Graphical abstract
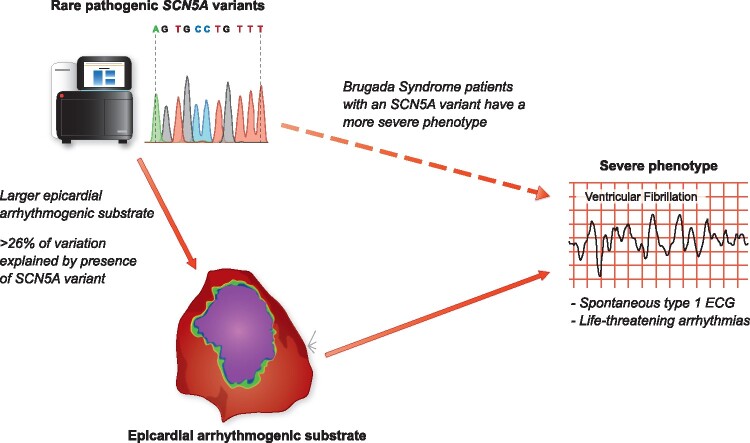
The presence of rare pathogenic SCN5A variants is associated with more severe phenotypes in Brugada syndrome patients, which may be at least partly explained by greater epicardial arrhythmogenic substrates in these patients.
This editorial refers to ‘Brugada syndrome genetics is associated with phenotype severity’†, by G. Ciconte et al., on page 1082.
Brugada syndrome (BrS) is a relatively rare arrhythmia disorder that can lead to sudden cardiac death (SCD) due to ventricular tachycardia/fibrillation (VT/VF), predominantly in adults. Despite being considered an inherited disease, the genetic basis of BrS appears to be considerably more complex than other genetic cardiac conditions such as long QT syndrome (LQTS) and hypertrophic cardiomyopathy (HCM) that have a more prominent Mendelian-like inheritance. Although rare variants in numerous genes encoding ion channels and related proteins have been implicated in BrS, recent re-evaluation of the evidence for these associations has downgraded all but one gene, SCN5A, to disputed status.1 Rare pathogenic variants in SCN5A, which encodes for the NaV1.5 sodium channel, are detected in only about 20% of BrS cases of European descent however (with diagnostic yields lower still in East Asian populations where BrS is much more prevalent). It is now believed that most BrS patients, particularly those without a rare SCN5A variant, are likely to have a complex aetiology comprising multiple genetic and non-genetic factors rather than as yet undiscovered Mendelian causes of disease. Genome-wide association studies (GWAS) have identified an unexpectedly strong contribution of common genetic variants to the BrS phenotype, with common non-coding variants at the SCN5A locus being particularly prominent.2
Genetic testing to identify rare pathogenic variants in SCN5A can be pursued in patients with BrS with a Class IIa recommendation.3 While the identification of a causative rare variant may be used for identifying at risk family members through cascade genetic screening, knowledge of the genetic status plays little role in the clinical management or prognosis for BrS patients.3 This is despite the fact that the presence of a rare pathogenic SCN5A variant has been associated with a more severe phenotype in BrS patients, including a higher occurrence of major arrhythmic events.4 While it is not atypical in cardiac disease for worse outcomes to be associated with the presence of rare pathogenic variants (HCM patients with pathogenic variants in sarcomere genes also have a poorer prognosis than those without5), the mechanism underlying this in BrS patients is not fully understood. Now, a new study by Ciconte and colleagues has investigated this issue through the analysis of a critical intermediate phenotype, the epicardial arrhythmogenic substrate area, in a cohort of BrS patients with and without SCN5A variants.6
In their paper, Ciconte and colleagues share with us their data on BrS patients referred to the San Donato hospital, Milan, Italy, because of their inferred high-risk features. From the 195 included patients, 49 (25%) appeared to harbour a pathogenic or likely pathogenic SCN5A variant. All patients had an implantable cardioverter defibrillator (ICD) and underwent endocardial and epicardial mapping and ablation to modify their arrhythmogenic substrate to the example of the pioneering study of Nademanee and colleagues in Bangkok, Thailand.7 Noteworthy, the San Donato experience with this procedure is now possibly one of the largest in the world. The mapping and ablation procedure described in the study is well structured and conducted systematically, including endocardial right ventricular (RV) mapping followed by epicardial mapping with a decapolar catheter to assess the area with wide, low voltage and fragmented electrograms and local late potentials (predominantly at the RV outflow tract [RVOT]). Subsequently, ajmaline was administered to evaluate enlargement of this area by additional sodium channel blockade, thereby defining the arrhythmogenic substrate and setting the target area for ablation. This protocol mirrors the protocol used in multiple centres.8 , 9 Although not specifically addressed in this paper, the target area would then be modified by the application of RF energy to, ideally, reach a non-inducible type-1 ECG after resolution of the initial ST-segment changes after epicardial ablation. This non-inducibility of the type-1 ECG is believed to mirror a (vast) reduction in arrhythmogenic risk.
