Abstract
With the advent of deep learning, convolutional neural networks (CNNs) have evolved as an effective method for the automated segmentation of different tissues in medical image analysis. In certain infectious diseases, the liver is one of the more highly affected organs, where an accurate liver segmentation method may play a significant role to improve the diagnosis, quantification, and follow-up. Although several segmentation algorithms have been proposed for liver or liver-tumor segmentation in computed tomography (CT) of human subjects, none of them have been investigated for nonhuman primates (NHPs), where the livers have a wide range in size and morphology. In addition, the unique characteristics of different infections or the heterogeneous immune responses of different NHPs to the infections appear with a diverse radiodensity distribution in the CT imaging. In this study, we investigated three state-of-the-art algorithms; VNet, UNet, and feature pyramid network (FPN) for automated liver segmentation in whole-body CT images of NHPs. The efficacy of the CNNs were evaluated on 82 scans of 37 animals, including pre and post-exposure to different viruses such as Ebola, Marburg, and Lassa. Using a 10-fold cross-validation, the best performance for the segmented liver was provided by the FPN; an average 94.77% Dice score, and 3.6% relative absolute volume difference. Our study demonstrated the efficacy of multiple CNNs, wherein the FPN outperforms VNet and UNet for liver segmentation in infectious disease imaging research.
Keywords: deep learning, infectious disease, segmentation, liver, nonhuman primate, CT, whole-body
1. Introduction
Effective volumetric segmentation of livers from computed tomography (CT) images is crucial for dose estimation, follow-up assessment, and evaluation of therapeutic response. The liver is a spatially complex, three-dimensional (3D) organ that usually shows similar density to the nearby organs on non-contrast CT scans. Manual delineation of liver is considered as the gold-standard, however the process is time-consuming and suffers from inter- and intra-observer variability. Specifically, for a large dataset, the manual process is quite unmanageable in a short period of time. Therefore, an effective and automated segmentation method is highly desirable to accurately segment the liver and allows faster processing of large data sets, while removing inter- and intra-observer biases and variation.
Among recent methods, a number of 2D and 3D convolutional neural network (CNN) based automated liver and liver-tumor segmentation methods were reported in the Liver Tumor Segmentation (LiTS) challenge [1] with a primary focus on liver tumor segmentation. Han et al. [2] proposed a combination of two UNet-like models in a 2.5 D framework, where the first model was used only for coarse liver segmentation and the second model was trained with the segmented liver region to segment both liver and tumors in a fine-tuned manner. Li et al. [3] developed another UNet based model in a hybrid densely connected fashion (H-DenseUNet) for liver and liver-tumor segmentation. Chlebus et al. [4] also employed a UNet like fully convolutional network architecture for liver tumor segmentation. The H-DenseUNet consisting of a 2D DenseUNet and a 3D counterpart aggregates the volumetric contexts in an auto-context mode [5]. Jin et al. [6] used a 3D hybrid residual attention-aware segmentation method RA-UNet for liver and liver-tumor segmentation, which has a basic 3D UNet structure with low-label and high-label feature fusion to extract the contextual information. On the other hand, Jiang et al. [7] proposed a 3D fully connected network structure, composed of multiple attention hybrid connection blocks (AHCBlocks) densely connected with both long and short skip connections and soft self-attention modules for liver-tumor segmentation. Although the above methods were effective in human livers and liver-lesion segmentation from contrast-enhanced CT scans, their performances haven’t been evaluated yet for animal models. Recently, Solomon et al. [8] proposed an atlas-based liver segmentation for NHPs to support quantitative image analysis of preclinical studies. However, the performance of the atlas-based method depends on the closest matching of a rigid registration process and suffers mis-classification if the shape and radiodensity of pathologic liver is not well represented or the field of view differs significantly in the atlas.
