Summary
Introduction of a C4 photosynthetic mechanism into C3 crops offers an opportunity to improve photosynthetic efficiency, biomass and yield in addition to potentially improving nitrogen and water use efficiency. To create a two‐cell metabolic prototype for an NADP‐malic enzyme type C4 rice, we transformed Oryza sativa spp. japonica cultivar Kitaake with a single construct containing the coding regions of carbonic anhydrase, phosphoenolpyruvate (PEP) carboxylase, NADP‐malate dehydrogenase, pyruvate orthophosphate dikinase and NADP‐malic enzyme from Zea mays, driven by cell‐preferential promoters. Gene expression, protein accumulation and enzyme activity were confirmed for all five transgenes, and intercellular localization of proteins was analysed. 13CO2 labelling demonstrated a 10‐fold increase in flux though PEP carboxylase, exceeding the increase in measured in vitro enzyme activity, and estimated to be about 2% of the maize photosynthetic flux. Flux from malate via pyruvate to PEP remained low, commensurate with the low NADP‐malic enzyme activity observed in the transgenic lines. Physiological perturbations were minor and RNA sequencing revealed no substantive effects of transgene expression on other endogenous rice transcripts associated with photosynthesis. These results provide promise that, with enhanced levels of the C4 proteins introduced thus far, a functional C4 pathway is achievable in rice.
Keywords: C4 photosynthesis, rice, metabolic engineering
Introduction
Installation of a C4 photosynthetic pathway in rice has been predicted to increase rice yields by up to 50% (Hibberd et al., 2008) and has been the focus of a large international consortium for more than a decade (https://c4rice.com/; von Caemmerer et al., 2012; Ermakova et al., 2020). Whereas a full compendium of genes required to engineer anatomical specialization for a full two‐cell C4 pathway in rice is still a way off, the genes and promoter sequences required for a metabolic prototype are currently available (Ermakova et al., 2020). An obstacle thus far in developing a metabolic prototype has been the difficulty of genotype engineering wherein multiple genes originating from distinct single gene transgenics are crossed to assemble a complete metabolic pathway (Karki et al., 2020). This has required screening of thousands of individuals across the initial transgenic generation stage and through multiple rounds of crosses to track the presence of all transgenes and confirm their expression in the appropriate cell‐type in segregating material (Karki et al., 2020; Lin et al., 2020). Modular cloning technologies such as Golden Gate (Engler et al., 2014) offer the opportunity to introduce a full suite of C4 enzymes into rice on a single construct, simplifying generation of lines and minimizing the potential for deleterious effects arising from transgene insertion.
Introduction of full C4 biochemistry into a C3 plant will require significant changes to chloroplast proteomes of mesophyll (M) and bundle sheath (BS) cells (Hernández‐Prieto et al., 2019; Majeran and van Wijk, 2009), but a minimal C4 cycle could be built by introducing just five enzymes from Zea mays (maize) into specific cells and compartments of the rice leaf (Ermakova et al., 2020; Karki et al., 2020). In this scenario, carbonic anhydrase (CA) and PEP carboxylase (PEPC) in the cytosol of M cells convert CO2 into bicarbonate and fix it into oxaloacetate. NADP‐malate dehydrogenase (MDH) inside the mesophyll chloroplasts then converts oxaloacetate into malate using NADPH produced by photosynthetic electron transport. After malate diffuses into BS cells, it is decarboxylated inside the chloroplast by NADP‐dependent malic enzyme (NADP‐ME). The released CO2 is refixed by ribulose‐1,5‐bisphosphate carboxylase oxygenase (Rubisco) and the residual pyruvate diffuses back to M cells to be regenerated into PEP by pyruvate orthophosphate dikinase (PPDK) inside the chloroplast. Individually, expression of these enzymes does not affect plant fitness or photosynthetic function (Karki et al., 2020). However, collectively this minimal cycle would contribute to establishing higher CO2 partial pressure around Rubisco in BS cells and could be beneficial even in plants with C3 leaf anatomy (Ermakova et al., 2020).
Previous attempts to introduce C4 metabolism into C3 species were focused primarily on using maize enzymes to replicate the single cell type C4 pathway that is found in species such as Hydrilla verticillata (Miyao et al., 2011). Stacking of four maize genes was achieved by crossing individual rice lines transformed with single genes of interest or by multiple re‐transformation of transgenic lines with additional gene constructs (Taniguchi et al., 2008). Although extractable activity of C4 cycle enzymes was unambiguously demonstrated in vitro for both single and multigene rice transgenics (Fukayama et al., 2001; Karki et al., 2020; Ku et al., 1999; Taniguchi et al., 2008; Tsuchida et al., 2001), expression of maize PEPC did not result in any 14C incorporation into C4 acids in vivo (Fukayama et al., 2003) and reports confirming C4 metabolic function using isotopic labelling (Arrivault et al., 2016; Hatch and Slack, 1966) have so far been missing.
Here we express five enzymes required to form a minimal NADP‐ME C4 cycle in appropriate cell types with the correct subcellular compartment in rice using a single construct. 13CO2 labelling and metabolomics revealed in vivo incorporation of CO2 into C4 acids (C4‐carboxylation) but no evidence of subsequent decarboxylation, indicating that the C4 cycle is partially functional. We show that physiological consequences of C4‐carboxylation in rice are minor with limited perturbation to photosynthetic light induction and no substantive effects on the global leaf transcriptome. Our results demonstrate that expression of a multigene construct is feasible in rice and provides a basis for the establishment of a C4 cycle in C3 species.
Results
A single construct for expression of five C4 enzyme transgenes
The coding sequences of five Z. mays genes encoding core enzymes of the C4 cycle were assembled in a single construct using the Golden Gate cloning system (see Materials and Methods; Engler et al., 2014). PEPC promoters from four different C4 grass species were used to drive M‐preferential gene expression, and the glycine decarboxylase P‐protein (GLDP) promoter from Flaveria trinervia was used for BS‐preferential expression (Engelmann et al., 2008; Gupta et al., 2020). The nucleotide sequence of the baculovirus envelope gp64 protein (AcV5 tag; Lawrence et al., 2003) was added as a tag to the N‐terminal end of ZmCA to aid in its detection in planta. Constructs were transformed into O. sativa spp. japonica cultivar Kitaake using stable agrobacterium‐mediated transformation and either hygromycin or bialaphos as selective agents. Two hygromycin‐resistant T0 plants (lines 1 and 29) and one bialaphos‐resistant T0 plant (line B6) were selected for further analysis based on the presence of transcripts from all five Z. mays transgenes. Homozygous plants of the three selected lines were identified in the T1 progeny, and seeds originating from those plants were used in all further experiments.
Expression, activity and localization of the C4 enzymes
Transgene transcript abundance was analysed by RNA sequencing. Figure 1a reveals some variation in transgene expression between the lines with line 1 having the highest levels of ZmCA transcripts but about two‐fold lower levels of ZmMDH transcripts in comparison with lines 29 and B6. Across all three transgenic lines, ZmCA transcript abundance was highest, followed by ZmMDH, ZmPPDK, ZmPEPC and ZmNADP‐ME, with ZmNADP‐ME transcripts detectable above background only in lines 29 and B6. Transcript abundance of the endogenous rice orthologs of the Z. mays transgenes did not change in response to transgene expression in the three transgenic lines.
Figure 1.

Expression of C4 enzymes in O. sativa. (a) Transcript abundance (in transcripts per million) of Z. mays transgenes and orthologous to them endogenous genes in wild‐type (WT) and three transgenic rice lines. CA, carbonic anhydrase; MDH, NADP‐malate dehydrogenase; NADP‐ME, NADP‐dependent malic enzyme; PEPC, PEP carboxylase; PPDK, pyruvate orthophosphate dikinase. Mean ± SD, n = 3 biological replicates. (b) Immunodetection of proteins in leaf extracts loaded on leaf area basis. Z. mays leaf extract dilution series was used for relative quantification; three plants from each transgenic line were analysed. Signal from RbcL (the large subunit of ribulose bisphosphate carboxylase oxygenase) was used as loading control. (c) Confocal micrographs of protein localization on leaf cross‐sections. Fluorescence signals are pseudo‐coloured: green ‐ protein of interest labelled with secondary antibodies conjugated with Alexa Fluor 488; magenta ‐ chlorophyll autofluorescence; blue ‐ calcofluor white‐stained cell walls. BS, bundle sheath; M, mesophyll. Scale bars = 20 µm. Localization of C4 enzymes in transgenic lines 1 and B6 is presented in Figure S1 and the summary of localization is presented in Table S1.
Accumulation of C4 enzymes was confirmed by immunoblotting of leaf protein extracts (Figure 1b). When compared to wild‐type (WT) plants, all transgenic lines accumulated higher levels of PEPC, MDH and PPDK. These abundances correspond to 20‐60% of levels in Z. mays (per leaf area) for MDH and 3‐5% for PEPC and PPDK. Consistent with the transcript abundance data, CA protein abundance was also highest in line 1. NADP‐ME protein was only just detectable in lines B6 and 29 and not detectable in line 1. In accordance with protein abundance, enzyme assays demonstrated significantly increased activities of PEPC, MDH and PPDK compared to WT in all three transgenic lines whereas NADP‐ME activity was only higher in lines B6 and 29 (Table 1). PEPC, MDH, NADP‐ME and PPDK activities were increased by up to three‐fold, 13‐fold, 15‐fold and seven‐fold compared to WT, reaching values that were up to 2%, 47%, 2% and 10% of those reported in maize, respectively. No increase in CA activity was detected in transgenic lines, likely due to the high‐activity levels of the endogenous chloroplast‐targeted CA isoform present in WT rice. Rubisco activity, chlorophyll content and leaf dry weight were all unaltered compared to WT (Table 1) and no growth phenotype was observed in the transgenic plants.
Table 1.
