Abstract
Bioluminescence imaging with luciferase-luciferin pairs is routinely used to monitor cellular functions. Multiple targets can be visualized in tandem using luciferases that process unique substrates, but only a handful of such orthogonal probes are known. Multiplexed studies require additional robust, light-emitting molecules. In this work, we report new luciferins for orthogonal imaging that comprise disubstituted cores. These probes were found to be bright emitters with various engineered luciferases. The unique patterns of light output also provided clues into enzyme-substrate interactions necessary for productive emission. Screening studies identified mutant luciferases that could preferentially process the disubstituted analogs, enabling orthogonal imaging with existing bioluminescent reporters. Further mutational analyses revealed the origins of substrate selectivity. Collectively, this work provides insights into luciferase-luciferin features relevant to bioluminescence and expands the number of probes for multicomponent tracking.
Graphical Abstract
INTRODUCTION
Bioluminescence imaging is a powerful technique for monitoring cellular events in live organisms.1–4 This technology features enzymes (luciferases) that catalyze the oxidation of small molecule substrates (luciferins), releasing visible light in the process. One of the most popular luciferase-luciferin pairs for in vivo imaging derives from the firefly: firefly luciferase (Fluc) and D-luciferin (D-luc, Figure 1A).5, 6 Fluc and D-luc can be introduced into a variety of cells, and the light produced can report on cell movements, proliferation, and other parameters.7–9 Since no excitation light is required, background signal is virtually nonexistent. Thus, bioluminescence can be preferred to traditional fluorescence imaging in tissues and whole organisms—environments where autofluorescence often diminishes sensitivity.2, 10, 11
Figure 1.
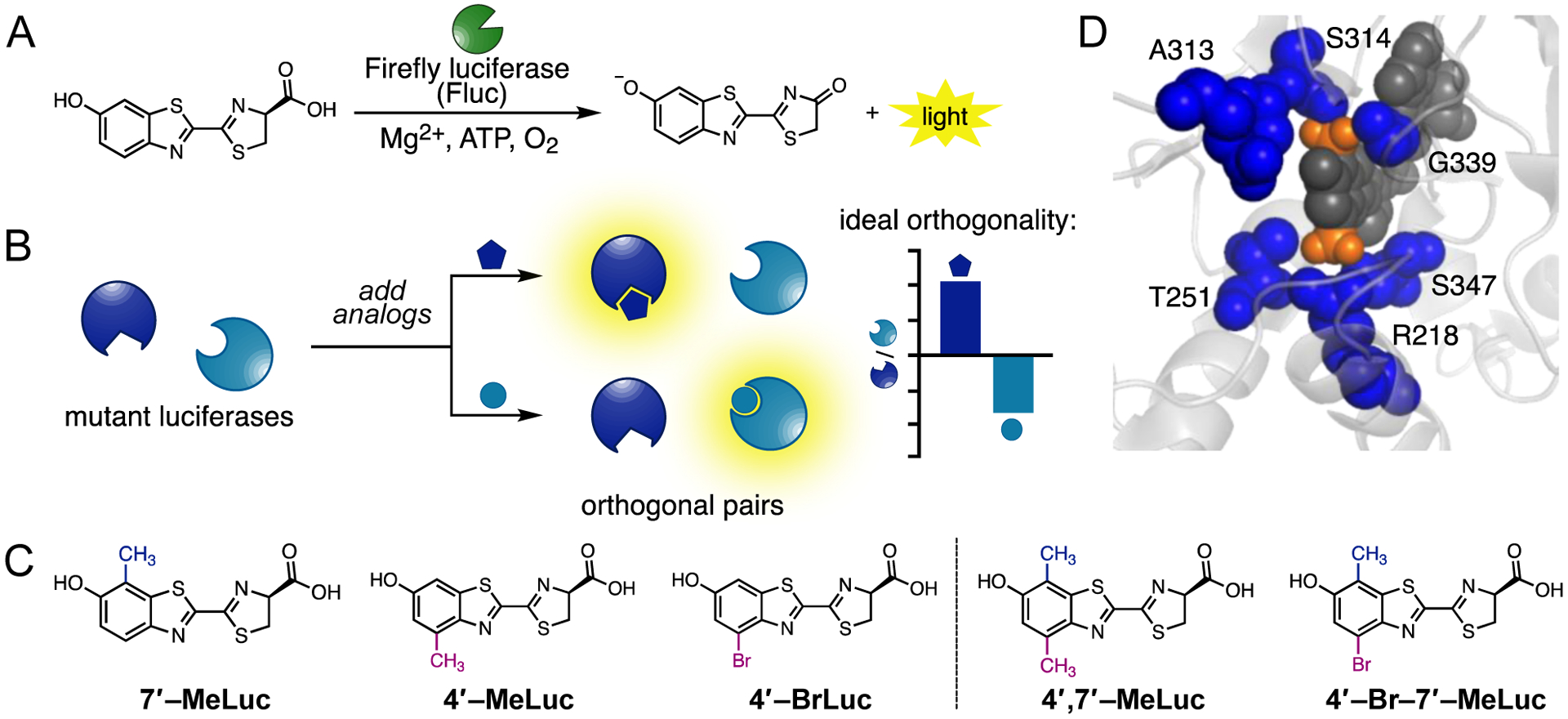
Multicomponent bioluminescence imaging with engineered luciferases and luciferins. (A) Fluc catalyzes the oxidation of D-luc, producing photons of light. (B) Mutant luciferases preferentially process sterically modified analogs. Orthogonal pairs comprise enzymes that are bright with one analog, while dim with another. Substrate preference can be graphically depicted as shown in the cartoon plot. (C) Disubstituted luciferins examined in this work (right), alongside the relevant singly modified analogs (left). (D) Overlay of 4’,7’-MeLuc (docked as AMP ester conjugate, dark gray) in the active site of Fluc (PDB: 4G36). Residues within 5 Å of the methyl substituents (orange) are shown in blue.
While versatile, bioluminescence has been largely limited to imaging only one or two targets at a time in heterogeneous environments.12 Several of the most popular luciferases use the same substrate, making them difficult to differentiate in vivo.6 Spectral resolution is possible,13–15 and red-shifted enzyme-substrate pairs have enabled two-component imaging in mice.15–18 Resolving multiple reporters by color alone remains challenging, though, due to the broad emission spectra of luciferases and the complexities associated with tissue absorption.19 To address the need for more unique and distinguishable probes, we and others have been developing orthogonal pairs: luciferin analogs that can be preferentially processed by engineered luciferases (Figure 1B).20–26 Orthogonal enzymes can be distinguished by substrate, even if they emit similar colors of light. The more structurally divergent the luciferases and luciferins, the more readily they can be differentiated.24
Accessing diverse luciferase and luciferin architectures is key to expanding the number of orthogonal probes. Inspiration can come from nature, where dozens of distinct light-emitting chemistries (and thus unique luciferase-luciferin pairs) have evolved. However, only a few naturally occurring bioluminescent pairs have been successfully re-purposed for imaging in mammalian hosts. The requisite luciferins are often difficult to isolate and characterize, and many are poorly bioavailable.6,9 Engineering efforts can provide more rapid access to orthogonal probes. Among the most fruitful approaches involves simultaneous modification of substrate architectures and enzyme active sites—a classic method for achieving selectivity.12,27, 28 Luciferase residues can be mutated to accommodate chemically distinct luciferins. Robust signal is thus observed when complementary enzymes and substrates interact. Parallel engineering of Fluc and D-luc, in particular, has greatly expanded the number of orthogonal tools for imaging in vitro and in vivo. There are now dozens of distinct D-luc analogs and complementary luciferases that can be used in cells and tissues. In most cases, perfect selectivity among the enzymes and substrates is not required for multicomponent bioluminescence.20 Rather, unique patterns of light emission (i.e., “barcodes”) are sufficient to resolve collections of luciferases and luciferins (Figure 1B).
While the number of orthogonal luciferases and luciferins has grown in recent years, most emit far less light than native bioluminescent tools. The probes can still be used to track bulk biological processes, but are less suitable for imaging rare events. Brighter, more structurally distinct luciferins are necessary for sensitive, multiplexed analyses. Unfortunately, there is often a tradeoff between orthogonality and light output. Our empirical observations suggest that large modifications to the luciferin core (e.g., morpholino appendages) are easier to discriminate, enabling rapid identification of distinct enzymes.20, 24 However, larger perturbations often present a barrier to efficient processing, resulting in poor turnover and low light production. Consequently, the most orthogonal probes are often orders of magnitude less bright than native bioluminescent pairs. Such drastic changes in light output can be difficult to recoup via traditional enzyme engineering.
