Abstract
Background
Circular RNA (circRNA) is a key regulator of cancer, and it has been proved to be involved in the regulation of cancer progression including non-small cell lung cancer (NSCLC). Circ-PITX1 was found to be a significantly upregulated circRNA in NSCLC, and its role and potential mechanism in NSCLC progression deserve further investigation.
Methods
The expression levels of circ-PITX1, microRNA (miR)-1248 and cyclin D2 (CCND2) were examined by quantitative real-time PCR (qRT-PCR). Cell proliferation, apoptosis, cell cycle process, migration and invasion were determined using cell counting kit 8 (CCK8) assay, colony formation assay, flow cytometry, wound healing assay and transwell assay. Xenograft models were built to explore the role of circ-PITX1 in NSCLC tumor growth in vivo. The glycolysis and glutamine metabolism of cells were assessed by detecting the consumptions of glucose and glutamine, cell extracellular acidification rate (ECAR), and the productions of lactate, α-ketoglutaric acid (α-KG) and ATP. The protein levels of hexokinase 2 (HK-2), glutaminase 1 (GLS1) and CCND2 were tested by Western blot (WB) analysis. Dual-luciferase reporter assay and RIP assay were employed to verify the interaction between miR-1248 and circ-PITX1 or CCND2.
Results
Circ-PITX1 was upregulated in NSCLC and its silencing could inhibit the proliferation, migration, invasion, cell cycle process, glycolysis, glutamine metabolism, and promote the apoptosis of NSCLC cells in vitro, as well as reduced tumor growth in vivo. In the terms of mechanism, we found that circ-PITX1 could act as a sponge of miR-1248, and miR-1248 could target CCND2. In addition, miR-1248 inhibitor reversed the inhibitory effect of circ-PITX1 knockdown on NSCLC progression. Similarly, CCND2 overexpression also reversed the suppressive effect of miR-1248 on NSCLC progression. Moreover, circ-PITX1 positively regulated CCND2 expression by sponging miR-1248.
Conclusion
Circ-PITX1 served as a sponge of miR-1248 to promote NSCLC progression by upregulating CCND2.
Keywords: non-small cell lung cancer, circ-PITX1, miR-1248, CCND2
Introduction
Non-small cell lung cancer (NSCLC) is a malignant tumor derived from bronchial mucosal epithelium or alveolar epithelium.1,2 At present, the etiology of NSCLC is unclear, but the risk factors such as environmental factors and genetic factors have been considered to affect the occurrence of NSCLC.3,4 Although many efforts have been made for NSCLC treatment, the metastasis and recurrence make the treatment of NSCLC particularly difficult.5,6 Therefore, exploring effective biomarkers is essential for treating NSCLC.
Circular RNA (circRNA) is a kind of non-coding RNA with circular structure, which is widely and stably expressed in eukaryotic cells.7 Due to its abundance and stability, circRNA has shown good application prospects in becoming a biomarker of human diseases, especially cancer.8,9 More importantly, circRNA has been found to have many binding sites of microRNA (miRNA), which can participate in the regulation of downstream targets by acting as competitive endogenous RNA (ceRNA) for miRNA.10,11 For example, circ-NCOR2 was highly repressed in papillary thyroid cancer, and it could enhance cancer cell growth and metastasis by the miR-615a-5p/MTA2 axis.12 Hsa_circ_0001944 was considered to be a potential biomarker for bladder cancer, which served as a miR-548 sponge to facilitate cell migration and invasion via PROK2.13 In addition, circ_0000376 was found to facilitate NSCLC cell glycolysis, viability and metastasis through regulating the miR-1182/NOVA2 network.14
Hsa_circ_0074027 is located at chr5 and derived from the PITX1 host gene, so it is also called circ-PITX1. Recently, circ-PITX1 had been found to promote the growth and invasion of glioma.15 In NSCLC, many studies confirmed that circ-PITX1 might be a potential biomarker for the prognosis of NSCLC, which was closely related to cell proliferation and metastasis.16–18 Therefore, it is worth continuing to explore more about the role and the underlying molecular mechanism of circ-PITX1 in NSCLC. Our study aims to reveal more functions of circ-PITX1 in NSCLC progression and to reveal new molecular mechanisms of circ-PITX1 using the circRNA/miRNA/mRNA axis.
Materials and Methods
Patient Samples
Forty-one paired NSCLC tumor tissues and adjacent normal tissues were collected from 41 NSCLC patients at the First People’s Hospital of Yunnan Province. All tissues were stored in liquid nitrogen. All patients signed written informed consent. This study was approved by the Ethics Committee of the First People’s Hospital of Yunnan Province and was carried out according to the guidelines of Declaration of Helsinki.
Cell Culture and Transfection
Human NSCLC cell lines (HCC827 and H1650) were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA), and normal bronchial epithelial cell line (BESA-2B) were grown in BEGM Bullet Kit (Lonza, Basel, Switzerland). All mediums contained 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Procell, Wuhan, China). All cells were obtained from ATCC (Manassas, VA, USA) and were cultured at 37°C with 5% CO2 incubator.
The small interference RNA (siRNA), overexpression vector and lentiviral short hairpin RNA (shRNA) of circ-PITX1 (si-circ-PITX1, circ-PITX1 and sh-circ-PITX1), miR-1248 mimic and inhibitor (miR-1248 and anti-miR-1248), pcDNA cyclin D2 (CCND2) overexpression plasmid, and their negative controls (si-NC, vector, sh-NC, miR-NC, anti-miR-NC and pcDNA) were designed and synthesized from General Biosystems (Anhui, China). Transfection could be performed using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). After 48 h, the cells were collected for function experiments.
Quantitative Real-Time PCR (qRT-PCR) and RNase R Assay
Trizol reagent (Invitrogen) was used to extract total RNA, and cDNA Synthesis Kit (Roche, Basel, Switzerland) was performed to synthesis cDNA. SYBR Green (Invitrogen) was mixed with the special primers for qRT-PCR. β-actin or U6 was used as the internal reference. The primer sequences were listed as below: circ-PITX1, F 5ʹ-GCGTCCCTGTGTATGTTGGA-3ʹ, R 5ʹ-GCCGGCAGAGTCTGTCTTAAA-3ʹ; PITX1, F 5ʹ-CTAGAGGCCACGTTCCAGAG-3ʹ; R 5ʹ-TGGTTACGCTCGCGCTTAC-3ʹ; miR-1248, F 5ʹ-GTCCACCTTCTTGTATAAGCACT-3ʹ, R 5ʹ-GCAGGGTCCGAGGTATTC-3ʹ; CCND2, F 5ʹ-ACCTTCCGCAGTGCTCCTA-3ʹ, R 5ʹ-CCCAGCCAAGAAACGGTCC-3ʹ; β-actin, F 5ʹ-GACCTCTATGCCAACACAGT-3ʹ, R 5ʹ-AGTACTTGCGCTCAGGAGGA-3ʹ; U6, F 5ʹ-CAGCACATATACTAAAATTGGAACG-3ʹ, R 5ʹ-ACGAATTTGCGTGTCATCC-3ʹ. The 2−ΔΔCT method was used to calculate relative expression.