The automated liver segmentation evaluated in this study using whole-body CT scans of these NHP animal models is challenging due to the diversity of the subjects that includes different species with a wide range of animal ages/sizes. In addition, the infected livers appear with heterogeneous radiodensity distribution, changes in size and shape (e.g. enlarged left lobe of the liver due to the infectious disease process), and the motion blur due to breathing artifact. Example images shown in Figure 1 demonstrate the challenges associated with this segmentation task. In this study, we employed three CNN methods; VNet [9], UNet [10], and feature pyramid network (FPN) [11] to investigate the efficacy of different CNNs for automated liver segmentation in animal models. To the best of our knowledge, this is the first study that uses CNN-based liver segmentation in nonhuman primates (NHPs) with a history of infectious diseases.
Figure 1.
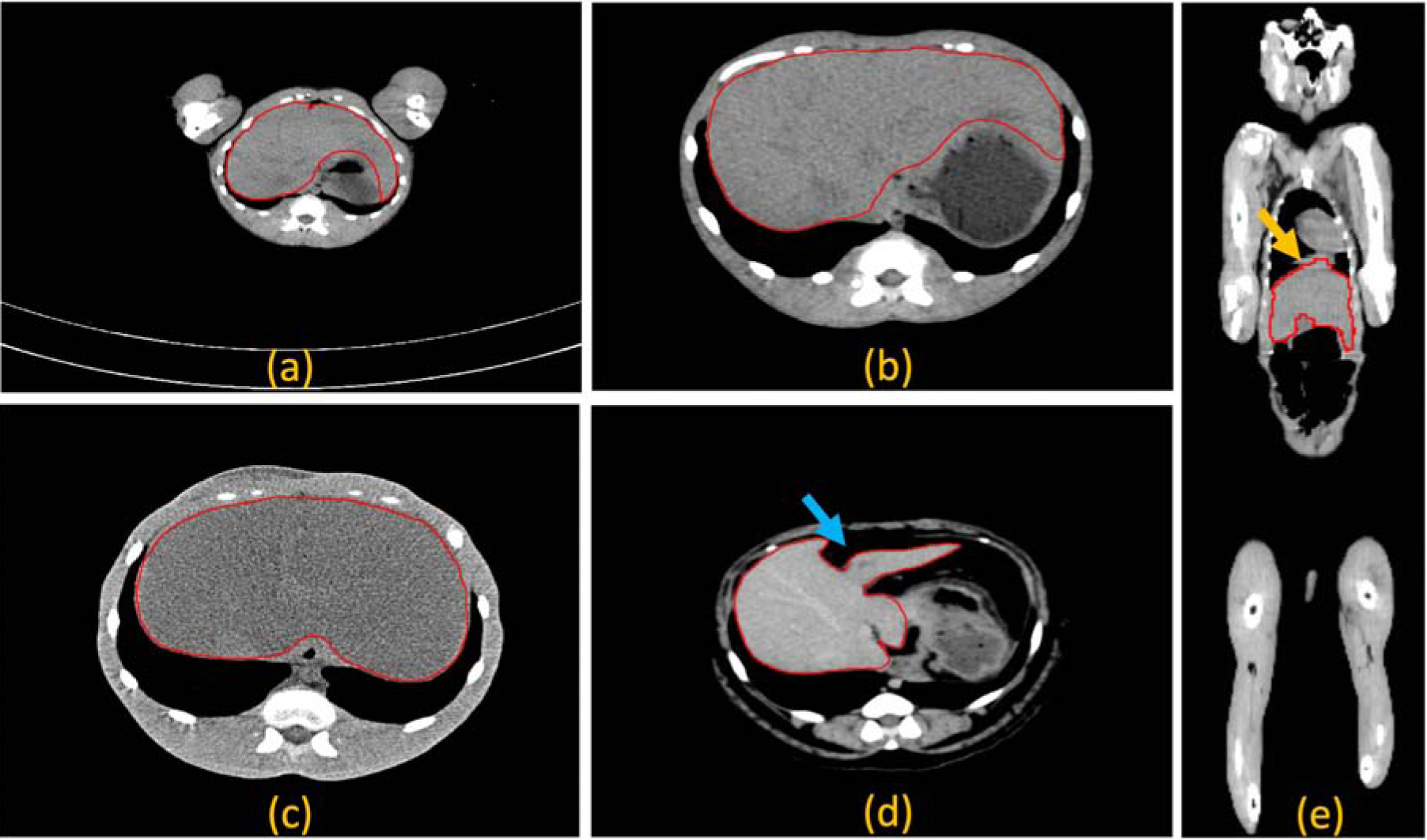
Challenges related to automated liver segmentation in virus infected nonhuman primates. The liver appears within a range of fields of view (a)-(b), heterogeneous radiodensity distribution due to infection (b)-(c), deformation due to the presence of intra-abdominal fat, indicated by the blue arrow (d), and motion blur due to breathing artifacts, indicated by the yellow arrow (e). The liver margins are shown by the red polygons.