The search for orthogonal probes would benefit from larger collections of diverse molecules that retain robust emission. Since luciferin modifications that boost orthogonality typically come at the expense of brightness, less perturbing substituents are often better starting points for enzyme engineering. In previous work, we identified luciferins that strike a balance between structural diversity (for orthogonality) and small size (for brightness). These molecules comprise bromo and methyl appendages at C4′ (e.g., 4’-BrLuc and e.g., 4’-MeLuc, respectively).24, 25 These analogs were uniquely processed by luciferases comprising mutations at residues 240, 247, and 347. Docking studies suggested that the engineered mutants harbored additional space for the luciferin appendages. The C4′-modified analogs were also readily differentiated from C7′-modified substrates, owing to their structurally divergent cores. The top orthogonal pairs emitted light on par with D-luc and Fluc and were amenable to orthogonal imaging in vitro, in cells, and even in whole animals.20, 24
We surmised that additional bright and orthogonal probes could be obtained by building on the success of the C4′ modified compounds and imparting additional minimal modifications to the ring. We were particularly attracted to the C7′ position, based on its synthetic accessibility and our previous success in installing alkyl groups at this site. Analogs with substituents at both C4′ and C7′ would likely be orthogonal to existing luciferins and could thus be used in tandem with several existing imaging tools, expanding the bioluminescent toolkit. Moreover, such disubstituted probes could provide valuable insight into enzyme-substrate interactions governing substrate selectivity.
Herein we show that two disubstituted luciferins are suitable probes for bioluminescence imaging and comprise a unique class of orthogonal substrates. The molecules were synthesized and found to be competent emitters in vitro and in cells. The luciferins were further screened with a collection of mutant luciferases, and enzymes that exhibited preference for the disubstituted analogs were identified. Importantly, the structural modifications to the luciferin core were subtle enough to maintain brightness, yet distinct enough to achieve orthogonality. The hybrid molecules could be differentiated from their singly modified counterparts and even the native substrate, D-luc. Biochemical analyses revealed the luciferase residues responsible for preferential substrate processing. Surprisingly, only a few mutations contributed to overall selectivity. Our results suggest that novel bioluminescent reactivity can be achieved by combining minimal structural perturbations to the luciferin core.
EXPERIMENTAL DETAILS
Structural analyses of luciferin analogs in the active sites of Fluc (PDB:4G36) and related mutant active sites were performed using AutoDock Vina.29 Mutant luciferases were generated using RosettaBackrub.30 The enzyme structures were prepared for docking by removing water molecules and co-crystallized ligands. The luciferase (Fluc and mutant) and luciferin analog input files were generated using AutoDock Tools.31 All nonpolar hydrogens were removed from the structures prior to docking. Residues in the Fluc and mutant active sites were kept rigid, and grid coordinates were generated to define the sampling space for conformational analysis (Table S1). Rotational and torsional degrees of freedom for the minimized ligands were defined in AutoDock Tools. Additional details are provided in the Supporting Information file.
RESULTS AND DISCUSSION
Design and synthesis of disubstituted luciferins.
We and others have shown that C4′-modified variants of D-luc can be processed by Fluc to release light.6, 32, 33 Compounds with a methyl (4′-MeLuc) or bromo substituent (4′-BrLuc, Figure 1C) are particularly robust and biocompatible, and rank among the brightest sterically modified luciferin analogs.20, 24 The bromo and methyl substituents do not drastically alter the electronic configuration of the luciferin and are straightforward to install, making them good starting points for bioluminescent probe development.22 In addition, both 4′-MeLuc and 4′-BrLuc can be readily discriminated from analogs bearing substituents at C7′ (e.g., 7′-MeLuc) using engineered luciferases. Indeed, pairs of C4′ and C7′-modified luciferins (and their cognate enzymes) have been used for multicomponent imaging both in vitro and in vivo.20, 24, While the origins of selectivity remain unknown, biochemical data suggest that mutants capable of processing 4′-MeLuc and 4′-BrLuc harbor modifications proximal to the C4′ position of bound substrates. Such mutations likely accommodate the extra bulk on the substrate.24
Given the success of C4′-modified compounds in bioluminescence imaging, we aimed to build on these scaffolds and expand the collection of orthogonal probes. We reasoned that appending another small group to the luciferin core would provide added structural diversity, without sacrificing brightness. We were particularly interested in installing a methyl substituent at C7′. C7′-Methyl modifications are synthetically tractable and biocompatible, and on their own, do not severely impede enzyme processing.20 To determine whether the combination of dual C4′- and C7′-substituents could boost orthogonality while maintaining robust emission, we set out to produce a new class of disubstituted bioluminescent emitters, with 4′,7′-MeLuc and 4′-Br-7′-MeLuc as the initial targets. Docking studies suggested that the analogs could be reasonably accommodated in the Fluc active site (Figures 1D and S1). Potential sites of steric clashing were observed, but such interactions would likely be advantageous for orthogonal luciferase development. Enzyme engineering can be used to overcome steric penalties and provide more substrate-specific luciferases.20, 34, 35
The disubstituted analogs were prepared using a general method previously reported by our laboratory.36, 37 This route features functionalized anilines and Appel’s salt (blue, Scheme 1) to access key cyanobenzothiazole intermediates. Cyanobenzothiazoles can be readily condensed with D-cysteine to afford luciferin architectures.21, 22, 35, 37 Following this overall approach, we began the synthesis of 4′,7′-MeLuc with commercially available 2,5-dimethyl-4-nitrophenol (1). Treatment of phenol 1 with acetic anhydride followed by palladium-catalyzed hydrogenation afforded aniline 3 (Scheme 1). Compound 3 was then incubated with Appel’s salt (4) to form dithiazole adduct 5. Thermolysis of 5 provided the key cyanobenzothiazole intermediate (6), albeit in small amounts. The low yield was likely due to the electron-deficient core of dithiazole 5. More electron-rich scaffolds (e.g., anisoles) are typically better substrates for the cyclization reaction.37 Indeed, substituting a methoxy protecting group for the acetyl unit boosted the overall yield of the thermolysis reaction (Scheme S1).
Scheme 1.
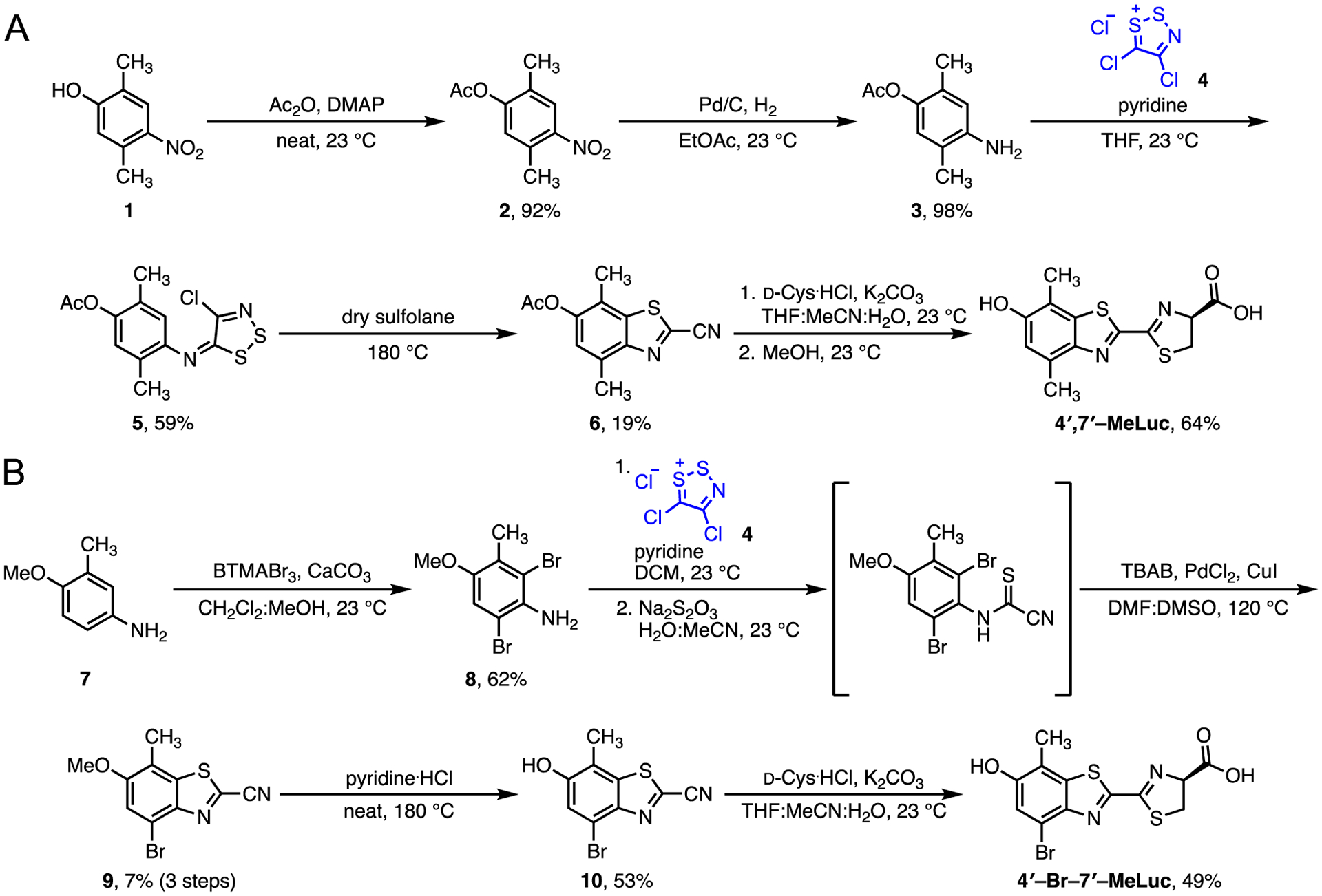
Synthesis of disubstituted luciferin analogs (A) 4′,7′-MeLuc and (B) 4′-Br-7′-MeLuc
The cyanobenzothiazole en route to 4′-Br-7′-MeLuc was synthesized using a similar approach. Aniline 7 was first reacted with benzyltrimethylammonium tribromide to provide dibrominated intermediate 8. The corresponding dithiazole adduct was then formed and fragmented in a single pot. The cyanothioformamide intermediate was prone to cyclization upon isolation, so the product was taken on crude. Palladium-mediated cyclization provided two regiosiomeric cyanobenzothiazoles that were separable by chromatography. The isomers were subjected to palladium-catalyzed dehalogenation to aid in structural assignment (Figure S2). The desired intermediate 9 was then deprotected to afford the corresponding cyanobenzothiazole 10. Intermediates 10 and 6 were both ultimately condensed with D-cysteine to afford the disubstituted luciferins, 4′,7′-MeLuc and 4′-Br-7′-MeLuc.