For RNase R assay, the extracted RNA needed to be incubated with RNase R (Epicenter, Madison, WI, USA) for an additional 15 min, and then the expression of circ-PITX1 and linear mRNA PITX1 was detected.
Cell Counting Kit 8 (CCK8) and Colony Formation Assays
Both assays were used to evaluate cell proliferation. For CCK8 assay, NSCLC cells were seeded into 96-well plates. After 48 h, the cells were incubated CCK8 solution (Dojindo, Kumamoto, Japan) for 4 h. The absorbance was measured at 450 nm under a microplate reader to calculate cell viability. For colony formation assay, NSCLC cells were seeded in 6-well plates (150 cells/well) and cultured for 14 days at 37°C. The colonies were then immobilized and stained by crystal violet (Beyotime, Shanghai, China). The colonies were photographed under a microscope and counted the colony number larger than 50 cells.
Cell Apoptosis and Cell Cycle Detection
Annexin V-FITC/propidium iodide (PI) Apoptosis Detection Assay Kit (Yeasen, Shanghai, China) and Cell cycle DNA Content Quantitation Assay (Cell Cycle) (Solarbio, Beijing, China) were used to analyze cell apoptosis and cell cycle process. NSCLC cells were harvested from 6-well plates after culturing 48 h. In the detection of cell apoptosis, the cell lysates were stained with Annexin V-FITC and PI. Flow cytometry (BD Biosciences, San Jose, CA, USA) was used to analyze cell apoptosis rate. In the detection of cell cycle, the cell lysates were fixed in 70% ethanol and then hatched with RNase A and PI. The cell cycle distribution was analyzed using flow cytometer.
Wound Healing Assay
When the NSCLC cells in 6-well plates reached 90% confluences, a 200 μL pipette tip was used to create a wound on the cell layer. After incubating for 24 h at 37°C, the wound was photographed under a microscope (40 ×) to analyze the wound healing area for evaluating cell migration index = (area at T0 – area at T24)/area at T0 ×100%.
Transwell Assay
24-well Transwell chambers pre-coated with Matrigel (Corning Inc., Corning, NY, USA) were used to detecting cell invasion. NSCLC cells were seeded into the upper chamber with serum-free medium coated. The medium containing 10% FBS was added to the lower chamber. Twenty-four hours later, the cells that invaded to the lower surface were fixed and stained by crystal violet. The number of invaded cells was counted under a microscope (100×).
Glycolysis Detection
Cell glycolysis was assessed by detecting glucose consumption and lactate production using the Glucose Assay Kit and Lactate Assay Kit (Biovision, San Francisco, CA, USA) according to the manufacturer’s instructions.
Detection of Cell Extracellular Acidification Rate (ECAR)
Glycolysis Rate Determination Kit was bought from Agilent (Santa Clara, CA, USA). HCC827 and H1650 cells were seeded into the cell-culture dish, and sequentially added Glucose, Oligomycin, and 2-DG. The ECAR of cells was analyzed by Seahorse XFe Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA).
Glutamine Metabolism
Glutamine consumption, α-ketoglutaric acid (α-KG) production and ATP production were determined to assess the glutamine metabolism of cells. According the protocol of manufacturer, the Glutamine Assay Kit, α-KG Assay Kit and ATP Assay Kit (Enzyme-linked Biotechnology, Shanghai, China) were used to detect the glutamine consumption, α-KG production and ATP production of cells.
Western Blot (WB) Analysis
Cells and tissues were treated with RIPA Lysis Buffer (Beyotime) to extract total protein. After quantifying, protein was subjected to SDS-PAGE gel, transferred to PVDF membranes, following by hatching with non-fat milk. The membranes were incubated with the primary antibodies targeting hexokinase 2 (HK-2, 1:1000, Abcam, Cambridge, MA, USA), glutaminase 1 (GLS1, 1:5000, Abcam), CCND2 (1:2000, Abcam) or β-actin (1:1000, Abcam), following by secondary antibody (Goat anti-Rabbit IgG, 1:50,000, Abcam). The protein blots were presented using an Enhanced Chemiluminescence Kit (Millipore, Billerica, MA, USA).
Dual-Luciferase Reporter Assay
The fragments of circ-PITX1 or CCND2 3ʹUTR including the putative binding sites or mutant sites of miR-1248 were cloned into the pMIR-REPORT vectors (Promega, Madison, WI, USA), generating the wild-type (WT) or mutate-type (MUT) vectors, respectively. The vectors were co-transfected with miR-1248 mimic or miR-NC into NSCLC cells and incubated for 48 h. Luciferase activity was assessed by Dual-Luciferase Reporter Assay System (Promega).
RIP Assay
MiR-1248 mimic or miR-NC was transfected into NSCLC cells for 48 h. Then, the cells were harvested for collecting the cell lysates. According to the instructions of RIP Kit (Millipore), cell lysates were hatched with magnetic beads coated with IgG antibody (RIP-IgG) or Ago2 antibody (RIP-Ago2). After extracting the RNA, circ-PITX1 and CCND2 enrichment was measured by qRT-PCR.
Xenograft Models
Ten BALB/c mice (male, Vital River, Beijing, China) were divided into 2 groups (n = 5). In the flank of mice, H1650 cells transfected with sh-NC or sh-circ-PITX1 were subcutaneously injected into mice. After 7 days, tumor length and width were detected every 5 days until 32 days to calculate tumor volume. All mice were sacrificed to obtain the tumor tissues. Animal experiments were approved by the Ethics Committee of the First People’s Hospital of Yunnan Province and performed in accordance with the guidelines of the National Animal Care and Ethics Institution.