2. Materials and Methods
2.1. Dataset
A total of 82 CT images were acquired from 37 nonhuman primates (rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques), ranging in weight from 3 kg to 14 kg. Pre and post-exposure scans at different time intervals for different studies were used for this project. These animals were exposed to either Ebola (EBOV), Marburg (MARV) or Lassa (LASV) viruses. The data set is summarized in Table 1. Sixty-four non-contrast and eighteen contrast-enhanced scans were obtained from using either a Philips Gemini 16-slice CT scanner or a Philips Precedence 16-slice CT scanner (Philips Healthcare, Cleveland, OH, USA) in helical scan mode. The whole-body CT scans acquired for this study covered either head to pelvis or head to knee of the animals. The scanner parameters were set at 140 kVp, 250 or 300 mAs/slice, 1-mm thickness with 0.5-mm increments.
Table 1.
Diversity in the dataset for automated liver segmentation.
| Virus types277 | # of animals (total 37) | # of scans (total 82) | Range of animal weights (kg) | Range of liver volumes (ml3) | ||||
|---|---|---|---|---|---|---|---|---|
| Rhesus | Cynomolgus | Pre | Post | min | max | min | max | |
| EBOV | 25 | 0 | 25 | 21 | 3.06 | 14.52 | 61.78 | 253.14 |
| LASV | 0 | 6 | 9 | 9 | 3 | 10 | 99.14 | 122.88 |
| MARV | 6 | 0 | 12 | 6 | 4.8 | 10 | 128.65 | 279.486 |
2.2. Ground-truth creation
The manual liver outlining was performed by a previously trained imaging fellow and monitored by a CT body radiologist. The contours were drawn in the axial plane using the 2D brush, Contour Copilot, and interpolation tools within MIM software (MIM Maestro version 6.9 Cleveland, Ohio). If breathing motion was observed (even if subjects were anesthetized and intubated), the contour was drawn halfway through the extents of the breathing artifact. The gallbladder was excluded if it was clearly visible. The 82 CT images and associated manual liver masks used in this study have been made publicly available for researchers to download at https://www.synapse.org/#!Synapse:syn22174612/wiki/604069.
2.3. Preprocessing
In the pre-processing step, first, all the CT images were resampled to 1 mm3 isotropic voxels using cubic interpolation. Second, the density values were thresholded to include the range [–40, 160] that excluded irrelevant organs and objects. Finally, the thresholded image intensities were rescaled between 0 to 1. An anisotropic diffusion filter [12] was applied to each image for noise reduction and boundary preservation.
2.4. CNN architecture
In order to measure the efficacy of the CNN-based method, we applied a 3D version of UNet [10] which is one of the most evaluated [1, 2, 3, 6, 4] state-of-the-art algorithms that has been successfully applied in many segmentation tasks in medical image analysis. In addition, VNet [9] is also applied as it has been effective in CT image analysis [13]. We also employed feature pyramid network, FPN [11], which has also been adopted in many object-recognition tasks [14, 15]. Recognizing objects at vastly different scales is a fundamental challenge in computer vision. A standard solution to this scale-variant issue is to use feature pyramids that enables a model to detect objects across a large range of scales by scanning the model over both positions and pyramid levels. Recently, FPN has been seen in various semantic [16, 17] and instance [18] segmentation tasks. The FPN consists of two pyramidal pathways; bottom-up and top-down. The top-down architecture with lateral connections builds the high-level semantic feature maps at all scales. The principle advantage of FPN is that it produces a multi-scale feature representation in which all levels are semantically strong, including the high-resolution levels [19]. For the bottom-up feature encoder, we have chosen to use ResNet50 [20] architecture with four resolution levels. Instead of five as in the original ResNet50 [20] structure, we chose four which is the maximum number of levels achievable by downsampling our input patches described in a later section. Following Kirillov et al. [16], we implemented the network overhead for semantic segmentation (shown by the red box in Figure 2) and then adjusted the filter parameters for our application. However, our primary focus in this work was to experiment with state-of-the-art deep learning segmentation methods and implement the most accurate method for the novel application of liver segmentation in CT scans of NHPs.
