Summary
Lysine is the main limiting essential amino acid (EAA) in the rice seeds, which is a major energy and nutrition source for humans and livestock. In higher plants, the rate‐limiting steps in lysine biosynthesis pathway are catalysed by two key enzymes, aspartate kinase (AK) and dihydrodipicolinate synthase (DHDPS), and both are extremely sensitive to feedback inhibition by lysine. In this study, two rice AK mutants (AK1 and AK2) and five DHDPS mutants (DHDPS1–DHDPS5), all single amino acid substitution, were constructed. Their protein sequences passed an allergic sequence‐based homology alignment. Mutant proteins were recombinantly expressed in Escherichia coli, and all were insensitive to the lysine analog S‐(2‐aminoethyl)‐l‐cysteine (AEC) at concentrations up to 12 mm. The AK and DHDPS mutants were transformed into rice, and free lysine was elevated in mature seeds of transgenic plants, especially those expressing AK2 or DHDPS1, 6.6‐fold and 21.7‐fold higher than the wild‐type (WT) rice, respectively. We then engineered 35A2D1L plants by simultaneously expressing modified AK2 and DHDPS1, and inhibiting rice LKR/SDH (lysine ketoglutaric acid reductase/saccharopine dehydropine dehydrogenase). Free lysine levels in two 35A2D1L transgenic lines were 58.5‐fold and 39.2‐fold higher than in WT and transgenic rice containing native AK and DHDPS, respectively. Total free amino acid and total protein content were also elevated in 35A2D1L transgenic rice. Additionally, agronomic performance analysis indicated that transgenic lines exhibited normal plant growth, development and seed appearance comparable to WT plants. Thus, AK and DHDPS mutants may be used to improve the nutritional quality of rice and other cereal grains.
Keywords: rice, lysine metabolism, aspartate kinase, dihydrodipicolinate synthase, lysine feedback insensitive, biosynthetic pathway modification
Introduction
Cereals play an important role in agriculture worldwide, especially rice (Oryza sativa L.), which contributes significantly to the global food pool and thereby helps to achieve food and nutritional security (Vasal, 2004). Lysine, the main limiting essential amino acid (EAA) in cereals, restricts the absorption and utilization of other amino acids and proteins, which leads to nutrient deficiency in humans and other animals (Cohen et al., 2014; Lee et al., 2001; Ufaz and Galili, 2008). Thus, enhancing the lysine content in cereal grains, especially rice, is a major interest of plant breeders aiming to improve the nutritional value of grains and prevent nutrient deficiency diseases such as kwashiorkor (Toride, 2004).
Owing to the importance of lysine, regulating the biosynthesis and catabolism of lysine is well characterized in plants (Wang et al., 2018). In higher plants, lysine biosynthesis involves a branch of aspartate metabolism, and two key enzymes, aspartate kinase (AK) and dihydrodipicolinate synthase (DHDPS), regulate lysine accumulation in this pathway. AK, the first limiting enzyme in this pathway, has been characterized at the molecular level in microorganisms and some higher plants (Azevedo and Lea, 2001; Matthews, 1999) including Arabidopsis, rice, barley, maize, carrot and coix (Azevedo et al., 1992; Frankard et al., 1997; Lea et al., 1992; Lugli et al., 2002; Teixeira et al., 1998; Wilson et al., 1991). However, there are at least two AK isoenzymes in plants; a lysine‐sensitive isoenzyme and a threonine‐sensitive isoenzyme (Azevedo et al., 1997). DHDPS catalyses the first reaction in lysine biosynthesis and is more important than AK for enhancing free lysine levels (Galili, 2002). DHDPS has also been characterized at molecular and biochemical levels in plants (Craciun et al., 2000; Frisch et al., 1991; Matthews, 1999), and it is also feedback‐inhibited by lysine or lysine analogs (Lee et al., 2001).
The first insight into feedback regulation of lysine biosynthesis was obtained by investigating the Escherichia coli (E. coli) gene dapA encoding a DHDPS enzyme that was ~20‐fold less sensitive to inhibition by lysine than a typical plant DHDPS (Lee et al., 2001; Shaul and Galili, 1992). An AK mutant gene (lysC) insensitive to lysine feedback was subsequently identified in bacteria (Kikuchi et al., 1999). A number of reports have focused on expressing dapA and/or mutated AK to enhance lysine and threonine content in plants (Galili and Amir, 2013). This approach proved successful, and more AK and/or DHDPS mutant genes are being sought in plants (Matthews, 1999). Previous studies identified natural mutants of lysine‐insensitive AK and/or DHDPS in Corynebacterium glutamicum, E. coli, barley and maize, and their sequences have single nucleotide substitutions (Matthews, 1999). In maize, two threonine‐overproducing, lysine‐insensitive AK mutants (Ask1‐LT19 and Ask2‐LT20) exhibited increased free threonine and lysine levels compared with wild‐type (WT) plants (Muehlbauer et al., 1994). In barley, the feedback characteristics of AK activity in mutant plants were changed so that lysine was half‐maximally inhibitory at 10 mm rather than 0.4 mm (Bright et al., 1982). Many attempts have been made to obtain lysine‐insensitive AK and DHDPS or lysine overproducing mutants of various plants by selecting plants resistant to the lysine analog S‐(2‐aminoethyl)‐cysteine (AEC), and some lysine overproducing mutants have been successfully generated for several species including tobacco, maize, carrot, rice and bulrush millet (Galili et al., 1994; Lee et al., 2001). However, lysine levels have only been modestly increased in these mutants. For example, rice plants were developed from resistant cells by treating with the lysine analog, and lysine content in rice seed was elevated up to 15% of total protein (Schaeffer and Sharpe, 1987; Schaeffer et al., 1997; Wenefrida et al., 2013). Unfortunately, mutants of AK and/or DHDPS that are insensitive to lysine feedback regulation have not yet been identified in rice.
The bi‐functional enzyme lysine‐ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH) was shown to play a pivotal role in the catabolism and accumulation of lysine via the lysine metabolic regulation pathway in plants (Azevedo and Lea, 2001). A series of studies have attempted to inhibit the activity of LKR/SDH, or combine overexpression of AK and/or DHDPS with interference of LKR/SDH to simultaneously enhance lysine biosynthesis and reduce lysine catabolism (Galili et al., 2017). In our previous studies, we expressed bacterial AK and DHDPS, and inhibited rice LKR/SDH activity, resulting in engineered and pyramid transgenic rice with ~25‐fold and ~60‐fold improvements in free lysine content in mature seeds (Long et al., 2013; Yang et al., 2016). Similar findings were also reported in plants using different strategies (Galili and Amir, 2013).
The lysine content has been successfully enhanced in cereals, Arabidopsis, tobacco, canola and soya bean (Wang et al., 2018). However, some unexpected effects on plant growth and/or development have been observed in lysine‐mutated and engineered plants, such as low germination rate, acute reduction in yield, low oil content, poor grain quality and dark‐brown seed phenotype (Jia et al., 2013; Wang et al., 2018; Yang et al., 2018, 2020). Thus, breeding crops with a high lysine content remains a major goal of plant breeders.
In the present study, in an attempt to increase lysine level, we modified the cDNAs encoding rice AK and DHDPS, and selected AK and DHDPS mutants that are less sensitive to lysine inhibition. Transgenic rice accumulated more free lysine in mature seeds harbouring mutated AK or DHDPS genes. We then identified two efficient mutants, and combined them with RNA interference (RNAi)‐mediated inhibition of LKR to construct polygenic transgenic rice that displayed a further enhancement in lysine accumulation in mature seeds.