Disubstituted analogs are functional light emitters.
With the new analogs in hand, we first measured their light outputs with native Fluc. Luciferin scaffolds that retain some level of activity with the enzyme are desirable, as a basal level of light emission is a necessary entry point for subsequent engineering. Light production with Fluc (even if minimal) also provides assurance that the molecules are capable of photon production. The disubstituted probes 4′-Br-7′-MeLuc and 4′,7′-MeLuc were incubated with recombinant Fluc and ATP, and photon production was measured over time. Dose-dependent light emission was observed over a range of concentrations (Figure 2A). The apparent Km values for both analogs were on par with the native substrate (D-luc, Table S2) and analogs bearing either a single methyl (7′-MeLuc) or bromo substituent (4′-BrLuc).20, 38 These results were encouraging, as luciferins with larger modifications at either C4′ or C7′ typically exhibit increased Km values (>100 μM) and correspondingly poor light outputs.20 The emission intensity for 4′-Br-7′-MeLuc was also 10-fold higher than 4′,7′-MeLuc, likely due to the more electron-rich nature of the scaffold.22
Figure 2.
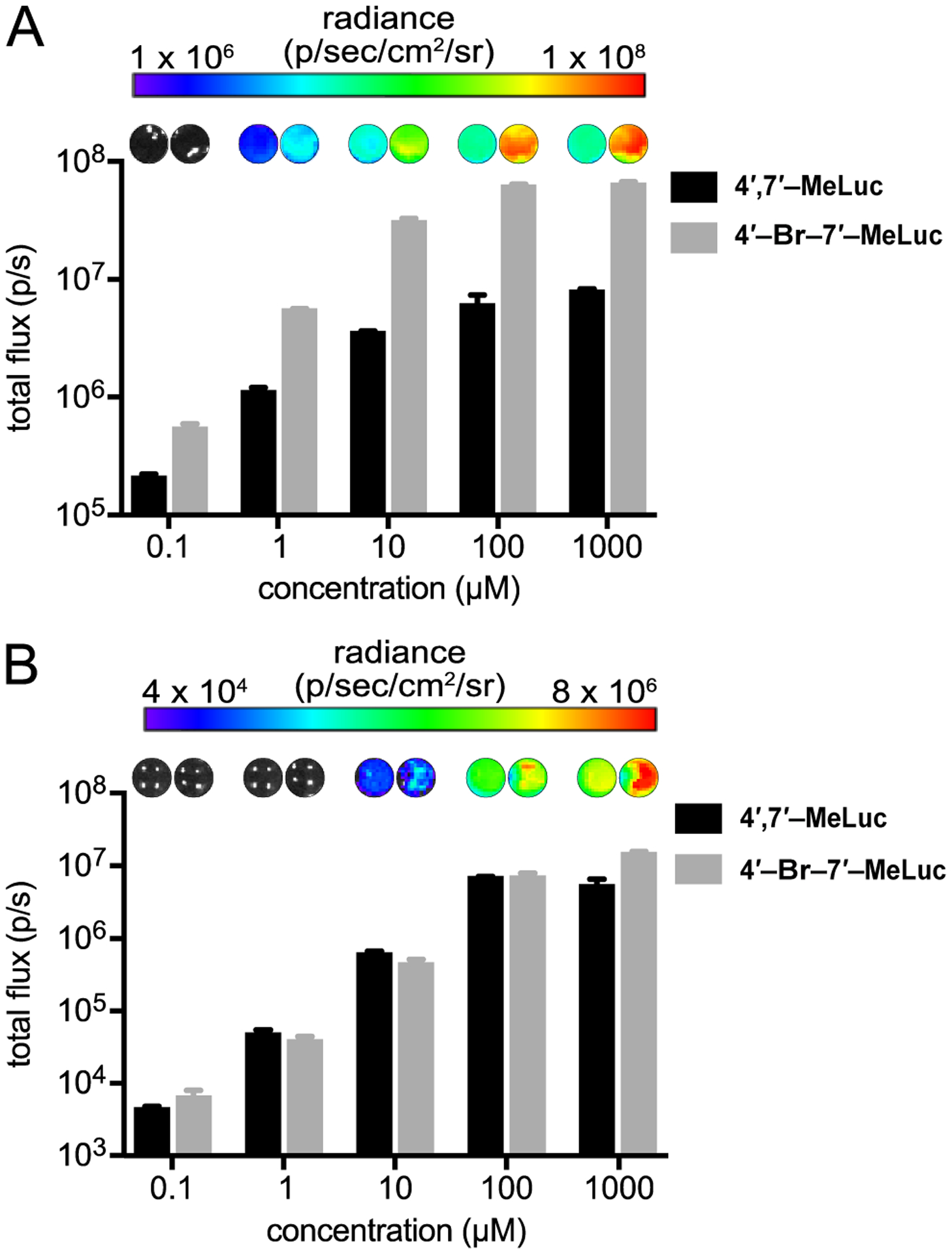
Disubstituted analogs produce light with Fluc. Analogs (0–1 mM) were incubated with (A) recombinant Fluc or (B) Fluc-expressing HEK293 cells. For (B), photon outputs were monitored over time and steady state emission values (t = 25 min) are shown. For (A)-(B), emission intensities are plotted as photon flux values on log scales. Error bars represent the standard error of the mean for n ≥ 3 experiments.
We also examined the disubstituted compounds in cultured cell assays to gauge their relative biocompatibility and cell permeability. When 4′,7′-MeLuc and 4′-Br-7′-MeLuc were incubated with Fluc-expressing HEK293 cells, photon production was observed (Figures 2B and S3). The analogs were well tolerated by cells at saturating doses (Figure S4). Light emission from the disubstituted compounds was ~10 to 100-fold reduced compared to D-luc and relevant singly modified scaffolds, including 4′-BrLuc.20, 22 However, the photon outputs were on par with those of other luciferin analogs employed in cells,20 suggesting that the disubstituted probes were in a useful range for imaging and good candidates for further engineering. Interestingly, the two disubstituted luciferins produced similar levels of light in cells. This result implied that 4′,7′-MeLuc is more cell permeable than 4′-Br-7′-MeLuc, as the latter luciferin generated more light in enzymatic assays.
Screen for robust and orthogonal luciferases.
We next aimed to identify enzymes that could selectively process the disubstituted analogs, enabling them to be easily discriminated from D-luc and other robust-emitting architectures (e.g., 4′-BrLuc and 7′-MeLuc). As noted above, collections of bright, orthogonal probes are rare. We previously developed a general approach to identifying orthogonal luciferase-luciferin pairs that relies on screening functional luciferases.20, 24 Enzyme-substrate pairs are designated as orthogonal if robust reactivity is observed when complementary partners interact, but diminished reactivity is observed in all other cases. It is important to note that perfect selectivity is not required for orthogonal imaging. For example, enzyme-substrate pairs that exhibit ten-fold differences in selectivity can be readily discriminated in multicomponent assays.39
To identify luciferases that could preferentially process the disubstituted analogs, we screened 4′-Br-7′-MeLuc and 4′,7′-MeLuc against a collection of 222 characterized mutants.24 This library includes enzymes with ~1–8 mutations per sequence (spread over 23 distinct sites) near the luciferin-binding site. Enzymes from the library were distributed across 96-well plates and incubated with the disubstituted analogs. Fluc was also included as a control. As shown in Figure 3A, a range of photon outputs was observed upon analog addition. The majority of mutant luciferases produced less light with the disubstituted probes compared to Fluc, although a few clusters of robust emitting pairs were observed. Mutants that were active with 4′,7′-MeLuc also appeared to be less bright with 4′-Br-7′-MeLuc, consistent with the in vitro results.