Concerning the impact of genotype on phenotypic expression in these patients, already at baseline a notable difference in conduction delay was apparent; patients with a pathogenic or likely pathogenic SCN5A variant had at mean 30 ms longer PR intervals, 10 ms longer filtered QRS complex durations, while the other parameters from signal averaged ECGs were also worse in the SCN5A variant positive patients. This resembles previous reports on conduction slowing in BrS.10 , 11 In their study, Ciconte et al. for the first time now also demonstrate that invasive epicardial measures of conduction delay, already apparent in all BrS cases, are exaggerated in patients with a pathogenic or likely pathogenic SCN5A variant, translating to larger substrates and longer electrogram durations. (Ultra)structural abnormalities are believed to play an important role in the occurrence of conduction delay and the arrhythmogenic BrS substrate.12 , 13 In patients with an SCN5A variant, such abnormalities likely conspire with the reduced sodium current to determine the ultimate characteristics and severity of the substrate. Interestingly, in patients without a current type-1 ECG, abnormal electrograms can also be seen at the epicardial RVOT. This resembles the notion that additional excitation failure at the RVOT epicardium is obligatory to reach a type-1 ECG, while such excitation failure is dependent on structural abnormalities coinciding with sufficiently reduced conductivity13 (and the latter can be initiated by, e.g., ajmaline).
While this study provides important insights on the relationship between genotype and phenotype in BrS, our understanding of these associations is still rudimentary. For the interpretation of SCN5A variants, a generic classification tool was applied – while the majority of variants were likely to have been correctly classified, gene-specific approaches using high throughput functional assays14 or quantitative case-control approaches15 may lead to improved classification and estimation of variant effect size rather than binary predictions. Limited phenotypic differences were observed in this cohort based on SCN5A variant classes and properties - a larger baseline (though not ajmaline-induced) substrate was noted for patients with non-missense variants but there were no significant differences based on topographical location. Larger cohorts are likely to be needed to further characterise the interaction between SCN5A variant class and phenotypic measures like the epicardial arrhythmogenic substrate. In addition, it is important to note that this San Donato cohort is (much) less severely affected compared to other cohorts:7 , 8 12% of patients had a previous aborted cardiac arrest, 22% had a spontaneous type-1 ECG and 38% had appropriate ICD therapy, while in 48% VT/VF was inducible (undoubtedly with considerable overlap between categories). Interestingly, the BrS substrate at the RV epicardium can also be seen in lower risk patients. Still, whether the ablation procedure in a sample of this population outweighs baseline risk is currently uncertain. It is interesting to note that, although not formally tested, based on the numbers provided (Table 1 and supplemental data in Ciconte et al.), a spontaneous type-1 ECG, inducible VT/VF and presence of an SCN5A variant appear to be associated with appropriate ICD therapy, while a family history of sudden death seems to have the opposite association. Similarly, the presence of an SCN5A variant appears to be associated with a spontaneous type-1 ECG and symptoms (aborted cardiac arrest, syncope and appropriate ICD therapy).
The genetic architecture of BrS is of course likely to be much more complex than the simple presence or absence of a rare pathogenic SCN5A variant and efforts to uncover the associations between the genetic status of BrS patients and their phenotype and clinical outcomes are dependent on furthering our understanding of BrS genetics. While most non-SCN5A Mendelian BrS associations have been disputed, it is likely that some novel, and likely infrequent, disease genes that harbour rare genetic variation remain to be discovered and these could at least partially define the genetic aetiology in some patients. The role of common variants identified by GWAS in determining phenotype also needs to be further explored. The SCN5A-SCN10A locus is the strongest association with BrS, where causal variants are likely to act through regulation of SCN5A gene expression.2 Other significant loci may also act indirectly on the dosage of the SCN5A transcript, further emphasising the central role of sodium channel function in BrS. Defining the collective contribution of common variants, through instruments like polygenic risk scores, and their interaction with rare pathogenic SCN5A variants will lead to an enhanced estimation of the overall genetic risk in BrS patients. Such insights may then enable more comprehensive and accurate investigations into genotype-phenotype associations in BrS, for both intermediate phenotypes like the epicardial arrhythmogenic substrate and clinical events and outcomes. While we are still at the early stages of being able to offer individualised prognosis for BrS patients, studies like the one by Ciconte and colleagues are beginning to illuminate the path between genetic risk factors and clinical outcomes.
