Dear editor,
RNA interference (RNAi) is a promising approach for developing insect‐resistant crops. In the first two proof‐of‐concept studies, DNA fragments derived from essential insect genes were constructed into plant expression cassettes as inverted repeats, enabling long double‐stranded RNAs (dsRNAs) to be transcribed in host plants (Baum et al., 2007; Mao et al., 2007). After ingestion, dsRNAs overexpressed in plants suppressed target gene expression via small interference RNA (siRNA)‐mediated RNAi in western corn rootworm (Diabrotica virgifera virgifera; Baum et al., 2007) and cotton bollworm (Helicoverpa armigera) (Mao et al., 2007), and thereby reduced their viability. Subsequent studies have mostly employed a similar approach and attempted to develop RNAi crops against various insect species (Liu et al., 2020). However, except for coleopterans that are generally susceptible to RNAi, most insects (e.g. lepidopterans, dipterans, hymenopterans and hemipterans) exhibit unpredictable responses to dsRNA‐induced RNAi, and this has become a hurdle for ubiquitous adaption of this strategy (Cooper et al., 2019). Potential factors influencing RNAi efficacy in insects involve dsRNA stability, cellular dsRNA absorption, core RNAi machinery integrity, systemic RNAi spread and target gene amenability (Cooper et al., 2019).
Although most studies on RNAi‐mediated insect‐resistant plants are based on dsRNA (Liu et al., 2020), limited studies have suggested that microRNA (miRNA)‐mediated RNAi is also practically applicable for insect‐resistant plant development. MiRNA‐mediated RNAi plants can be classified into four types based on the origin of miRNA effectors: artificially designed miRNAs (Guo et al., 2014), natural insect miRNAs (Jiang et al., 2017), natural plant miRNAs (Wamiq and Khan, 2018) and modified insect pre‐miRNAs (Bally et al., 2020).
Our previous study demonstrated that striped stem borer (SSB; Chilo suppressalis), a destructive insect pest of rice (Oryza sativa), is refractory to dsRNA‐induced RNAi but relatively susceptible to artificial miRNA (amiRNA)‐induced RNAi. Transgenic rice expressing SSB endogenous miRNA candidates with unknown functions retarded the growth of SSB larvae, without showing significant lethality (Jiang et al., 2017).
In this study, we successfully engineered highly SSB‐resistant rice (named csu260) expressing amiRNA of SSB endogenous miRNA csu‐novel‐miR260 through amiRNA expression technology. Csu‐novel‐miR260 negatively regulates ecdysteroid biosynthesis in SSB by suppressing the expression of disembodied (dib) that encodes a cytochrome P450 enzyme catalysing the C22‐hydroxylation of 2, 22‐dideoxy‐3‐dehydroecdysone (Figure 1a; He et al., 2017). Two representative homozygous csu260 rice lines (csu260‐16 and csu260‐18) with a single‐copy insertion were obtained from 28 independent primary transformants.
Figure 1.
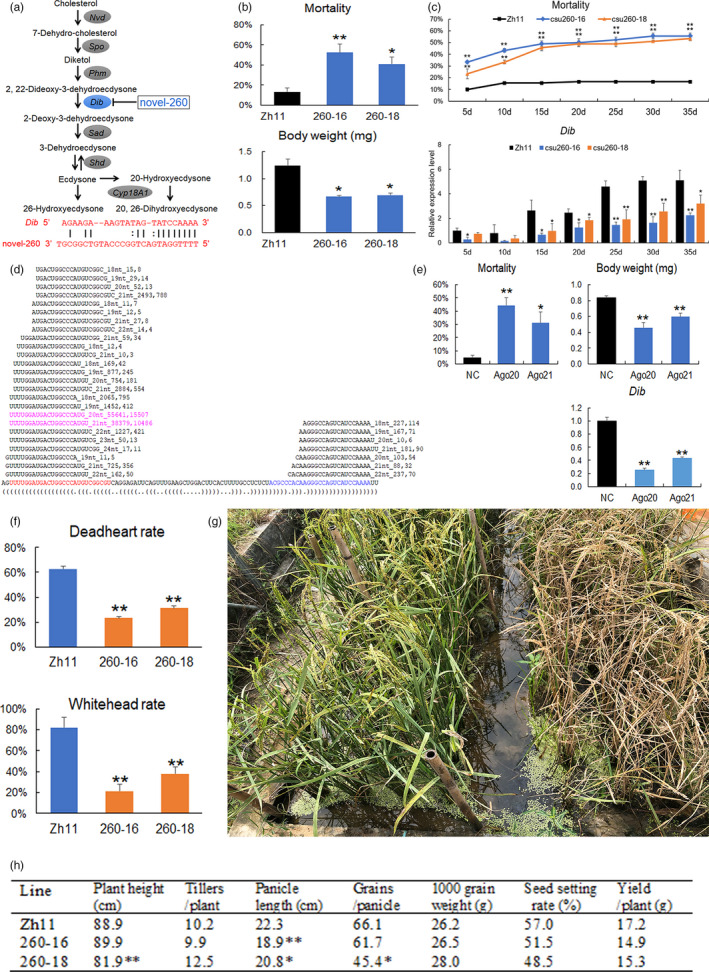
Overexpressing amiRNA of csu‐novel‐260 confers high resistance against striped stem borer (SSB). (a) The biosynthetic pathway of ecdysteroids and putative target site of csu‐novel‐260 in 5′ UTR of dib (modified from He et al., 2017). (b) Five‐day stemcutting feeding assay. (c) Thirty‐five day consecutive stemcutting feeding assay. (d) Small RNA sequencing of csu260‐16 and csu260‐18 rice. The bottom sequence is the amiRNA precursor of csu‐novel‐260. The red and blue bases in the amiRNA precursor are the positions of amiRNA and amiRNA*, respectively. Sequences above the amiRNA precursor of csu‐novel‐260 are the sequenced amiRNAs (threshold of read number ≥ 10 in csu260‐16) matching the amiRNA precursor. The two sequences in pink are the two most abundant amiRNA sequences. The three numbers at the end of each amiRNA sequence are their length, and read counts of csu260‐16 and csu260‐18. (e) Agomir feeding assay. (f) Field assessment of SSB resistance in the tillering stage (deadheart rate) and the mature stage (whitehead rate) under manual infestation conditions. (g) Field performance under manual infestation in the maturity stage. Left: csu260‐18; right: Zh11 control. (h) Agronomic assessment of csu260‐16 and csu260‐18 under field conditions. Data in (b), (c), (e) and (f) are presented as mean ± SE. * and ** indicate statistically significant difference at P < 0.05 and P < 0.01, respectively, compared with the WT control, according to Student’s t‐test