Figure 3.
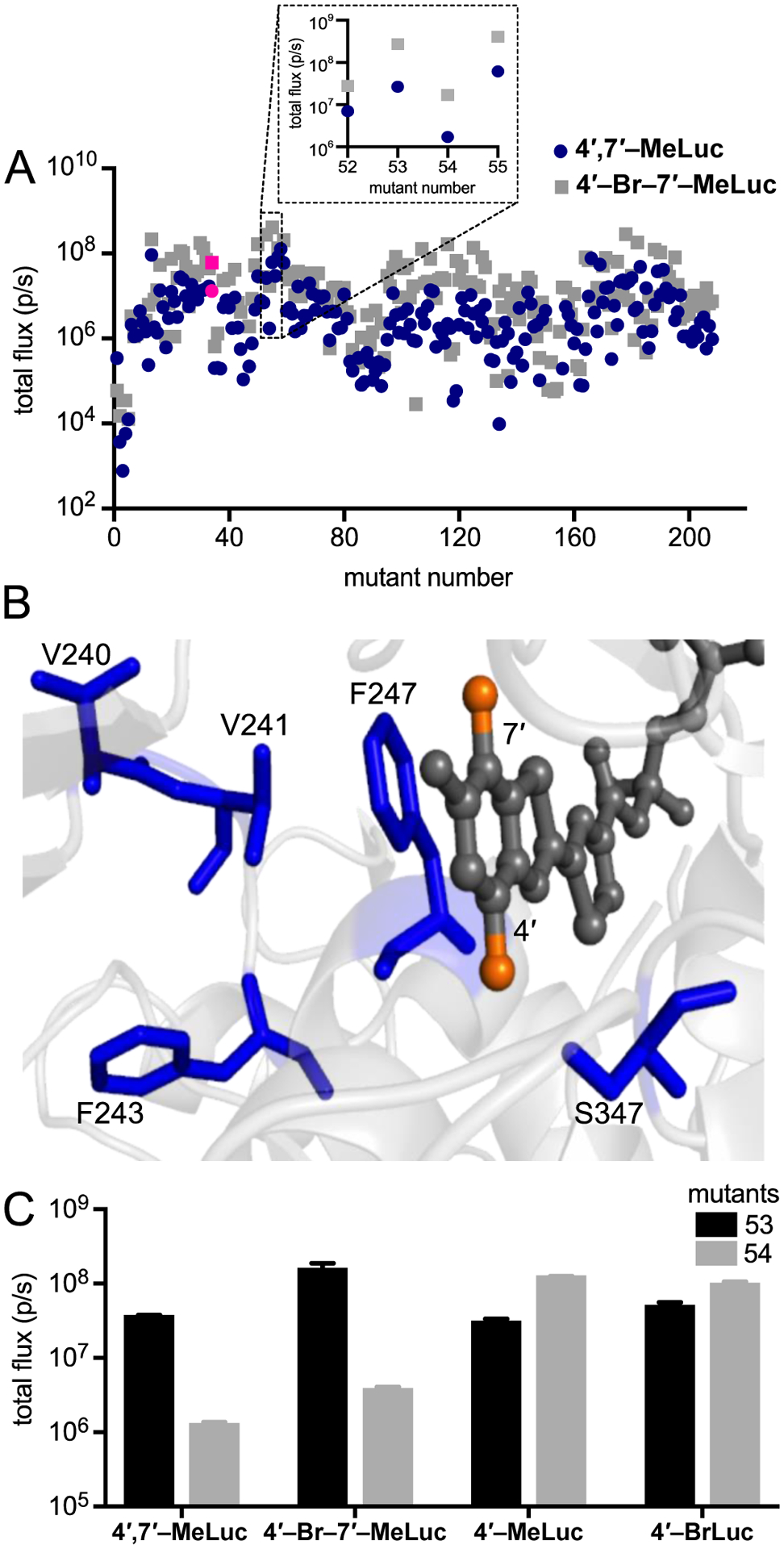
Light emission of luciferin analogs incubated with a panel of mutant luciferases. (A) Mutant luciferase-expressing bacteria were lysed, plated in 96-well black plates, and incubated with 4′,7′-MeLuc (blue circles, 250 μM) or 4′-Br-7′-MeLuc (gray squares, 250 μM). Fluc-expressing bacteria were included in the screen, and the corresponding light emission values are highlighted in pink. The inset highlights a subset of mutants further analyzed in this work. (B) Residues of interest for orthogonal probe development. Analog 4′,7′-MeLuc is shown (as AMP ester conjugate, dark gray) bound in the Fluc active site (PDB: 4G36). The methyl groups are colored orange, while the sites targeted in mutants 53 and 54 are shown in blue. (C) Light emission from a subset of mutants and analogs. Saturating doses of 4′,7′-MeLuc (250 μM), 4′-Br-7′-MeLuc (250 μM), 4′-MeLuc (100 μM), or 4′-BrLuc (100 μM) were incubated with lysed bacterial cells expressing mutants 53 or 54. For each replicate in (A) and (C), a single lysate was split among the wells and treated with the various analogs. Emission intensities are shown as photon flux values on log scales. Error bars represent the standard error of the mean for n ≥ 3 experiments.
The luciferases that produced the most light with the disubstituted analogs were clustered in wells 52–55. These enzymes harbored mutations at residues V240, V241, F243, F247, and S347 (Figure 3B). F247 and S347 are known to modulate the binding and light emission of the native substrate, D-luc. Moreover, mutations at these positions have been shown to disrupt D-luc processing and promote the utilization of substrate analogs.38, 40 Residues 240, 241, and 243 have been less extensively characterized in terms of their effects on light emission or substrate selectivity. These residues are proximal to key amino acids in the active site, though, and can potentially alter substrate recognition. For example, V241 is located near F247 in the luciferin binding site and likely influences the positioning (and pi-stacking ability) of F247. Bulkier residues at V241 have also been speculated to force F247 closer to the benzothiazole ring of D-luc.41 Smaller residues at this position could presumably open up more space for luciferin analogs.
We were particularly drawn to two mutants from the screening analysis: mutant 53 (V240I, F243M, F247Y, and S347G) and mutant 54 (V241A, F247L, S347A, Figure S5). These enzymes harbored mutations in similar regions of the luciferin-binding pocket, but exhibited drastically different outcomes relevant to orthogonal substrate usage. Mutant 53 was previously shown to selectively process 4′-BrLuc over C7′-modified luciferins, enabling two-component imaging both in cells and in vivo.24 When mutant 53 was treated with disubstituted analogs 4′,7′-MeLuc and 4′-Br-7′-MeLuc, similar levels of robust light emission were observed (Figure 3C). Previous docking studies suggested that the mutations present in mutant 53 (e.g. S347G) create space for bulky substituents (e.g., C4′-bromo groups).24 Such mutations might be indispensable for processing C4′-brominated or methylated compounds, regardless of the C7′ substitutent.24, 25 Mutant 53 did not process all C4′-modified luciferins to a similar extent, though, as shown in Figures 4 and S6. When luciferins bearing larger C4′ appendages were screened, light emission decreased as the substituent size increased. The disubstituted analogs were preferentially processed compared to a range of singly modified compounds (indicated by the upward black bars in Figures 4 and S6). They were also preferred over C7′ compounds and D-luc, suggesting that they might be useful orthogonal probes in combination with these known bright-emitting tools.
Figure 4.