Figure 2.
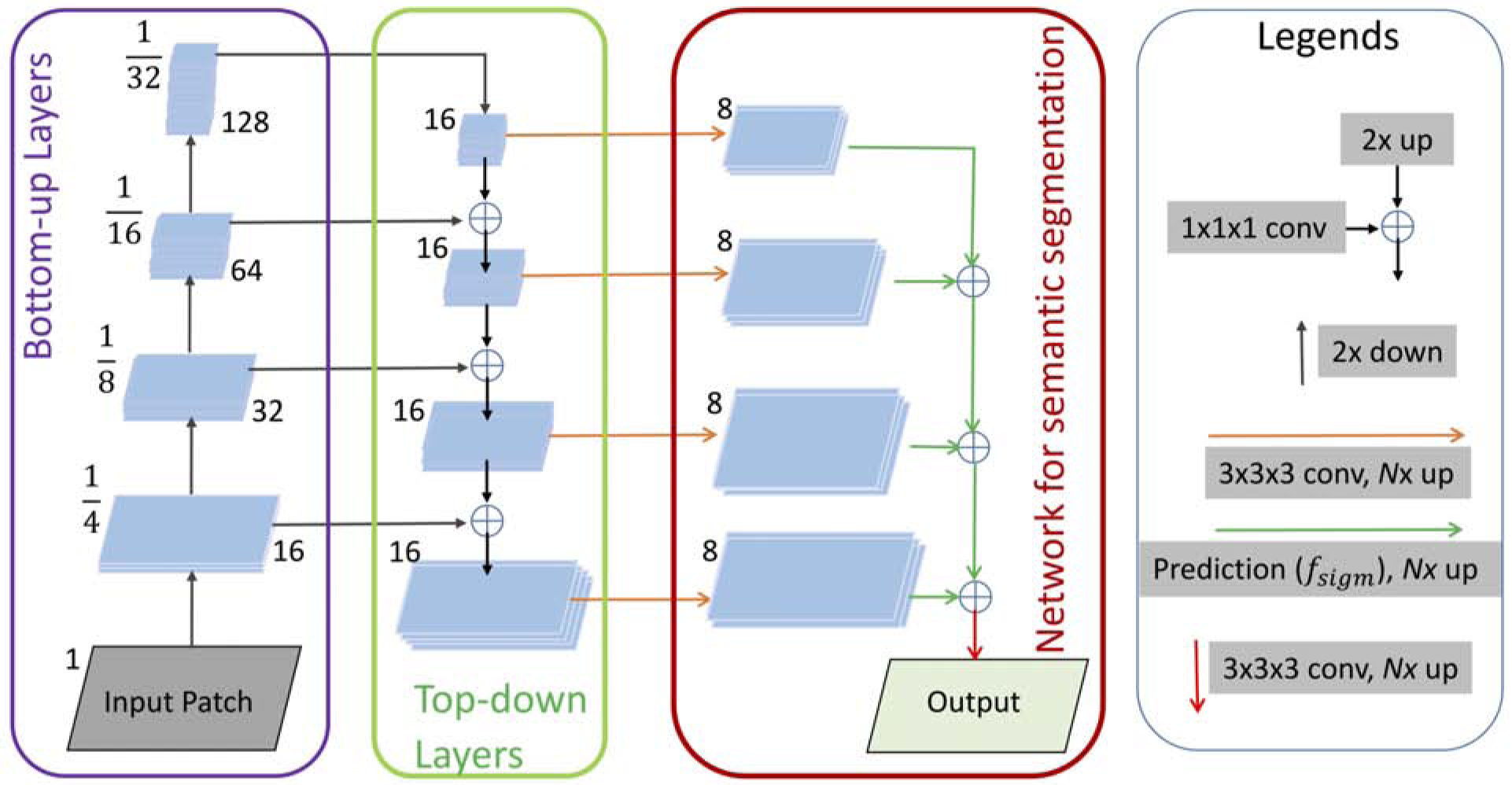
The schematic diagram of the proposed FPN for liver segmentation.
