genes based on their higher abundance in flower, was performed by qRT-PCR. The results showed that 27 HcSnRK genes significantly respond to ABA treatment, while 28, 24 and 28 HcSnRK genes significantly responded to IAA, ethylene, MeJA, and their corresponding inhibitors, respectively. Similarly, HbSnRK2.5, 2.7, and 2.10 from Hevea brasiliensis were also significantly up-regulated under ABA, ethylene, and MeJA treatment. Alike, HbSnRK2.8, 2.9 up-regulated under ABA and MeJA treatment, whilst, HbSnRK2.2 down-regulated under ABA and MeJA treatment, however, HbSnRK2.6 significantly up-regulated under MeJA, down-regulated under ethylene and do not respond to ABA treatment. In the present study, HcSnRK2.4 significantly up-regulated under IAA and down-regulated under PCIB, while HcSnRK2.6 up-regulated under MeJA and down-regulated under ASA treatment. Furthermore, HcSnRK2.2 and HcSnRK2.9 showed significant differential expression under ABA, IAA, ethylene, MeJA, and their corresponding inhibitor treatments. Previous studies verified that ABA, IAA, ethylene, and MeJA have a significant effect on the regulation of floral aroma. These results implied that HcSnRK2.2 and HcSnRK2.9 maybe involved in multiple hormone metabolism pathways to regulate the metabolism of floral fragrance.
Tissue-specific expression pattern of HcSnRK genes in different tissue permits different regulation of tissue development and alternate means of metabolic regulation. The transcriptome data of different varieties (strong, moderate, and almost no floral fragrance variety), different tissues (rhizome, leaf, and flower), and different flower development stages (bud stage, full-bloom stage, and fade stage) were used to analyze the expression pattern of HcSnRK genes. The 23 HcSnRK genes showed high expression in strong floral fragrance variety, 22 HcSnRK genes were highly expressed in the flower, whereas, 23 HcSnRK genes showed their preferential expression at the full-bloom stage of the flower. It was also observed that 15 HcSnRK genes have extremely high expression during the senescence stage indicating their possible role in the regulation of flower aging. Moreover, the expression pattern of HcSnRK2.2 and HcSnRK2.9 was similar to the emission of floral substances, suggesting that these two genes might play an important role in the regulation of floral aroma synthesis. The differential expression pattern of SnRK genes has been found in many species. In apple, MdCIPK4, 9, 15, and 32 were highly expressed in the flower and MdCIPK 29 show relatively high expression in fruit implied their different biological functions in respective tissues (Niu et al., 2018). In Brassica napus L., BnCIPK9 was tissue-specific and developmental stage-specific expressed in seed, and overexpression of BnCIPK9 reduced oil synthesis in the transgenic plant (Guo et al., 2018). The virus-induced gene silencing of HcSnRK2.2 and HcSnRK2.9 confirm their role in floral scent regulation. The silencing of HcSnRK2.2 and HcSnRK2.9 genes did not alter the flowering process, however, resulted in the emission of the low amount of floral volatile and decreases the expression pattern of key genes involved in the biosynthesis of floral scent (Fig. 9). The decrease in the emission of volatile compounds might be because of their involvement in the hormone signaling especially in ABA and ethylene signaling pathway. This is the first report regarding the role of HcSnRK genes in the regulation of the floral scent biosynthetic pathway. The function of SnRK family in ABA-dependent and independent pathway have been extensively studied, however, their role in floral scent pathway needs to be elucidated further.
Conclusion
In brief, we identified the SnRK gene family in Hedychium coronarium; analyzed expression profiles based on three different transcriptome data, and screened numerous key candidate genes for functional characterization. Through virus-induced gene silencing, we find out the functional involvement of HcSnRK2.2 and HcSnRK2.9 in floral scent formation. Our findings will bring new insights into the function of HcSnRK genes in secondary metabolism
Supplemental Information
Table S1: Sequences information of 20 SnRK Motifs.
Table S2:Ka/Ks ratios of paralogous HcSnRK genes.
Table S3: Cis-acting regulatory elements numbers found in the promoter region of HcSnRK genes.
Table S4: Gene IDs of SnRK genes from Arabidopsis and rice.