A five‐day stemcutting feeding assay was performed in triplicate with 15 first‐instar SSB larvae for each replicate. It showed that csu260‐16 and csu260‐18 exhibited significant lethality against and retarded the growth of feeding SSB larvae compared with those fed the wild‐type (WT) recipient variety Zhonghua11 (Zh11) (Figure 1b). Then, a consecutive stemcutting feeding assay was performed in triplicate with 30 first‐instar SSB larvae for each replicate. It confirmed that csu260‐16 and csu260‐18 rice caused 55.6% and 53.3% larval mortality, respectively, at 35 days after feeding, whereas the mortality of the control larvae was less than 20% (Figure 1c). Correspondingly, the stem–loop quantitative polymerase chain reaction analysis showed consistently lower dib expression in the csu260‐fed larvae than in the control larvae throughout the feeding assay (Figure 1c).
We used small RNA sequencing to characterize the amiRNA expression in csu260‐16 and csu260‐18 (Figure 1d). The sequenced amiRNA reads were normalized as reads per million, and the normalized amiRNA abundance was 1.8‐fold higher in csu260‐16 than in csu260‐18. Consistently, csu260‐16 showed higher and more stable insect resistance than csu260‐18, and dib expression in SSB larvae fed csu260‐16 was lower than that in larvae fed csu260‐18 (Figure 1b, c). These results suggest that amiRNA dose is crucial for insect resistance of amiRNA plants. We used the osa‐MIR528 precursor as a backbone for constructing amiRNA expression vectors (Jiang et al., 2017). Small RNA sequencing showed that amiRNAs in csu260 rice are preferentially processed into 20‐ and 21‐nt long sequences, similar to mature osa‐MIR528 (21 nt), although the amiRNA sequence of csu‐novel‐260 originally constructed into amiRNA expression vector is 27‐nt long. The predominant 20 and 21 nt amiRNAs in cus260 rice possess the intact 5′ end of original csu‐novel‐260, but they lacked 7mer and 6mer at the 3′ end, respectively (Figure 1d). MiRNAs generally function effectively as long as the ‘seed sequence’ (positions 2–8 at the 5′ end of miRNA) matches between miRNA and the 3′ untranslated region (UTR) of messenger RNA (Brennecke et al., 2005). An in vitro feeding assay was performed using 100 nmol synthetic miRNA mimics (agomirs) with the same sequences as the predominant 20 (Ago20) and 21 nt (Ago21) sequences. The agomirs were delivered via five‐day germinated Zh11 seeds, which soaked in agomir solutions for 30 min before the assay. The result confirmed the efficacy of these trimmed amiRNAs (Figure 1e), indicating that the length of natural miRNAs would not be a major consideration when constructing the amiRNA expression vectors.
Moreover, we evaluated SSB resistance under field conditions, resembling practical rice production. Because SSB infestation causes ‘deadheart’ in the tillering stage and ‘whitehead’ in the heading stage, we conducted two independent field assessments of SSB resistance in these two stages. In both field assessments of SSB resistance, each of csu260‐16, csu260‐18, and Zh11 was grown in three plots as three replicates. The deadheart rate of csu260‐16 and csu260‐18 decreased by 62.9% and 50.4%, respectively, compared with that of the WT control, 35 days after the manual inoculation of 30 second‐instar SSB larvae per plant in the early tillering stage (Figure 1f). While the whitehead rate of the two csu260 lines decreased by 74.6% and 54.0%, respectively, compared with that of the WT control, 40 days after manual infestation with 1.4 SSB adult moths per plant in the early heading stage (Figure 1f, g). These results indicate that csu260 rice substantially prevented yield loss even under severe SSB infestation.