2.5. Training CNNs
Training parameters:
All the networks were trained using input patches of size 64×64×64 voxels. The input patches were extracted randomly from both liver and non-liver areas with equal numbers. Patches were called liver or non-liver patches based on the center voxel. The corresponding manual segmentation mask was blurred with a 3D Gaussian filter with sigma equal to 1 × 1 × 1 mm3. We set the mini-batch size to 90. The parameter updates were performed using the Adam optimizer with a learning rate of 5 × 10−5 and decay rate of 5×10−5. The model was trained for 30 epochs, whereas the input patches were reshuffled before each epoch.
Loss function:
Segmentation of any organ is often seen as a voxel-wise classification task, where high class-imbalance (sample ratio of different classes) has a significantly negative impact on the performance. Considering the variation of class-ratios (foreground/background) among the subjects and hard-to-detect boundaries with the nearby organs, we chose focal [21] as the loss function which is defined as;
Where, gi is the probability of voxel, i to the foreground, and pi is the ground-truth. The parameter, α is a weighting factor to adjust the class imbalance and (1 − gi)σ is called the modulating factor that gives more weight for hard-to-detect samples and less weight for easy-to-detect samples in training. Empirically, α = 0.3, and σ = 2 was found the best fit in this liver segmentation task.
2.6. Post-processing
The output of the CNN is a probability map image at 1 mm3 resolution. This probability map image was resampled to the original image size. Similar to the preprocessing step, a Gaussian filter was used to smooth the resampled probability map image. Later, an empirically chosen threshold of value 0.5 was used to suppress the low probability values and set higher probability values as the liver.
2.7. Quantitative Evaluation
To validate the effectiveness and robustness of our approach, we used four metrics; Dice coefficient, sensitivity, relative absolute volume difference (RAVD), and average surface distance (ASD). For the last two metrics, the smaller the value is, the better the segmentation. The above metrics are commonly used to measure the segmentation performance [22] and defined as follows:
where S is the automated segmentation and M is the manual mask of the liver.
where TP and FN stand for True Positive and False Negative, respectively.
where VS and VM indicate liver volumes by automated and manual segmentation, respectively.
where , are the number of voxels on the surface of the detected liver and manually outlined liver, d(pD,SM) measures the minimum distance between the detected voxels and surface of the manual mask and d(pM,SD) measures the opposite.
2.8. Statistical Analysis
Mean, standard deviation, median, maxima, and minima were used for descriptive statistics of the above metrics; Dice coefficient, Sensitivity, RAVD, and ASD. A Wilcoxon signed-rank test which is a non-parametric version of the paired T-test was used to measure the statistical significance (p < 0.05) between the performances of two models. All the statistical analysis was performed in Python using SciPy and NumPy tools.