Table S5: List of primers used in the experimentations.
Abbreviations
- SnRK
Snf1-Related protein kinase
- VIGS
Virus-induced gene silencing
- qRT-PCR
Quantitative real-time polymerase chain reaction
- MAPK
Mitogen-activated protein kinase
- CDPK
Calcium-dependent protein kinases
- GSK3
Glycogen synthase kinase 3
- SNF1
Sucrose non-fermenting 1
- ABA
Abscisic acid
- SAPK
Stress/ABA-activated protein kinase
- bZIP
Basic region-leucine zipper
- CIPK
Calcineurin B-like interacting protein kinases
- NDGA
Nordihydroguaiaretic acid
- IAA
Indole-3-Acetic Acid
- PCIB
2-(4-chlorophenoxy)-isobutyric acid
- ET
Ethylene
- 1-MCP
1-Methylcyclopropene
- MeJA
Methyl jasmonate
- ASA
Acetylsalicylic acid
- GSDS
Gene Structure Display Server
- MEME
Multiple Expectation Maximization for Motif Elicitation
- Ka
Non-synonymous substitutions
- Ks
Synonymous substitutions
- PDMS
Polydimethylsiloxane
- GC-MS
Gas chromatography-mass spectrometry
- BSMV
Barley stripe mosaic virus
- GFP
Green fluorescence protein
Funding Statement
This work was supported by the Key-Areas Research and Development Program of Guangdong Province (Grant no. 2020B0202022007) to Yanping Fan, National Natural Science Foundation of China (Grant no. 31770738), the People’s Livelihood Science and Technology projects of Guangzhou (Grant no. 201903010054) to Yanping Fan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Chutian Wang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Farhat Abbas performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Yiwei Zhou analyzed the data, prepared figures and/or tables, and approved the final draft.
Yanguo Ke performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Xinyue Li, Yuechong Yue and Yunyi Yu performed the experiments, prepared figures and/or tables, and approved the final draft.
Rangcai Yu and Yanping Fan conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Raw data are available in the Supplemental Files.
References
- Abbas et al. (2019).Abbas F, Ke Y, Yu R, Fan Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta. 2019;249:71–93. doi: 10.1007/s00425-018-3006-7. [DOI] [PubMed] [Google Scholar]
- Abbas et al. (2017).Abbas F, Ke Y, Yu R, Yue Y, Amanullah S, Jahangir MM, Fan Y. Volatile terpenoids: multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta. 2017;246:803–816. doi: 10.1007/s00425-017-2749-x. [DOI] [PubMed] [Google Scholar]
- Abbas et al. (2021).Abbas F, Ke Y, Zhou Y, Yu Y, Waseem M, Ashraf U, Wang C, Wang X, Li X, Yue Y, Yu R, Fan Y. Genome-Wide Analysis Reveals the Potential Role of MYB Transcription Factors in Floral Scent Formation in Hedychium coronarium. Frontiers in Plant Science. 2021;12:623742. doi: 10.3389/fpls.2021.623742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran-Delalande et al. (2012).Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant and Cell Physiology. 2012;53:659–672. doi: 10.1093/pcp/pcs022. [DOI] [PubMed] [Google Scholar]
- Baba, Rigó & Ayaydin (2018).Baba AI, Rigó G, Ayaydin F. Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: AtCRK1 regulates responses to continuous light. International Journal of Molecular Sciences. 2018;19(5):1282. doi: 10.3390/ijms19051282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González & Sheen (2008).Baena-González E, Sheen J. Convergent energy and stress signaling. Trends in Plant Science. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey et al. (2009).Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel, Grieco & Jope (2015).Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacology and Therapeutics. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, Barbier-Brygoo & Laurière (2004).Boudsocq M, Barbier-Brygoo H, Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. Journal of Biological Chemistry. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Boudsocq et al. (2007).Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Molecular Biology. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- Chan et al. (2017).Chan A, Carianopol C, Tsai AY, Varatharajah K, Chiu RS, Gazzarrini S. SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. Journal of Experimental Botany. 2017;68:4219–4231. doi: 10.1093/jxb/erx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler (2011).Chandler J. The hormonal regulation of flower development. Journal of Plant Growth Regulation. 2011;30:242–254. doi: 10.1007/s00344-010-9180-x. [DOI] [Google Scholar]