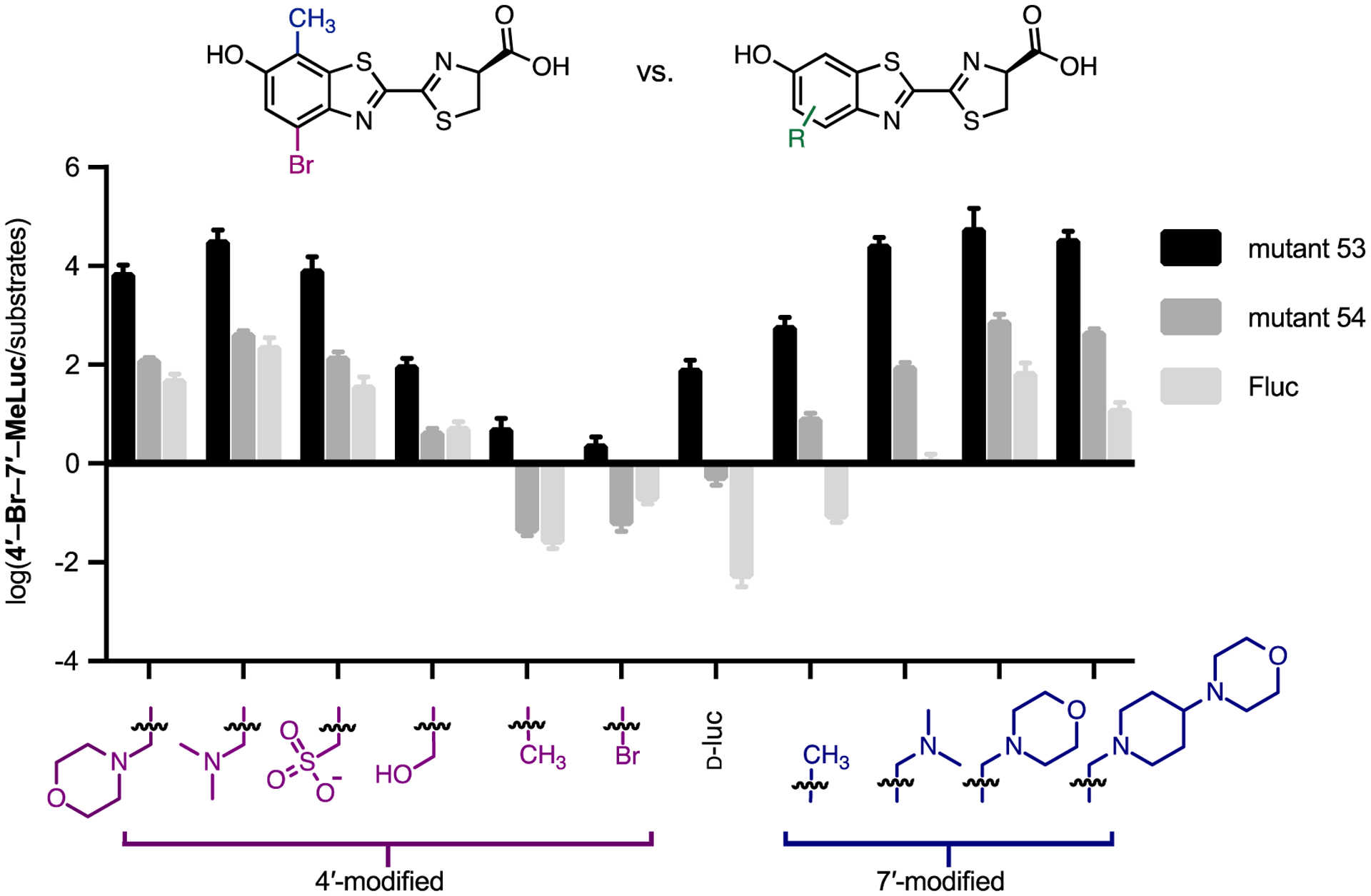
Orthogonal substrate usage observed between mutants 53 and 54. Mutant 53 produces more light with disubstituted luciferins than singly modified analogs. Bacteria expressing mutant 53, mutant 54, or Fluc were lysed and incubated with 4’-Br-7’-MeLuc or a singly modified luciferin at saturating doses (100–250 μM). The light output for each enzyme-substrate reaction was measured. Comparative emission values are plotted as the ratio of disubstituted luciferin activity over the activity of each substrate shown, on a log scale. Error bars represent the standard error of the mean for n ≥ 3 experiments. For each replicate, a single lysate was split among three wells and treated with the various analogs.
Mutant 54 was also previously shown to selectively process C4′-modified luciferins, including 4′-BrLuc and 4′-MeLuc.24 This enzyme was similarly hypothesized to create space for C4′ modifications. However, unlike mutant 53, light emission was diminished (>50-fold) when mutant 54 was treated with the disubstituted analogs. As shown in Figures 4 and S7, 4′-BrLuc, 4′-MeLuc, and D-luc all emitted more light with mutant 54 than the disubstituted analogs (indicated by the downward gray bars). Similar trends in substrate use were observed at physiological temperatures (Figure S7). We were motivated to pursue these results further, both to learn more about the features governing enzyme-substrate specificity and to exploit the reactivity differences for orthogonal pair development (vide infra).
Mutational origin of selectivity for disubstituted luciferins.
The initial enzyme screen implicated luciferase residues responsible for orthogonal reactivity between the di- and mono-substituted luciferins. To examine their impacts in more detail, a library was constructed comprising mutations from both mutants 53 and 54 (Figure 5A). Each site was allowed to code for either the native or mutated residue. The final library was constructed using synthetic gene assembly, followed by circular polymerase extension cloning (CPEC).42, 43 The library was transformed into bacteria and screened in two batches: (1) with 4′,7′-MeLuc, 4′-MeLuc, and D-luc, and (2) 4′-Br-7′-MeLuc, 4′-BrLuc, and D-luc. The native luciferin (D-luc) was included in both screens to normalize the data sets and examine whether similar patterns of selectivity emerged. The enzymes were distributed across 96-well plates and incubated with the various analogs. Clear differences in photon output were observed when the luciferins were incubated with different library members (Figures 5B and S8).
Figure 5.
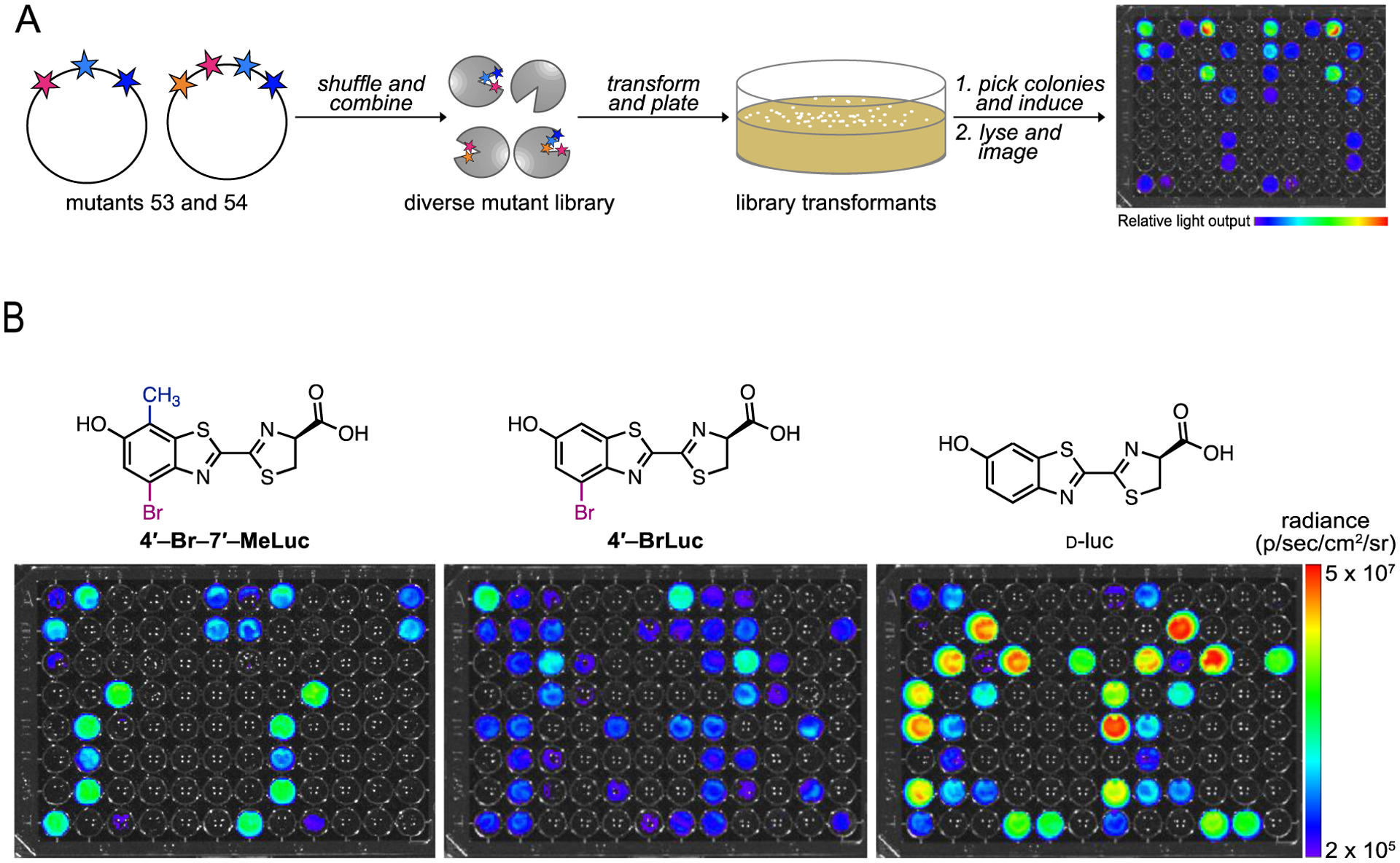
Screen to identify key residues involved in analog selectivity. (A) Library screening workflow. (B) Sample bioluminescence images from library screens with 4′-Br-7′-MeLuc, 4′-BrLuc, and D-luc. Selective colonies were selected for sequencing analysis. For each replicate, a single lysate was split among the wells and treated with the compounds shown.
