One of the important goals of crop breeding is yield improvement. Among the yield indices, the tiller angle is tightly associated with enhancing photosynthetic efficiency and facilitating enhanced planting density (Sakamoto et al., 2006; Wang and Li, 2008). Rice plants with erect tillers, leaves and panicles allow a high‐density planting system for high yields but are more susceptible to the occurrence of sheath blight disease causing yield reduction. Therefore, the antagonistic relationship between crop yield and immunity pathways makes crop breeding extremely difficult (Ning et al., 2017). In our previous studies, we found that overexpression of loose plant architecture 1 (LPA1) reduced the tiller and lamina joint angle but increased resistance to sheath blight disease through activation of PIN1a‐mediated auxin distribution, suggesting the breeding potential of LPA1 in high‐density planting systems (Liu et al., 2016; Sun et al., 2019). To further analyse the mechanism of tiller angle and sheath blight regulation, we performed a yeast two‐hybrid selection and identified G‐protein γ subunit DEP1 (dense and erect panicle 1, Os09g26999) as a novel interactor of LPA1. The heterotrimeric G proteins, comprising α, β and γ subunits, are key players in the transmission of extracellular signals via membrane‐spanning G‐protein‐coupled receptors to intracellular effectors (Gilman, 1987), and panicle erectness is controlled by a dominant allele of DEP1, which reduces the length of the inflorescence internode (Huang et al., 2009). Further analysis indicated that DEP1 interacted with both full‐length LPA1 and its N‐terminal region (indeterminate domain, IDD) (Figure 1a). Furthermore, coimmunoprecipitation (co‐IP) and split‐GFP assays confirmed that LPA1 interacted with DEP1 in the nucleus (Figure 1b,c).
Figure 1.
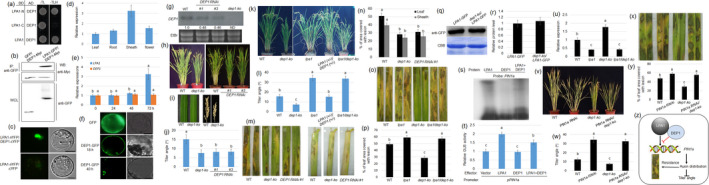
DEP1 controls the tiller angle and resistance to sheath blight disease in rice. (a) The interaction between LAP1 and DEP1 was analysed in a yeast two‐hybrid system. Interaction between activating domain (AD)‐DEP1 and binding domain (BD)‐LPA1 (full length), BD‐LPA1‐N (N‐terminal), or BD‐LAP1‐C (C‐terminal) was analysed. (b) Coimmunoprecipitation (co‐IP) was performed to analyse the interaction between DEP1 and LPA1 in tobacco leaves. DEP1‐Myc + LPA1‐GFP or DEP1‐Myc + GFP were transfected into tobacco leaves. Proteins immunoprecipitated using anti‐GFP were detected using anti‐Myc antibody. The levels of DEP1‐Myc, GFP and LPA1‐GFP from whole‐cell lysates (WCL) were detected using anti‐Myc and anti‐GFP antibodies, respectively. (c) Reconstitution of GFP fluorescence from LPA1‐nYFP + DEP1‐cCFP and LPA1‐nYFP + cCFP. Bars = 10 μm. (d) Expression patterns of DEP1 in the leaf, root, sheath and flower tissues were examined. Data indicate average ± standard error (SE) (n =3). (e) Expressions of LPA1 and DEP1 were tested after 0, 24, 48 and 72 h of R. solani AG1‐IA inoculation. Data indicate average ± standard error (SE) (n =3). (f) Free GFP and DEP1‐GFP signals were detected in the protoplasts. GFP signal was checked at 18 and 48 h. Bars = 10 μm. (g) DEP1 expression level in wild type (WT), DEP1 RNAi lines (#1 and #3) and dep1‐ko was analysed by northern blot analysis. EtBr staining was used as a loading control. The numbers shown at the bottom of the blot indicate the relative density of each band. ND: not detected. (h) Shown are 3‐month‐old WT, dep1‐ko and DEP1 RNAi lines (#1 and #3) plants. (i) The leaf and panicle of WT and dep1‐ko were photographed. (j) Tiller angles of plants from panel (h) are shown. Data indicate average ± SE (n> 10). (k) Shown are 3‐month‐old WT, dep1‐ko, lpa1, DEP1 (+/‐)/LPA1 (+/‐) and lpa1/dep1‐ko. (l) Tiller angles of the lines from panel (k) are calculated. Data