- Chen et al. (2019).Chen H, Yue Y, Yu R, Fan Y. A Hedychium coronarium short chain alcohol dehydrogenase is a player in allo-ocimene biosynthesis. Plant Molecular Biology. 2019;101:297–313. doi: 10.1007/s11103-019-00904-z. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2013).Chen JJ, Ting CW, Wu YC, Hwang TL, Cheng MJ, Sung PJ, Wang TC, Chen JF. New Labdane-type diterpenoids and anti-inflammatory constituents from Hedychium coronarium. International Journal of Molecular Sciences. 2013;14:13063–13077. doi: 10.3390/ijms140713063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2011).Chen X, Gu Z, Xin D, Hao L, Liu C, Huang J, Ma B, Zhang H. Identification and characterization of putative CIPK genes in maize. Journal of Genetics and Genomics. 2011;38:77–87. doi: 10.1016/j.jcg.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Cheong et al. (2007).Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. The Plant Journal. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- Cna’ani et al. (2015).Cna’ani A, Spitzer-Rimon B, Ravid J, Farhi M, Masci T, Aravena-Calvo J, Ovadis M, Vainstein A. Two showy traits, scent emission and pigmentation, are finely coregulated by the MYB transcription factor PH4 in petunia flowers. New Phytologist. 2015;208:708–714. doi: 10.1111/nph.13534. [DOI] [PubMed] [Google Scholar]
- Coello, Hey & Halford (2011).Coello P, Hey SJ, Halford NG. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. Journal of Experimental Botany. 2011;62:883–893. doi: 10.1093/jxb/erq331. [DOI] [PubMed] [Google Scholar]
- Colina et al. (2019).Colina F, Amaral J, Carbó M, Pinto G, Soares A, Cañal MJ, Valledor L. Genome-wide identification and characterization of CKIN/SnRK gene family in Chlamydomonas reinhardtii. Scientific Reports. 2019;9:350. doi: 10.1038/s41598-018-35625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui et al. (2020).Cui Y, Su Y, Wang J, Jia B, Wu M, Pei W, Zhang J, Yu J. Genome-wide characterization and analysis of CIPK gene family in two cultivated allopolyploid cotton species: sequence variation, association with seed oil content, and the role of GhCIPK6. International Journal of Molecular Sciences. 2020;21(3):863. doi: 10.3390/ijms21030863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo et al. (2006).D’Angelo C, Weinl S, Batistic O, Pandey GK, Cheong YH, Schültke S, Albrecht V, Ehlert B, Schulz B, Harter K, Luan S, Bock R, Kudla J. Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. The Plant Journal. 2006;48:857–872. doi: 10.1111/j.1365-313X.2006.02921.x. [DOI] [PubMed] [Google Scholar]
- Das et al. (2013).Das A, Lee SH, Hyun TK, Kim SW, Kim JY. Plant volatiles as method of communication. Plant Biotechnology Reports. 2013;7:9–26. doi: 10.1007/s11816-012-0236-1. [DOI] [Google Scholar]
- Dudareva et al. (2013).Dudareva N, Klempien A, Muhlemann JK, Kaplan I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytologist. 2013;198:16–32. doi: 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- Dudareva et al. (2006).Dudareva N, Negre F, Nagegowda DA, Orlova I. Plant volatiles: recent advances and future perspectives. Critical Reviews in Plant Sciences. 2006;25:417–440. doi: 10.1080/07352680600899973. [DOI] [Google Scholar]
- Fan et al. (2007).Fan YP, Wang XR, Yu RC, Yang P. Analysis on the aroma components in several species of Hedychium. Acta Horticulturae Sinica. 2007;34:231. [Google Scholar]
- Finn et al. (2016).Finn RD, Coggill P, Eberhardt RY, Eddy SR. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, Verslues & Zhu (2011).Fujii H, Verslues PE, Zhu J-K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita et al. (2009).Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant and Cell Physiology. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Gazzarrini et al. (2004).Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Developmental Cell. 2004;7:373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2018).Guo Y, Huang Y, Gao J, Pu Y, Wang N, Shen W, Wen J, Yi B, Ma C, Tu J, Fu T, Zou J, Shen J. CIPK9 is involved in seed oil regulation in Brassica napus L. and Arabidopsis thaliana (L.) Heynh. Biotechnology for Biofuels. 2018;11:124. doi: 10.1186/s13068-018-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford & Hey (2009).Halford NG, Hey SJ. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochemical Journal. 2009;419:247–259. doi: 10.1042/bj20082408. [DOI] [PubMed] [Google Scholar]