Results
Selection of target sites for modification in rice AK and DHDPS enzymes
Previous studies showed that feedback inhibition of the E. coli AKIII enzyme is a major regulatory point in lysine biosynthesis, and AKIII mutants with T344M (LysC1), S345L (LysC2) or G323D (LysC12) amino acid substitutions are insensitive to lysine feedback inhibition (Figure S1A; Kikuchi et al., 1999). Following comparison of the amino acid sequences of various AK enzymes in natural rice and Arabidopsis, and native and mutant E. coli strains (Figure S1A), we concluded that amino acid residues 448–452, corresponding to the residues 344–348 in E. coli AKIII, are conserved in rice AK proteins and correspond to the lysine‐binding domain (Figure S1A). Changes in this domain may affect the lysine‐insensitive AK characteristics. Therefore, Thr (T) and Ser (S) residues at positions 448 and 449 in rice AK enzyme were selected for substitution (Figure 1a).
Figure 1.

Sequence modification of rice AK and DHDPS genes, and AEC resistance testing in E. coli cells expressing WT and modified AK and DHDPS genes. (a) Sequence modification of rice AK. (b) Sequence modification of rice DHDPS. (c) AEC (12mM concentration) resistance testing in E. coli cells expressing AK. (d) AEC (12mM concentration) resistance testing in E. coli cells expressing DHDPS. Blue font indicates modified nucleotides, and red font indicates modified amino acids. AEC, S‐(2‐aminoethyl)‐cysteine.
Similarly, from the sequence alignment results of feedback‐insensitive DHDPS mutants in E. coli, tobacco, maize and soya bean (Matthews, 1999), it was concluded that the conserved region (residues 124–133) in rice DHDPS defines the lysine‐binding domain, and its mutation might cause the loss of lysine‐mediated inhibition of DHDPS activity (Figure S1B). Thus, five amino acids, Asn (N), Ile (I), Lys (K), Thr (T) and Val (V) at positions 124, 125, 129, 133 and 133 in the natural rice DHDPS enzyme, were selected for single amino acid substitution (Figure 1b).
Single amino acid mutation in rice AK and DHDPS
The cDNAs encoding AK or DHDPS were cloned from rice. To obtain feedback‐insensitive AK and DHDPS by overlapping PCR, specific primers were designed for nucleotide modification (Table S1). According to the selected site, two modified AK clones (AK1 and AK2) and five modified DHDPS clones (DHDPS1–5) were successfully generated via point mutations, and all mutations clustered within the predicted lysine‐binding domain (Figure 1A‐B). For AK modifications, mutation of AK1 at 448th codon (nucleotides 1343 and 1344) caused a Thr (T) to Met (M) substitution, and mutation of AK2 at 449th codon (nucleotides 1345 and 1346) resulted in a Ser (S) to Leu (L) substitution (Figure 1a). For DHDPS modifications, mutation of DHDPS1–5 resulted in G to A, A to T or C to T substitutions at nucleotides 371, 374, 385, 397 and 398, respectively, in the DHDPS coding region. Each nucleotide change created a single amino acid substitution: Ser (S) to Asn (N) at position 124 (DHDPS1), Asn (N) to Ile (I) at position 125 (DHDPS2), Glu (E) to Lys (K) at position 129 (DHDPS3), Ala (A) to Thr (T) at position 133 (DHDPS4) and Ala (A) to Val (V) at position 133 (DHDPS5) (Figure 1b).
In addition, the results of allergenic assessment showed that rice AK protein and its modified proteins passed the 8‐mer and 80‐mer searches, but had a small hit with a Par h I allergen (44.7% identity) in the full FASTA search (Table S2). Their E value had <3.9E‐07, and the allergen come from Parthenium hysterophorus and different from rice species (Table S2). The rice DHDPS and its modified proteins showed no significant alignment with any known allergen. As well as, these protein sequences identity were not likely to be cross‐reactive with other allergens. Thence, bioinformatics results predicted that rice AK and DHDPS proteins and their modified proteins had no potential allergenicity.
AK and DHDPS mutants are resistant to feedback inhibition by lysine in Escherichia coli
S‐(2‐aminoethyl)‐l‐cysteine (AEC) can substitute lysine to elicit feedback inhibition of key enzymes AK and DHDPS in the aspartate metabolism pathway (Shaver et al., 1996; Xu et al., 2016). Escherichia coli BL21 cells expressing modified (AK1, AK2 or DHDPS1–5) or unmodified AK and DHDPS from rice were cultured on medium containing 12 mm AEC. Colonies harbouring modified AK or DHDPS genes grew normally, and better than those harbouring unmodified genes and blank controls, although the growth states differed (Figure 1c,d). Cells harbouring unmodified genes grew poorly because expression was feedback‐inhibited by the lysine analog AEC (Figure 1c,d). These results indicate reduced sensitivity of modified AK and DHDPS to lysine‐mediated feedback inhibition compared with cells expressing native rice AK and DHDPS.
Functions of mutated AK and DHDPS in transgenic rice
We generated nine constructs with single target gene, including two for unmodified controls with the native rice AK or DHDPS genes and seven with their modified genes (Figure S2A; Table S3), where the rice endosperm‐specific glutelin 1 (Gt1) promoter were used to drive the expression of target genes in rice. These constructs were used for Agrobacterium‐mediated transformation to generate transgenic rice. The leaves of transgenic plants were collected for PCR analysis to confirmed target gene insertion, and more than 10 individual transgenic lines per construct were identified. Both northern and Western blotting analyses showed that rice AK and DHDPS genes were highly expressed in developing seeds at 15 days after fertilization (DAF) in transgenic rice containing modified or unmodified AK and DHDPS, and expression levels were higher than those in untransformed wild‐type (WT) rice (Figure 2a–d).
Figure 2.

AK and DHDPS expression in developing rice seeds, and free lysine content in monofactorial transgenic and WT rice. (a, b) Northern blot analyses of rice AK and DHDPS expression in 15 DAF developing seeds. (c, d) Western blot analyses of rice AK and DHDPS expression in 15 DAF developing seeds. (e) Free lysine content (mg/g dry seed weight) in modified and unmodified AK and DHDPS transgenic and WT rice. Error bars represent SD for three biological replicates, and * and ** indicate significant difference between transgenic and WT plants with P < 0.05 and P < 0.01, respectively. The white lines represent the images from discontinuous lanes in a film or different films.
Preliminary analysis showed that the free lysine content was increased in most transgenic mature seeds, especially in transgenic lines harbouring modified AK2 and DHDPS1 (Figure 2e). The level of free lysine was significantly increased up to 6.6‐fold and 21.7‐fold in AK2 and DHDPS1 transgenic lines, respectively (Figure 2e). The free lysine content was also increased in AK (1.6‐ to 1.9‐fold), AK1 (2.0‐ to 2.1‐fold), DHDPS (2.0‐ to 4.4‐fold), DHDPS2 (1.0‐ to 5.8‐fold), DHDPS3 (2.5‐ to 2.6‐fold), DHDPS4 (1.0‐ to 5.0‐fold) and DHDPS5 (3.1‐ to 9.5‐fold) transgenic lines (Figure 2e). The results revealed that increasing the expression of endogenous AK or DHDPS genes can enhance free lysine levels in rice, and modification of AK2 (Ser to Leu substitution at amino acid residue 449 in rice AK) and DHDPS1 (Ser to Asn substitution at amino acid residue 124 in rice DHDPS) can significantly boost free lysine levels in mature rice seeds compared with either unmodified AK or DHDPS transgenic rice or wild‐type rice.
Regulation of lysine metabolism by simultaneous expression of mutated AK2 and DHDPS1 in rice
Based on previous successful strategies related to lysine catabolism to manipulate free lysine accumulation in plants, we investigated whether lysine levels could be further elevated in transgenic rice by simultaneously expressing mutated AK2 and DHDPS1 (Figure S2B,C; Table S3). Constructs were generated for expressing native (35ADL) or modified (35A2D1L) rice AK and DHDPS genes, and for inhibiting expression of the LKR/SDH gene by LKR‐RNAi, all driven by the CaMV 35S promoter, in an attempt to simultaneously stimulate lysine biosynthesis and diminish lysine catabolism (Figure S2B,C). Over 40 independent transgenic lines were obtained using the two constructs, and transgenic lines (both 35ADL and 35A2D1L) were randomly chosen as representative lines for subsequent analysis.