Statistical Analysis
All data were presented as mean ± standard deviation from at least three repeats. The difference of targets expression in paired NSCLC tumor tissues and adjacent normal tissues uses the paired-t test. And the analysis between the two groups in the other results uses unpaired-t test. One-way analysis of variance followed by Tukey post hoc test was employed to compare the differences among multi-groups. The statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad, La Jolla, CA, USA). P < 0.05 indicated as significant difference.
Results
Circ-PITX1 Was Highly Expressed in NSCLC Tissues and Cells
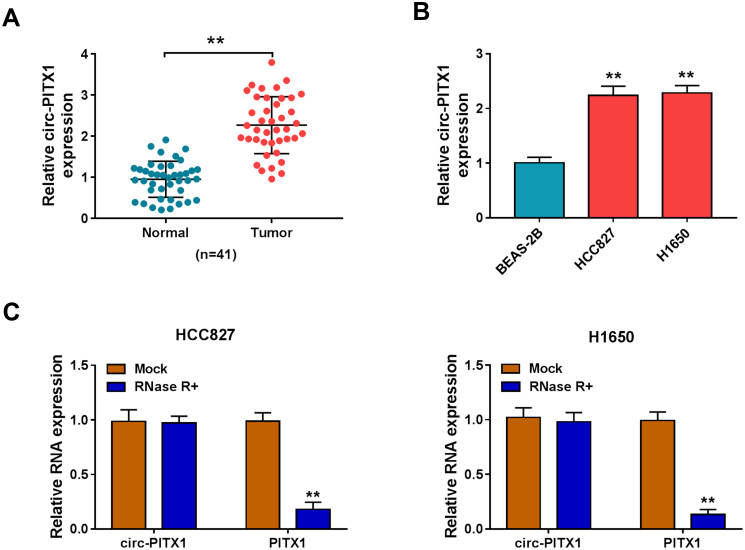
In 41 paired NSCLC tumor tissues and adjacent normal tissues, we found that circ-PITX1 had notably increased expression in NSCLC tumor tissues (Figure 1A). Similarly, the expression of circPITX1 was significantly higher in NSCLC cell lines (HCC827 and H1650) than in BESA-2B cells (Figure 1B). Subsequently, the stability of circ-PITX1 was assessed by RNase R assay, and the results presented that RNase R could digest linear mRNA PITX1, while had not effect on circ-PITX1 (Figure 1C).
Figure 1.
The expression of circ-PITX1 in NSCLC tissues and cells. (A) The expression of circ-PITX1 in 41 paired NSCLC tumor tissues (Tumor) and adjacent normal tissues (Normal) was determined by qRT-PCR. (B) QRT-PCR was performed to measure circ-PITX1 expression in BEAS-2B cells and NSCLC cell lines (HCC827 and H1650). (C) RNase R assay was used to evaluate the stability of circ-PITX1. **P < 0.01.
Circ-PITX1 Played an Oncogenic Role in NSCLC
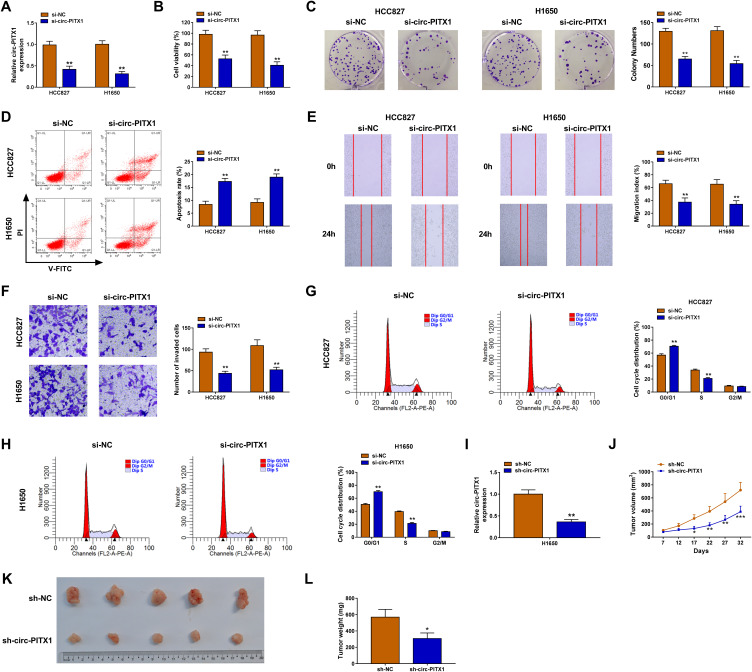
To investigate the biological roles of circ-PITX1 in NSCLC cells, the siRNA of circ-PITX1 was designed for the loss-of-function experiment. The significant decrease of circ-PITX1 expression confirmed that transfection of si-circ-PITX1 could effectively inhibit circ-PITX1 expression (Figure 2A). Using the CCK8 assay and colony formation assay, we found that circ-PITX1 knockdown could suppress the viability and colony number of HCC827 and H1650 cells (Figure 2B and C). The detection results of cell apoptosis indicated that circ-PITX1 silencing also enhanced the apoptosis rate of HCC827 and H1650 cells (Figure 2D). Moreover, wound healing assay and transwell assay suggested that the migration and invasion of HCC827 and H1650 cells also were inhibited by circ-PITX1 silencing (Figure 2E and F). In addition, we also evaluated the cell cycle process of NSCLC cells. The results showed that the cell number in G0/G1 phase was markedly increased and in S phase was obviously decreased in the presence of si-circ-PITX1, indicating that circ-PITX1 knockdown induced cell cycle arrest (Figure 2G and H). In addition, we also built the circ-PITX1 overexpression vector. The significant high expression of circ-PITX1 confirmed the successful transfection of the circ-PITX1 overexpression vector (Supplementary Figure 1A). In contrast, we found that overexpressed circ-PITX1 could promote the viability, colony number, migration, invasion, and suppress apoptosis of HCC827 and H1650 cells (Supplementary Figure 1B–F). Therefore, these data confirmed that circ-PITX1 had a positive role in NSCLC progression. Furthermore, the subcutaneous xenograft tumors were constructed to investigate the effect of circ-PITX1 knockdown on NSCLC tumorigenesis in vivo. After transfected with sh-circ-PITX1 into H1650 cells, we confirmed that circ-PITX1 was indeed decreased (Figure 2I). Then, the transfected H1650 cells were injected into nude mice. The detection of tumor volume confirmed that circ-PITX1 knockdown did inhibit the tumor volume of NSCLC (Figure 2J). By comparing tumor sizes, we found that the tumors of the circ-PITX1 knockdown group were significantly smaller than the control group (Figure 2K), and the tumor weight of circ-PITX1 knockdown group also was reduced compared to the control group (Figure 2L). All data suggested that circ-PITX1 played a tumor promoter role in NSCLC.
