Common wheat has a large genome with three subgenomes (A, B and D), making it challenging to create mutations at multiple genomic sites simultaneously. The CRISPR/Cas9 system offers a game‐changing tool for editing crop genomes (Chen et al., 2019). Three main strategies have been developed to produce multiple single‐guide RNAs (sgRNAs), including the conventional multiplex system with tandem repeats of separate U3 or U6 promoters (TRSP), the tRNA‐processing system (Xie et al., 2015) and the ribozyme‐processing system (Gao and Zhao, 2014). Although CRISPR/Cas9‐mediated genome editing was previously achieved by biolistic (Wang et al., 2014, 2018) and Agrobacterium transformation (Zhang et al., 2019a), a most efficient CRISPR/Cas9 system for multiplex editing in wheat remains elusive. To address this important question, we designed three multiplex editing constructs corresponding to these three systems, based on the pBUE411 vector (Figure 1a). For the TRSP system, wheat Pol III promoters, TaU3, TaU6.3 and TaU6.1 (Zhang et al., 2019a), were used to drive sgRNA expression independently. For the tRNA system, a TaU3 promoter was also used to express the tRNA‐sgRNA cassettes in a single transcript unit. For the ribozyme system, a Pol II promoter, Cestrum yellow leaf curling virus (CmYLCV) promoter (Cermak et al., 2017), was employed for expressing hammerhead ribozyme (HH)–sgRNA–hepatitis delta virus (HDV) ribozyme cassettes in a single transcript unit. A longer sgRNA scaffold was applied in three vectors to optimize the sgRNA structure (Dang et al., 2015). Wheat codon‐optimized Cas9 was driven under a maize (Zea mays) ubiquitin promoter (Ubip). Three genes, TaDA1, TaPDS and TaNCED1, were selected for simultaneous editing. The sgRNA for TaDA1 could target its homoeologous genes on A and B chromosomes, while the sgRNAs for TaPDS and TaNCED1 were designed to target all three homoeologous genes, respectively (Zhang et al., 2019b). In total, three sgRNAs could target 8 genomic sites in common wheat (Figure 1b). The sgRNA cassettes in the vectors were all arranged in the same order for close comparison. These T‐DNA vectors were introduced into hexaploid wheat Fielder via Agrobacterium tumefaciens‐mediated transformation.
Figure 1.
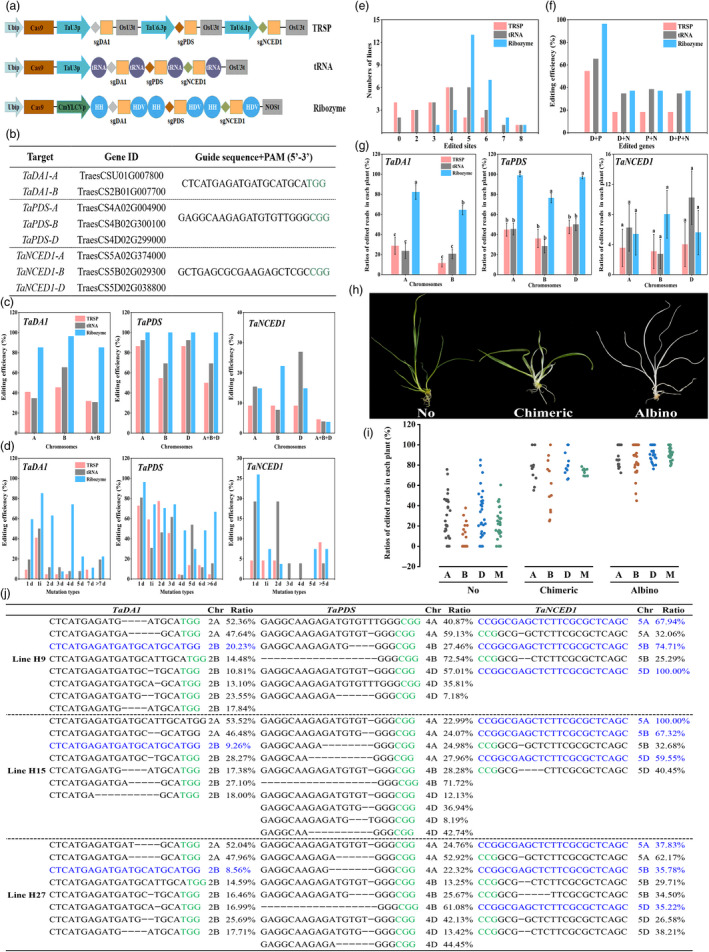
Multiplex genome editing mediated by CRISPR/Cas9 system in common wheat. (a) Illustration of vectors for three distinct multiplex CRISPR/Cas9 systems used in this study. The protospacers and sgRNA scaffolds are indicated by diamonds and orange, respectively. (b) Target gene IDs and sequences of the target sites. The PAM is labelled in green. (c) Editing efficiencies of homoeologous genes at different subgenomes in three multiplex editing systems. A + B, plants with genes edited at both A and B chromosomes. A + B + D, plants with genes simultaneously edited at A, B and D chromosomes. (d) Editing efficiencies of mutation types. The number of nucleotide insertion (i) and deletion (d) is indicated for each mutation type. (e) Number of lines with different numbers of edited sites. (f) Multiplex gene editing efficiencies in three editing