indicate average ± SE (n > 10). (m) Leaves and sheath from the WT, dep1‐ko and DEP1 RNAi #1 had been inoculated with R. solani AG1‐IA. Each experiment was performed in triplicate. (n) The percentage of lesions in the leaves and sheath shown in (m) was examined. Data indicate average ± SE (n > 10). (o) Leaves from the WT, lpa1, dep1‐ko and lpa1/dep1‐ko plants were inoculated with R. solani AG1‐IA. (p) The percentage of lesions in the leaves from panel (o) was examined. Data indicate average ± SE (n > 10). (q) LPA1‐GFP protein levels in LPA1‐GFP and dep1‐ko/LPA1‐GFP transgenic plants were examined. The proteins stained with Coomassie brilliant blue (CBB) were used as the loading control. (r) The band density shown in panel (q) was calculated. Data indicate average ± SE (n =3). (s) An electrophoretic mobility shift assay (EMSA) was conducted to evaluate the affinities of LPA1 and DEP1 to the PIN1a promoter. (t) Transient expression of p35S:LPA1 alone or p35S:LPA1 and p35S:DEP1 together with a construct including the GUS gene under the control of 1.5 kb PIN1a promoter in protoplast cells. A luciferase gene driven by the 35S promoter was used as an internal control to normalize GUS expression. Error bars represent ± SE (n = 6). (u) Expression level of PIN1a in WT, lpa1, dep1‐ko and lpa1/dep1‐ko plants. Data indicate average ± SE (n =3). (w) Shown are 3‐month‐old WT, PIN1a RNAi, dep1‐ko and PIN1a RNAi/dep1‐ko plants. (v) Tiller angles of the lines from panel (u). Data indicate average ± SE (n > 10). (x) Leaves from the WT, PIN1a RNAi, dep1‐ko and PIN1a RNAi/dep1‐ko plants were inoculated with R. solani AG1‐IA and were photographed after infection. (y) The percentage of lesions in the leaves shown in panel (x) was examined. Data indicate average ± SE (n > 10). (z) Schematic diagram showing LAP1‐DEP1 regulating PIN1a transcription, and its mediated auxin distribution on sheath blight resistance and tiller angle. Different letters above the columns indicate a statistically significant difference between groups. The lesion areas in the leaves and sheath were calculated after 3 days following inoculation.
Through qPCR, we found that DEP1 expression in sheaths is high compared with that in leaves, roots and flowers (Figure 1d). Rhizoctonia solani inoculation induced LPA1, but not DEP1 (Figure 1e), and DEP1‐GFP was localized at the plasma membrane and nucleus (Figure 1f). To analyse DEP1 function in the japonica rice cultivar Dongjin, a DEP1 knockout mutant dep1‐ko (An et al., 2003) (PFG_3A‐02648) with the T‐DNA inserted into the first intron, and DEP1 RNAi lines were used. Northern blot results confirmed that DEP1 expression was suppressed by about 50% in two RNAi lines (#1 and #3) while it was not detected in dep1‐ko (Figure 1g). Compared with wild type, dep1‐ko and DEP1 RNAi plants exhibited a narrow tiller angle, similar shape of leaves and a short panicle (Figure 1h,i,j).
It has previously been shown that lpa1 causes a wider tiller angle (Liu et al., 2016; Wu et al., 2013). Further genetic studies showed that lpa1 plants were similar to lpa1/dep1‐ko plants exhibiting a wider tiller angle. However, the tiller angle of plants that were heterozygous for both genes (LPA1 (+/‐)/DEP1 (+/‐)) was similar to that of wild‐type plants (Figure 1k,l). In addition, overexpression of LPA1 has been shown to increase resistance to rice sheath blight (Sun et al., 2019). Interestingly, dep1‐ko and DEP1 RNAi plants were less susceptible to sheath blight compared with wild‐type plants (Figure 1m,n). Upon further examination, we discovered that lpa1 and lpa1/dep1‐ko plants exhibited similar symptoms and were more susceptible, while dep1‐ko plants were significantly less susceptible to sheath blight than wild‐type plants (Figure 1o,p).