Enzyme activity and leaf parameters determined on wild‐type (WT) O. sativa and three transgenic lines expressing enzymes of the C4 metabolic pathway. Z. mays enzyme activity rates are given for comparison
| Parameter | O. sativa | Z. mays | |||
|---|---|---|---|---|---|
| WT | Line 1 | Line B6 | Line 29 | ||
| Enzyme activity | |||||
| PEPC, µmol m‐2 s‐1 | 2.40 ± 0.23a | 7.15 ± 0.32b | 6.14 ± 0.66b | 7.01 ± 1.02b | 315.20 |
| MDH, µmol m‐2 s‐1 | 11.18 ± 1.59a | 104.01 ± 14.02b | 147.93 ± 27.07b | 152.87 ± 34.08b | 323.93 |
| NADP‐ME, µmol m‐2 s‐1 | 0.14 ± 0.03a | 0.23 ± 0.05a | 0.96 ± 0.05c | 0.60 ± 0.03b | 48.66 |
| PPDK, µmol m‐2 s‐1 | 0.31 ± 0.10a | 4.75 ± 0.33c | 3.53 ± 0.27b | 3.66 ± 0.14b | 37.81 |
| CA, mol m‐2 s‐1 bar‐1 | 15.74 ± 0.96a | 15.01 ± 1.60a | 15.67 ± 1.58a | 16.22 ± 2.84a | 9.71 |
| Rubisco, µmol m‐2 s‐1 | 83.59 ± 5.63a | 74.12 ± 7.40a | 74.45 ± 6.13a | 65.00 ± 11.45a | 47.68 |
| LMA, g (dry weight) m‐2 | 57.62 ± 0.22a | 59.52 ± 0.15a | 69.52 ± 0.55a | 54.76 ± 0.33a | |
| Chlorophyll (a + b), mmol m‐2 | 0.53 ± 0.02a | 0.53 ± 0.03a | 0.52 ± 0.04a | 0.55 ± 0.04a | |
| Chlorophyll a/b | 4.57 ± 0.08a | 4.44 ± 0.12a | 4.48 ± 0.06a | 4.65 ± 0.10a | |
Mean ± SE, n = 4 biological replicates. Significance was evaluated by one‐way ANOVA and Tukey’s post hoc test, letters indicate significant differences between the groups (α> 0.05).
Abbreviations: CA, carbonic anhydrase; LMA, leaf mass per area; MDH, malate dehydrogenase; NADP‐ME, NADP‐dependent malic enzyme; PEPC, PEP carboxylase; PPDK, pyruvate orthophosphate dikinase; Rubisco, ribulose bisphosphate carboxylase oxygenase.
To determine whether the C4 enzymes accumulated in the correct cellular compartment, immunolocalization was performed with specific antibodies against the Z. mays enzymes or, in the case of CA, the AcV5 tag, using thin leaf cross‐sections and laser confocal microscopy (Figure 1c, Figure S1, Table S1). PEPC was correctly compartmentalized in the cytosol of M cells in lines 29 and B6 but was also present in BS cells in line 1; MDH was present in both cell‐types in all three lines rather than being M cell‐specific, as it would in a C4 plant; CA‐AcV5 was correctly localized to M cells in line 29 but was in both cell‐types in the other two lines; and NADP‐ME could not be detected in any of the lines. PPDK accumulated in both cell‐types in all three transgenic lines, as it does in maize (Majeran et al., 2005). Given these accumulation patterns, we predicted that carboxylation by PEPC would be possible in M cells of all three lines, as would conversion of oxaloacetate to malate by MDH. Metabolic consequences beyond those steps could not be predicted, but the inability to detect NADP‐ME in BS cells of the transgenic lines suggested that decarboxylation of malate was unlikely to occur.
Detection of C4‐carboxylation
13CO2 labelling experiments were performed to assess whether there was any C4 pathway flux in transgenic lines. Leaves from WT and transgenic plants were supplied with 13CO2 under ambient growth conditions using a custom‐designed labelling chamber (Figure S2; see Method S2). Isotopomers of C4 pathway intermediates and Calvin–Benson cycle (CBC) intermediates were quantified by reverse‐phase liquid chromatography, coupled to tandem mass spectrometry (LC‐MS/MS), after both 13CO2‐pulse and 13CO2‐pulse/12CO2 chase labelling experiments.
When WT plants were pulse‐labelled with 13CO2, 13C enrichment rose rapidly in 3‐phosphoglycerate (3PGA) and other CBC intermediates ‐ dihydroxyacetone phosphate (DHAP) and ribulose‐1,5‐bisphosphate (RuBP) ‐ (up to 90% 13C enrichment after 600 s), and almost as rapidly in the photorespiratory intermediate 2‐phosphoglycolate (2PG), but there was little 13C enrichment in C4 acids, malate and aspartate (Figure 2; Figure S3). These labelling patterns are typical for C3 species, as shown previously in Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum) and cassava (Manihot esculenta) (Arrivault et al., 2019; Hasunuma et al., 2009; Ma et al., 2014; Szecowka et al., 2013). By contrast, 13C enrichment of malate rose faster in the three transgenic rice lines than in WT (Figure 2), with the m1 isotopomer as well as the m2 and m3 isotopomers being more abundant than in WT (Figure 3). Line 29 had the highest 13C enrichment in malate (Figure 2, Figure 3). Similar results were observed for aspartate where line B6 had the highest enrichment (Figure 2, Figure 3). Calculation of 13C enrichment half times confirmed the faster rise in 13C enrichment of malate and aspartate in all three transgenic lines (Table S2).
Figure 2.

13C enrichment (%) during 13CO2‐pulse labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway. 3PGA, 3‐phosphoglycerate; DHAP, dihydroxyacetone phosphate; PEP, phosphoenolpyruvate; RuBP, ribulose‐1,5‐bisphosphate; 2PG, 2‐phosphoglycolate. The x‐axes show the pulse labelling time on a log scale. Values from the 30 s time point are mean ± SD, n = 3‐4 biological replicates. Values at all other time points are from individual samples or means of two biological replicates. Abundances of individual isotopomers are shown in Figure 3 and Figure S9. The original data are presented in Data S1.
Figure 3.
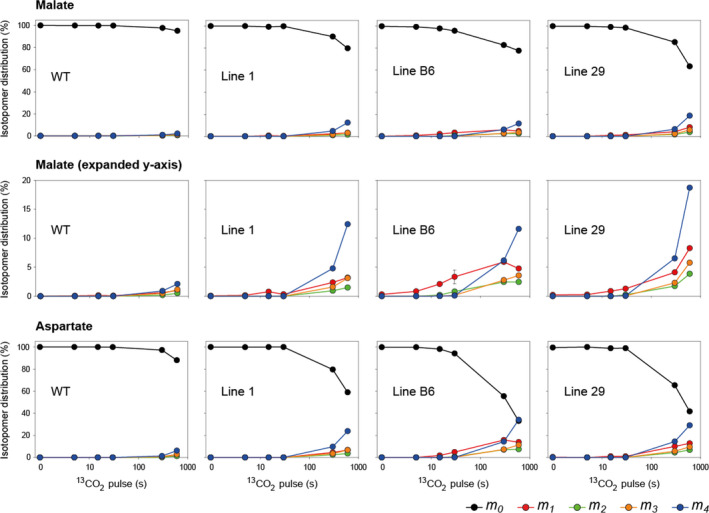
Isotopomer distribution (%) of malate and aspartate during 13CO2‐pulse labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway. The relative abundance of each isotopomer (mn ) for a given metabolite is represented, where n is the number of 13C atoms incorporated. The x‐axes show the pulse labelling time on a log scale. Values from the 30 s time point are mean ± SD, n = 3‐4 biological replicates. Values at all other time points are from individual samples or means of two biological replicates. Isotopomer abundances for further metabolites are shown in Figure S9. The original data are presented in Data S1.
Rates of 13C accumulation, estimated from the initial slope of net 13C accumulation plotted against 13C‐pulse labelling time, provided a proxy for minimum 13C fluxes (Figure S4; Data S1). 13C accumulation rates were similar in the three transgenic lines and approximatively 10‐fold higher than in WT for malate and three‐fold higher for aspartate (Table S3). The summed rates of 13C accumulation in malate and aspartate in the transgenic lines were equivalent to up to 330 nmol 13C equivalents g‐1 FW h‐1, which is about 1.7% of a typical photosynthesis rate in maize under similar conditions (~190 µmol CO2 g‐1 FW h‐1) (Arrivault et al., 2016). Moreover, total measured metabolite pool sizes indicated that all three transgenic lines had significantly more malate (up to 50%) and less aspartate than the WT (Figure S5; Table S4).
The kinetics of 13C enrichment in PEP and 3PGA differs between C3 and C4 plants (Arrivault et al., 2019; Arrivault et al., 2016) and the ratio of [13C enrichment in PEP]: [13C enrichment in 3PGA], termed hereafter the PEP:3PGA enrichment ratio, can provide evidence for the operation of C4 cycle (Figure S6). In WT rice the enrichment rose more slowly in PEP than 3PGA, similar to cassava (Arrivault et al., 2019). In maize, enrichment rose slower in PEP than 3PGA (data from (Arrivault et al., 2016) is replotted in Figure S6) reflecting a large flux of unlabelled C from C4 cycle intermediates into PEP (see legend of Figure S6). The kinetics of the PEP:3PGA enrichment ratio in the three transgenic rice lines broadly resembled that of WT rice, suggesting that PEP was mostly labelled from 3PGA via the reversible reactions catalysed by phosphoglyceromutase and enolase (Furbank and Leegood, 1984). However, the enrichment ratio was slightly lower at the first time points suggesting the operation of C4 cycle at a low level (Figure S6, significant for lines B6 and 29). Calculation of 13C enrichment half times confirmed the slightly slower rise in enrichment in PEP in lines B6 and 29 compared to WT rice (Table S2).
No consistent evidence of loss of label from malate or aspartate in the transgenic lines and no increase in 13C labelling of 3PGA or other CBC intermediates were found during the pulse/chase labelling (Figures S7 and S8). These results were corroborated by the pulse labelling kinetics and the rates of 13C accumulation in RuBP, DHAP, pyruvate and 2PG that were broadly similar between the transgenic and WT plants (Figure 2, Figure S9). The transgenic lines had more PEP and RuBP than the WT plants, but there were no significant differences in 3PGA, DHAP, pyruvate and 2PG content (Figure S5, Table S4). Movement of the 13C label to the citric acid cycle in mitochondria was evident from the 13C labelling of citrate being readily detectable in the three transgenic lines but not in WT rice (Figure 2, Figure S9). Calculation of half times confirmed the faster rise in 13C enrichment of citrate in all three transgenic lines (Table S2). This enrichment was low in percentage terms and no significant differences in total citrate amount were detected (Figure S5, Table S4).