systems: D, TaDA1; P, TaPDS; and N, TaNCED1. (g) Ratios of edited reads for homoeologous genes at different subgenomes in each plant. Data are presented as the mean ± SE. n = 22, 26, 27 for TRSP, tRNA and ribozyme systems, respectively. Different letters above the columns indicate groups with significant differences (ANOVA, P < 0.05). (h) Representative lines with no, chimeric and albino phenotype with TaPDS editing. (i) Ratios of edited reads for TaPDS at different chromosomes and their means in plants with no, chimeric and albino phenotype. M, mean value for ratios of TaPDS‐A, TaPDS‐B and TaPDS‐D in each plant. Each dot represents the ratios of edited reads in a single plant. A total of 31, 12 and 32 T0 transgenic plants were collected with no, chimeric and albino phenotype, respectively, and used in this analysis. (j) Examples of genotyping results for three T0 mutants generated by the ribozyme system. The alleles for each target gene were quantified by Hi‐TOM NGS. The PAM is labelled in green, and the wild type alleles are shown in blue. Chr, chromosome.
A total of 22, 26 and 27 T0 plants were generated from the transformed calli of TRSP, tRNA and ribozyme systems, respectively. The genotype of each plant was characterized by Hi‐TOM sequencing of the PCR amplicons with primers flanking each target site (Liu et al., 2019). The editing efficiency of individual genes was first analysed. Edits of TaDA1‐A and TaDA1‐B were detected in all three systems, and the ribozyme system was most effective (Figure 1c). The superior editing ability of the ribozyme system was also observed at TaPDS where mutations could cause albino phenotype. Impressively, 22 out of 27 plants showed albino phenotype in the ribozyme system, whereas only 5 plants displayed albino phenotype in either the TRSP or tRNA system. Sequencing results supported the observation as the ribozyme system achieved the highest efficiency, up to 100.00% (Figure 1c). Although fewer plants showed albino phenotype, the editing efficiencies in TRSP and tRNA systems still reached to 86.36% and 92.31%, respectively. For TaNCED1, all three vectors exhibited low activity, and the tRNA and ribozyme systems resulted in higher gene editing rates than the TRSP system (Figure 1c). The three systems showed similar mutation profiles for individual genes where small deletions and 1bp insertions predominated (Figure 1d).
The ability to target multiple genomic sites was further analysed for three systems. Compared with the TRSP system, more plants with over 4 edited sites were identified in the tRNA and ribozyme systems (Figure 1e), and the ribozyme system generated the highest simultaneous editing rates. The efficiencies of simultaneous editing in three genes in the tRNA and ribozyme systems were 34.62% and 37.04%, respectively, which were about twofold higher than that in the TRSP system (Figure 1f). Thus, the tRNA and ribozyme systems are more effective than TRSP, and the ribozyme system appeared to be most robust. The high editing efficiency of the ribozyme system might partly result from the use of the Pol II promoter, CmYLCV, for sgRNA expression.