Figure 2.
Circ-PITX1 silencing inhibited NSCLC progression in vitro. HCC827 and H1650 cells were transfected with si-NC or si-circ-PITX1. (A) The expression of circ-PITX1 was detected by qRT-PCR. CCK8 assay (B), colony formation assay (C), flow cytometry (D), wound healing assay (E) and transwell assay (F) were used to measure the viability, colony number, apoptosis, migration and invasion of cells. (G and H) The cell cycle process was analyzed using flow cytometry. (I) H1650 cells were transfected with sh-NC or sh-circ-PITX1. The expression of circ-PITX1 was measured by qRT-PCR to confirm the transfection efficiency of sh-circ-PITX1. (J–L) H1650 cells transfected with sh-NC or sh-circ-PITX1 were injected into nude mice. (J) Tumor volume was measured in each group. (K) After 32 days, the tumors were removed and photographed. (L) Tumor weight was detected in each group were measured. *P < 0.05, **P < 0.01, ***P < 0.001.
Circ-PITX1 Silencing Restrained the Glycolysis and Glutamine Metabolism of NSCLC Cells
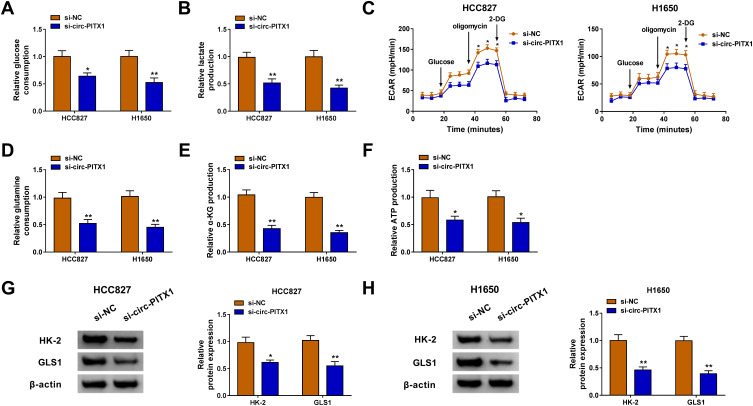
Glycolysis and glutamine metabolism are important sources of energy gain in cancer cells, and their inhibition is an important direction in the treatment of cancer.19,20 By detecting the glucose consumption and lactate production, we assessed the glycolysis of NSCLC cells. The results revealed that knockdown of circ-PITX1 could significantly reduce the glucose consumption and lactate production of HCC827 and H1650 cells (Figure 3A and B), indicating that the glycolysis of NSCLC could be inhibited by circ-PITX1 silencing. By detecting the ECAR of HCC827 and H1650 cells, we confirmed that circ-PITX1 knockdown could inhibit the glycolysis ability of NSCLC cells (Figure 3C). Furthermore, the glutamine consumption, α-KG production and ATP production of HCC827 and H1650 cells also were remarkably repressed by circ-PITX1 silencing (Figure 3D–F), which showed that circ-PITX1 downregulation inhibited the glutamine metabolism of NSCLC cells. In addition, we also uncovered that the protein levels of glycolysis rate-limiting enzyme HK-2 and glutamine hydrolase GLS1 in HCC827 and H1650 cells also were markedly decreased after circ-PITX1 knockdown (Figure 3G and H).
Figure 3.
Circ-PITX1 silencing restrained the glycolysis and glutamine metabolism of NSCLC cells in vitro. HCC827 and H1650 cells were transfected with si-NC or si-circ-PITX1. (A–C) The glucose consumption, lactate production and ECAR were measured to assess cell glycolysis. (D–F) Glutamine consumption, α-KG production and ATP production were determined using corresponding Assay Kits, respectively. (G and H) WB analysis was performed to assess the protein expression of HK-2 and GLS1. *P < 0.05, **P < 0.01.
Circ-PITX1 Could Sponge miR-1248
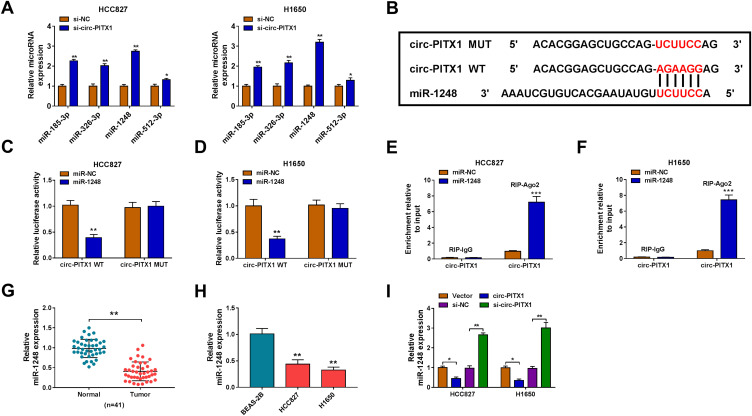
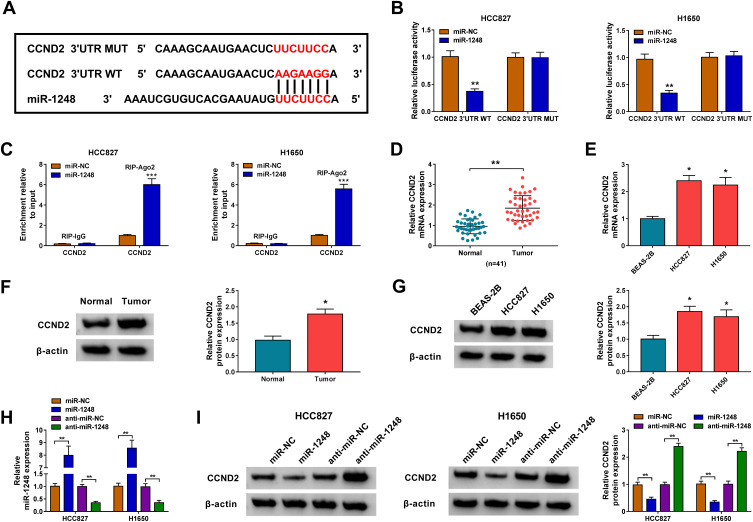
To explore the mechanism of circ-PITX1, the Circular RNA Interactome software was used to predict the targeted miRNAs of circ-PITX1. Four miRNAs with high scores and reported to be highly expressed in NSCLC were selected as candidate miRNAs (miR-185-3p, miR-326-3p, miR-1248 and miR-512-3p). Under the condition of knocking down circ-PITX1, we tested the expression of each candidate miRNA, and the results showed that the expression of miR-1248 was the most significantly increased among them (Figure 4A), so we chose miR-1248 for this study. As exhibited in Figure 4B, the binding sites between miR-1248 and circ-PITX1 were shown. Then, the circ-PITX1 WT vector and circ-PITX1 MUT vector were constructed for dual-luciferase reporter assay. The data indicated that miR-1248 overexpression had an inhibitory effect on the luciferase activity of the circ-PITX1 WT vector, but had no effect on the luciferase activity of the circ-PITX1 MUT vector (Figure 4C and D). Meanwhile, the results of RIP assay also showed that the overexpression of miR-1248 could make circ-PITX1 enriched in RIP-Ago2 compared with that in RIP-IgG (Figure 4E and F). Additionally, the significant low expression of miR-1248 was found in tumor tissues and cell lines of NSCLC (Figure 4G and H), which was contrary to the circ-PITX1 expression trend. To further confirm the regulation of circ-PITX1 on miR-1248, we measured miR-1248 expression in HCC827 and H1650 cells transfected with circ-PITX1 overexpression vector and siRNA, we found that miR-1248 expression could be inhibited by circ-PITX1 overexpression, while promoted by circ-PITX1 knockdown (Figure 4I).