Effects of enhanced C4‐carboxylation on C3 photosynthesis
To determine whether the altered metabolism observed in transgenic lines had a fitness cost, we carried out comprehensive gas‐exchange and fluorescence analysis. At steady‐state, when leaves were adapted to photosynthetic photon flux density (PPFD) of 1500 µmol m‐2 s‐1, no significant differences in net CO2 assimilation rate, stomatal conductance to H2O or the ratio between intercellular and ambient pCO2 were detected between the transgenic lines and WT (Table 2). The response of CO2 assimilation rate, stomatal conductance and Photosystem II (PSII) electron transport rate (ETR) to varying intercellular pCO2 for the three transgenic lines were similarly not significantly different from WT (Figure 4a), nor were the maximum carboxylation rate of Rubisco or the rate of photosynthetic electron transport (Table 2). The rate of respiration in the dark was not affected in the transgenic rice lines (Table 2).
Table 2.
Gas‐exchange and fluorescence parameters of wild‐type (WT) O. sativa and three transgenic lines expressing enzymes of C4 metabolic pathway
| Parameter | WT | Line 1 | Line B6 | Line 29 |
|---|---|---|---|---|
| Rd, µmol CO2 m‐2 s‐1 | 1.05 ± 0.05a | 1.11 ± 0.22a | 1.12 ± 0.10a | 1.05 ± 0.04a |
| FV/FM | 0.79 ± 0.01b | 0.78 ± 0.01b | 0.75 ± 0.01a | 0.77 ± 0.01ab |
| Ci/Ca | 0.74 ± 0.01a | 0.75 ± 0.02a | 0.75 ± 0.02a | 0.76 ± 0.02a |
| A, µmol CO2 m‐2 s‐1 | 29.0 ± 1.2a | 26.5 ± 2.7a | 23.7 ± 2.3a | 22.2 ± 1.9a |
| gs_H2O, mol H2O m‐2 s‐1 | 0.60 ± 0.05a | 0.55 ± 0.05a | 0.48 ± 0.05a | 0.45 ± 0.03a |
| V cmax , µmol CO2 m‐2 s‐1 | 132 ± 12a | 111 ± 20a | 95 ± 18a | 88 ± 13a |
| J, µmol e‐ m‐2 s‐1 | 144 ± 8a | 140 ± 16a | 121 ± 17a | 119 ± 8a |
| J/V cmax | 1.11 ± 0.06a | 1.32 ± 0.11a | 1.32 ± 0.12a | 1.39 ± 0.12a |
| TPU, µmol CO2 m‐2 s‐1 | 10.6 ± 0.4a | 11.0 ± 1.0a | 9.4 ± 1.1a | 9.5 ± 0.6a |
Leaf dark respiration rates (Rd) and the maximum quantum efficiency of Photosystem II (FV/FM) were determined after 40‐min dark adaptation. Ratio between intercellular and ambient pCO2 (Ci/Ca), net CO2 assimilation rates (A) and stomatal conductance to H2O (gs_H2O) were determined in steady‐state conditions at 1500 µmol m‐2 s‐1 PPFD and Ca = 37 Pa. Maximum carboxylation rate allowed by Rubisco (V cmax ), rate of photosynthetic electron transport based on NADPH requirement (J), and triose phosphate use (TPU) were determined by fitting the A‐Ci response curves (Figure 4a). Mean ± SE, n = 5 biological replicates for WT, n = 4 otherwise. Statistical analysis was performed using one‐way ANOVA and Tukey’s post hoc test, letters indicate significant differences between the groups (α > 0.05).
Figure 4.
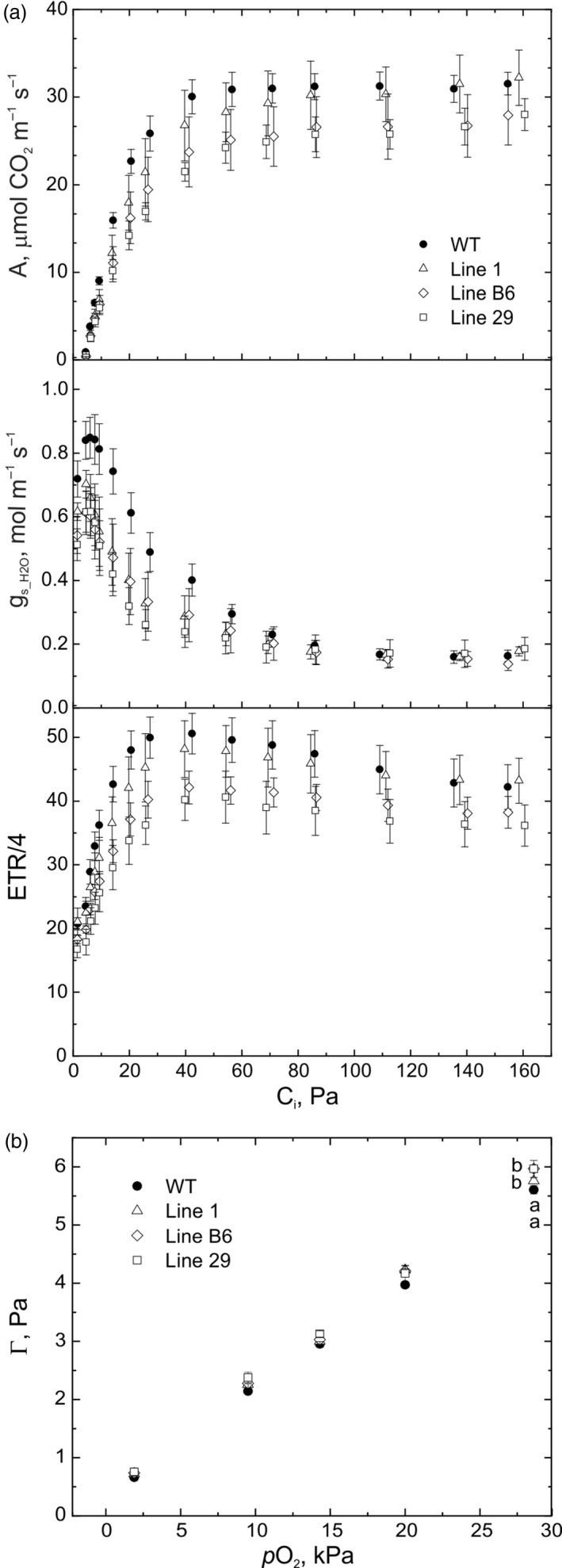
Gas‐exchange and fluorescence analysis of wild‐type (WT) O. sativa and three transgenic lines expressing enzymes of the C4 metabolic pathway. (a) A, net CO2 assimilation rate; gs_H2O, leaf stomatal conductance to water vapour; ETR/4, light‐driven electron transport rate through PHotosystem II divided to four, measured at different intercellular pCO2 (Ci), PPFD of 1500 µmol m‐2 s‐1 and ambient pO2 (20 kPa). Parameters determined by fitting the A‐Ci response curves and statistical analysis are provided in Table 2. (b) Leaf CO2 compensation point (Г). Mean ± SE, n = 5 biological replicates for WT, n = 4 otherwise. Statistical analysis was performed using one‐way ANOVA and Tukey’s post hoc test, letters indicate significant differences between the groups (α > 0.05) .
Since engagement of the C4 cycle in transgenic rice was predicted to influence the CO2 compensation point (Ermakova et al., 2020), this parameter was determined at different atmospheric O2 levels (Figure 4b). At pO2 between 1.9 and 20 kPa, the CO2 compensation points of transgenic lines did not differ from WT. However, at pO2 of 28.6 kPa, above the ambient level, lines B6 and 29 had a significantly higher CO2 compensation point compared to WT rice and line 1 (Figure 4b). In addition, the maximal dark‐adapted quantum efficiency of PSII, which is often decreased in plants subjected to various stresses (Murchie and Lawson, 2013), was significantly lower in line B6 (Table 2). These results pointed to a potentially lower resilience of the transgenic plants to stress conditions.
To further explore these effects, we studied the induction of photosynthesis over the first 20 min of illumination on plants grown under normal (400 µmol photons m−2 s−1) and low irradiance (200 µmol photons m−2 s−1). No significant differences in CO2 assimilation rate, stomatal conductance, ΦPSII (the effective quantum yield of PSII) and non‐photochemical quenching (NPQ, a measure of the absorbed light energy that is actively dissipated as heat in the PSII antennae) were found between the plants grown under normal light (Figure S10). However, when grown at a lower irradiance, all three transgenic lines had a slower induction of CO2 assimilation rate and stomatal conductance during the first 5 min of illumination (Figure 5). After 10 min of illumination, lines B6 and 29 still showed reduced CO2 assimilation rates whereas after 15 min neither of the transgenic lines differed from WT. Stomatal conductance in plants of line 29 was still lower than in WT after 15 min of illumination but after 20 min this difference was greatly diminished (Figure 5). Importantly, the ratio of intercellular and ambient pCO2 did not differ between the lines and WT indicating that stomatal conductance did not impose direct limitations to the assimilation rate. Although differences in ΦPSII were not significant, NPQ was significantly higher in all three transgenic lines after 5 min of illumination and in line B6 NPQ remained significantly higher also after 20 min of induction (Figure 5).
Figure 5.