PCR analysis of the chosen transgenic lines demonstrated the stable integration of the transgenic cassettes in the genome of rice plants (Figure S2D–F). Rice AK and DHDPS expressions were elevated at both transcript and protein levels in 15 DAF developing seeds produced by 35ADL or 35A2D1L transgenic plants (Figure 3a–g) relative to their WT. Meanwhile, expression levels of LKR/SDH were inhibited or down‐regulated dramatically as expected in all transgenic lines according to qRT‐PCR and Western blot analyses (Figure 3a–g). These results revealed the successful expression of the introduced genes in the selected transformants.
Figure 3.
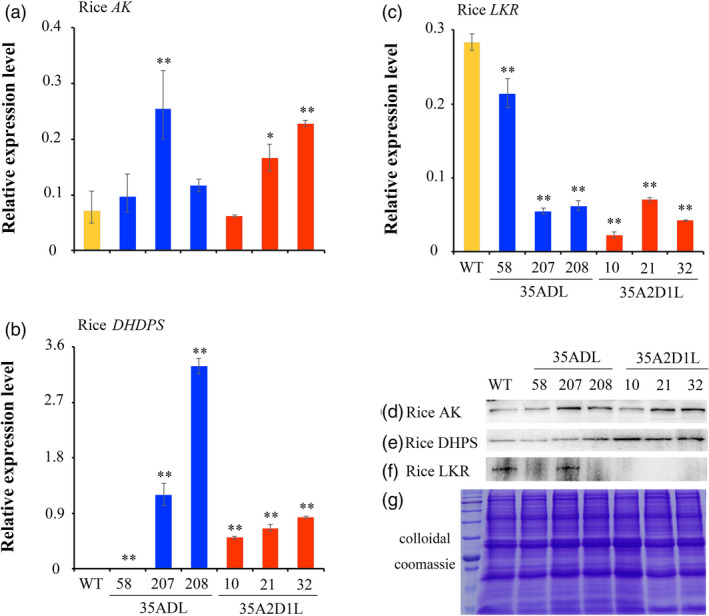
AK, DHDPS and LKR expression in developing rice seeds of polygenic transgenic and WT rice. (a–c) qRT‐PCR analyses of rice AK, DHDPS and LKR genes in 15 DAF developing seeds of transgenic and WT rice; (d–g) Western blot analyses of rice AK, DHDPS and LKR expression in 15 DAF developing seeds of transgenic and WT rice. Error bars represent SD for three biological replicates, and * and ** indicate significant difference between transgenic and WT plants with P < 0.05 and P < 0.01, respectively.
Enhancement of free lysine content in 35A2D1L transgenic rice
To be nutritionally useful, increased lysine content in transgenic rice should be retained in seed endosperm. Thus, after natural growth and development, mature seeds were harvested and analysed, and all 35ADL transgenic lines displayed a 1.5‐ to 13.9‐fold increase in free lysine content compared with WT seeds (Figure 4a). Surprisingly, 35A2D1L transgenic lines retained significantly elevated levels of free lysine in their seeds, reaching 1544.77 ± 7.20 µg/g dry seed weight, a 58‐fold increase over the 26.41 ± 3.79 µg/g dry seed weight in WT seeds, even higher than achieved by our 35R construct generated in previous work (Figure 4a). The 35R lines are elite high lysine lines expressing bacterial AK and DHDPS genes driven by the 35S promoter (Table S3; Long et al., 2013). As well as, free lysine content remained higher in transgenic mature seeds at T5 generation compared to WT rice (Table S4). Additionally, the increase in free lysine content in transgenic rice seeds was associated with a significant increase in the content of all measurable free amino acids, especially in 35A2D1L transgenic lines, with levels 2.7‐ to 7.6‐fold higher than in WT seeds (Figure 4b).
Figure 4.
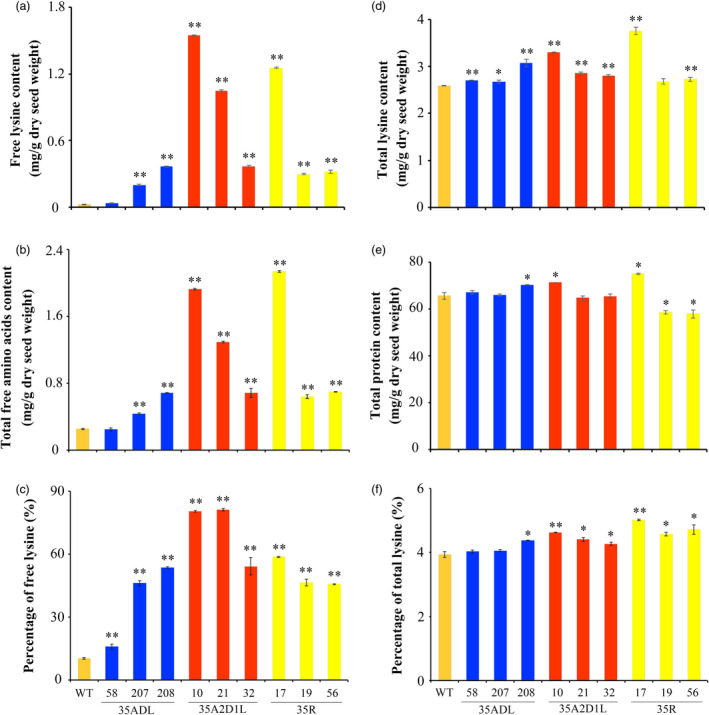
Lysine, total amino acids and protein content in mature seeds of transgenic and WT rice. (a) Free lysine content (mg/g dry seed weight). (b) Total free amino acids content (mg/g dry seed weight). (c) Proportion of free lysine among total free amino acids (weight basis). (d) Total lysine content (mg/g dry seed weight). (e) Total protein content (mg/g dry seed weight). (f) Proportion of total lysine among total proteins (weight basis). Error bars represent SD of three biological replicates, and * and ** indicate significant difference between transgenic and WT plants with P < 0.05 and P < 0.01, respectively.
The total lysine and protein contents were determined in mature seeds from WT and transgenic plants to assess whether the elevated free lysine caused changes in total lysine. As shown in Figure 4, both 35ADL and 35A2D1L transgenic lines displayed significantly higher total lysine content compared with WT rice, and it increased by 27.6% in the case of the 35A2D1L‐10 transgenic line containing the highest free lysine level (Figure 4d). The total protein content varied among different transgenic lines, and changes were correlated with the observed increase in free lysine and total lysine levels (Figure 4e). The 35A2D1L‐10 transgenic line yielded the highest total lysine and total protein (+4.62%), 17.4% higher than WT plants (Figure 4f).
Thus, expression of modified AK2 and DHDPS1 genes driven by the 35S promoter combined with LKR interference greatly increased the free lysine content in mature seeds, and enhanced the total lysine content.
Impact of regulating lysine metabolism on other amino acids in 35A2D1L transgenic rice
The composition of other free amino acids (FAAs) was examined to investigate the effects of regulating lysine metabolism on their abundance in rice. Compared with WT plants, levels of most FAAs were increased, but the proportion of each amino acid among total FAAs was reduced due to the great increase in the content of total FAAs (Figure 5; Figure S3). This may be due to the increased accumulation of free lysine, since the percentage of free lysine among total FAAs was 54.1%–81.0% in 35A2D1L transgenic lines, but only 10.3% in WT plants (Figure 4c; Figure S3). These results implied that the increase in total measurable FAA levels in 35A2D1L transgenic lines is mainly due to the accumulation of free lysine in the endosperm. Asp and Glu, two major substrates of the aspartate family pathway, were elevated in 35A2D1L‐10 containing the highest free lysine content, but there was no increase in 35A2D1L‐21 and 35A2D1L‐32 transgenic lines (Figure 5). Besides, the change of free asparagine, one of the most abundant free amino acids in most cereal grains, was similar as that of Asp in 35A2D1L transgenic rice (Figure 5).