Figure 4.
Circ-PITX1 could sponge miR-1248. (A) After transfected with si-NC or si-circ-PITX1 into HCC827 and H1650 cells, the expression of candidate miRNAs (miR-185-3p, miR-326-3p, miR-1248 and miR-512-3p) was measured by qRT-PCR. (B) The sequences of binding sites and mutated sites between miR-1248 and circ-PITX1 were presented. Dual-luciferase reporter assay (C and D) and RIP assay (E and F) were used to verify the interaction between miR-1248 and circ-PITX1. (G) QRT-PCR was performed to test the expression of miR-1248 in 41 paired NSCLC tumor tissues (Tumor) and adjacent normal tissues (Normal). (H) The expression of miR-1248 in BEAS-2B cells and NSCLC cell lines (HCC827 and H1650) was measured by qRT-PCR. (I) HCC827 and H1650 cells were transfected with vector, circ-PITX1, si-NC or si-circ-PITX1. The expression of miR-1248 was detected by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001.
Circ-PITX1 Regulated NSCLC Progression by Sponging miR-1248
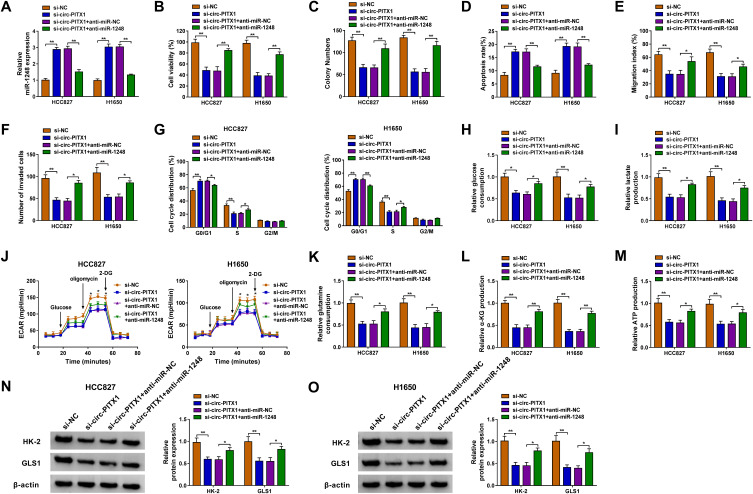
To investigate whether circ-PITX1 could sponge miR-1248 to regulate NSCLC progression, we co-transfected with si-circ-PITX1 and anti-miR-1248 into HCC827 and H1650 cells. By detecting the expression of miR-1248, we discovered that the promotion effect of circ-PITX1 silencing on miR-1248 expression was effectively reversed by miR-1248 inhibitor, which confirmed that the transfection of both was successful (Figure 5A). Then, we found that the inhibitory effect of circ-PITX1 knockdown on the viability and colony number, as well as the increasing effect on the apoptosis rate in HCC827 and H1650 cells could be reversed by miR-1248 inhibitor (Figure 5B–D). Moreover, miR-1248 inhibitor also could overturn the suppressive effect of circ-PITX1 silencing on the migration, invasion and cell cycle process of HCC827 and H1650 cells (Figure 5E–G). Furthermore, the inhibitory effect of circ-PITX1 downregulation on the glucose consumption, lactate production, ECAR, glutamine consumption, α-KG production and ATP production of in HCC827 and H1650 cells could also be eliminated by inhibiting miR-1248 expression (Figure 5H–M). Meanwhile, miR-1248 inhibitor also abolished the decreasing effect of circ-PITX1 knockdown on HK-2 and GLS1 protein expression in HCC827 and H1650 cells (Figure 5N and O).
Figure 5.
Circ-PITX1 regulated NSCLC progression by sponging miR-1248. HCC827 and H1650 cells were transfected with si-NC, si-circ-PITX1, si-circ-PITX1+ anti-miR-NC or si-circ-PITX1 + anti-miR-1248. (A) The expression of miR-1248 was measured by qRT-PCR. The viability, colony number, apoptosis, migration and invasion of cells were determined using CCK8 assay (B), colony formation assay (C), flow cytometry (D), wound healing assay (E) and transwell assay (F). (G) Flow cytometry also was used to detect cell cycle process. (H–J) Cell glycolysis was assessed by detecting the glucose consumption, lactate production and ECAR. (K–M) Glutamine consumption, α-KG production and ATP production were evaluated by the corresponding Assay Kits, respectively. (N and O) The protein expression of HK-2 and GLS1 was assessed using WB analysis. *P < 0.05, **P < 0.01.