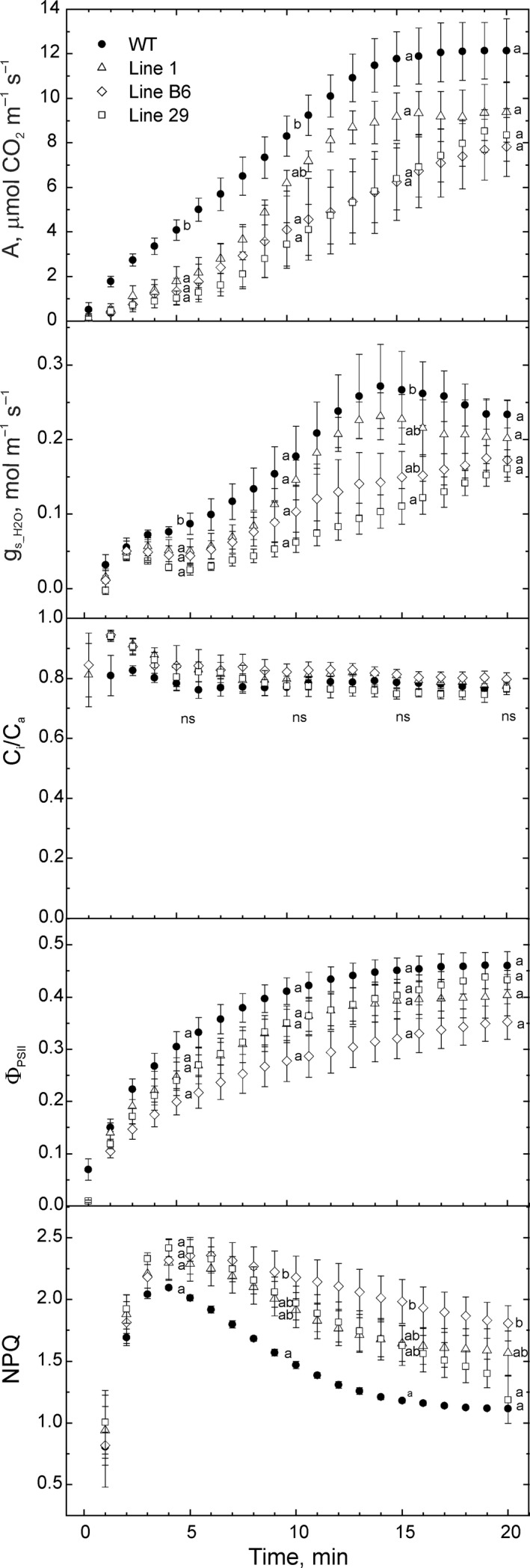
Gas‐exchange and fluorescence analysis of wild‐type (WT) O. sativa and three transgenic lines expressing enzymes of the C4 metabolic pathway grown at lower irradiance of about 200 µmol m‐2 s‐1 during the first 20 min after dark‐to‐light transition. Measurements were done on 40‐min dark‐adapted leaves at PPFD of 500 µmol m‐2 s‐1 and ambient pO2 (20 kPa). A, net CO2 assimilation rate; gs_H2O, leaf stomatal conductance to water vapour; Ci/Ca, ratio between intercellular and ambient pCO2; ΦPSII, quantum efficiency of Photosystem II; NPQ, non‐photochemical quenching. Mean ± SE, n = 5 biological replicates for WT, n = 4 otherwise (α> 0.05). Statistical analysis was performed at 5, 10, 15 and 20 min after the beginning of illumination using one‐way ANOVA and Tukey’s post hoc test, letters indicate significant differences between the groups (α > 0.05); ns, not significant.
Differentially regulated genes
Given that overexpression of functional C4 enzymes, and the concomitant establishment of C4‐carboxylation, produced minimal effects on the physiological response of the C3 leaf we sought to determine whether the lack of phenotypic response was attributable to compensatory mechanisms arising through altered gene expression. In addition, we investigated whether alteration of the expression of genes encoding enzymes or transporters in related biochemical pathways might help to explain the lack of a phenotypic perturbation or provide insight into why a fully operational C4 cycle was not established. To address these questions, WT and transgenic plants were subject to transcriptome sequencing and differential expression analysis.
In total, only 58 genes (0.2% of all genes) were consistently differentially expressed when transgenic lines were compared to WT control (Figure S11). Thus, the transcriptome phenotype is minimally perturbed, mirroring the subtle physiological phenotype. Although several genes that are differentially expressed lack functional annotations, none of the detected genes with functional annotations encode proteins that are known to transport or convert metabolites of the core C4 cycle. Thus, compensatory changes to the transcriptome that could divert malate from being decarboxylated in BS chloroplast did not occur.
Although changes to the expression of genes encoding C4 cycle‐related enzymes and transporters were not detected, shared changes in gene expression of several photosynthesises‐related genes were observed (Table S5). These include genes whose products function in the regulation of stomatal movement, photomorphogenesis, the circadian clock, thioredoxin(‐like) genes and multiple genes involved in the production of xanthophyll. Collectively these may contribute to the moderate effect on CO2 assimilation that was observed. Moreover, it is noteworthy that the rice ortholog of the Arabidopsis gene PROTEIN DISULFIDE ISOMERASE 6, which is an attenuator of D1 protein synthesis facilitating photoinhibition in response to high light (Wittenberg et al., 2014), is upregulated in the transgenic lines. Enhanced susceptibility to photoinhibition may also help to explain the enhanced NPQ and slightly decreased maximum PSII efficiency observed in the transgenic lines.
Discussion
Multiple C4 enzymes can be functionally expressed in rice from a single construct
Engineering C4 photosynthesis into C3 plants is proposed to be a major way to increase radiation, nitrogen and water‐use efficiencies and consequently crop yield (Hibberd et al., 2008; Schuler et al., 2016). In this study, we used a single construct to introduce five enzymes of the C4 pathway into rice, exploiting previously reported cell‐preferential promoters to drive expression of maize cDNA sequences (Engelmann et al., 2008; Gupta et al., 2020). Less than half of the screened events showed expression of all five transgenes (Figure S12), possibly due to deletion of key regulatory elements by recombination events either in agrobacteria or during the T‐DNA integration. Protein levels (Figure 1b) and extractable enzyme activity (Table 1) in homozygous segregants from three independent homozygous lines reflected relative transcript levels (Figure 1a) and were consistent with previous reports of expression from cDNAs encoding C4 enzymes (Fukayama et al., 2001; Taniguchi et al., 2008). However, protein levels were substantively lower than those seen when genomic gene sequences were expressed in rice (Miyao et al., 2011) suggesting that intron sequences or other untranslated regions may elevate levels of transcripts, and hence protein, in the context of the rice leaf. In addition, considerable variation in gene and protein expression between the three transgenic lines demonstrated that genomic context and selective down‐regulation of some transgenes in the T‐DNA may affect expression patterns.
Of the five C4 enzymes, NADP‐ME showed the least consistent and lowest protein accumulation levels in all three transgenic lines. The NADP‐ME cDNA sequence was driven by the GLDP promoter shown to be active in bundle sheath and vascular cells (Engelmann et al., 2008). Although the strength of this promoter relative to the M‐preferential PEPC promoter is unknown, line B6 did have appreciable NADP‐ME expression at the transcript level, detectable NADP‐ME protein by immunoblotting, and an almost seven‐fold increase in enzyme activity above the albeit very low basal value in WT (Figure 1, Table 1). However, the level of increased activity was insufficient to detect decarboxylation of malate in vivo.
Operation of a partial C4 cycle in rice
Labelling of C4 acids—malate and aspartate—was consistently higher in the three independent transgenic lines than in WT rice plants (Figure 2, Figure 3), indicating substantially increased fixation of CO2 by PEPC in lines expressing the Z. mays PEPC enzyme. Rates of 13C accumulation in malate were similar in the three transgenic lines and approximatively 10‐fold higher than in WT (Table S3). This increase in estimated flux was larger than the 2.5‐ to three‐fold increase in PEPC activity and resembled the 10‐fold increase in MDH expression in these lines (Table 1). This over‐proportional increase in flux indicates that Z. mays PEPC can operate efficiently in vivo in rice, perhaps more efficiently than the endogenous rice PEPC due to the altered kinetic properties of the C4 enzyme, that is, insensitivity to inhibition by post‐translational modification and product‐inhibition by malate (Endo et al., 2008). Nonetheless, the estimated flux at PEPC in the transgenic rice lines was only about 2% of that required during photosynthesis in maize and thus corresponding to the in vitro PEPC activity (Table 1). Thus, a considerably larger increase in PEPC expression will likely be required to establish C4 photosynthesis in rice.
The pulse‐chase experiments provided no compelling evidence for C4 acid decarboxylation by the introduced Z. mays NADP‐ME or for re‐fixation of CO2 into CBC intermediates in the transgenic lines. It should be noted that the 13CO2 pulse was kept short (30 s) so that the m1 isotopomers of malate and aspartate were the most abundant labelled isotopomers (Hatch, 1971). With such a short pulse, 13C enrichments for malate and aspartate were less than 2% in the transgenic lines and likely too low to detect their decarboxylation. Another confounding factor is that there was already substantial labelling of the CBC intermediates by the end of the 30 s pulse (around 30%), so movement of 13C from the weakly labelled pools of C4 acids into CBC intermediates would have been difficult to detect above this background. However, a slight delay in labelling of PEP in the transgenic lines, potentially caused by a flux of unlabelled C from pyruvate into PEP at early times during a pulse (Figure S6, see also Arrivault et al., 2016; Hatch, 1971; Hatch and Slack, 1966), indicated that a C4 cycle may be operating, albeit at very low rates. This label disequilibrium between 3PGA and PEP may provide a proxy for flux around the C4 cycle, as suggested by the significant changes in lines B6 and 29, which had the largest increase in NADP‐ME activity. Consistently higher 13C labelling of citrate in all three transgenic lines (Figure 2, Figure S9) could be explained by movement of 13C‐labelled C4 acids (malate, oxaloacetate or fumarate) from the cytosol into the mitochondria and the citric acid cycle. Although small in absolute terms, entry of organic acids into the citric acid cycle could lead to futile cycling of C4 acids and have an adverse impact on operation of a photosynthetic C4 cycle.