Figure 5.

The content of other measurable free amino acids in mature seeds of transgenic and WT rice. Data are presented as μg/g dry seed weight. Error bars represent SD of three biological replicates, and * and ** indicate significant difference between transgenic and WT plants with P < 0.05 and P < 0.01, respectively.
In addition, we also compared the total amounts of each measurable amino acid in seeds from 35A2D1L polygenic transgenic and WT rice (Figure S4). Surprisingly, the total content of essential amino acids, including threonine (Thr), valine (Val), methionine (Met), isoleucine (Ile), leucine (Leu), tyrosine (Tyr), phenylalanine (Phe) and histidine (His), was also increased significantly in the 35A2D1L‐10 transgenic line (Figure S4). We also calculated the amino acid score (AAS) for polygenic transgenic lines and WT plants (Table S5) based on the protein reference pattern recommended for school‐age children and adolescents (WHO, 2007). The AAS for 35A2D1L was 0.89–0.96, almost within the recommended range. These results indicated that the balance of other amino acids was surprisingly elevated in the 35A2D1L transgenic line in concert with the increase in lysine content, compared with other transgenic rice and WT controls.
Agronomic performance of 35A2D1L transgenic rice
Major agronomic traits were examined in the field trial, and transgenic lines exhibited normal plant growth and development for most traits as their wild type. There were no statistically significant differences in 1000‐grain weight, seed setting rate, grains per panicle, effective tiller number or plot yield between transgenic rice and WT plants (Figure 6c–g). These results indicated no obvious effect on yield upon regulating lysine metabolism by modifying rice AK and DHDPS. Interestingly, there were no obvious changes in seed chalkiness or appearance among rice lines (Figure 6b). Taken together, the above results suggested that engineering lysine metabolism in 35A2D1L transgenic lines increases the free lysine content and results in desirable trait improvements, especially in the case of the 35A2D1L‐10 transgenic line.
Figure 6.

Major agronomic traits and appearance of mature seeds from transgenic and WT rice. (a) Aspartate‐derived lysine metabolism pathway in higher plants. The red lines indicate allosteric product feedback inhibitions, while blue arrow indicates allosteric activation. (b) Appearance of mature brown rice. (c) Effective tiller number, (d) Grain number per panicle. (e) Seed setting rate (%). (f) 1000‐grain weight (g). (g) Plot grain yield (kg/m2). Error bars represent SD of three biological replicates, and no significant difference was found between transgenic and WT plants.
Discussion
The endosperm of the maize opaque‐2 mutant exhibited higher lysine content than normal WT kernels, providing the first insight into lysine metabolism in crops (Mertz et al., 1964). Subsequently, a series of mutants with high lysine content were bred or selected in Arabidopsis, tobacco, soya bean, rapeseed, wheat and barley (Galili and Amir, 2013). However, a high lysine rice mutant is still lacking due to the limitation of rice germplasm resources (Sun and Liu, 2004). Genetic engineering has been employed to increase the lysine content by regulating lysine metabolism, which provides an effective strategy for breeding high lysine rice (Long et al., 2013; Yang et al., 2017; Yang et al., 2016). Mutation of AK and DHDPS can generate lysine feedback insensitivity, but natural and engineered lysine feedback‐insensitive AK and DHDPS mutants are not yet available for rice. In the present study, we cloned rice AK and DHDPS genes and modified the corresponding enzymes to generate lysine feedback‐insensitive mutants (Figure 1). We found that the free lysine content of mature seeds was up to 21‐fold higher in transgenic lines containing modified AK and DHDPS driven by the rice Gt1 promoter, relative to WT controls (Figure 2).
On the other hand, allergenic assessments are one stage in the process of biosafety assessments of genetically modified products. The use of bioinformatics for allergenic assessment of novel proteins in allergen databases is recommended by the FAO/WHO, the European Food Safety Authority, and the US Environmental Protection Agency (Fard et al., 2015). In this study, matching the 80 amino acids (domain) and 8 amino acids (epitope), in general, showed no similarity between our interest proteins (rice AK and DHDPS and their modified versions) and allergen proteins. Although AllergenOnline and Allermatch indicated that AK and its modified proteins shared 44.7% sequence identity with a Par h I allergen protein from Parthenium hysterophorus, the AK and its modified protein had <50% sequence identity to allergen proteins and their E value had <3.9E‐07. These results suggested no significant alignment between the AK (and its modified proteins) protein and allergen proteins in full sequence matching. Therefore, as expected, we obtained high lysine rice via the generation of lysine feedback‐insensitive mutants, which could make up for the blank of expressing endogenous lysine biosynthetic genes.
Regulation of the biosynthesis and catabolism of lysine is well characterized in plants (Galili et al., 2016; Wang et al., 2018). AK and DHDPS are key enzymes that participate in the lysine biosynthesis pathway (Galili et al., 2016). Various studies have aimed to enhance the lysine content in seeds by expressing AK and/or DHDPS feedback‐insensitive to lysine under constitutive or seed‐specific regulation, and marked accumulations of lysine have been achieved in Arabidopsis, barely, canola and soya bean (Wang et al., 2018). For example, a 2.5‐fold increase in free lysine was reported for barley seeds expressing a maize lysine feedback‐insensitive dhps gene under the control of the CaMV 35S or Gt1 promoter, relative to WT plants (Lee et al., 2001). However, there was no increase in free lysine in mature seeds of 35S transgenic rice expressing bacterial lysine feedback‐insensitive AK and DHDPS genes under the control of the CaVM 35S promoter (Long et al., 2013; Yang et al., 2017). In the present study, endogenous rice AK and DHDPS genes were modified, resulting in two AK transgenic line (AK1 and AK2) and five DHDPS transgenic lines (DHDPS1–5) (Figure 1). The free lysine content was increased up to 21.7‐fold in mature seeds from modified DHDPS1 transgenic lines (Figure 2), compared with a 4.0‐fold increase reported in a previous study (Lee et al., 2001; Long et al., 2013). These results indicated that single amino acid mutation of AK and DHDPS can be effective for enhancing the free lysine content, as demonstrated most effectively in AK2 and DHDPS1 transgenic lines.
Previous studies reported a 2.5‐fold increase in free lysine in seeds expressing a maize lysine feedback‐insensitive DHDPS gene under the control of the CaMV 35S or Gt1 promoter, relative to WT plants (Lee et al., 2001), and similar results have been reported in other studies (Lee et al., 2001; Yang et al., 2016). This suggests that transgenic manipulation of AK and DHDPS under the control of a constitutive promoter may be more effective than using a seed‐specific promoter in terms of improving lysine biosynthesis. One explanation might be that lysine biosynthesis plays a more active role in regulating the accumulation of lysine in leaves, while lysine catabolism is the major regulatory factor in seeds (Long et al., 2013; Ufaz and Galili, 2008). Moreover, the LKR/SDH expression could be induced via the accumulation of lysine to balance amino acid flow (Stepansky et al., 2006; Sun and Liu, 2004; Zhu and Galili, 2003). It has been another important strategy by inhibiting or down‐regulating LKR/SDH enzyme activity for improving lysine content in plants (Hournard et al., 2007; Reyes et al., 2009). Furthermore, the expression of feedback‐insensitive AK and/or DHDPS genes combined with inhibiting the lysine catabolism pathway explosively increased free lysine content in Arabidopsis, maize and rice (Frizzi et al., 2008; Yang et al., 2016; Zhu and Galili, 2003). Therefore, simultaneous modulation of lysine biosynthesis and catabolism should be a relatively effective strategy for enhancing lysine content in plants. Indeed, in the present study, we generated the 35A2D1L transgenic line with both CaMV 35S‐driven expression of mutant rice AK2 and DHDPS1 and RNAi‐mediated inhibition of rice LKR/SDH expression, and the free lysine content was increased ~58‐fold in mature seeds, relative to WT plants (Figure 4), as good or even better than the excellent high lysine rice line 35R (Yang et al., 2016).