We analyzed the frequency of mutated residues that correlated with preferential luciferin processing. The top 1–10% of mutants that preferred each compound were analyzed via Sanger sequencing (Tables S4–11). We observed sequence convergence among mutants that exhibited a 10-fold or greater preference for the disubstituted analogs over D-luc. These mutants comprised F247Y and S347G/A (Figure 6). These same mutations were present in mutants that preferred the disubstituted scaffold 4′,7′-MeLuc over 4′-MeLuc, although they were observed less frequently. Mutants that preferred D-luc or the singly modified luciferins 4′-MeLuc, and 4′-BrLuc primarily comprised a single mutation (F247L) along with S347 (the native residue). Sequencing results suggested that V240, V241, and F243 contribute little to analog selectivity, since they had minimal impact on activity. Overall, these results are in accord with previous observations relevant to luciferin processing. F247 and S347 are known to impact the binding of D-luc and various analogs.21, 38, 40 We’ve also previously observed that S347G appears to favor analogs with C4′ substituents, likely by creating additional space in the active site.24
Figure 6.
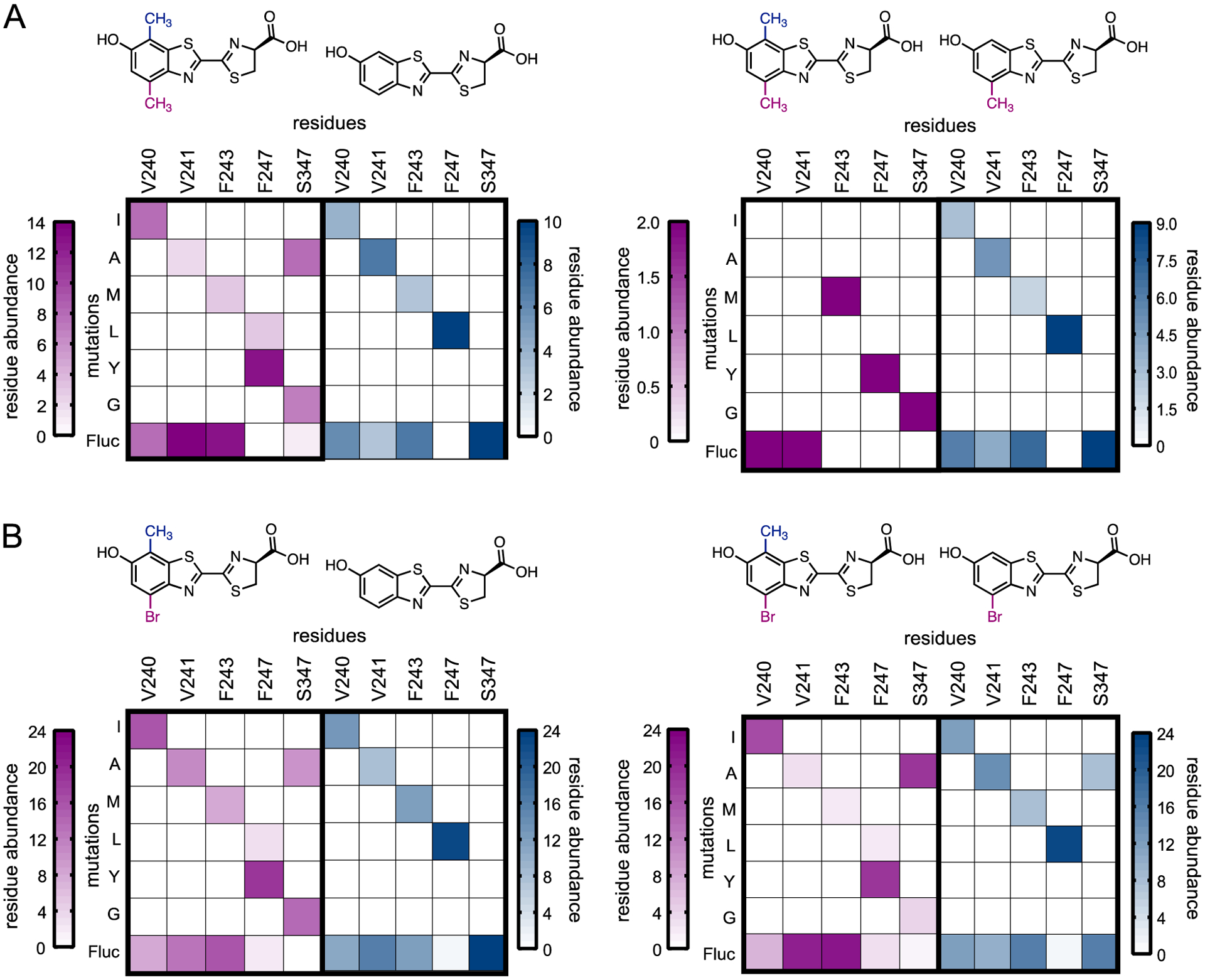
Substrate selectivity of frequently occurring mutants. (A) Frequency of residues observed in mutants that prefer 4’,7’-MeLuc over D-luc and 4’-MeLuc, and vice versa. (B) Frequency of residues observed in mutants that prefer 4’-Br-7’-MeLuc over D-luc and 4’-BrLuc, and vice versa. The pink heat maps represent mutants that preferred 4’,7’-MeLuc or 4’-Br-7’-MeLuc, while the blue heat map displays mutants that preferred either D-luc, 4’-BrLuc or 4’-MeLuc.
The library screens suggested that F247Y and S347G/A were the key drivers of selective processing for disubstituted analogs 4′,7′-MeLuc and 4′-Br-7′-MeLuc. However, it was unclear whether both mutations were necessary, or the extent to which the mutations were influencing selectivity. To uncover the roles of the specific mutations and gain additional sequence-function information, we created single point mutants (F247Y, S347G/A) and evaluated their selectivities for various analogs. Additional point mutants (V240I, V241A, F243M, F247L) and double mutants (F247Y/S347G, F247Y/S347A, F243M/S347G) were included in the analyses, along with mutants from the initial screen (mutants 53–54, and Fluc).
The mutants were expressed and incubated with the hybrid analogs, the singly modified compounds (4′-BrLuc, 4′-MeLuc) or D-luc, and light emission was measured (Figures S9–11). Not surprisingly, single mutants comprising V240I, V241A, or F243M were ~10–100-fold less selective for the disubstituted analogs compared to other luciferins. These results corroborate our earlier screening data, where variation at sites 240, 241, and 243 was observed. Interestingly, mutant F247Y preferentially processed the singly modified luciferins and D-luc over the disubstituted analogs. Only when this mutation was combined with S347G/A did preference for the hybrid analogs begin emerge. The F247Y/S347G double mutant was particularly selective for the hybrid luciferins over the native luciferin, D-luc. The selectivity was on par with the parental enzyme identified from the initial screen (mutant 53), confirming that the other residues (V240I and F243M) do not contribute substantially to orthogonal substrate usage. Biochemical analyses further revealed that the F247Y/S347G double mutant exhibited higher affinity for the disubstituted analogs compared to D-luc (Table S3/Figures S12–13).
Disubstituted analogs are orthogonal to existing probes.
The residue analyses revealed combinations of probes that would be useful for orthogonal bioluminescence imaging. As mentioned above, cell-compatible and robust architectures for multicomponent imaging have been difficult to identify. Combinations of enzymes and substrates must be minimally cross-reactive and, ideally, exhibit bright emission. Compatibility with native Fluc/D-luc is also desirable, due to the popularity and availability of this reporter pair.
Our data indicated that the disubstituted luciferins would be good candidates for orthogonal imaging, in combination with Fluc/D-luc and other robust sets. Mutant F247Y/S347G was selective for the hybrid analogs, while Fluc preferentially processed D-luc. To assess whether they could be used as orthogonal pairs, the luciferins were incubated with mutant F247Y/S347G or Fluc in bacterial cell lysate. Photon outputs were measured and orthogonality was assessed (Figure 7A). F247Y/S347G exhibited a >10-fold preference for the disubstituted analogs, while Fluc exhibited >100-fold selectivity for D-luc. Such fold differentials are sufficient for orthogonal imaging.39
Figure 7.