Acknowledgements
C.R.B. is supported by the Dutch Heart Foundation (CVON Predict2 project), the Netherlands Organization for Scientific Research (VICI fellowship, 016.150.610) and Fondation Leducq (17CVD02). R.W. is supported by an Amsterdam Cardiovascular Sciences fellowship.
Conflict of interest: none declared.
Footnotes
† doi:10.1093/eurheartj/ehaa942.
Contributor Information
Pieter G Postema, Department of Clinical Cardiology, Amsterdam UMC, AMC Heart Center, Amsterdam, The Netherlands.
Roddy Walsh, Department of Experimental Cardiology, Amsterdam UMC, AMC Heart Center, Amsterdam, The Netherlands.
Connie R Bezzina, Department of Experimental Cardiology, Amsterdam UMC, AMC Heart Center, Amsterdam, The Netherlands.
The opinions expressed in this article are not necessarily those of the Editors of the European Heart Journal or of the European Society of Cardiology.
References
- 1. Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, Garcia J, Care M, Sturm AC, Novelli V, Ackerman MJ, Ware JS, Hershberger RE, Wilde AAM, Gollob MH, On behalf of the National Institutes of Health Clinical Genome Resource Consortium National Institutes of Health Clinical Genome Resource Consortium. Reappraisal of reported genes for sudden arrhythmic death. Circulation 2018;138:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud J-B, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, Guicheney P, Fressart V, Leenhardt A, Antzelevitch C, Bartkowiak S, Borggrefe M, Schimpf R, Schulze-Bahr E, Zumhagen S, Behr ER, Bastiaenen R, Tfelt-Hansen J, Olesen MS, Kääb S, Beckmann BM, Weeke P, Watanabe H, Endo N, Minamino T, Horie M, Ohno S, Hasegawa K, Makita N, Nogami A, Shimizu W, Aiba T, Froguel P, Balkau B, Lantieri O, Torchio M, Wiese C, Weber D, Wolswinkel R, Coronel R, Boukens BJ, Bézieau S, Charpentier E, Chatel S, Despres A, Gros F, Kyndt F, Lecointe S, Lindenbaum P, Portero V, Violleau J, Gessler M, Tan HL, Roden DM, Christoffels VM, Le Marec H, Wilde AA, Probst V, Schott J-J, Dina C, Redon R. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet 2013;45:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Marec HL, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Hear Rhythm 2011;8:1308–1339. [DOI] [PubMed] [Google Scholar]
- 4. Yamagata K, Horie M, Aiba T, Ogawa S, Aizawa Y, Ohe T, Yamagishi M, Makita N, Sakurada H, Tanaka T, Shimizu A, Hagiwara N, Kishi R, Nakano Y, Takagi M, Makiyama T, Ohno S, Fukuda K, Watanabe H, Morita H, Hayashi K, Kusano K, Kamakura S, Yasuda S, Ogawa H, Miyamoto Y, Kapplinger JD, Ackerman MJ, Shimizu W. Genotype-phenotype correlation of SCN5A mutation for the clinical and electrocardiographic characteristics of probands with Brugada syndrome: a Japanese Multicenter Registry. Circulation 2017;135:2255–2270. [DOI] [PubMed] [Google Scholar]
- 5. Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, Caleshu CA, Helms AS, Colan SD, Girolami F, Cecchi F, Seidman CE, Sajeev G, Signorovitch J, Green EM, Olivotto I, For the SHaRe Investigators Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 2018;138:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciconte G, Monasky MM, Santinelli V, Micaglio E, Vicedomini G, Anastasia L, Negro G, Borrelli V, Giannelli L, Santini F, de Innocentiis C, Rondine R, Locati ET, Bernardini A, Mazza BC, Mecarocci V, Ćalović Ž, Ghiroldi A, D'Imperio S, Benedetti S, Di Resta C, Rivolta I, Casari G, Petretto E, Pappone C. Brugada syndrome genetics is associated with phenotype severity. Eur Heart J 2021;42:1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nademanee K, Veerakul G, Chandanamattha P, Chaothawee L, Ariyachaipanich A, Jirasirirojanakorn K, Likittanasombat K, Bhuripanyo K, Ngarmukos T. Prevention of ventricular fibrillation episodes in Brugada syndrome by catheter ablation over the anterior right ventricular outflow tract epicardium. Circulation 2011;123:1270–1279. [DOI] [PubMed] [Google Scholar]