Great achievements have been made for high lysine maize within long‐term studies (Azevedo and Arruda, 2010). However, the opaque‐2 mutant and Quality Protein Maize showed abnormal endosperm and starch property (Galili and Amir, 2013). A commercial high lysine maize line (LY038) was designed to express the bacterial CordapA gene in the embryo, and presented 19‐fold increment in free lysine content and 79% increment in total lysine content in the seeds (Lucas et al., 2007). Nevertheless, no phenotypic trait of the transgenic lines was reported. In our study, the free lysine level in the seeds of 35A2D1L‐10 transgenic line was 58.5‐fold and 39.2‐fold higher than that in WT or 35ADL transgenic rice with native AK and DHDPS overexpressed, respectively. Moreover, no obvious negative effect was found in these transgenic lines compared with their wild type. Therefore, these advantages will come right for the potential commercialization of such high lysine transgenic rice.
Although the free lysine content of the 35A2D1L transgenic rice seeds was greatly increased, the total lysine content only increased 27%, lower than that of the high lysine crops which expressing lysine‐rich proteins (Liu et al., 2015; Liu et al., 2016; Wong et al., 2015; Yu et al., 2005). It implied that different genetic engineering strategies may have different effects on enhancing the contents of free lysine and total lysine. Generally, there were obvious negative effects by overexpressing lysine rice proteins in plants (Liu et al., 2016; Wong et al., 2015; Yang et al., 2020). Thus, to avoid the obvious adverse effects, it could be widely considered to enhance the lysine content by modulating the lysine metabolism pathway in higher plants.
Due to different amino acids share the same metabolic trunk and/or acting as a synthetic substrate or intermediate, regulating metabolism of a certain amino acid may affect the accumulation of other amino acids (Figure 6a). In this study, the content of Met, the intermediate of the aspartate pathway, was significantly increased in the seeds of 35A2D1L transgenic rice relative to that of the WT (Figure 5). However, the contents of Thr and Gly, which are also derived from the aspartate pathway, were not affected in the seeds of 35A2D1L transgenic lines (Figure 5). This may be due to the overexpression of AK2 mutant gene leading to the increase of upstream products of Met. Interestingly, the leucine and isoleucine levels were decreased in the transgenic rice seeds (Figure 5), which might be resulted from the substrate competition. Asp can be produced in plant seeds either from Glu by Asp aminotransferase or from Asn by Asn synthetase (Long et al., 2013). In the current study, the free Asn level was elevated in 35A2D1L‐10 transgenic rice seeds containing the highest free lysine content, but there was no increase of Asn level in 35A2D1L‐32 and 35ADL‐207 transgenic lines, and even decrease in 35A2D1L‐21, 35ADL‐58 and 35ADL‐208 transgenic lines (Figure 5). One possible reason could be that the involvement of Asp aminotransferase in the production of Asp or Asp may participate in other metabolic pathways. Additionally, the contents of essential amino acids were significantly increased or not affected in the transgenic rice (Figure S4). These results suggested that a complex regulatory network exists in modulating the overall content of these beneficial amino acids.
Notably, transgenic and mutant plants with high lysine levels exhibited some negative effects on agronomic and economic traits, characterized by retarded seed germination in Arabidopsis (Zhu and Galili, 2003), hardened endosperm in Quality Protein Maize (Gibbon et al., 2003), wrinkled seeds in soya bean (Falco et al., 1995), low oil content in rapeseed (Wang et al., 2018) and altered morphology and increased chalkiness in rice (Lee et al., 2001; Wong et al., 2015). In previous studies, we bred high free lysine transgenic rice lines HFL and 35R, and whose mature endosperm had a dark‐brown appearance (Yang et al., 2017; Yang et al., 2018). These findings indicate that the aspartate family may play multiple roles in plant metabolism, and their biological functions might differ among species. As shown in Figure 6, in this study, there was no obvious change in certain agronomic traits or the appearance of seeds, and the dark‐brown seed phenotype was not evident in the 35A2D1L transgenic rice. Previous studies showed that elevated 2‐aminoadipate from lysine catabolism may play a key role in the dark‐brown phenotype of rice endosperm (Yang et al., 2018). As shown in Figure S5, when the bacterial AK and DHDPS highly expressed in 35R transgenic rice, the expression levels of rice native AK and DHDPS were not different from WT plants (Long et al., 2013). These results suggest that the regulation of bacterial AK and DHDPS in lysine metabolism may differ from that of rice endogenous AK and DHDPS, and upstream lysine biosynthesis and downstream LKR catabolism may be relatively coordinated, resulting in insufficient 2‐aminoadipate accumulation to generate the dark‐brown phenotype. These regulatory mechanisms had been investigated using LKR knockout mutants and tryptophan decarboxylase (TDC) overexpressing transgenic lines according to our recent works on the connection between lysine metabolism and other potential pathways (Yang et al., 2018, 2020).
Recently, the rapid development of genome editing, including base editing via CRISPR/Cas system (Gao, 2018), has provided an important tool for precise modification of the target enzymes without foreign transgene(s). In this report, we have successfully created high lysine rice without introduction of foreign genes before the fully developed CRISPR/Cas technology. Overall, our results are promising and indicate that molecular genetic modification of the lysine synthetic pathway is a viable approach for increasing free lysine levels in important crop species such as rice.
Experimental procedures
Cloning and sequence modification of AK and DHDPS
Based on the cDNA sequence of rice AK (AK073189) and DHDPS (L77616) from the NCBI database, gene‐specific primers were designed (Table S1). Rice AK and DHDPS cDNAs were obtained by RT‐PCR amplification and DNA sequencing was performed to confirm construction and modification. Specific sequence modifications were performed by the overlapping PCR technique, and specific primers were designed for nucleotide modifications (Table S1).
Allergenic assessment of candidate proteins
The allergen databases AllergenOnline (http://www.allergenonline.org/) and Allermatch (http://www.allermatch.org) were used to identify any potential sequence matches to allergen proteins. The rice original protein sequences and their modified protein sequences were searched for full FASTA, 80‐mer and 8‐mer, as suggested by the FAO/WHO guidelines for genetically modified foods (FAO and WHO, 2001). The full FASTA alignment recommendation applies to sequences of less than 80 amino acids in the FAO/WHO guidelines. And the studies showed that the efficiency of E value <3.9E‐07 in the full FASTA alignment was the same as that of sequence consistency >35% in sequence similarity ratio of 80‐mer by analysing a large number of known allergens and non‐allergens (Andre et al., 2009). Therefore, in this study, E < 3.9e‐07 was used as the judgment threshold for the full FASTA alignment.
The structural database of allergenic proteins (SDAP, http://fermi.utmb.edu/ SDAP/) provides rapid, cross‐referenced access to their sequences, structures and IgE epitopes that might indicate allergenic cross‐reactivity with other sequences. This alignment was performed in December 2019.
Transgene constructs and rice transformation
Wuxiangjing 9 (WXJ9, WT), an elite japonica rice cultivar from China, was used for transformation in this work. AK and DHDPS genes and their mutated variants were designed and subcloned into the binary vector pSB130 (Liu, 2002), as shown in Figure S1. Nine transgene constructs were used to analyse the effects of modified AK and DHDPS in rice (Figure S1). These constructs were used for overexpression of rice AK, DHDPS and their mutated variants AK1 and AK2, and DHDPS1–5 (amino acid sequence information for mutants is shown in Figure 1), driven by the rice endosperm‐specific glutelin Gt1 promoter.