- Halford et al. (2004).Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Zhang Y, Paul MJ. Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. Journal of Experimental Botany. 2004;55:35–42. doi: 10.1093/jxb/erh019. [DOI] [PubMed] [Google Scholar]
- Hrabak et al. (2003).Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiology. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2015).Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im et al. (2014).Im JH, Cho YH, Kim GD, Kang GH, Hong JW, Yoo SD. Inverse modulation of the energy sensor Snf1-related protein kinase 1 on hypoxia adaptation and salt stress tolerance in Arabidopsis thaliana. Plant, Cell and Environment. 2014;37:2303–2312. doi: 10.1111/pce.12375. [DOI] [PubMed] [Google Scholar]
- Iqbal et al. (2017).Iqbal N, Khan NA, Ferrante A, Trivellini A, Francini A, Khan M. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Frontiers in Plant Science. 2017;8:475. doi: 10.3389/fpls.2017.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong et al. (2015).Jeong EY, Seo PJ, Woo JC, Park CM. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biology. 2015;15:110. doi: 10.1186/s12870-015-0503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo & Choi (2015).Jo BS, Choi SS. Introns: the functional benefits of introns in genomes. Genomics & inform. 2015;13:112–118. doi: 10.5808/GI.2015.13.4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar et al. (2014).Kanwar P, Sanyal SK, Tokas I, Yadav AK, Pandey A, Kapoor S, Pandey GK. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium. 2014;56:81–95. doi: 10.1016/j.ceca.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Ke et al. (2019).Ke Y, Abbas F, Zhou Y, Yu R, Yue Y, Li X, Yu Y, Fan Y. Genome-wide analysis and characterization of the Aux/IAA family genes related to floral scent formation in Hedychium coronarium. International Journal of Molecular Sciences. 2019;20:3235. doi: 10.3390/ijms20133235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi et al. (2004).Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. The Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2018).Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch et al. (2012).Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander D, Garcia-Hernandez M, Karthikeyan AS. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan et al. (2013).Lan JB, Yu RC, Yu YY, Fan YP. Molecular cloning and expression of Hedychium coronarium farnesyl pyrophosphate synthase gene and its possible involvement in the biosynthesis of floral and wounding/herbivory induced leaf volatile sesquiterpenoids. Gene. 2013;518:360–367. doi: 10.1016/j.gene.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2015).Lee HJ, Park YJ, Seo PJ, Kim JH, Sim HJ, Kim SG, Park CM. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. The Plant Cell. 2015;27:3425–3438. doi: 10.1105/tpc.15.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot et al. (2002).Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, Doerks & Bork (2012).Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Research. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2010).Li G, Peng F, Zhang L, Shi X, Wang Z. Cloning and characterization of a SnRK1-encoding gene from Malus hupehensis Rehd. and heterologous expression in tomato. Molecular Biology Reports. 2010;37:947–954. doi: 10.1007/s11033-009-9734-9. [DOI] [PubMed] [Google Scholar]
- Librado & Rozas (2009).Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Livak & Schmittgen (2001).Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lumba et al. (2012).Lumba S, Tsuchiya Y, Delmas F, Hezky J, Provart NJ, Lu QShi, McCourt P, Gazzarrini S. The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biology. 2012;10:8. doi: 10.1186/1741-7007-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2018).Ma N, Ma C, Liu Y, Shahid MO, Wang C, Gao J. Petal senescence: a hormone view. Journal of Experimental Botany. 2018;69:719–732. doi: 10.1093/jxb/ery009. [DOI] [PubMed] [Google Scholar]
- Mai, Wang & Yang (2011).Mai YX, Wang L, Yang HQ. A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis. Journal of Integrative Plant Biology. 2011;53:480–492. doi: 10.1111/j.1744-7909.2011.01050.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer et al. (2011).Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann, Klempien & Dudareva (2014).Muhlemann JK, Klempien A, Dudareva N. Floral volatiles: from biosynthesis to function. Plant, Cell & Environment. 2014;37:1936–1949. doi: 10.1111/pce.12314. [DOI] [PubMed] [Google Scholar]