3. Results
We performed subject-wise 10-fold cross-validation using images from 37 animals. In other words, all pre and post scans of the test subjects/animals were left out from the training. Animal images were randomized before splitting into 10 folds. The example images shown in Figure 3 are the the probability maps provided by the CNNs. Figure 4 shows example images of the segmented liver for a comparison among VNet, UNet, and FPN. As seen in the CT images, when the livers appear with an iso-dense distribution with the surrounding tissues like muscles, kidney, stomach, and the partial volume effect of the gallbladder, the segmentation algorithms mostly suffer in finding a clear boundary between the tissues. In Figure 4 as example, we observed that the proposed FPN is better than VNet and UNet in these regions. Architecturally, FPN provides multiple predictions: one for each up-sampling layer. Although the FPN has the lateral connections between the bottom-up pyramid and the top-down pyramid as UNet, the FPN applies a 1×1 convolutional layer before adding them whereas UNet only copies and appends the features. This allows the FPN to use any suitable architecture for the bottom-up pyramid, in this case, ResNet50, extracts strong semantic features than simple down-sampling. Intutively, the above characteristics of FPN offer better generalizability than the other two networks. Therefore, the FPN has a better performance in handling the variation of the livers that appears in our dataset.
Figure 3.
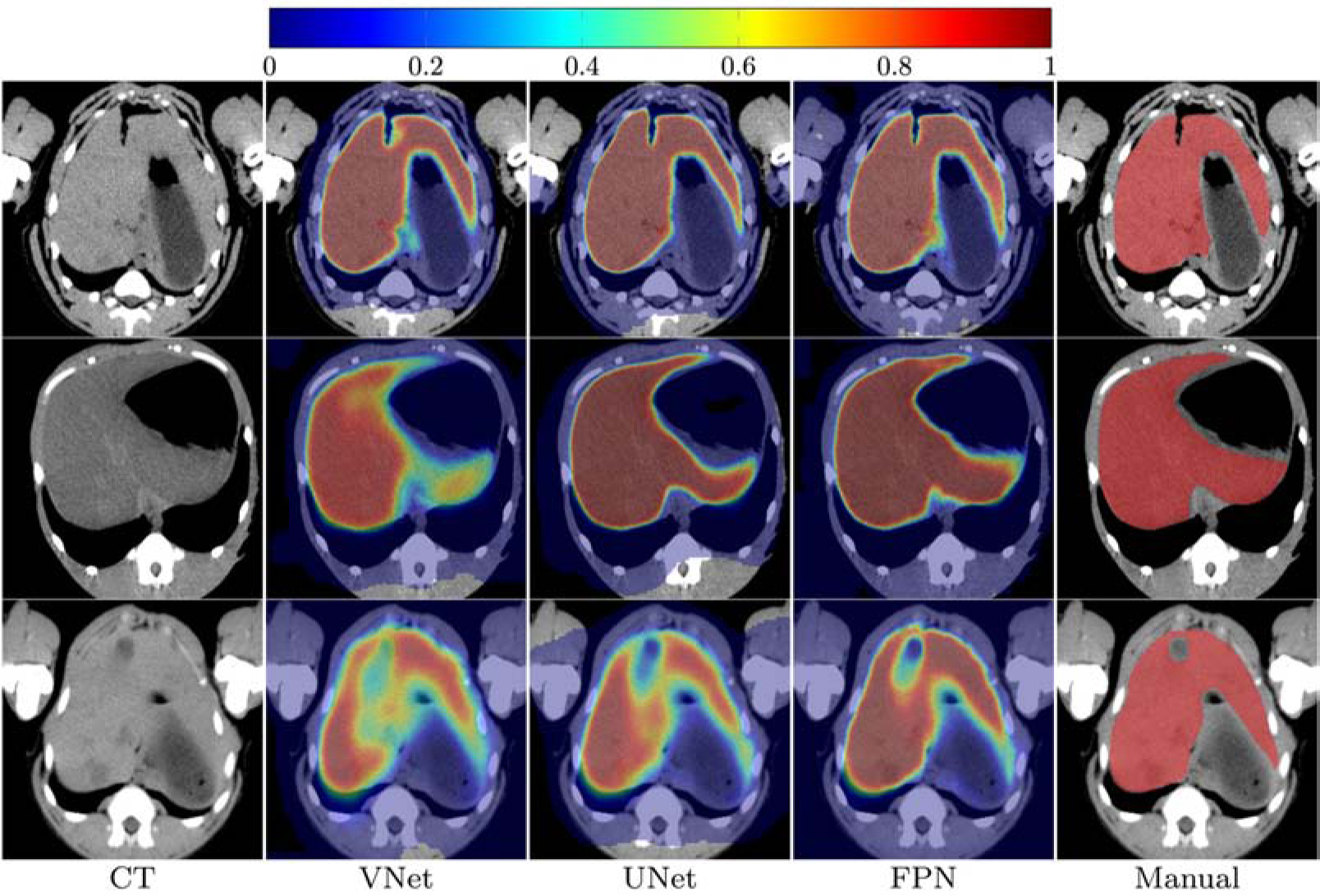
Example images of liver segmentation; column-wise from left to right, original CT images, the probability maps provided by VNet, UNet, FPN, and manual mask, respectively. Note: All the images are cropped and zoomed for better visualization.