Additionally, two transgene constructs (35ADL and 35A2D1L; Figure S1) were applied to breed high lysine rice by expressing rice AK (or mutated AK2) and DHDPS (or mutated DHDPS1) genes, and simultaneously down‐regulating the endogenous rice LKR/SDH gene, via three cassettes driven by the CaMV 35S promoter. Detailed information on the above constructs is included in Table S3. Rice calli from mature embryos were used as explants for Agrobacterium‐mediated transformation according to our previously published procedure (Liu et al., 1998). Stably transformed plants were regenerated after screening by PCR (Figure S2) with transgene‐specific primers (Table S1). All selected transgenic lines were homozygous for the transgenes and were grown in a greenhouse at the Chinese University of Hong Kong or in paddy fields at Yangzhou University (Yangzhou, Jiangsu Province, China).
AEC resistance in Escherichia coli‐expressing modified AK and DHDPS
AEC (S‐(2‐aminoethyl)‐l‐cysteine), a toxic lysine analog structurally similar to lysine, can compete with lysine for incorporation into proteins, and only mutants overproducing lysine or with defective AEC uptake can survive in the presence of this compound (Galili, 1995). Herein, 12 mm AEC was added to the growing medium of E. coli BL21 cells expressing modified or unmodified AK and DHDPS. These constructs were cloned by digesting the pET‐30a vector (Novagen, Darmstadt, Germany) with restriction enzymes Bam HI and Sac I, then ligating the PCR‐amplified and digested modified and unmodified AK or DHDPS gene. PCR amplification was performed using an ABI PRISM dRhodamine Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and analysed using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, ABI) as described in the user manual.
Analysis of RNA and protein expressions
Total RNAs were isolated from developing seeds at 15 days after flowering (DAF) using TRIzolRNA (Invitrogen, Carlsbad, CA) and purified by treatment with DNase I. First‐strand cDNA was generated using Perfect Real Time PrimeScript RT reagent (TaKaRa, Japan). The rice actin gene was used as an internal control to measure the relative expression levels of rice AK and DHDPS, bacterial AK and DHDPS, and rice LKR/SDH genes. Gene‐specific primers used for qRT‐PCR are listed in Table S1. The absolute values (M values) of the slope difference between the reference gene and the target genes were calculated for AK, DHDPS and LKR, respectively. Because the M value (0.040, 0.045 and 0.058, respectively) calculated based on these experiments was less than 0.1, the data were analysed according to the ΔΔcycle threshold (C t) method (Livak and Schmittgen, 2001) using actin as the CT normalizer. ΔC t = C t(target) − C t(actin), while ΔΔC t = ΔC t(test sample) − ΔC t(control sample). Relative gene expression was analysed by the ΔΔC t method.
For northern blot analysis, developing total RNA was separated on a 1% agarose/formaldehyde gel and transferred to a nylon membrane, and hybridization and detection were performed as described previously (Long et al., 2013). Total seed proteins were extracted from developing seeds as previously described (Long et al., 2013). Proteins were detected by Western blotting using antibodies specific for rice AK, DHDPS and LKR, or bacterial AK and DHDPS.
Grain component analyses
Mature seeds were shelled, milled and processed into flour for grain component analysis (Zhu et al., 2010). The contents of free and total amino acids, both based on the dry rice seeds, were determined as previously described (Yang et al., 2016). Briefly, the free amino acids were extracted from rice flours by ultrasound assisted, and the total amino acids were isolated by hydrolysed rice flours with 6 N HCl. These treated samples were then dissolved in Na‐S™ buffer and finally filtered through a 0.45‐μm nylon membrane syringe filter (Pall Life Sciences, Port Washington, NY, USA) for injection and analysis by HPLC (high‐performance liquid chromatography) using an L8900 amino acid analyser (Hitachi, Tokyo, Japan). The measurement of seed total protein contents was performed according to a previous report (Zhou et al., 2020). Crude protein content was estimated by Kjeldahl method via a nitrogen determiner (Foss Tecator Kjeltec 2300; Tecator AB, Hoganas, Sweden), and the nitrogen content was converted to protein content by multiplying the former by 5.95. Three biological replicates were designed for each sample.
Field trials and agronomic trait investigation
Transgenic and non‐transgenic WT plants were grown at in the experimental field in Yangzhou University (Yangzhou, Jiangsu Province, China) with permission for small‐scale field trials under genetically modified safety supervision during the summer of years 2013–2019. A three randomized block design was adopted, and each plot contained six rows, with 10 plants per row and a spacing of 15 cm between plants and 15 cm between rows. Rice plants were grown under the same climate and management conditions. Major agronomic traits and plot yield were investigated after maturity, and mature seeds were harvested for grain component analysis.
Statistical analysis
Results are presented as the mean ± standard deviation (SD). Comparison of multiple transgenic and WT plants was performed by t‐test analysis using SPSS 17.0 for windows (SPSS Inc., Chicago, IL, USA). Asterisks * and ** indicate statistical significance between transgenic and WT plants at P < 0.05 and P < 0.01, respectively.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
Q.Y., W.Y., H.W. and C.Z. and performed the experiments; Q.Y, S.S.S. and Q.L. designed the experiments; Q.Y., W.Y., S.S.S. and Q.L. analysed the data; Q.Y. and Q.L. wrote the paper. All authors have commented on the manuscript and approved the final manuscript.
Supporting information
Figure S1 Sequence Alignments of the putative lysine‐binding domain from native AK and DHDPS and their mutants.
Figure S2 Transgene constructs for expressing modified or unmodified AK and DHDPS in rice and PCR analyses of transgenic plants.
Figure S3 Comparison of the proportion (by weight) of other individual free amino acids among total measurable free amino acids in mature seeds of transgenic and WT rice.
Figure S4 The contents of total essential amino acids in mature seeds of transgenic and WT rice.
Figure S5 The expression of AK, DHDPS and LKR in developing rice seeds of transgenic and WT plants.
Table S1 Primers used in this study.
Table S2 Sequence identity after full searching of candidate proteins in allergen databases of AllergenOnline and Allermatch (E value < 1).
Table S3 The information of chimeric genes for production of transgenic rice used in this study.
Table S4 Free lysine content in mature seeds of transgenic and WT rice.
Table S5 Proposed scores for essential amino acids in transgenic and WT rice.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31801322 and 31825019) to Q.Y. and Q.L., the Ministry of Agriculture of China (2016ZX08001006 and 2016ZX08009003‐004) and the Government of Jiangsu Province (BE2018357 and PAPD) to Q.L., and the Bill and Melinda Gates Foundation, the State Key Laboratory of Agrobiotechnology and the Lo Kwee Seong and Lee Hysan Foundations to S.S.S.
Yang, Q.‐Q. , Yu, W.‐H. , Wu, H.‐Y. , Zhang, C.‐Q. , Sun, S. S.‐M. and Liu, Q.‐Q. (2021) Lysine biofortification in rice by modulating feedback inhibition of aspartate kinase and dihydrodipicolinate synthase. Plant Biotechnol. J., 10.1111/pbi.13478
Contributor Information
Samuel Sai‐Ming Sun, Email: ssun@cuhk.edu.hk.
Qiao‐Quan Liu, Email: qqliu@yzu.edu.cn.