- 8. Nademanee K, Hocini M, Haïssaguerre M. Epicardial substrate ablation for Brugada syndrome. Hear Rhythm 2017;14:457–461. [DOI] [PubMed] [Google Scholar]
- 9. Haanschoten DM, Elvan A, Postema PG, Smit JJJ, Adiyaman A, Bekke RT, Asaad N, Aanhaanen WTJ, Misier ARR, Delnoy PPHM, Crijns HJGM, Wilde AAM. Catheter ablation in highly symptomatic Brugada patients: a Dutch case series. Clin Res Cardiol 2020;109:560–569. [DOI] [PubMed] [Google Scholar]
- 10. Smits JPP, Eckardt L, Probst V, Bezzina CR, Schott JJ, Remme CA, Haverkamp W, Breithardt G, Escande D, Schulze-Bahr E, LeMarec H, Wilde AAM. Genotype-phenotype relationship in Brugada syndrome: electrocardiographic features differentiate SCN5A-related patients from non-SCN5A-related patients. J Am Coll Cardiol 2002;40:350–356. [DOI] [PubMed] [Google Scholar]
- 11. Postema PG, Dessel P. V, Kors JA, Linnenbank AC, Herpen G. V, Ritsema van Eck HJ, Geloven N. V, Bakker J. D, Wilde AAM, Tan HL. Local depolarization abnormalities are the dominant pathophysiologic mechanism for type 1 electrocardiogram in brugada syndrome a study of electrocardiograms, vectorcardiograms, and body surface potential maps during ajmaline provocation. J Am Coll Cardiol 2010;55:789–797. [DOI] [PubMed] [Google Scholar]
- 12. Nademanee K, Raju H, Noronha S. V D, Papadakis M, Robinson L, Rothery S, Makita N, Kowase S, Boonmee N, Vitayakritsirikul V, Ratanarapee S, Sharma S, Wal A. V D, Christiansen M, Tan HL, Wilde AA, Nogami A, Sheppard MN, Veerakul G, Behr ER. Fibrosis, connexin-43, and conduction abnormalities in the Brugada syndrome. J Am Coll Cardiol 2015;66:1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoogendijk MG, Potse M, Linnenbank AC, Verkerk AO, Ruijter H. D, Amersfoorth S. V, Klaver EC, Beekman L, Bezzina CR, Postema PG, Tan HL, Reimer AG, Wal A. V D, Harkel AT, Dalinghaus M, Vinet A, Wilde AAM, Bakker J. D, Coronel R. Mechanism of right precordial ST-segment elevation in structural heart disease: excitation failure by current-to-load mismatch. Hear Rhythm 2010;7:238–248. [DOI] [PubMed] [Google Scholar]
- 14. Glazer AM, Wada Y, Li B, Muhammad A, Kalash OR, O’Neill MJ, Shields T, Hall L, Short L, Blair MA, Kroncke BM, Capra JA, Roden DM. High-throughput reclassification of SCN5A variants. Am J Hum Genet 2020;107:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walsh R, Lahrouchi N, Tadros R, Kyndt F, Glinge C, Postema PG, Amin AS, Nannenberg EA, Ware JS, Whiffin N, Mazzarotto F, Škorić-Milosavljević D, Krijger C, Arbelo E, Babuty D, Barajas-Martinez H, Beckmann BM, Bézieau S, Bos JM, Breckpot J, Campuzano O, Castelletti S, Celen C, Clauss S, Corveleyn A, Crotti L, Dagradi F, Asmundis C. D, Denjoy I, Dittmann S, et al. Enhancing rare variant interpretation in inherited arrhythmias through quantitative analysis of consortium disease cohorts and population controls. Genet Med 2020. 10.1038/s41436-020-00946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]