Figure 4.
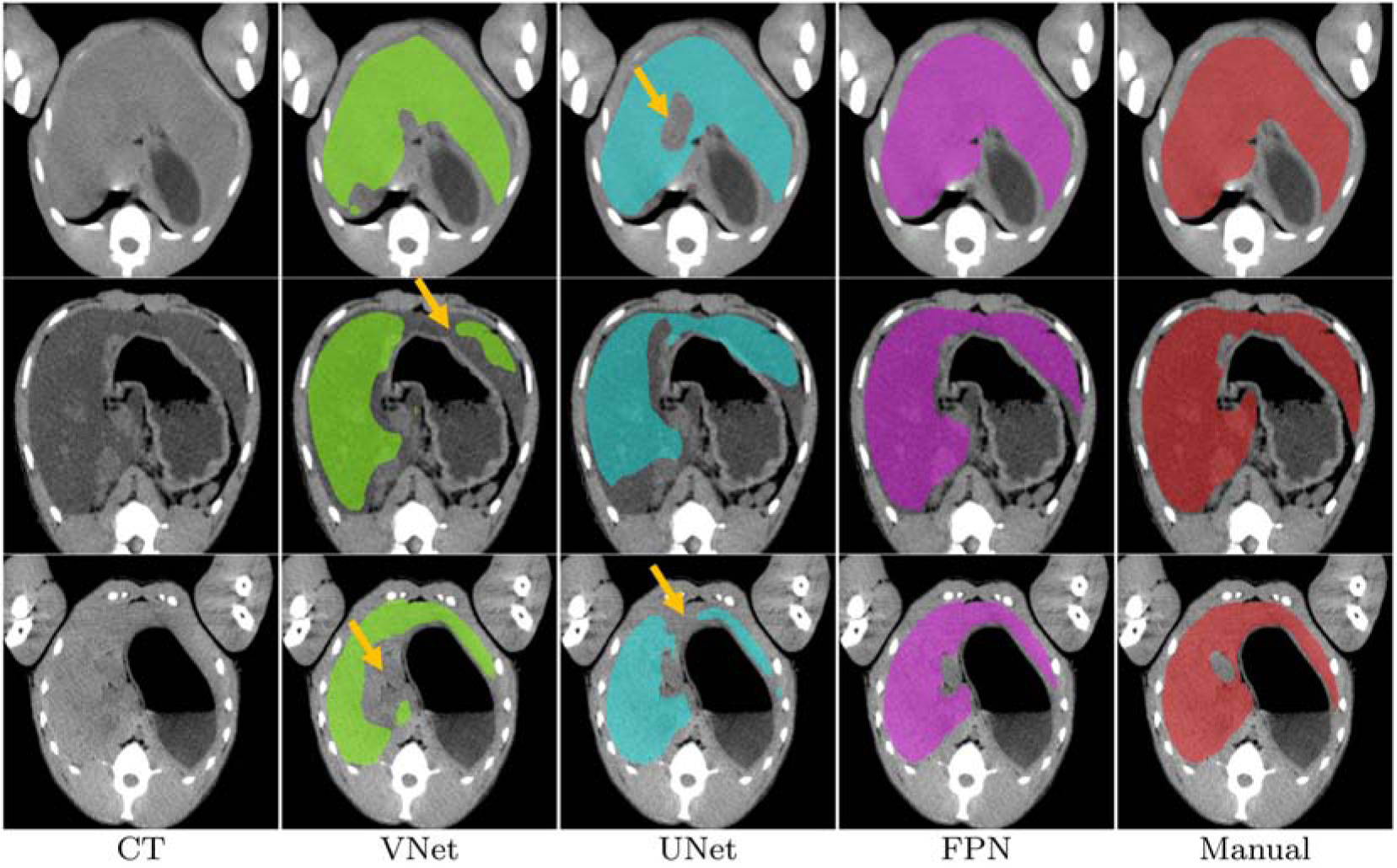
State-of-the-art comparison. From left to right, original CT image, segmented liver by VNet, UNet, FPN, and the manual liver mask. Example images are shown to demonstrate where FPN outperforms VNet and UNet (marked by the yellow arrows). Note: All the images are cropped and zoomed for better visualization.