References
- Andre, S. , Gary, B. and Scott, M. (2009) The use of E‐scores to determine the quality of protein alignments. Regul. Toxicol. Pharm. 54, S26–S31. [DOI] [PubMed] [Google Scholar]
- Azevedo, R.A. and Arruda, P. (2010) High‐lysine maize: the key discoveries that have made it possible. Amino Acids, 39, 979–989. [DOI] [PubMed] [Google Scholar]
- Azevedo, R.A. , Arruda, P. , Turner, W.L. and Lea, P.J. (1997) The biosynthesis and metabolism of the aspartate derived amino acids in higher plants. Phytochemistry, 46, 395–419. [DOI] [PubMed] [Google Scholar]
- Azevedo, R.A. and Lea, P.J. (2001) Lysine metabolism in higher plants. Amino Acids, 20, 261–279. [DOI] [PubMed] [Google Scholar]
- Azevedo, R.A. , Smith, R.J. and Lea, P.J. (1992) Aspartate kinase regulation in maize: evidence for co‐purification of threonine‐sensitive aspartate kinase and homoserine dehydrogenase. Phytochemistry, 31, 3731–3734. [Google Scholar]
- Bright, S.W.J. , Muflin, B.J. and Rognes, S.E. (1982) Threonine accumulation in the seeds of a barley mutant with an altered aspartate kinase. Biochem. Genet. 20, 229–243. [DOI] [PubMed] [Google Scholar]
- Cohen, H. , Israeli, H. , Matityahu, I. and Amir, R. (2014) Seed‐specific expression of a feedback‐insensitive form of CYSTATHIONINE‐γ‐SYNTHASE in Arabidopsis stimulates metabolic and transcriptomic responses associated with desiccation stress. Plant Physiol. 166, 1575–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun, A. , Jacobs, M. and Vauterin, M. (2000) Arabidopsis loss of function mutant in the lysine pathway points out complex regulation mechanisms. FEBS Lett. 487, 234–238. [DOI] [PubMed] [Google Scholar]
- F.A.O. and W.H.O. (2001) Evaluation of Allergenicity of Genetically Modified Foods. Rome, Italy: FAO/WHO. [Google Scholar]
- Falco, S.C. , Guida, T. , Locke, M. , Mauvais, J. , Sanders, C. , Ward, R.T. and Webber, P. (1995) Transgenic canola and soybean seeds with increased lysine. Nat. Biotechnol. 13, 577–582. [DOI] [PubMed] [Google Scholar]
- Fard, N.A. , Minuchehr, Z. and Rahgozar, M. (2015) Novel genetically modified foods and allergenicity assessment of them, case study: Tarom GM rice. Curr. Nutr. Food Sci. 11, 11–15. [Google Scholar]
- Frankard, V. , Vauterin, M. and Jacobs, M. (1997) Molecular characterization of an Arabidopsis thaliana cDNA coding for a monofunctional aspartate kinase. Plant Mol. Biol. 34, 233–242. [DOI] [PubMed] [Google Scholar]
- Frisch, D.A. , Tommey, A.M. , Gengenbach, B.G. and Somers, D.A. (1991) Direct genetic selection of a maize cDNA for dihydrodipicolinate synthase in an Escherichia coli dapA‐auxotroph. Mol. Gen. Genet. 228, 287–293. [DOI] [PubMed] [Google Scholar]
- Frizzi, A. , Huang, S. , Gilbertson, L.A. , Armstrong, T.A. , Luethy, M.H. and Malvar, T.M. (2008) Modifying lysine biosynthesis and catabolism in corn with a single bifunctional expression/silencing transgene cassette. Plant Biotechnol. J. 6, 13–21. [DOI] [PubMed] [Google Scholar]
- Galili, G. (1995) Regulation of lysine and threonine synthesis. Plant Cell, 7, 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, G. (2002) New insights into the regulation and function significance of lysine metabolism in plants. Annu. Rev. Plant Biol. 53, 27–43. [DOI] [PubMed] [Google Scholar]
- Galili, G. and Amir, R. (2013) Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol. J. 11, 211–22. [DOI] [PubMed] [Google Scholar]
- Galili, G. , Amir, R. and Fernie, A.R. (2016) The regulation of essential amino acid synthesis and accumulation in plants. Annu. Rev. Plant Biol. 67, 153–178. [DOI] [PubMed] [Google Scholar]
- Galili, G. , Shaul, O. , Karchi, H. and Perl, A. (2017) Synthesis and accumulation of the essential amino acids lysine and threonine in seeds. In Seed Development and Germination ( Kigel, J. , ed), pp. 811–831. New York: Routledge. [Google Scholar]
- Galili, G. , Shaul, O. and Perl, A. (1994) Transgenic plants overproducing threonine and lysine. U.S. Patent No. 5,367,110.
- Gao, C.X. (2018) The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 19, 275–276. [DOI] [PubMed] [Google Scholar]
- Gibbon, B.C. , Wang, X. and Larkins, B.A. (2003) Altered starch structure is associated with endosperm modification in Quality Protein Maize. Proc. Natl. Acad. Sci. USA, 100, 15329–15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houmard, N.M. , Mainville, J.L. , Bonin, C.P. , Huang, S. , Luethy, M.H. and Malvar, T.M. (2007) High‐lysine corn generated by endosperm‐specific suppression of lysine catabolism using RNAi. Plant Biotechnol. J. 5, 605–614. [DOI] [PubMed] [Google Scholar]
- Jia, M. , Wu, H. , Clay, K.L. , Jung, R. , Larkins, B.A. and Gibbon, B.C. (2013) Identification and characterization of lysine‐rich proteins and starch biosynthesis genes in the opaque2 mutant by transcriptional and proteomic analysis. BMC Plant Biol. 13, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Kojima, H. and Tanaka, T. (1999) Mutational analysis of the feedback sites of lysine‐sensitive aspartokinase of Escherichia coli . FEMS Microbiol. Lett. 173, 211–215. [DOI] [PubMed] [Google Scholar]
- Lea, P.J. , Blackwell, R.D. and Azevedo, R.A. (1992) Analysis of barley metabolism using mutant genes. In Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology( Shewry, P.R. , ed.), pp. 181–208. Wallingford: CAB International. [Google Scholar]
- Lee, S.I. , Kim, H.U. , Lee, Y.H. , Suh, S.C. , Lim, Y.P. , Lee, H.Y. and Kim, H.I. (2001) Constitutive and seed‐specific expression of a maize lysine‐feedback insensitive dihydrodipicolinate synthase gene leads to increased free lysine levels in rice seeds. Mol. Breed. 8, 75–84. [Google Scholar]
- Liu, Q.Q. (2002) Genetically engineering rice for increased lysine. Dissertation, Yangzhou University. [Google Scholar]
- Liu, C. , Li, S. , Yue, J. , Xiao, W. , Zhao, Q. , Zhu, D. and Yu, J. (2015) Microtubule‐associated protein SBgLR facilitates storage protein deposition and its expression leads to lysine content increase in transgenic maize endosperm. Int. J. Mol. Sci. 16, 29772–29786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q.Q. , Zhang, J.L. , Wang, Z.Y. , Hong, M.M. and Gu, M.H. (1998) A highly efficient transformation system mediated by agrobacterium tumefaciens in rice (Oryza sativa L.). Acta Photophysiol. Sin. 24, 259–271. [Google Scholar]
- Liu, X. , Zhang, C. , Wang, X. , Liu, Q. , Yuan, D. , Pan, G. , Sun, S.S.M. et al. (2016) Development of high‐lysine rice via endosperm‐specific expression of a foreign LYSINE RICH PROTEIN gene. BMC Plant Biol. 16, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2‐ΔΔC T method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Long, X.H. , Liu, Q.Q. , Chan, M.L. , Wang, Q. and Sun, S.S.M. (2013) Metabolic engineering and profiling of rice with increased lysine. Plant Biotechnol. J. 11, 490–501. [DOI] [PubMed] [Google Scholar]
- Lucas, D.M. , Taylor, M.L. , Hartnell, G.F. , Nemeth, M.A. , Glenn, K.C. , Davis, S.W. (2007) Boiler performance and carcass characteristic when fed diet containing lysine maize (LY038 or LY038 x MON810), control or conventional reference maize. Poult. Sci. 86, 2152–2161. [DOI] [PubMed] [Google Scholar]