C4‐carboxylation has minimal effects on C3 leaf fitness
Previous studies examining physiological phenotypes in rice overexpressing individual enzymes of the C4 cycle have struggled to find effects (Giuliani et al., 2019; Miyao et al., 2011; Taniguchi et al., 2008), except for rice expressing the Z. mays NADP‐ME cDNA driven by the Arabidopsis chlorophyll a/b‐binding protein promoter (Takeuchi et al., 2000; Tsuchida et al., 2001), which severely inhibited photosynthesis and growth. Indeed, this has led authors to propose that high levels of activity and expression of C4 cycle enzymes in a C3 leaf is neutral under normal growth conditions (Karki et al., 2020). Fukayama et al. (2003) proposed that in rice with high‐level expression of maize PEPC, flux through PEPC in vivo was low, as PEP levels were limiting. Similarly, MDH overexpression alone had little physiological effect, potentially because of low availability of oxaloacetate in chloroplasts (Kandoi et al., 2018). Conceivably, concurrent overexpression of five C4 enzymes was required to detect 13C flux into malate, as well as the photosynthetic phenotype seen here. Synthesis of malate in the transgenic rice lines produced here presumably occurred in chloroplasts and consumed NADPH produced by electron transport. This additional consumption of NADPH could potentially inhibit CO2 fixation and deplete reducing power required for other reactions. All three transgenic lines showed an extended photosynthetic dark‐to‐light induction (Figure 5), and the severity of this phenotype was greater in lines B6 and 29 showing higher CO2 flux to C4 acids (Figure 3). This extended induction phase could be related to slower opening of stomata (Lawson and Blatt, 2014), autocatalytic build‐up of metabolites (Walker, 1973) or delayed light‐activation of enzymes (Anderson et al., 1978). Although enzymes of the C4 cycle establish guard cell turgor (Santelia and Lawson, 2016), stomatal conductance is not the primary cause of this phenomenon since the Ci/Ca ratio was similar in all plants (Figure 5). Depletion of reducing power, however, could down‐regulate activity of the chloroplast thioredoxin reductase that uses NADPH to activate CBC enzymes and ATP synthase specifically during the dark‐to‐light transition and under low light (Nikkanen et al., 2018; Yoshida and Hisabori, 2016). This reduced activation could contribute to the build‐up of ΔpH and result in the slower NPQ relaxation observed in the transgenic lines (Figure 5). Additionally, reduced FV/FM and the strongest NPQ phenotype during the induction seen in line B6 (Table 2, Figure 5) could be a result of the enhanced NADP‐ME abundance seen in this line, leading to a mild version of the chlorotic phenotype reported by Tsuchida et al. (2001).
in C3 plants, chloroplastic MDH is part of the malate shuttle; it contributes to photoprotection by oxidizing stromal electron acceptors (Scheibe and Stitt, 1988) and enables regulation of gene expression by equilibrating the redox state between cellular compartments (Dietz et al., 2016). C3 MDH is regulated by thioredoxin m and thus is active only when NAPDH accumulates and the thioredoxin system is pushed into a more reduced state (Collin et al., 2003; Scheibe, 1987). In C4 plants, the activation state of MDH is dynamically regulated by the ratio of NADPH to NADP+ (Ashton and Hatch, 1983; Rebeille and Hatch, 1986). The higher malate efflux from chloroplasts may potentially contribute to the differential regulation of multiple genes involved in light sensing, stomatal regulation and the circadian clock observed in the RNA profiles of the transgenic lines (Table S5). In summary, taken together these results indicate that the C4 photosynthetic enzymes expressed in the transgenic lines are functional in vivo and catalyse a partial C4 pathway.
Concluding comments
For the first time, we demonstrate that a partial C4 pathway can be established in rice by transformation with a single construct harbouring coding sequences for five enzymes of C4 metabolism. Whilst expression levels of these enzymes require improvement, the cell‐specific expression patterns were largely appropriate for two‐cell C4 photosynthesis and the observed photosynthetic phenotypes of the transgenic plants were consistent with C4‐carboxylation occurring in vivo. These results suggest that a full C4 metabolic pathway may be achievable in rice.
Materials and methods
Generation of transgenic plants
Two constructs, EC18089 and EC18089B (Figure S12), were assembled in a plant binary vector pAGM4723 using the Golden Gate MoClo Plant Parts Kit (Engler et al., 2014). The first expression module was occupied either by the hygromycin phosphotransferase gene (hpt) in EC18089 or bialaphos resistance gene (bar) in EC18089B, in both cases the Z. mays ubiquitin promoter. Other modules were identical in the two constructs. The second module was occupied by the coding sequence of ZmPEPC (GRMZM2G083841) driven by the Panicum miliaceum PEPC promoter + 5’‐UTR (Gupta et al., 2020). The third module was occupied by the coding sequence of ZmMDH (GRMZM2G129513) under the control of the Z. mays PEPC promoter (Matsuoka et al., 1994). The fourth module was occupied by the coding sequence of ZmNADP‐ME (GRMZM2G085019) under the control of the F. trinervia GLDP promoter (Engelmann et al., 2008). The fifth module was occupied by the coding sequence of ZmPPDK (GRMZM2G097457) driven by the Setaria viridis PEPC promoter + 5’‐UTR (Gupta et al., 2020). The sixth module was occupied by the coding sequence of ZmCA (GRMZM2G348512) driven by the Urochloa maxima PEPC promoter + 5’‐UTR (Gupta et al., 2020). All coding and promoter sequences were domesticated for the Golden Gate cloning system (Engler et al., 2014). The bacterial terminator tNos was used in all modules. Both constructs were verified by sequencing and transformed into Agrobacterium tumefaciens strain AGL1 for stable rice transformation (Toki et al., 2006) as described in detail in Method S1.
T0 plants were analysed for insertion copy number of the hpt or bar genes by droplet digital PCR (iDNA genetics, UK). T0 plant of line 1 carried three copies of hpt, whilst T0 plants of lines 29 and B6 had one copy of hpt and bar, respectively. Analysis of hpt copy number in the T1 progeny of line 1 suggested that the three insertions segregated as a single genetic locus. Plants containing homozygous insertions were identified in T1 progenies of the three transgenic lines and seeds of those plants were thereafter used in all experiments. From a total of 47 independent transformation events, 25 were examined, and of these, less than half showed expression of all five genes at the transcript level (Figure S13).
Plant growth conditions
Rice plants were grown in a controlled environment chamber (Model PGC Flex, Conviron, Winnipeg, MB, Canada) under ambient CO2 partial pressure, 16‐h photoperiod, 28 °C day, 22 °C night and 60% humidity. Irradiance of 400 μmol photons m−2 s−1 (if not stated otherwise) was supplied by a mixture of fluorescent tubes (Master TL5 HO 54W/840, Philips Lighting, The Netherlands) and halogen incandescent globes (42W 2800K warm white clear glass 630 lumens, CLA, Brookvale, Australia). Plants were individually grown in 1‐L pots in a soil mix composed by 80% peat/10% perlite/10% vermiculite (pH 5.6‐5.8) mixed with 5 g of slow‐release fertilizer (Osmocote, Evergreen Garden Care, Australia) supplied once at the beginning of the growth cycle. All pots were kept at field water capacity. Z. mays cv. B73 plants were grown in a controlled environment chamber using the same settings except that the light was supplied by 1000W red sunrise 3200K lamps (Sunmaster Growlamps, Solon, OH).
For 13CO2 labelling experiments, rice plants were grown in 1‐L pots in a 2:1 mixture of peat substrate and medium‐sized grain quartz sand (Einheitserdewerke Werkverband e.V, Sinntal‐Altengronau, Germany), containing 0.66 mL L‐1 Plantacote Depote 4M (Wilhelm Haug GmbH & Co. KG, Ammerbuch, Germany) and 0.66 mL L‐1 Fetrilon Combi (COMPO EXPERT GmbH, Muenster, Germany) fertilizer. Plants were grown in a controlled environment chamber with a 16‐h photoperiod and an irradiance of 350 μmol photons m−2 s−1 provided by LED lights, day/night temperatures of 26 °C/22 °C and constant 70% humidity. Pots were submerged in water. The plants were vegetatively propagated by detaching and re‐planting tillers at 35 days after sowing (DAS) and used for 13CO2 labelling experiments at 60 and 61 DAS. An additional 13CO2 labelling experiment was performed at 79 DAS.
RT‐PCR and RNA sequencing
Leaf discs were collected from the mid‐distal leaf blade portion of the youngest fully expanded leaf from the central stem of 4‐week‐old rice plants, frozen in liquid N2 and stored at −80 °C. Frozen samples were homogenized using a Qiagen TissueLyser II (Qiagen, Venlo, The Netherlands). RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Venlo, The Netherlands). DNA from the samples was removed using an Ambion TURBO DNA free kit (Thermo Fisher Scientific, Tewksbury, MA) and RNA quality was determined using a NanoDrop (Thermo Fisher Scientific, Tewksbury, MA). For RT‐PCR, 200 ng of total RNA were reverse transcribed into cDNA using an RT2 HT First Strand cDNA synthesis kit (Qiagen, Venlo, The Netherlands). Primers listed in Table S6 were used for RT‐PCR and amplicons were visualized in 1% agarose gels.
For RNA sequencing, leaf discs from three plants were pooled together as one biological replicate and three biological replicates per genotype were analysed. cDNA synthesis, library preparation and sequencing were performed by BGI (https://www.bgi.com/). The raw sequence reads are available from EBI array express under the accession number E‐MTAB‐9129. Trimmomatic v0.39 (Bolger et al., 2014) was used to trim off the sequencing adapters and remove low quality bases using the following settings: LEADING:20 TRAILING:20 SLIDINGWINDOW:5:20 HEADCROP:1 MINLEN:35. The most recent version of the complete set of O. sativa transcripts (Osativa_323_v7.0.transcript.fa) was obtained from Phytozome V13 (Goodstein et al., 2011). The transcript sequences with correct 5’UTR and 3’UTR sequences (as per promoters and terminators used in the construct) corresponding to ZmCA, ZmPEPC, ZmMDH, ZmPPDK and ZmME were added to this file so that the transgenes could be simultaneously quantified with the rice transcriptome. The quality filtered trimmed reads were mapped to this modified reference transcriptome using Salmon (Patro et al., 2017). Prior to differential expression testing, the read counts for multiple isoforms of the same gene were summed to produce a single transcript abundance estimate per gene locus. These counts were then used as input to test for differential expression using DESeq2 v3.9 (Love et al., 2014). Genes were considered differentially expressed between WT and transgenic plants if the Benjamini–Hochberg adjusted P‐value was ≤0.01. The read counts for genes of the WT and transgenic rice lines are provided in Data S2. Accession numbers for endogenous rice genes that are orthologous to C4 transgenes and were used for comparison in Figure 1a are: OsKitaake01g256600.1.p (CA), OsKitaake08g251400.2.p (MDH), OsKitaake01g064200.1.p (NADP‐ME), OsKitaake02g105200.1.p (PEPC), OsKitaake05g157000.1.p (PPDK).
Enzyme assays and immunodetection
Enzyme activities were determined using leaf extracts from fresh or frozen leaf tissue. For PPDK activity, a leaf disc was taken from the portion of leaf illuminated with the LI‐6800 (LI‐COR Biosciences, Lincoln, NE) during gas‐exchange measurements and immediately ground in a glass homogenizer in extraction buffer (Ashton et al., 1990; Voznesenskaya et al., 2003). PEPC activity was determined from the same fresh leaf extract (Pengelly et al., 2010). Additional leaf discs were frozen in liquid N2 and stored at −80 °C to later determine activities of Rubisco (Pengelly et al., 2010), NADP‐ME (Pengelly et al., 2012), MDH (Johnson and Hatch, 1970; Tsuchida et al., 2001) and CA (von Caemmerer et al., 2004). All enzyme activities were assayed at 25 °C and expressed per unit leaf surface area. Leaf chlorophyll content was determined spectrophotometrically in 80% acetone buffered with 25 mm 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (Hepes)‐KOH (Porra et al., 1989).