The phenotype caused by gene editing depends on the genotype in individual plants. Therefore, we investigated the ratio of mutated reads at each target site in each plant through Hi‐TOM sequencing (Liu et al., 2019). No significant differences were detected at all targeted sites between the TRSP and tRNA systems, but the ratios of edited reads were significantly increased in the ribozyme system except for TaNCED1 (Figure 1g). Ratios of edited reads in the ribozyme system were about threefold higher for TaDA1 and twofold higher for TaPDS than those in the TRSP and tRNA systems, respectively. The results suggested that the ribozyme system greatly decreased the proportions of unedited reads at multiplex chromosomes and therefore increased the probability of the loss‐of‐function phenotype in T0 generation. This might explain the discrepancy between the high editing efficiency and less albino phenotype caused by TaPDS mutation in the TRSP and tRNA systems. Although over 86.00% of the plants carried edited TaPDS in the TRSP and tRNA systems, the editing ratios in most plants were not enough to display albino phenotype. To further quantify the relationship between ratios of the edited reads and observable phenotype, we compared the ratios of edited reads for TaPDS‐A, TaPDS‐B and TaPDS‐D among three groups of plants as no albino, chimeric and albino phenotype, respectively (Figure 1h). Positive correlation between the phenotype and editing ratio was observed (Figure 1i). The lowest average ratio for plants with albino phenotype was 80.59%, indicating an editing threshold for displaying loss‐of‐function phenotype. Wild type alleles were not detected at TaPDS in 6 lines of the ribozyme system despite different levels of chimerism (Figure 1j). These results collectively revealed that the phenotype caused by targeted mutagenesis occurred only when the ratios of edited homoeologous genes achieved a higher level at all homoeologous chromosomes simultaneously.
In summary, we compared three multiplex CRISPR/Cas9 systems for simultaneous genome editing at 8 target sites in common wheat. The tRNA and ribozyme systems were more effective than the TRSP system in multiplex genome editing. Furthermore, the ribozyme system could significantly increase the ratios of edited homoeologous genes at multiplex chromosomes in individual plants and therefore generated more plants with loss‐of‐function phenotypes. The ribozyme system established in our study would greatly aid fundamental and translational research in wheat.
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
J.L. and S.Z. constructed the vectors, analysed the data and wrote the manuscript. R.Z. designed the sgRNAs and performed Hi‐TOM sequencing. G.S. and W.L. collected samples and extracted DNA. J.G. performed the wheat transformation. Y.Q. analysed the data and revised the manuscript. Y.L. and G.L supervised the project.
Acknowledgements
We appreciated Qixin Sun’s group of China Agricultural University for providing us with the pBUE411 vector. This work was funded by grants from Agricultural Variety Improvement Project of Shandong Province (2019LZGC015, 2019LZGC001), the Ministry of Science and Technology of China (2016YFD0100500), the National Natural Science Foundation of China (31701428) and the Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2019G02). The work was also supported by the start‐up funds from University of Maryland, College Park.
Li, J. , Zhang, S. , Zhang, R. , Gao, J. , Qi, Y. , Song, G. , Li, W. , Li, Y. and Li, G. (2021) Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J., 10.1111/pbi.13508
Contributor Information
Yulian Li, Email: liyulian01@163.com.
Genying Li, Email: lgy111@126.com.
References
- Čermák, T. , Curtin, S.J. , Gil‐Humanes, J. , Čegan, R. , Kono, T.J.Y. , Konečná, E. , Belanto, J.J. et al. (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29, 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Dang, Y. , Jia, G. , Choi, J. , Ma, H. , Anaya, E. , Ye, C. , Shankar, P. et al. (2015) Optimizing sgRNA structure to improve CRISPR‐Cas9 knockout efficiency. Genome Biol. 16, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. and Zhao, Y. (2014) Self‐processing of ribozyme‐flanked RNAs into guide RNAs in vitro and in vivo for CRISPR‐mediated genome editing. J. Integr. Plant Biol. 56, 343–349. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X. , Zhang, H. , Song, L. , Li, Y. , Gao, C. et al. (2019) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Pan, Q. , He, F. , Akhunova, A. , Chao, S. , Trick, H. and Akhunov, E. (2018) Transgenerational CRISPR‐Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 1, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. . (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl. Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Hua, L. , Gupta, A. , Tricoli, D. , Edwards, K. , Yang, B. and Li, W. (2019a) Development of an Agrobacterium‐delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 17, 1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Zhang, R. , Gao, J. , Gu, T. , Song, G. , Li, W. , Li, D. et al. (2019b) Highly efficient and heritable targeted mutagenesis in wheat via the Agrobacterium tumefaciens‐mediated CRISPR/Cas9 system. Int. J. Mol. Sci. 20, 4257. [DOI] [PMC free article] [PubMed] [Google Scholar]