- Niu et al. (2018).Niu L, Dong B, Song Z, Meng D. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica) International Journal of Molecular Sciences. 2018;19(7):2131. doi: 10.3390/ijms19072131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou & Nei (1987).Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai et al. (2013).Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T, Yamada Y. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant and Cell Physiology. 2013;54:e6–e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz et al. (2003).Schmelz EA, Engelberth J, Alborn HT, O’Donnell P, Sammons M, Toshima H, Tumlinson 3rd JH. Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10552–10557. doi: 10.1073/pnas.1633615100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2016).Tang J, Lin J, Li H, Li X, Yang Q, Cheng ZM, Chang Y. Characterization of CIPK family in asian pear (Pyrus bretschneideri Rehd) and co-expression analysis related to salt and osmotic stress responses. Frontiers in Plant Science. 2016;7:1361. doi: 10.3389/fpls.2016.01361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2020).Tang RJ, Wang C, Li K, Luan S. The CBL-CIPK Calcium signaling network: unified paradigm from 20 years of discoveries. Trends in Plant Science. 2020;25:604–617. doi: 10.1016/j.tplants.2020.01.009. [DOI] [PubMed] [Google Scholar]
- Tena, Boudsocq & Sheen (2011).Tena G, Boudsocq M, Sheen J. Protein kinase signaling networks in plant innate immunity. Current Opinion in Plant Biology. 2011;14:519–529. doi: 10.1016/j.pbi.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson et al. (1997).Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi et al. (2009).Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. The Plant Journal. 2009;58:778–790. doi: 10.1111/j.1365-313X.2009.03812.x. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2015).Wang L, Hu W, Sun J, Liang X, Yang X, Wei S, Wang X, Zhou Y, Xiao Q, Yang G, He G. Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Scientific Reports. 2015;237:33–45. doi: 10.1016/j.plantsci.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2019).Wang Y, Yan H, Qiu Z, Hu B, Zeng B, Zhong C, Fan C. Comprehensive analysis of SnRK gene family and their responses to salt stress in Eucalyptus grandis. International Journal of Molecular Sciences. 2019;20(11):2786. doi: 10.3389/fpls.2017.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu & Zhang (2015).Xu J, Zhang S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science. 2015;20:56–64. doi: 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Yoshida et al. (2006).Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. Journal of Biological Chemistry. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- Yuan et al. (2011).Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLOS ONE. 2011;6:e26468. doi: 10.1371/journal.pone.0026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Yu & Fan (2014).Yue Y, Yu R, Fan Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta. 2014;240:745–762. doi: 10.1007/s00425-014-2127-x. [DOI] [PubMed] [Google Scholar]
- Yue, Yu & Fan (2015).Yue Y, Yu R, Fan Y. Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genomics. 2015;16:470. doi: 10.1186/s12864-015-1653-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2020).Zhang Y, Ye Y, Jiang L, Lin Y. Genome-wide characterization of Snf1-Related Protein Kinases (SnRKs) and expression analysis of SnRK1.1 in strawberry. Gene. 2020;11(4):427. doi: 10.3390/genes11040427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2016).Zhu K, Chen F, Liu J, Chen X, Hewezi T, Cheng ZM. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Scientific Reports. 2016;6:28225. doi: 10.1038/srep28225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Sequences information of 20 SnRK Motifs.
Table S2:Ka/Ks ratios of paralogous HcSnRK genes.
Table S3: Cis-acting regulatory elements numbers found in the promoter region of HcSnRK genes.
Table S4: Gene IDs of SnRK genes from Arabidopsis and rice.
Table S5: List of primers used in the experimentations.
Data Availability Statement
The following information was supplied regarding data availability:
Raw data are available in the Supplemental Files.