For western blotting, aliquots of the leaf protein extracts used for enzyme activity measurements were supplemented with 2% (w/v) sodium dodecyl sulphate, separated by polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane according to (Ermakova et al., 2019). Proteins were probed with antisera against Z. mays PEPC (1:10,000 dilution, Karki et al., 2020), Z. mays PPDK (1:20,000 dilution, Karki et al., 2020), Z. mays MDH (1:5000 dilution, Karki et al., 2020), Z. mays NADP‐ME (1:5000 dilution, Karki et al., 2020; Sonawane et al., 2018), AcV5 tag (ab49581, Abcam, Cambridge, UK, 1:3000 dilution) and tobacco Rubisco (prepared by S.M. Whitney, 1:10,000). Secondary goat anti‐rabbit horseradish peroxidase‐conjugated (Biorad, Hercules, CA) or goat anti‐mouse horseradish peroxidase‐conjugated (Agrisera, Vännäs, Sweden) antibodies were used at a 1:5000 dilution. Chemiluminescence signal from the membranes was obtained with a Western Lightning Ultra kit (Perkin Elmer, Waltham, MA) and detected by a ChemiDoc MP imaging system (Biorad, Hercules, CA). Results were analysed in Image Lab software (Biorad, Hercules, CA). Details of immunolocalization of C4 enzymes performed on leaf sections are in Method S3. Proteins were visualized under a Leica SP8 laser scanning confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) at 488 nm excitation and 546 to 600 nm emission. Simultaneously, fluorescence from calcofluor white‐stained cell walls was detected at 434 to 445 nm following excitation at 405 nm. Chlorophyll autofluorescence was captured using 633 nm excitation and 650 to 742 nm emission. Images were analysed using LAS X software (Leica Microsystems GmbH, Wetzlar, Germany).
Leaf gas‐exchange analysis
Leaf‐atmosphere CO2 and H2O exchange and chlorophyll fluorescence measurements were conducted with two portable photosynthesis systems LI‐6800 equipped with a Multiphase flashTM fluorometer circular chamber (6800‐01A) as described in detail Method S4. The LI‐COR leaf chamber was set at PPFD of 1500 µmol photons m−2 s−1 (90% red/10% blue), leaf temperature of 25 °C, leaf‐to‐air vapour‐pressure deficit of 1.0 kPa and the airflow rate of 500 µmol s−1. Multiphase flash of 10,000 µmol photons m‐2 s‐1 (ramp 25%) was applied to leaves to transiently close all PSII reaction centres and monitor the maximum fluorescence in the light (F'M) and the steady‐state fluorescence (FS). Photochemical yield of PSII was determined as ΦPSII = (F'M‐FS)/F'M (Genty et al., 1989). The light‐driven electron transport rate through PSII was determined as ETR = α*PPFD*β*ΦPSII, where α is the leaf absorbance of photosynthetic quanta (0.843 according to Björkman and Demmig, 1987) and β is the fraction of photons absorbed by PSII (0.5 according to Maxwell and Johnson, 2000).
For dark‐to‐light induction measurements, leaves were dark‐adapted for 40 min and then enclosed in the dark fluorometer chamber for 10 minutes to measure dark respiration rate, the minimum PSII fluorescence (F0) and the maximum PSII fluorescence (FM). The maximum quantum yield of PSII was calculated as FV/FM = (FM‐F0)/FM. Afterwards, the leaf was illuminated with actinic light of 500 µmol photons m‐2 s−1 to monitor photosynthetic induction. Non‐photochemical quenching was estimated as NPQ = (FM‐F'M)/F'M.
Fitting of the A‐Ci response curves
The maximum leaf carboxylation rate allowed by Rubisco, the rate of photosynthetic electron transport and triose phosphate use were obtained by fitting the A‐Ci response curves (Ca steps from 0 to 190 Pa) at atmospheric pO2 using the fitting routine (Sharkey et al., 2007). Leaf mesophyll conductance to CO2 diffusion (gm) of 6.7 µmol CO2 m‐2 s‐1 Pa‐1 was previously determined for rice (von Caemmerer and Evans, 2015). For A‐Ci response curves recorded at different O2 levels, the least square regression method was applied to the initial slope (for Ci ≤ 9 Pa) to calculate the CO2 compensation point (Γ, Pa).
13CO2 labelling and quenching procedure
Labelling chambers were custom‐designed as shown in Figure S2. Details of chambers and the labelling procedure are described in detail in Method S2. Two sets of experiments were conducted: (i) 13CO2‐pulse labelling (0, 5, 15, 30, 300 and 600 s) was performed to determine the extent of 13C incorporation into C4 acids and other intermediates, and (ii) pulse‐chase labelling (30 s 13CO2‐pulse/0, 5, 15, 30, 60, 90 and 120 s 12CO2 chase) to detect movement of label out of C4 acids and into CBC intermediates. To gain a more complete overview of the temporal kinetics of 13C incorporation into each metabolite, we analysed single samples at multiple time points. Unlabelled samples (0) were also collected from leaves placed in the labelling chamber and flushed with the unlabelled air mixture for 1 min. The order of the 13CO2‐pulse labelling and pulse‐chase labelling times was randomized.
Metabolite analyses and calculation of total pool size, 13C enrichment and isotopomer distributions
Frozen samples were homogenized using a ball mill (Tesch, Haan, Germany) at liquid nitrogen temperature. Chemicals used for quantification were from Sigma‐Aldrich (St. Louis, Missouri, USA), Roche (Basel, Switzerland) or Merck (Darmstadt, Germany). For LC‐MS/MS analysis, samples were extracted with chloroform–methanol as described in Arrivault et al. (2016). Isotopomers were measured by reverse‐phase LC‐MS/MS (malate, aspartate, PEP, RuBP, 3PGA, DHAP and 2PG; Arrivault et al., 2016) or anion‐exchange LC‐MS/MS (with modifications as described in Figueroa et al., 2016; pyruvate and citrate; Lunn et al., 2006) with authentic standards for accurate metabolite quantification. Total amounts of malate, aspartate, citrate, RuBP, DHAP and 2PG were calculated by summing isotopomers. The total amounts of PEP, pyruvate and 3PGA were determined enzymatically in freshly prepared trichloroacetic acid extracts as described in (Merlo et al., 1993), except for a modified assay buffer containing 50 mm Hepes‐KOH, pH 7.5, 200 mm KCl, 40 mm MgCl2 used for the determination of PEP and pyruvate. Net accumulation of 13C in each metabolite was calculated by multiplying the 13C enrichment by the number of C atoms in the molecule (n) and by the total amount of the metabolite. Net accumulation was plotted against 13C‐pulse labelling time (Figure S4) and the initial slope provided a proxy for minimum 13C fluxes (see Data S1 for calculation steps); these are a minimum estimate because some 13C may exit the metabolite pool during the pulse. Estimation of flux at PEPC is described in Data S1.
Statistics and reproducibility
One‐way ANOVA and Tukey post hoc test for the pairwise comparisons of means (α = 0.05) was applied to all leaf traits determined in the present study using OriginPro 2018b software (OriginLab Corp., Northampton, MA). At least three biological replicates were used for each genotype for each measurement. At least ten sets of plants were grown for various experiments. Gas‐exchange, immunodetection of C4 enzymes, 13CO2 labelling and enzyme activity assays were partially replicated on different sets of plants.
Conflict of interest
Authors declare no competing interests.
Authors contribution
ABC, JAL, JEL, MS, SvC and RTF designed research. ME, SA, RG, FD, HAC, DV, HI, RF, MG, GLB performed research. ME, SA, RG, SK, JEL, MS, SvC and RTF analysed data. ML contributed research tools. ME, SA, RG, JAL, SK, JEL, MS, SvC and RTF wrote the paper.
Supporting information
Figure S1. Confocal micrographs of C4 enzymes localization on leaf cross‐sections of Z. mays and O. sativa wild‐type (WT) and three transgenic O. sativa lines expressing C4 metabolic enzymes
Figure S2. 13CO2 labelling apparatus and quenching procedure
Figure S3. 13C enrichment (%) of wild‐type (WT) rice during (A) 13CO2‐pulse labelling and (B) pulse‐chase labelling
Figure S4. Net accumulation of 13C (nmol 13C equivalents g‐1 FW) during 13CO2‐pulse labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S5. Metabolite amounts (nmol g‐1 FW) of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S6. Relative labelling of PEP and 3PGA during 13CO2‐pulse labelling of wild‐type (WT) rice, three transgenic lines expressing enzymes of the C4 metabolic pathway, maize and cassava
Figure S7. Pulse‐chase labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S8. Isotopomer distribution (%) during pulse‐chase labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S9. Isotopomer distribution (%) during 13CO2‐pulse labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S10. Gas‐exchange and fluorescence analysis of wild‐type (WT) O. sativa and three transgenic lines expressing enzymes of the C4 metabolic pathway during a dark‐to‐light shift
Figure S11. Number of differentially regulated genes in the three transgenic O. sativa lines expressing the C4 metabolic pathway compared to WT
Figure S12. Constructs used for stable rice transformation
Figure S13. RT‐PCR detection of Z. mays gene transcripts in T0 O. sativa lines transformed with the gene construct for C4 metabolic pathway expression
Table S1. Summary of C4 enzymes localization from the confocal images on Figure 1c and Figure S1.
Table S2. Estimation of 13C enrichment half times in different metabolites in wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway during 13CO2‐pulse labelling.
Table S3. Estimation of minimum 13C fluxes using slopes of 13C accumulation (nmol 13C equivalents g‐1 FW h‐1) as a proxy in wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway during 13CO2‐pulse labelling.
Table S4. Metabolite amounts of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway.
Table S5. Genes differentially regulated in all three transgenic O. sativa lines expressing the enzymes of C4 metabolic pathway.
Table S6. Primers used for RT‐PCR.
Method S1. Generation of transgenic rice plants.
Method S2. 13CO2 labelling and quenching procedure.
Method S3. Immunolocalization of C4 enzymes on leaf sections.
Method S4. Leaf gas‐exchange analysis.
Data S1. (separate file). Isotopomer and metabolite amounts, 13C enrichments and relative isotopomer abundances in wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway.