CCND2 Was Targeted by miR-1248
Then, we used the Starbase tool to predict the targets of miR-1248, and found that miR-1248 could bind to the 3ʹUTR of CCND2 (Figure 6A). The further experiments revealed that the luciferase activity of CCND2 3ʹUTR WT vector rather than the CCND2 3ʹUTR MUT vector could be decreased by miR-1248 overexpression (Figure 6B), and CCND2 could be markedly enriched in RIP-Ago2 in HCC827 and H1650 cells overexpressed miR-1248 (Figure 6C). Importantly, we discovered a remarkably highly expressed CCND2 in NSCLC tumor tissues and cells in the mRNA level and protein level (Figure 6D–G). To explore the regulation of miR-1248 on CCND2, miR-1248 mimic and inhibitor were transfected into HCC827 and H1650 cells. The significantly high and low expression of miR-1248 confirmed the transfection effectiveness of mimic and inhibitor (Figure 6H). The detection results of CCND2 protein level indicated that miR-1248 overexpression could obviously inhibit CCND2 expression, while its inhibitor had an opposite effect (Figure 6I).
Figure 6.
CCND2 was targeted by miR-1248. (A) The sequences of binding sites and mutated sites between CCND2 3ʹUTR and miR-1248 were exhibited. The interaction between CCND2 and miR-1248 was confirmed using dual-luciferase reporter assay (B) and RIP assay (C). (D–G) The mRNA and protein expression levels of CCND2 in 41 paired NSCLC tumor tissues (Tumor) and adjacent normal tissues (Normal), as well as in BEAS-2B cells and NSCLC cell lines (HCC827 and H1650) were measured by qRT-PCR and WB analysis. (H) The transfection efficiencies of miR-1248 mimic and inhibitor were evaluated by measuring miR-1248 expression in HCC827 and H1650 cells using qRT-PCR. (I) HCC827 and H1650 cells were transfected with miR-NC, miR-1248, anti-miR-NC or anti-miR-1248. The protein expression of CCND2 was examined by WB analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
Overexpressed CCND2 Overturned the Inhibition of miR-1248 on NSCLC Progression
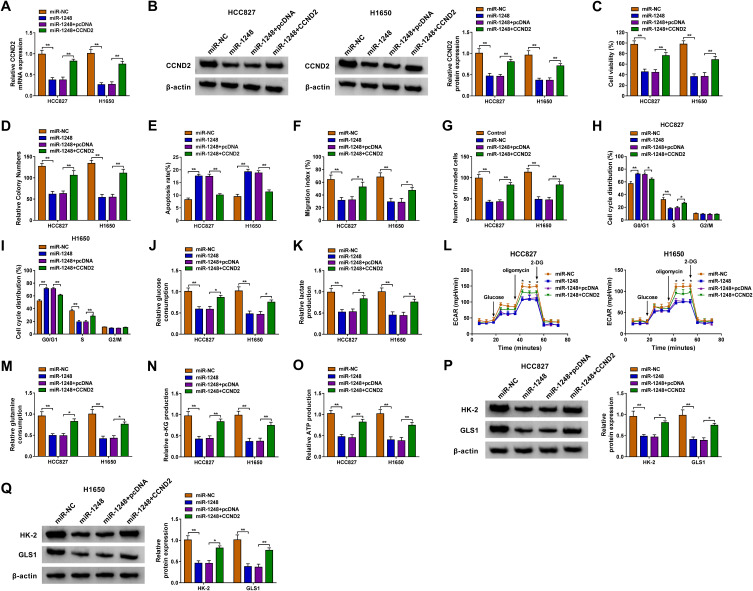
For illuminating whether miR-1248 regulated NSCLC progression by targeting CCND2, miR-1248 mimic and pcDNA CCND2 overexpression plasmid were co-transfected into HCC827 and H1650 cells. The detection of CCND2 mRNA and protein expression showed that the addition of pcDNA CCND2 overexpression plasmid could eliminate the inhibitory effect of miR-1248 mimic on CCND2 expression (Figure 7A and B). Then, we measured the biological functions of NSCLC cells. The results indicated that miR-1248 overexpression could repress cell viability, colony number, and enhance apoptosis in HCC827 and H1650 cells. However, these effects could be reversed by CCND2 overexpression (Figure 7C–E). Furthermore, the inhibition effect of miR-1248 mimic on the migration, invasion and cell cycle process also could be recovered by CCND2 overexpression (Figure 7F–I). Similarly, miR-1248 also suppressed the glucose consumption, lactate production, ECAR, glutamine consumption, α-KG production and ATP production of HCC827 and H1650 cells, while CCND2 overexpression also overturned these effects (Figure 7J–O). At the same time, overexpressed CCND2 also promoted the protein levels of HK-2 and GLS1 reduced by miR-1248 in HCC827 and H1650 cells (Figure 7P and Q).
Figure 7.
MiR-1248 and CCND2 overexpression regulated NSCLC progression. HCC827 and H1650 cells were transfected with miR-NC, miR-1248, miR-1248 + pcDNA or miR-1248 + CCND2. (A and B) The mRNA and protein expression of CCND2 was tested using qRT-PCR and WB analysis. CCK8 assay (C), colony formation assay (D), flow cytometry (E), wound healing assay (F) and transwell assay (G) were performed to detect the viability, colony number, apoptosis, migration and invasion of cells. (H and I) Flow cytometry was employed to measure the cell cycle process. (J–L) Cell glycolysis was determined using analyzing the glucose consumption, lactate production and ECAR of cells. (M–O) The corresponding Assay Kits were used to determine the glutamine consumption, α-KG production and ATP production, respectively. (P and Q) The protein expression of HK-2 and GLS1 was evaluated by WB analysis. *P < 0.05, **P < 0.01.
Circ-PITX1 Regulated CCND2 Expression via Sponging miR-1248
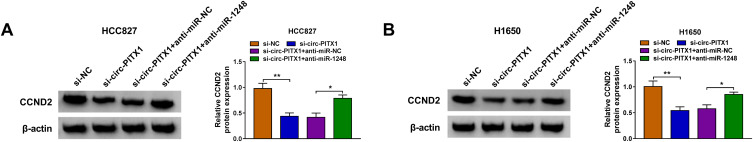
Based on the above studies, we found that circ-PITX1 could sponge miR-1248, and miR-1248 could target CCND2. To determine whether circ-PITX1 sponged miR-1248 to indirectly regulate CCND2, we detected CCND2 protein expression in HCC827 and H1650 cells co-transfected with si-circ-PITX1 and anti-miR-1248. As presented in Figure 8A and B, circ-PITX1 knockdown could markedly inhibit CCND2 expression, while this effect could be reversed by miR-1248 inhibitor. Therefore, we confirmed that circ-PITX1 positively regulated CCND2 by targeting miR-1248.