Even though DEP1 interacts with LPA1, but Western blot analysis showed that LPA1‐GFP protein levels were similar in LPA1‐GFP and dep1‐ko/LPA1‐GFP, a genetic combination by crossing LPA1‐GFP and dep1‐ko plants (Figure 1q,r). LPA1 activates PIN1a via promoter binding, which increases planting density and resistance to sheath blight disease (Sun et al., 2019). Therefore, we further tested the role of DEP1 in LPA1‐mediated PIN1a activation via the EMSA and transient assays. EMSA result indicated that DEP1 inhibits the binding of LPA1 to the PIN1a promoter (Figure 1s). The transient assay by co‐transformed with p35S:LPA1, p35S:DEP1 or p35S:LPA1 together with p35S:DEP1 and a vector expressing the beta‐glucuronidase gene (GUS) under the control of pPIN1a promoter in protoplast cells revealed that co‐expression of DEP1 reduced the ability of LPA1 to stimulate the relative GUS activity (Figure 1t), and qPCR results also showed that PIN1a expression level was higher in dep1‐ko than in lpa1, lpa1/dep1‐ko and wild‐type plants (Figure 1u). Further genetic studies demonstrated that PIN1 RNAi plants were similar to PIN1 RNAi/dep1‐ko plants exhibiting a wider tiller angle (Figure 1v,w). In addition, PIN1a RNAi and PIN1 RNAi/dep1‐ko plants were more susceptible, while dep1‐ko was less susceptible to sheath blight compared with wild‐type plants (Figure 1x,y).
Taken together, our analyses revealed that DEP1 interacts with LPA1 to regulate PIN1a expression and that down‐regulation of DEP1 enhanced planting density by decreasing the tiller angle and at the same time promoted rice resistance to sheath blight disease (Figure 1z). In addition, DEP1 inhibited LPA1‐dependent activation of PIN1a transcription via interacts with the N‐terminal region of LPA1, which is the IDD domain region, a known DNA‐binding domain (Kozaki et al., 2004). Our data suggest that the interaction between DEP1 and the IDD domain inhibits the DNA‐binding ability of LPA1, thereby suppressing PIN1a expression, leading to an increase in planting density and resistance to sheath blight disease in rice.
Conflict of interests
The authors declare no conflict of interest.
Authors’ contributions
JML and YHX designed the experiments. JML, QM, CYX and ZYW performed the experiments. DPL and YXZ manipulated plant materials. JML, QM and YHX analysed data. JML and YHX wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Support Program for Science and Technology Innovation Talents of Shenyang (RC190489), Natural Science Foundation of Liaoning Province (2020‐YQ‐05), the Support Plan for Innovative Talents in Colleges and Universities of Liaoning Province (LR2017037) and Natural Science Foundation of Zhejiang Province (LQ19C020004).
Miao Liu, J. , Mei, Q. , Yun Xue, C. , Yuan Wang, Z. , Pin Li, D. , Xin Zhang, Y. and Hu Xuan, Y. (2021) Mutation of G‐protein γ subunit DEP1 increases planting density and resistance to sheath blight disease in rice. Plant Biotechnol. J., 10.1111/pbi.13500
References
- An, S. , Park, S. , Jeong, D.H. , Lee, D.Y. , Kang, H.G. , Yu, J.H. , Hur, J. et al. (2003) Generation and analysis of end sequence database for T‐DNA tagging lines in rice. Plant Physiol. 133, 2040–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman, A.G. (1987) G proteins: transducers of receptor‐generated signals. Annu. Rev. Biochem. 56, 615–649. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Qian, Q. , Liu, Z. , Sun, H. , He, S. , Luo, D. , Xia, G. et al. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. [DOI] [PubMed] [Google Scholar]
- Kozaki, A. , Hake, S. and Colasanti, J. (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32, 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.M. , Park, S.J. , Huang, J. , Lee, E.J. , Xuan, Y.H. , Je, B.I. , Kumar, V. et al. (2016) Loose Plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C‐22‐hydroxylated and 6‐deoxo brassinosteroids in rice. J. Exp. Bot. 67, 1883–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, Y. , Liu, W. and Wang, G.L. (2017) Balancing immunity and yield in crop plants. Trend. Plant Sci. 22, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T. , Morinaka, Y. , Ohnishi, T. , Sunohara, H. , Fujioka, S. , Ueguchi‐Tanaka, M. , Mizutani, M. et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Li, T.Y. , Li, D.D. , Wang, Z.Y. , Li, S. , Li, D.P. , Han, X. et al. (2019) Overexpression of Loose Plant Architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN‐FORMED 1a in rice. Plant Biotechnol. J. 17, 855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Li, J. (2008) Rice, rising. Nat. Genet. 40, 1273–1275. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Tang, D. , Li, M. , Wang, K. and Cheng, Z. (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 161, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]