To evaluate agronomic performance in the field, each of cus260‐16, csu260‐18 and Zh11 was grown in three plots as three replicates. The result demonstrated that the most important trait yield per plant of two csu260 lines was not significantly different from that of the control. However, panicle length of cus260‐16 and plant height, panicle length and grains per panicle of csu260‐18 showed significant differences from those of the control (Figure 1h). Agronomic variations in transgenic lines are usually caused by somatic mutations occurring in transformation procedure. Therefore, most agronomic variations may be recovered via backcrosses with the WT and subsequent self‐pollinations.
Our results demonstrated that amiRNA‐induced RNAi is more applicable for controlling SSB than dsRNA‐induced RNAi. Because endogenous miRNAs naturally regulate their gene targets in insects, their use theoretically avoids certain factors that cause inefficient RNAi, such as refractory target genes and relevant RNAi machinery deficiency (Cooper et al., 2019). Moreover, critical siRNA and miRNA components associated with their biogenesis and function are significantly different (Cooper et al., 2019), potentially causing variations among insects in terms of small RNA stability, cellular uptake and intercellular transport between siRNA and miRNA. These factors could explain why SSB is resistant to dsRNA‐induced RNAi but relatively susceptible to miRNA‐induced RNAi. Taken together, amiRNA‐induced RNAi can be an alternative approach for controlling these insect pests insensitive to dsRNA‐induced RNAi, which would broaden the applicability of RNAi‐mediated insect pest management.
Author contributions
H.C., F.L. and Y.L. designed the study; X.Z., Z.W., H.L., Z.K. and W.M. performed the experiments; H.C., X.Z. and Z.Z. analysed the data and prepared the figures; H.C. and X.Z. wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This project was financially supported by the National Natural Science Foundation of China (Grant No. 31772178) and the National Special Key Project for Transgenic Breeding (Grant No. 2016ZX0810002‐002).
Zheng, X. , Weng, Z. , Li, H. , Kong, Z. , Zhou, Z. , Li, F. , Ma, W. , Lin, Y. and Chen, H. (2021) Transgenic rice overexpressing insect endogenous microRNA csu‐novel‐260 is resistant to striped stem borer under field conditions. Plant Biotechnol J., 10.1111/pbi.13504
References
- Bally, J. , Fishilevich, E. , Doran, R.L. , Lee, K. , de Campos, S.B. , German, M.A. , Narva, K.E. et al. (2020) Plin‐amiR, a pre‐microRNA‐based technology for controlling herbivorous insect pests. Plant Biotechnol. J. 18, 1925–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, J.A. , Bogaert, T. , Clinton, W. , Heck, G.R. , Feldmann, P. , Ilagan, O. , Johnson, S. et al. (2007) Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Brennecke, J. , Stark, A. , Russell, R.B. and Cohen, S.M. (2005) Principles of microRNA–target recognition. PLoS Biol. 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A.M.W. , Silver, K. , Zhang, J. , Park, Y. and Zhu, K.Y. (2019) Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 75, 18–28. [DOI] [PubMed] [Google Scholar]
- Guo, H. , Song, X. , Wang, G. , Yang, K. , Wang, Y. , Niu, L. , Chen, X. et al. (2014) Plant‐generated artificial small RNAs mediated aphid resistance. PLoS One, 9, e97410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K. , Sun, Y. , Xiao, H. , Ge, C. , Li, F. and Han, Z. (2017) Multiple miRNAs jointly regulate the biosynthesis of ecdysteroid in the holometabolous insects, Chilo suppressalis . RNA, 23, 1817–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Wu, H. , Liu, H. , Zheng, J. , Lin, Y. and Chen, H. (2017) The overexpression of insect endogenous small RNAs in transgenic rice inhibits growth and delays pupation of striped stem borer (Chilo suppressalis). Pest Manag. Sci. 73, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Jaouannet, M. , Dempsey, D.A. , Imani, J. , Coustau, C. and Kogel, K.H. (2020) RNA‐based technologies for insect control in plant production. Biotechnol. Adv. 39, 107463. [DOI] [PubMed] [Google Scholar]
- Mao, Y.B. , Cai, W.J. , Wang, J.W. , Hong, G.J. , Tao, X.Y. , Wang, L.J. , Huang, Y.P. et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Wamiq, G. and Khan, J.A. (2018) Overexpression of ghr‐miR166b generates resistance against Bemisia tabaci infestation in Gossypium hirsutum plants. Planta, 247, 1175–1189. [DOI] [PubMed] [Google Scholar]