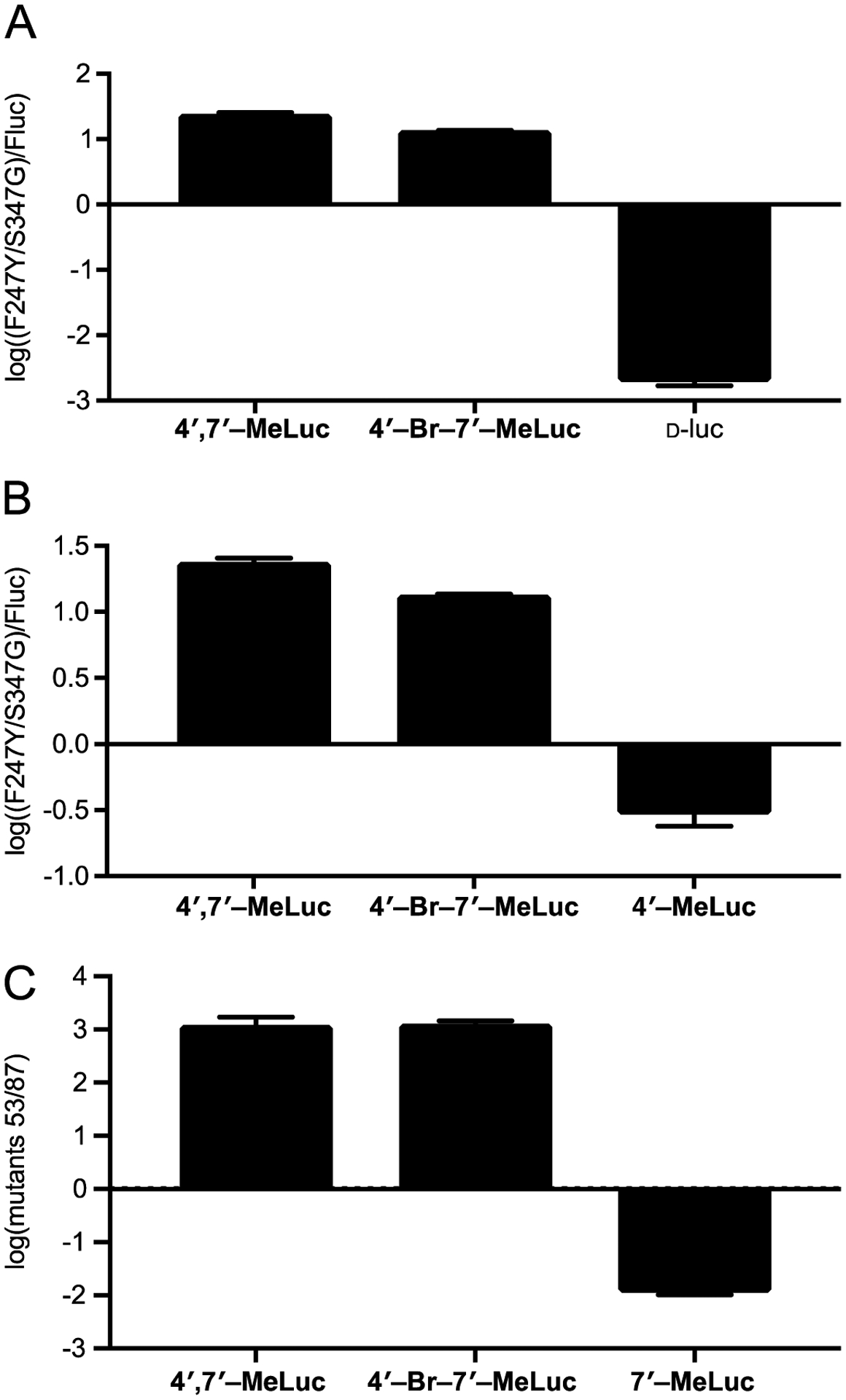
Disubstituted luciferins are orthogonal to robust light-emitting luciferins, including (A) D-luc, (B) 4′-MeLuc, and (C) 7′-MeLuc. Mutants F247Y/S347G and 53 selectively processes the disubstituted analogs, while Fluc and mutant 87 prefer other luciferins. Bacterial cells expressing (A)-(B) mutant F247Y/S347G or Fluc or (C) mutant 53 or 87 were lysed and plated over a 96-well plate. Comparative emission values are plotted as the ratio of one luciferase over the other, on a log scale, for each substrate. Error bars represent the standard error of the mean for n ≥ 3 experiments. For each replicate, a single lysate was split among the wells and treated with each analog.
We also examined whether 4′,7′-MeLuc and 4′-Br-7′-MeLuc could be used in combination with monosubstituted luciferins, including 4′-BrLuc, 4′-MeLuc, and 7′-MeLuc. As shown in Figures 7B and S14, the disubstituted probes were less orthogonal to 4′-MeLuc and 4′-BrLuc. Cross-reactivities among the compounds and complementary enzymes resulted in diminished orthogonality. Since the disubstituted analogs exhibited similar enzyme preferences as singly C4′-modified luciferins, we surmised that they might be more readily differentiated from analogs comprising a single C7′ appendage. To test this hypothesis, we selected luciferase mutants 53 and 54, in addition to mutant 87 (R218K, F250Y, S314T, S316T) for analysis. Mutants 53 and 54 were previously shown to be selective for C4′-modified luciferins, while mutant 87 is known to preferentially process 7′-MeLuc (Figure S15).24 The three mutants were individually expressed and then incubated with 4′,7′-MeLuc, 4′-Br-7′-MeLuc, or 7′-MeLuc. Light outputs were measured for all enzyme-substrate combinations. Excitingly, both disubstituted luciferins were orthogonal to 7′-MeLuc, with >10-fold differences in light output observed with the relevant luciferases (Figures 7C and S16). These results indicate that both 4′,7′-MeLuc and 4′-Br-7′-MeLuc are suitable for multicomponent imaging with existing probe sets.
Conclusions.
Multicomponent bioluminescence imaging requires collections of bright and structurally diverse luciferins. Analogs with more subtle structural modifications are typically better tolerated by luciferase enzymes (and provide more photons), but remain difficult to differentiate from other light-emitting substrates. Luciferin features most beneficial for orthogonality (e.g., large modifications to the core) are often detrimental to enzyme processing. Consequently, the most substrate-specific probes tend to give off the least light.
In this work, new luciferins were constructed by combining minimal structural components of singly modified analogs to create disubstituted versions. These luciferins were viable light emitters with Fluc, enabling orthogonal probe development via parallel engineering. Screening of the analogs with a collection of mutants revealed luciferases that could efficiently process the disubstituted analogs. We further identified mutants that could differentiate the hybrids from D-luciferin and monosubstituted analogs. To determine the enzyme residues responsible for selectivity, we evaluated a panel of individual mutants with various luciferin probes. Light emission data revealed unique patterns of reactivity for the disubstituted luciferins, with residues F247Y and S347G/A underlying substrate preference.
Our results provide further evidence that subtle changes to luciferins and luciferases can elicit sufficient orthogonality. Using minimally modified scaffolds, we were able to identify enzymes that preferentially processed disubstituted analogs over related architectures. Only a handful of luciferase mutations contributed to the overall selectivity. The hybrid probes reported here can be readily applied to multicomponent imaging and add to the growing number of bioluminescent tools. This work also informs on how to design robust and substrate-selective imaging agents via parallel engineering. Further applications of the method will expand the number of probes that can be used in tandem.
Supplementary Material
ACKNOWLEDGMENTS
We thank Krysten Jones for providing mammalian cell lines, along with Zi Yao and Carly Brennan for assisting with compound screening and bioluminescence kinetics measurements, respectively. We also thank members of the Weiss and Martin laboratories for reagents and equipment. Last, we thank members of the Prescher laboratory for helpful discussions and manuscript edits.
Funding Sources
This work was supported by the U. S. National Institutes of Health (R01GM107630 to J.A.P.). S.J.W. was supported by the National Science Foundation via the BEST IGERT program (DGE-1144901) and Graduate Research Fellowship under Grant No. DGE-1839285. C.S.H. was supported by the University of California, Irvine Undergraduate Research Opportunities Program (UROP) Fellowship under Grant No. GF10968.
ABBREVIATIONS
- D-luc
D-luciferin
- Fluc
Firefly luciferase
Footnotes
The following files are available free of charge.
Supporting Methods, Schemes S1, Figures S1–14, and Tables S1–10 (PDF)
The authors declare no competing financial interest.