Data S2. (separate file). The gene read counts obtained by RNA sequencing for wild‐type (WT) and transgenic rice lines expressing the enzymes of the C4 metabolic pathway.
Acknowledgements
This work was funded by a C4 Rice Project grant from Bill & Melinda Gates Foundation to the University of Oxford (2015–2019; OPP1129902), Max Planck Society (SA, HI, RF, MG, JEL, MS) and Australian Research Council (DP150101037 to ML, JEL, MS and CE140100015). Work in the SK laboratory was supported by the European Union’s Horizon 2020 research and innovation programme under grant agreement number 637765; SK is a Royal Society University Research Fellow. We thank Riya Kuruvilla, Xueqin Wang and Soumi Bala for technical support and Robert Sharwood and Spencer Whitney for the gifts of antibodies. We thank the Australian Plant Phenomics Facility supported under the National Collaborative Research Infrastructure Strategy of the Australian Government and CSIRO Black Mountain MicroImaging Centre.
Ermakova, M. , Arrivault, S. , Giuliani, R. , Danila, F. , Alonso‐Cantabrana, H. , Vlad, D. , Ishihara, H. , Feil, R. , Guenther, M. , Borghi, G. L. , Covshoff, S. , Ludwig, M. , Cousins, A. B. , Langdale, J. A. , Kelly, S. , Lunn, J. E. , Stitt, M. , von Caemmerer, S. and Furbank, R. T. (2021) Installation of C4 photosynthetic pathway enzymes in rice using a single construct. Plant Biotechnol. J., 10.1111/pbi.13487
References
- Anderson, L.E. , Nehrlich, S.C. and Champigny, M.‐L. (1978) Light modulation of enzyme activity. Activation of the light effect mediators by reduction and modulation of enzyme activity by thiol‐disulfide exchange. Plant Physiol. 61, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault, S. , Alexandre Moraes, T. , Obata, T. , Medeiros, D.B. , Fernie, A.R. , Boulouis, A. , Ludwig, M. et al.(2019) Metabolite profiles reveal interspecific variation in operation of the Calvin‐Benson cycle in both C4 and C3 plants. J. Exp. Bot. 70, 1843–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault, S. , Obata, T. , Szecówka, M. , Mengin, V. , Guenther, M. , Hoehne, M. , Fernie, A.R. et al. (2016) Metabolite pools and carbon flow during C4 photosynthesis in maize: 13CO2 labeling kinetics and cell type fractionation. J. Exp. Bot. 68:283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton, A.R. , Burnell, J.N. , Furbank, R.T. , Jenkins, C.L. and Hatch, M.D. (1990) Enzymes of C4 photosynthesis. In Methods in plant biochemistry ( Lea, P. , ed.), pp. 39–72. London: Academic press. [Google Scholar]
- Ashton, A.R. and Hatch, M.D. (1983) Regulation of C4 photosynthesis: Regulation of activation and inactivation of NADP‐malate dehydrogenase by NADP and NADPH. Arch. Biochem. Biophys. 227, 416–424. [DOI] [PubMed] [Google Scholar]
- Björkman, O. and Demmig, B. (1987) Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77 K among vascular plants of diverse origins. Planta 170, 489–504. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer, S. and Evans, J.R. (2015) Temperature responses of mesophyll conductance differ greatly between species. Plant, Cell Environ. 38, 629–637. [DOI] [PubMed] [Google Scholar]
- von Caemmerer, S. , Quick, W.P. and Furbank, R.T. (2012) The development of C4 rice: current progress and future challenges. Science 336, 1671–1672. [DOI] [PubMed] [Google Scholar]
- von Caemmerer, S. , Quinn, V. , Hancock, N.C. , Price, G.D. , Furbank, R.T. and Ludwig, M. (2004) Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant, Cell Environ. 27, 697–703. [Google Scholar]
- Collin, V. , Issakidis‐Bourguet, E. , Marchand, C. , Hirasawa, M. , Lancelin, J.‐M. , Knaff, D.B. and Miginiac‐Maslow, M. (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J. Biol. Chem. 278, 23747–23752. [DOI] [PubMed] [Google Scholar]
- Dietz, K.‐J. , Turkan, I. and Krieger‐Liszkay, A. (2016) Redox‐ and reactive oxygen species‐dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 171, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T. , Mihara, Y. , Furumoto, T. , Matsumura, H. , Kai, Y. and Izui, K. (2008) Maize C4‐form phosphoenolpyruvate carboxylase engineered to be functional in C3 plants: mutations for diminished sensitivity to feedback inhibitors and for increased substrate affinity. J. Exp. Bot. 59, 1811–1818. [DOI] [PubMed] [Google Scholar]
- Engelmann, S. , Wiludda, C. , Burscheidt, J. , Gowik, U. , Schlue, U. , Koczor, M. , Streubel, M. et al. (2008) The gene for the P‐subunit of glycine decarboxylase from the C4 species Flaveria trinervia: Analysis of transcriptional control in transgenic Flaveria bidentis (C4) and Arabidopsis (C3). Plant Physiol. 146, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. , Youles, M. , Gruetzner, R. , Ehnert, T.‐M. , Werner, S. , Jones, J.D.G. , Patron, N.J. et al. (2014) A golden gate modular cloning toolbox for plants. ACS Synthetic Biol. 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Ermakova, M. , Danila, F.R. , Furbank, R.T. and von Caemmerer, S. (2020) On the road to C4 rice: advances and perspectives. Plant J. 101, 940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova, M. , Lopez‐Calcagno, P.E. , Raines, C.A. , Furbank, R.T. and von Caemmerer, S. (2019) Overexpression of the rieske FeS protein of the cytochrome b6f complex increases C4 photosynthesis in Setaria viridis . Commun. Biol. 2: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, C.M. , Feil, R. , Ishihara, H. , Watanabe, M. , Kölling, K. , Krause, U. , Höhne, M. et al. (2016) Trehalose 6–phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 85, 410–423. [DOI] [PubMed] [Google Scholar]
- Fukayama, H. , Hatch, M.D. , Tamai, T. , Tsuchida, H. , Sudoh, S. , Furbank, R.T. and Miyao, M. (2003) Activity regulation and physiological impacts of maize C4‐specific phosphoenolpyruvate carboxylase overproduced in transgenic rice plants. Photosynth. Res. 77, 227–239. [DOI] [PubMed] [Google Scholar]
- Fukayama, H. , Tsuchida, H. , Agarie, S. , Nomura, M. , Onodera, H. , Ono, K. , Lee, B.‐H. et al. (2001) Significant accumulation of C4‐specific pyruvate, orthophosphate dikinase in a C3 plant, rice. Plant Physiol. 127, 1136–1146. [PMC free article] [PubMed] [Google Scholar]
- Furbank, R.T. and Leegood, R.C. (1984) Carbon metabolism and gas exchange in leaves of Zea mays L. Interaction between C3 and C4 pathways during photosynthetic induction. Planta 62, 457–462. [DOI] [PubMed] [Google Scholar]
- Genty, B. , Briantais, J.M. and Baker, N.R. (1989) The relationship between the quantum yield of photosynthetic electron‐transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta, 990, 87–92. [Google Scholar]
- Giuliani, R. , Karki, S. , Covshoff, S. , Lin, H.‐C. , Coe, R.A. , Koteyeva, N.K. , Evans, M.A. et al. (2019) Transgenic maize phosphoenolpyruvate carboxylase alters leaf–atmosphere CO2 and 13CO2 exchanges in Oryza sativa . Photosynth. Res. 142:153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein, D.M. , Shu, S. , Howson, R. , Neupane, R. , Hayes, R.D. , Fazo, J. , Mitros, T. et al. (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S.D. , Levey, M. , Schulze, S. , Karki, S. , Emmerling, J. , Streubel, M. , Gowik, U. et al. (2020) The C4Ppc promoters of many C4 grass species share a common regulatory mechanism for gene expression in the mesophyll cell. Plant J. 101, 204–216. [DOI] [PubMed] [Google Scholar]
- Hasunuma, T. , Harada, K. , Miyazawa, S.‐I. , Kondo, A. , Fukusaki, E. and Miyake, C. (2009) Metabolic turnover analysis by a combination of in vivo13C‐labelling from 13CO2 and metabolic profiling with CE‐MS/MS reveals rate‐limiting steps of the C3 photosynthetic pathway in Nicotiana tabacum leaves. J. Exp. Bot. 61, 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, M.D. (1971) The C4 pathway of photosynthesis. Evidence for an intermediate pool of carbon dioxide and the identity of the donor C 4 ‐dicarboxylic acid. Biochem. J. 125, 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, M.D. and Slack, C.R. (1966) Photosynthesis by sugar‐cane leaves: A new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Prieto, M.A. , Foster, C. , Watson‐Lazowski, A. , Ghannoum, O. and Chen, M. (2019) Comparative analysis of thylakoid protein complexes in the mesophyll and bundle sheath cells from C3, C4 and C3–C4 Paniceae grasses. Physiol. Plant. 166, 134–147. [DOI] [PubMed] [Google Scholar]
- Hibberd, J.M. , Sheehy, J.E. and Langdale, J.A. (2008) Using C4 photosynthesis to increase the yield of rice ‐ rationale and feasibility. Curr. Opin. Plant Biol. 11, 228–231. [DOI] [PubMed] [Google Scholar]
- Johnson, H.S. and Hatch, M.D. (1970) Properties and regulation of leaf nicotinamide‐adenine dinucleotide phosphate‐malate dehydrogenase and malic enzyme in plants with C4‐dicarboxylic acid pathway of photosynthesis. Biochem. J. 119, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoi, D. , Mohanty, S. and Tripathy, B.C. (2018) Overexpression of plastidic maize NADP‐malate dehydrogenase (ZmNADP‐MDH) in Arabidopsis thaliana confers tolerance to salt stress. Protoplasma 255, 547–563. [DOI] [PubMed] [Google Scholar]
- Karki, S. , Lin, H. , Danila, F.R. , AbuJamous, B. , Giuliani, R. , Emms, D.M. et al. (2020) A role for neutral variation in the evolution of C4 photosynthesis. bioRxiv, 104299. 10.1101/2020.05.19.104299 [DOI]
- Ku, M.S.B. , Agarie, S. , Nomura, M. , Fukayama, H. , Tsuchida, H. , Ono, K. , Hirose, S. et al. (1999) High‐level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat. Biotechnol. 17, 76–80. [DOI] [PubMed] [Google Scholar]
- Lawrence, S.D. , Novak, N.G. and Slack, J.M. (2003) Epitope tagging: a monoclonal antibody specific for recombinant fusion proteins in plants. Biotechniques 35, 488–492. [DOI] [PubMed] [Google Scholar]