Figure 8.
Circ-PITX1 regulated CCND2 expression via sponging miR-1248. HCC827 and H1650 cells were transfected with si-NC, si-circ-PITX1, si-circ-PITX1 + anti-miR-NC or si-circ-PITX1 + anti-miR-1248. (A and B) The protein expression of CCND2 was measured by WB analysis. *P < 0.05, **P < 0.01.
Discussion
In recent years, with the deepening of circRNA research, researchers have found that the expression of circRNA is particularly important for the progression of many diseases, so it has great potential in the diagnosis and treatment of diseases.21,22 In NSCLC, many circRNAs have been identified as potential biomarkers for NSCLC, such as circ_0000735,23 circ_102179,24 circ_103993,25 and circ_0020123.26 In previous studies, circ-PITX1 was found to be highly expressed in NSCLC and was related to NSCLC progression.16–18 Consistent with previous research results, we also found significantly high expression of circ-PITX1 in the tissues and cells of NSCLC. Functional experiments revealed that circ-PITX1 silencing could repress the proliferation, migration, invasion, cell cycle process, glycolysis, glutamine metabolism, and enhance the apoptosis of NSCLC cells. More importantly, our results also proposed that downregulated circ-PITX1 had an inhibitory effect on NSCLC tumorigenesis in vivo. These data indicated that circ-PITX1 had the oncogenic role in NSCLC, suggesting that circ-PITX1 knockdown might be an important measure for NSCLC treatment.
Circ-PITX1 has been reported to target miR-362-3p, miR-335-3p and miR-185-3p to regulate NSCLC progression.16–18 To find a new mechanism of circ-PITX1, we used bioinformatics analysis for prediction and found that circ-PITX1 could sponge miR-1248. MiR-1248 has been found to be involved in regulating the progression of a variety of cancers. For instance, Du et al suggested that miR-1248 was upregulated in esophageal squamous cell carcinoma, which could improve cancer cell proliferation and metastasis.27 However, Zhang et al reported that miR-1248 functioned a tumor suppressor in gastric cancer.28 Therefore, miR-1248 might have different roles in different cancers. However, it was worth noting that miR-1248 also was significantly low in NSCLC,29 and our findings were consistent with it. In addition, we discovered that miR-1248 expression was negatively regulated by circ-PITX1 in vitro and in vivo. Using the rescue experiments, we showed that circ-PITX1 promoted NSCLC progression by acting as a sponge of miR-1248. This is a new discovery.
In previous studies, miR-1248 had been reported to target C3A (lung squamous carcinoma), TYMS (NSCLC) and TRIM24 (NSCLC).29–31 In this study, CCND2 was found to be a target of miR-1248. CCND2 is a recognized oncogene which encodes proteins belonging to a highly conserved cyclin family.32,33 The significant high expression of CCND2 has been found in many cancers, including breast cancer,34 colorectal cancer,35 and cervical cancer.36 Therefore, CCND2 has been considered as a cancer biomarker. In many studies, inhibition of CCND2 had been shown to hinder NSCLC proliferation and metastasis.37–39 Further experiments revealed that the anti-cancer effect of miR-1248 in NSCLC could be reversed by CCND2 overexpression, indicating that miR-1248 targeted CCND2, an oncogene, to inhibit NSCLC progression. Additionally, the positive regulatory effect of circ-PITX1 on CCND2 expression also fully confirmed the hypothesis that circ-PITX1 sponged miR-1248 to regulate CCND2 in NSCLC.
In general, our results found a new mechanism for circ-PITX1 to regulate the progression of NSCLC. Our data showed that circ-PITX1 could promote the proliferation, metastasis, cell cycle, glycolysis and glutamine metabolism of NSCLC via regulating the miR-1248/CCND2 axis. Our findings perfected the role of circ-PITX1 in NSCLC and provided new evidence for circ-PITX1 to become a biomarker of NSCLC.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Skrickova J, Kadlec B, Venclicek O, Merta Z. Lung cancer. Cas Lek Cesk. 2018;157(5):226–236. [PubMed] [Google Scholar]
- 2.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 3.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr SR, Akerley W, Cannon-Albright LA. Genetic contribution to nonsquamous, non-small cell lung cancer in nonsmokers. J Thorac Oncol. 2018;13(7):938–945. doi: 10.1016/j.jtho.2018.03.024 [DOI] [PubMed] [Google Scholar]
- 5.Kim CG, Kim KH, Pyo KH, et al. Hyperprogressive disease during PD-1/PD-L1 blockade in patients with non-small-cell lung cancer. Ann Oncol. 2019;30(7):1104–1113. doi: 10.1093/annonc/mdz123 [DOI] [PubMed] [Google Scholar]
- 6.Cruz C, Afonso M, Oliveiros B, Pego A. Recurrence and risk factors for relapse in patients with non-small cell lung cancer treated by surgery with curative intent. Oncology. 2017;92(6):347–352. doi: 10.1159/000458533 [DOI] [PubMed] [Google Scholar]
- 7.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 9.Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. doi: 10.1186/s12967-018-1593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luan S, Fu P, Wang X, Gao Y, Shi K, Guo Y. Circular RNA circ-NCOR2 accelerates papillary thyroid cancer progression by sponging miR-516a-5p to upregulate metastasis-associated protein 2 expression. J Int Med Res. 2020;48(9):300060520934659. doi: 10.1177/0300060520934659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin M, Lu S, Wu Y, et al. Hsa_circ_0001944 promotes the growth and metastasis in bladder cancer cells by acting as a competitive endogenous RNA for miR-548. J Exp Clin Cancer Res. 2020;39(1):186. doi: 10.1186/s13046-020-01697-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Li C, Liu H, Niu Q, Gao J. Circ_0000376, a Novel circRNA, promotes the progression of non-small cell lung cancer through regulating the miR-1182/NOVA2 network. Cancer Manag Res. 2020;12:7635–7647. doi: 10.2147/CMAR.S258340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian L, Guan J, Wu Y, Wang Q. Upregulated circular RNA circ_0074027 promotes glioblastoma cell growth and invasion by regulating miR-518a-5p/IL17RD signaling pathway. Biochem Biophys Res Commun. 2019;510(4):515–519. doi: 10.1016/j.bbrc.2019.01.140 [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z, Yin J, Peng G, Long X. Circ_0074027 contributes to the progression of non-small cell lung cancer via microRNA-362-3p/clathrin heavy chain axis. Anticancer Drugs. 2021;32(1):1–10. doi: 10.1097/CAD.0000000000000990 [DOI] [PubMed] [Google Scholar]