References
- [1].Prescher JA, Contag CH (2010) Guided by the light: visualizing biomolecular processes in living animals with bioluminescence, Curr. Opin. Chem. Biol 14, 80–89. [DOI] [PubMed] [Google Scholar]
- [2].Thorne N, Inglese J, Auld DS . (2010) Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology, Chem. Biol 17, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Paley MA, Prescher JA (2014) Bioluminescence: a versatile technique for imaging cellular and molecular features, MedChemComm 5, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yeh H-W, Ai H-W (2019) Development and applications of bioluminescent and chemiluminescent reporters and biosensors, Annu. Rev. Anal. Chem 12, 129–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Contag CH, Spilman SD, Contag PR, Oshiro M, Eames B, Dennery P, Stevenson DK, Benaron DA . (1997) Visualizing gene expression in living mammals using a bioluminescent reporter, Photochem. Photobiol 66, 523–531. [DOI] [PubMed] [Google Scholar]
- [6].Rathbun CM, Prescher JA (2017) Bioluminescent probes for imaging biology beyond the culture dish, Biochemistry 56, 5178–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kim J, Urban K, Cochran E, Lee S, Ang A, Rice B, Bata A, Campbell K, Coffee R, Gorodinsky A, Lu Z, Zhou H, Kishimoto TK, Lassota P (2010) Non-invasive detection of a small number of bioluminescent cancer cells in vivo, PLOS ONE 5, e9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fraga H (2008) Firefly luminescence: a historical perspective and recent developments, Photochem. Photobiol. Sci 7, 146–158. [DOI] [PubMed] [Google Scholar]
- [9].Viviani VR (2002) The origin, diversity, and structure function relationships of insect luciferases, Cell Mol. Life Sci 59, 1833–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rumyantsev KA, Turoverov KK, Verkhusha VV . (2016) Near-infrared bioluminescent proteins for two-color multimodal imaging, Sci. Rep 6, 36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Troy T, Jekic-McMullen D, Sambucetti L, Rice B (2004) Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models, Mol. Imaging 3, 9–23. [DOI] [PubMed] [Google Scholar]
- [12].Williams SJ, Prescher JA (2019) Building biological flashlights: orthogonal luciferases and luciferins for in vivo imaging, Acc. Chem. Res 52, 3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Branchini BR, Southworth TL, Fontaine DM, Kohrt D, Florentine CM, and Grossel MJ . (2018) A firefly luciferase dual color bioluminescence reporter assay using two substrates to simultaneously monitor two gene expression events, Sci. Rep 8, 5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Masahiro N, and Nagai T (2016) Five color variants of bright luminescent protein for real-time multicolor bioimaging, Nat. Commun 7, 13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mezzanotte L, Qye I, Kaijzel E, Branchini B, Roda A, and Löwik C (2011) Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase, PLoS ONE 6, e19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zambito G, Hall MP, Wood MG, Gaspar N, Ridwan Y, Stellari FF, Shi C, Kirkland TA, Encell LP, Löwik C, Mezzanotte L (2020) Red-shifted click beetle luciferase mutant expands the multicolor bioluminescence palette dor deep tissue imaging, iScience 24, 101986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stowe CL, Burley TA, Allan H, Vinci M, Kramer-Marek G, Ciobota DM, Parkinson GN, Southworth TL, Agliardi G, Hotblack A, Lythgoe MF, Branchini BR, Kalber TL, Anderson JC, and Pule MA (2019) Near-infrared dual bioluminescence imaging in mouse models of cancer using infraluciferin, eLife 8, e45801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aswendt M, Vogel S, Schäfer C, Jathoul A, Pule M, and Hoehn M (2019) Quantitative in vivo dual-color bioluminescence imaging in the mouse brain. Neurophotonics, 6, 025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH (2005) Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo, J. Biomed. Opt 10, 041210. [DOI] [PubMed] [Google Scholar]
- [20].Jones KA, Porterfield WB, Rathbun CM, McCutcheon DC, Paley MA, Prescher JA (2017) Orthogonal luciferase-luciferin pairs for bioluminescence imaging, J. Am. Chem. Soc 139, 2351–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mofford DM, Reddy GR, Miller SC (2014) Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over D-luciferin, J. Am. Chem. Soc 136, 13277–13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Steinhardt RC, Rathbun CM, Krull BT, Yu J, Yang Y, Nguyen BD, Kwon J, McCutcheon DC, Jones KA, Furche F, Prescher JA (2016) Brominated luciferins are versatile bioluminescent probes, ChemBioChem 18, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iwano S, Sugiyama M, Hama H, Watakabe A, Hasegawa N, Kuchimaru T, Tanaka KZ, Takahashi M, Ishida Y, Hata J, Shimozono S, Namiki K, Fukano T, Kiyama M, Okana H, Kizaka-Kondoh S, McHugh TJ, Yamamori T, Hioki H, Maki S, Miyawaki A (2018) Single-cell bioluminescence imaging of deep tissue in freely moving animals, Science 359, 935–939. [DOI] [PubMed] [Google Scholar]
- [24].Rathbun CM, Porterfield WB, Jones KA, Sagoe MJ, Reyes MR, Hua CT, Prescher JA (2017) Parallel screening for rapid identification of orthogonal bioluminescent tools, ACS Cent. Sci 3, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rathbun CM, Ionkina AA, Yao Z, Jones KA, Porterfield WB Prescher JA (2019) Rapid multicomponent bioluminescent imaging via substrate unmixing, bioRxiv, 811026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Su Y, Walker JR, Park Y, Smith TP, Liu LX, Hall MP, Labanieh L, Hurst R, Wang DC, Encell LP, Namdoo K, Zhang F, Kay MA, Casey KM, Majzner RG, Cochran JR, Mackall CL, Kirkland TA, Lin MZ (2020) Novel NanoLuc substrates enable bright two-population bioluminescence imaging in animals, Nat. Methods 17, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Belshaw PJ, Schoepfer JG, Liu KQ, Morrison KL, Schreiber SL (1995) Rational design of orthogonal receptor-ligand combinations, Angew. Chem. Int. Ed. Engl 34, 2129–2132. [Google Scholar]
- [28].Koh JT (2002) Engineering selectivity and discrimination into ligand-receptor interfaces, Chem. Biol 9, 17–23. [DOI] [PubMed] [Google Scholar]
- [29].Trott O, Olsen AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lauck F, Smith CA, Friedland GF, Humphris EL, Kortemme T (2010) RosettaBackrub-a web server for flexible backbone protein structure modeling and design, Nucleic Acids Res. 38, W569–W575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility, J. Comput. Chem 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Miller SC, Mofford DM, Adams ST (2018) Lessons learned from luminous luciferins and latent luciferases, ACS Chem. Biol 13, 1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kaskova ZM, Tsarkova AS, Yampolsky IV (2016) 1001 Lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine, Chem. Soc. Rev 45, 6048–6077. [DOI] [PubMed] [Google Scholar]
- [34].Liu MD, Warner EA, Morrissey CE, Fick CW, Wu TS, Ornelas MY, Ochoa GV, Zhang BS, Rathbun CM, Porterfield WB, Prescher JA, Leconte AM (2018) Statistical coupling analysis-guided library design for the discovery of mutant luciferases, Biochemistry 57, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang BS, Jones KA, McCutcheon DC, Prescher JA (2018) Pyridone luciferins and mutant luciferases for bioluminescence imaging, ChemBioChem 19, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].McCutcheon DC, Paley MA, Steinhardt RC, Prescher JA . (2012) Expedient synthesis of electronically modified luciferins for bioluminescence imaging, J. Am. Chem. Soc 134, 7604–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].McCutcheon DC, Porterfield WB, Prescher JA (2015) Rapid and scalable assembly of firefly luciferase substrates, Org. Biomol. Chem 13, 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Branchini BR, Southworth TL, Murtiashaw MH, Boije H, Fleet SE (2003) A mutagenesis study of the putative luciferin binding site residues of firefly luciferase, Biochemistry 42, 10429–10436. [DOI] [PubMed] [Google Scholar]
- [39].Yao Z, Zhang BS, Steinhardt RC, Mills JH, Prescher JA (2020) Multicomponent bioluminescence imaging with a π-extended luciferin, J. Am. Chem. Soc 142, 14080–14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Harwood KR, Mofford DM, Reddy GR, Miller SC (2011) Identification of mutant firefly luciferases that efficiently utilize aminoluciferins, Chem. Biol 18, 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Branchini BR, Ablamsky DM, Rosenman JM, Uzasci L, Southworth TL, Zimmer M (2007) Synergistic mutations produce blue-shifted bioluinescence in firefly luciferase, Biochemistry 48, 13847–13855. [DOI] [PubMed] [Google Scholar]
- [42].Ness JE, Kim S, Gottman A, Pak R, Krebber A, Borchert TV, Govindarajan S, Mundorff EC, Minshull J (2002) Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine. independently, Nat. Biotechnol 20, 1251–1255. [DOI] [PubMed] [Google Scholar]
- [43].Quan J, Tian J (2011) Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries, Nat. Protoc 6, 242–251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