- Lugli, J. , Campbell, A. , Gaziola, S.A. , Smith, R.J. , Lea, P.J. and Azevedo, R.A. (2002) Enzymes of lysine metabolism from Coix lacryma‐jobi seeds. Plant Physiol. Bioch. 40, 25–32. [Google Scholar]
- Matthews, B.F. (1999) Lysine, threonine and methionine biosynthesis. In Plant Amino Acids: Biochemistry and Biotechnology ( Singh, B.K. , ed.), pp. 205–225. New York, NY: Marcel Dekker. [Google Scholar]
- Mertz, E.T. , Bates, L.S. and Nelson, O.E. (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science, 3629, 279–280. [DOI] [PubMed] [Google Scholar]
- Muehlbauer, G.J. , Gengenbach, B.G. , Somers, D.A. and Donovan, C.M. (1994) Genetic and amino‐acid analysis of two maize threonine‐overproducing, lysine‐insensitive aspartate kinase mutants. Theor. Appl. Genet. 89, 767–774. [DOI] [PubMed] [Google Scholar]
- Reyes, A.R. , Bonin, C.P. , Houmard, N.M. , Huang, S. and Malvar, T.M. (2009) Genetic manipulation of lysine catabolism in maize kernels. Plant Mol. Biol. 69, 81–89. [DOI] [PubMed] [Google Scholar]
- Schaeffer, G.W. and Sharpe, F.T. (1987) Increased lysine and seed storage protein in rice plants recovered from calli selected with inhibitory levels of lysine plus threonine and S‐(2‐aminoethyl) cysteine. Plant Physiol. 84, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, G.W. , Sharpe, F.T. and Sicher, R.C. (1997) Fructose 1,6‐bisphosphate aldolase activity in leaves of a rice mutant selected for enhanced lysine. Phytochemistry, 46, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Shaul, O. and Galili, G. (1992) Increased lysine synthesis in tobacco plants that express high‐levels of bacterial dihydrodipicolinate synthase in their chloroplasts. Plant J. 2, 203–209. [Google Scholar]
- Shaver, J.M. , Bittel, D.C. , Sellner, J.M. , Frisch, D.A. , Somers, D.A. and Gengenbach, B.G. (1996) Single‐amino acid substitutions eliminate lysine inhibition of maize dihydrodipicolinate synthase. Proc. Natl. Acad. Sci. USA, 93, 1962–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepansky, A. , Less, H. , Angelovici, R. , Aharon, R. , Zhu, X. and Galili, G. (2006) Lysine catabolism, an effective versatile regulator of lysine level in plants. Amino Acids, 30, 121–125. [DOI] [PubMed] [Google Scholar]
- Sun, S.S.M. and Liu, Q.Q. (2004) Transgenic approaches to improve the nutritional quality of plant proteins. In Vitro Cell Dev. 40, 155–162. [Google Scholar]
- Teixeira, C.M.G. , Gaziola, S.A. , Lugli, J. and Azevedo, R.A. (1998) Isolation, partial purification and characterization of isoenzymes of aspartate kinase from rice seeds. J. Plant Physiol. 153, 281–289. [Google Scholar]
- Toride, Y. (2004) Protein Sources for the Animal Feed Industry, pp. 161–165. Bangkok, Thailand: FAO Expert Consultation and Workshop. [Google Scholar]
- Ufaz, S. and Galili, G. (2008) Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 147, 954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasal, S.K. (2004) The Role of High Lysine Cereals in Animal and Human Nutrition in Asia. Protein Sources for the Animal Feed Industry, pp. 167–184. Rome: FAO. [Google Scholar]
- Wang, W. , Xu, M. , Wang, G. and Galili, G. (2018) New insights into the metabolism of aspartate‐family amino acids in plant seeds. Plant Reprod. 31, 203–211. [DOI] [PubMed] [Google Scholar]
- Wenefrida, I. , Utomo, H.S. and Linscombe, S.D. (2013) Mutational breeding and genetic engineering in the development of high grain protein content. J. Agric. Food Chem. 61, 11702–11710. [DOI] [PubMed] [Google Scholar]
- Wilson, B.J. , Gray, A.C. and Matthews, B.F. (1991) Bifunctional protein in carrot contains both aspartokinase and homoserine dehydrogenase activities. Plant Physiol. 97, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.W. , Liu, Q. and Sun, S.S.M. (2015) Biofortification of rice with lysine using endogenous histones. Plant Mol. Biol. 87, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, United Nations University (2007) Protein and Amino Acid Requirements in Human Nutrition, p. 935. Rome, Italy: . WHO. [PubMed] [Google Scholar]
- Xu, J. , Han, M. , Ren, X. and Zhang, W. (2016) Modification of aspartokinase III and dihydrodipicolinate synthetase increases the production of l‐lysine in Escherichia coli . Biochem. Eng. J. 114, 79–86. [Google Scholar]
- Yang, Q.Q. , Wu, H.Y. , Li, Q.F. , Duan, R.X. , Zhang, C.Q. , Sun, S.S.M. and Liu, Q.Q. (2017) Characterization of agronomy, grain physicochemical quality, and nutritional property of high‐lysine 35R transgenic rice with simultaneous modification of lysine biosynthesis and catabolism. J. Agric. Food Chem. 65, 4296–4304. [DOI] [PubMed] [Google Scholar]
- Yang, Q.Q. , Zhang, C.Q. , Chan, M.L. , Zhao, D.S. , Chen, J.Z. , Wang, Q. , Li, Q.F. et al. (2016) Biofortification of rice with the essential amino acid lysine: molecular characterization, nutritional evaluation, and field performance. J. Exp. Bot. 67, 4285–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q.Q. , Zhao, D.S. and Liu, Q.Q. (2020) Connections between amino acid metabolisms in plants: lysine as an example. Front Plant Sci. 11, 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q.Q. , Zhao, D.S. , Zhang, C.Q. , Wu, H.Y. , Li, Q.F. , Gu, M.H. , Sun, S.S.M. et al. (2018) A connection between lysine and serotonin metabolism in rice endosperm. Plant Physiol. 176, 1965–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Peng, P. , Zhang, X. , Zhao, Q. , Zhu, D. , Sun, X. , Liu, J. et al. (2005) Seed‐specific expression of the lysine‐rich protein gene sb401 significantly increases both lysine and total protein content in maize seeds. Food Nutr. Bull. 26, 427–431. [PubMed] [Google Scholar]
- Zhou, L. , Lu, Y. , Zhang, Y. , Zhang, C. , Zhao, L. , Yao, S. , Sun, X. et al. (2020) Characteristics of grain quality and starch fine structure of japonica rice kernels following preharvest sprouting. J. Cereal Sci. 95, 103023. [Google Scholar]
- Zhu, X. and Galili, G. (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell, 15, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.J. , Liu, Q.Q. , Sang, Y.J. , Gu, M.H. and Shi, Y.C. (2010) Underlying reasons for waxy rice flours having different pasting properties. Food Chem. 120, 94–100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sequence Alignments of the putative lysine‐binding domain from native AK and DHDPS and their mutants.
Figure S2 Transgene constructs for expressing modified or unmodified AK and DHDPS in rice and PCR analyses of transgenic plants.
Figure S3 Comparison of the proportion (by weight) of other individual free amino acids among total measurable free amino acids in mature seeds of transgenic and WT rice.
Figure S4 The contents of total essential amino acids in mature seeds of transgenic and WT rice.
Figure S5 The expression of AK, DHDPS and LKR in developing rice seeds of transgenic and WT plants.
Table S1 Primers used in this study.
Table S2 Sequence identity after full searching of candidate proteins in allergen databases of AllergenOnline and Allermatch (E value < 1).
Table S3 The information of chimeric genes for production of transgenic rice used in this study.
Table S4 Free lysine content in mature seeds of transgenic and WT rice.
Table S5 Proposed scores for essential amino acids in transgenic and WT rice.