- 17.Yu C, Ying J, Yu K, Shen W, Jiang M. Circ_0074027 contributes to nonsmall cell lung cancer progression by upregulating CUL4B expression through miR-335-5p. Cancer Biother Radiopharm. 2020. doi: 10.1089/cbr.2020.3579 [DOI] [PubMed] [Google Scholar]
- 18.Gao P, Wang Z, Hu Z, Jiao X, Yao Y. Circular RNA circ_0074027 indicates a poor prognosis for NSCLC patients and modulates cell proliferation, apoptosis, and invasion via miR-185-3p mediated BRD4/MADD activation. J Cell Biochem. 2020;121(3):2632–2642. doi: 10.1002/jcb.29484 [DOI] [PubMed] [Google Scholar]
- 19.Li C, Zhang G, Zhao L, Ma Z, Chen H. Metabolic reprogramming in cancer cells: glycolysis, glutaminolysis, and Bcl-2 proteins as novel therapeutic targets for cancer. World J Surg Oncol. 2016;14(1):15. doi: 10.1186/s12957-016-0769-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akins NS, Nielson TC, Le HV. Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Curr Top Med Chem. 2018;18(6):494–504. doi: 10.2174/1568026618666180523111351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMed. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680. doi: 10.1002/1878-0261.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, Xu X, Liu M, Cui W, Peng G. Downregulation of Hsa_circ_0000735 inhibits the proliferation, migration, invasion, and glycolysis in non-small-cell lung cancer by targeting miR-940/BMPER axis. Onco Targets Ther. 2020;13:8427–8439. doi: 10.2147/OTT.S253474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou ZF, Wei Z, Yao JC, et al. CircRNA_102179 promotes the proliferation, migration and invasion in non-small cell lung cancer cells by regulating miR-330-5p/HMGB3 axis. Pathol Res Pract. 2020;216(11):153144. doi: 10.1016/j.prp.2020.153144 [DOI] [PubMed] [Google Scholar]
- 25.Lv YS, Wang C, Li LX, Han S, Li Y. Effects of circRNA_103993 on the proliferation and apoptosis of NSCLC cells through miR-1271/ERG signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(16):8384–8393. doi: 10.26355/eurrev_202008_22635 [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Zhao L, Wang Y. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by sponging miR-590-5p to regulate THBS2. Cancer Cell Int. 2020;20:387. doi: 10.1186/s12935-020-01444-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du F, Guo T, Cao C. Restoration of UPK1A-AS1 expression suppresses cell proliferation, migration, and invasion in esophageal squamous cell carcinoma cells partially by sponging microRNA-1248. Cancer Manag Res. 2020;12:2653–2662. doi: 10.2147/CMAR.S239418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Wang M, Zang X, et al. CircHN1 affects cell proliferation and migration in gastric cancer. J Clin Lab Anal. 2020;34(10):e23433. doi: 10.1002/jcla.23433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T, Li M, Li H, Shi P, Liu J, Chen M. Downregulation of circEPSTI1 represses the proliferation and invasion of non-small cell lung cancer by inhibiting TRIM24 via miR-1248 upregulation. Biochem Biophys Res Commun. 2020;530(1):348–354. doi: 10.1016/j.bbrc.2020.06.106 [DOI] [PubMed] [Google Scholar]
- 30.Li G, Guo X. LncRNA STARD13-AS blocks lung squamous carcinoma cells growth and movement by targeting miR-1248/C3A. Pulm Pharmacol Ther. 2020;64:101949. doi: 10.1016/j.pupt.2020.101949 [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Tian S, Yin Z, et al. MicroRNA-binding site SNPs in deregulated genes are associated with clinical outcome of non-small cell lung cancer. Lung Cancer. 2014;85(3):442–448. doi: 10.1016/j.lungcan.2014.06.010 [DOI] [PubMed] [Google Scholar]
- 32.Hung CS, Wang SC, Yen YT, Lee TH, Wen WC, Lin RK. Hypermethylation of CCND2 in lung and breast cancer is a potential biomarker and drug target. Int J Mol Sci. 2018;19(10):3096. doi: 10.3390/ijms19103096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cameron SR, Nandi S, Kahn TG, Barrasa JI, Stenberg P, Schwartz YB. PTE, a novel module to target polycomb repressive complex 1 to the human cyclin D2 (CCND2) oncogene. J Biol Chem. 2018;293(37):14342–14358. doi: 10.1074/jbc.RA118.005010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Chen P, Yang J. miR-451a suppresses the development of breast cancer via targeted inhibition of CCND2. Mol Cell Probes. 2020;54:101651. doi: 10.1016/j.mcp.2020.101651 [DOI] [PubMed] [Google Scholar]
- 35.Li WC, Wu YQ, Gao B, Wang CY, Zhang JJ. MiRNA-574-3p inhibits cell progression by directly targeting CCND2 in colorectal cancer. Biosci Rep. 2019;39(12):BSR20190976. doi: 10.1042/BSR20190976 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Zhang H, Xue B, Wang S, Li X, Fan T. Long noncoding RNA TP73 antisense RNA 1 facilitates the proliferation and migration of cervical cancer cells via regulating microRNA607/cyclin D2. Mol Med Rep. 2019;20(4):3371–3378. doi: 10.3892/mmr.2019.10572 [DOI] [PubMed] [Google Scholar]
- 37.Yao Y, Zhou Y, Fu X. miR6713p is downregulated in nonsmall cell lung cancer and inhibits cancer progression by directly targeting CCND2. Mol Med Rep. 2019;19(3):2407–2412. doi: 10.3892/mmr.2019.9858 [DOI] [PubMed] [Google Scholar]
- 38.He X, Chen SY, Yang Z, et al. miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J Exp Clin Cancer Res. 2018;37(1):230. doi: 10.1186/s13046-018-0882-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Shu H, Guo S. MiR-646 suppresses proliferation and metastasis of non-small cell lung cancer by repressing FGF2 and CCND2. Cancer Med. 2020;9(12):4360–4370. doi: 10.1002/cam4.3062 [DOI] [PMC free article] [PubMed] [Google Scholar]