- Lawson, T. and Blatt, M.R. (2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164, 1556–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H.C. , Arrivault, S. , Coe, R.A. , Karki, S. , Covshoff, S. , Bagunu, E. et al. (2020) A partial C4 photosynthetic biochemical pathway in rice. Front Plant Sci, 1581. 10.3389/fpls.2020.564463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E. , Feil, R. , Hendriks, J.H.M. , Gibon, Y. , Morcuende, R. , Osuna, D. , Scheible, W.‐R. et al. (2006) Sugar‐induced increases in trehalose 6‐phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 397, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, F. , Jazmin, L.J. , Young, J.D. and Allen, D.K. (2014) Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc. Natl Acad. Sci. USA, 111, 16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran, W. , Cai, Y. , Sun, Q. and van Wijk, K.J. (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell, 17, 3111–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran, W. and van Wijk, K.J. (2009) Cell‐type‐specific differentiation of chloroplasts in C4 plants. Trends Plant Sci. 14, 100–109. [DOI] [PubMed] [Google Scholar]
- Matsuoka, M. , Kyozuka, J. , Shimamoto, K. and Kano‐Murakami, Y. (1994) The promoters of two carboxylases in a C4 plant (maize) direct cell‐specific, light‐regulated expression in a C3 plant (rice). Plant J. 6, 311–319. [DOI] [PubMed] [Google Scholar]
- Maxwell, K. and Johnson, G.N. (2000) Chlorophyll fluorescence ‐ a practical guide. J. Exp. Bot. 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Merlo, L. , Geigenberger, P. , Hajirezaei, M. and Stitt, M. (1993) Changes of carbohydrates, metabolites and enzyme activities in potato tubers during development, and within a single tuber along astolon‐apexgradient. J. Plant Physiol. 142, 392–402. [Google Scholar]
- Miyao, M. , Masumoto, C. , Miyazawa, S.‐I. and Fukayama, H. (2011) Lessons from engineering a single‐cell C4 photosynthetic pathway into rice. J. Exp. Bot. 62, 3021–3029. [DOI] [PubMed] [Google Scholar]
- Murchie, E.H. and Lawson, T. (2013) Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J. Exp. Bot. 64, 3983–3998. [DOI] [PubMed] [Google Scholar]
- Nikkanen, L. , Toivola, J. , Trotta, A. , Diaz, M.G. , Tikkanen, M. , Aro, E.‐M. and Rintamäki, E. (2018) Regulation of cyclic electron flow by chloroplast NADPH‐dependent thioredoxin system. Plant Direct 2, e00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R. , Duggal, G. , Love, M.I. , Irizarry, R.A. and Kingsford, C. (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nat. Methods, 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly, J.J.L. , Sirault, X.R.R. , Tazoe, Y. , Evans, J.R. , Furbank, R.T. and von Caemmerer, S. (2010) Growth of the C4 dicot Flaveria bidentis: photosynthetic acclimation to low light through shifts in leaf anatomy and biochemistry. J. Exp. Bot. 61, 4109–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly, J.J.L. , Tan, J. , Furbank, R.T. and von Caemmerer, S. (2012) Antisense reduction of NADP‐malic enzyme in flaveria bidentis reduces flow of CO2 through the C4 Cycle. Plant Physiol. 160, 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J. , Thompson, W.A. and Kriedemann, P.E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 975, 384–394. [Google Scholar]
- Rebeille, F. and Hatch, M.D. (1986) Regulation of NADP‐malate dehydrogenase in C4 plants: Relationship among enzyme activity, NADPH to NADP ratios, and thioredoxin redox states in intact maize mesophyll chloroplasts. Arch. Biochem. Biophys. 249, 171–179. [DOI] [PubMed] [Google Scholar]
- Santelia, D. and Lawson, T. (2016) Rethinking guard cell metabolism. Plant Physiol. 172, 1371–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe, R. (1987) NADP+‐malate dehydrogenase in C3‐plants: Regulation and role of a light‐activated enzyme. Physiol. Plant. 71, 393–400. [Google Scholar]
- Scheibe, R. and Stitt, M. (1988) Comparison of NADP‐malate dehydrogenase activation, Qa reductionand O2 evolution in spinach leaves. Plant Physiol. Biochem. 26, 473–481. [Google Scholar]
- Schuler, M.L. , Mantegazza, O. and Weber, A.P.M. (2016) Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 87, 51–65. [DOI] [PubMed] [Google Scholar]
- Sharkey, T.D. , Bernacchi, C.J. , Farquhar, G.D. and Singsaas, E.L. (2007) Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant, Cell Environ. 30, 1035–1040. [DOI] [PubMed] [Google Scholar]
- Sonawane, B.V. , Sharwood, R.E. , Whitney, S. and Ghannoum, O. (2018) Shade compromises the photosynthetic efficiency of NADP‐ME less than that of PEP‐CK and NAD‐ME C4 grasses. J. Exp. Bot. 69, 3053–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szecowka, M. , Heise, R. , Tohge, T. , Nunes‐Nesi, A. , Vosloh, D. , Huege, J. , Feil, R. et al. (2013) Metabolic fluxes in an Illuminated Arabidopsis Rosette. Plant Cell, 25, 694–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, Y. , Akagi, H. , Kamasawa, N. , Osumi, M. and Honda, H. (2000) Aberrant chloroplasts in transgenic rice plants expressing a high level of maize NADP‐dependent malic enzyme. Planta 211, 265–274. [DOI] [PubMed] [Google Scholar]
- Taniguchi, Y. , Ohkawa, H. , Masumoto, C. , Fukuda, T. , Tamai, T. , Lee, K. , Sudoh, S. et al. (2008) Overproduction of C4 photosynthetic enzymes in transgenic rice plants: an approach to introduce the C4‐like photosynthetic pathway into rice. J. Exp. Bot. 59, 1799–1809. [DOI] [PubMed] [Google Scholar]
- Toki, S. , Hara, N. , Ono, K. , Onodera, H. , Tagiri, A. , Oka, S. and Tanaka, H. (2006) Early infection of scutellum tissue with Agrobacterium allows high‐speed transformation of rice. Plant J. 47, 969–976. [DOI] [PubMed] [Google Scholar]
- Tsuchida, H. , Tamai, T. , Fukayama, H. , Agarie, S. , Nomura, M. , Onodera, H. , Ono, K. et al. (2001) High level expression of C4‐specific NADP‐malic enzyme in leaves and impairment of photoautotrophic growth in a C3 plant, rice. Plant Cell Physiol. 42, 138–145. [DOI] [PubMed] [Google Scholar]
- Voznesenskaya, E.V. , Edwards, G.E. , Kiirats, O. , Artyusheva, E.G. and Franceschi, V.R. (2003) Development of biochemical specialization and organelle partitioning in the single‐cell C4 system in leaves of Borszczowia aralocaspica (Chenopodiaceae). Am. J. Bot. 90, 1669–1680. [DOI] [PubMed] [Google Scholar]
- Walker, D.A. (1973) Photosynthetic induction phenomena and the light activation of ribulose diphosphate carboxylase. New Phytol. 72, 209–235. [Google Scholar]
- Wittenberg, G. , Levitan, A. , Klein, T. , Dangoor, I. , Keren, N. and Danon, A. (2014) Knockdown of the Arabidopsis thaliana chloroplast protein disulfide isomerase 6 results in reduced levels of photoinhibition and increased D1 synthesis in high light. Plant J. 78, 1003–1013. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. and Hisabori, T. (2016) Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl Acad. Sci. USA, 113, E3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Confocal micrographs of C4 enzymes localization on leaf cross‐sections of Z. mays and O. sativa wild‐type (WT) and three transgenic O. sativa lines expressing C4 metabolic enzymes
Figure S2. 13CO2 labelling apparatus and quenching procedure
Figure S3. 13C enrichment (%) of wild‐type (WT) rice during (A) 13CO2‐pulse labelling and (B) pulse‐chase labelling
Figure S4. Net accumulation of 13C (nmol 13C equivalents g‐1 FW) during 13CO2‐pulse labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S5. Metabolite amounts (nmol g‐1 FW) of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S6. Relative labelling of PEP and 3PGA during 13CO2‐pulse labelling of wild‐type (WT) rice, three transgenic lines expressing enzymes of the C4 metabolic pathway, maize and cassava
Figure S7. Pulse‐chase labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S8. Isotopomer distribution (%) during pulse‐chase labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S9. Isotopomer distribution (%) during 13CO2‐pulse labelling of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway
Figure S10. Gas‐exchange and fluorescence analysis of wild‐type (WT) O. sativa and three transgenic lines expressing enzymes of the C4 metabolic pathway during a dark‐to‐light shift
Figure S11. Number of differentially regulated genes in the three transgenic O. sativa lines expressing the C4 metabolic pathway compared to WT
Figure S12. Constructs used for stable rice transformation
Figure S13. RT‐PCR detection of Z. mays gene transcripts in T0 O. sativa lines transformed with the gene construct for C4 metabolic pathway expression
Table S1. Summary of C4 enzymes localization from the confocal images on Figure 1c and Figure S1.
Table S2. Estimation of 13C enrichment half times in different metabolites in wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway during 13CO2‐pulse labelling.
Table S3. Estimation of minimum 13C fluxes using slopes of 13C accumulation (nmol 13C equivalents g‐1 FW h‐1) as a proxy in wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway during 13CO2‐pulse labelling.
Table S4. Metabolite amounts of wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway.
Table S5. Genes differentially regulated in all three transgenic O. sativa lines expressing the enzymes of C4 metabolic pathway.
Table S6. Primers used for RT‐PCR.
Method S1. Generation of transgenic rice plants.
Method S2. 13CO2 labelling and quenching procedure.
Method S3. Immunolocalization of C4 enzymes on leaf sections.
Method S4. Leaf gas‐exchange analysis.
Data S1. (separate file). Isotopomer and metabolite amounts, 13C enrichments and relative isotopomer abundances in wild‐type (WT) rice and three transgenic lines expressing enzymes of the C4 metabolic pathway.
Data S2. (separate file). The gene read counts obtained by RNA sequencing for wild‐type (WT) and transgenic rice lines expressing the enzymes of the C4 metabolic pathway.