In order to understand the effect of virus infections on the liver, we separately evaluated the segmentation performance of the pre- and post-exposure scans for all three algorithms. The average scores of these metrics can be found in Table 2. More detail statistics on the above scores can be found in Figure 5.
Table 2.
Statistics on the quantitative scores using VNet, UNet, and FPN. The best scores are in bold
| Scans278 | Metrics | Dice Coefficient (%) | Sensitivity (%) | RAVD(%) | ASD (mm) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VNet | UNet | FPN | VNet | UNet | FPN | VNet | UNet | FPN | VNet | UNet | FPN | ||
| Pre | Mean | 92.50 | 92.55 | 94.77 | 89.13 | 89.41 | 94.45 | 8.14 | 7.79 | 3.60 | 1.57 | 1.11 | 0.49 |
| Std. | 2.48 | 3.21 | 1.43 | 4.82 | 5.87 | 2.65 | 5.01 | 6.23 | 3.20 | 4.32 | 1.76 | 0.20 | |
| Median | 92.88 | 93.92 | 94.80 | 89.38 | 91.13 | 94.81 | 7.15 | 5.84 | 2.58 | 0.62 | 0.60 | 0.45 | |
| Max | 96.13 | 95.84 | 96.58 | 96.15 | 97.93 | 98.41 | 18.32 | 27.52 | 16.90 | 26.89 | 9.90 | 1.38 | |
| Min | 86.46 | 82.81 | 90.75 | 79.11 | 71.42 | 86.21 | 0 | 0.18 | 0.14 | 0.36 | 0.35 | 0.30 | |
| Post | Mean | 91.88 | 91.36 | 93.19 | 88.84 | 88.19 | 92.44 | 8.13 | 7.80 | 4.53 | 1.49 | 1.16 | 0.90 |
| Std. | 3.00 | 3.05 | 5.71 | 4.82 | 5.92 | 8.61 | 5.70 | 6.66 | 8.63 | 3.19 | 1.20 | 1.62 | |
| Median | 92.77 | 92.05 | 94.30 | 90.22 | 89.53 | 94.73 | 6.06 | 6.12 | 2.13 | 0.71 | 0.83 | 0.55 | |
| Max | 95.83 | 95.34 | 96.52 | 96.24 | 96.05 | 98.07 | 23.66 | 24.86 | 50.52 | 19.41 | 6.67 | 10.38 | |
| Min | 82.17 | 82.91 | 62.34 | 72.45 | 72.80 | 46.59 | 0.68 | 0.11 | 0.07 | 0.37 | 0.45 | 0.32 | |
| Overall | Mean | 92.23 | 92.03 | 94.08 | 89.00 | 88.87 | 93.57 | 8.14 | 7.80 | 4.01 | 1.54 | 1.13 | 0.67 |
| Std. | 2.74 | 3.20 | 3.93 | 5.23 | 5.92 | 6.12 | 5.32 | 6.42 | 6.22 | 3.86 | 1.54 | 1.10 | |
| Median | 92.81 | 93.01 | 94.63 | 89.81 | 90.56 | 94.75 | 6.75 | 5.97 | 2.27 | 0.65 | 0.68 | 0.49 | |
| Max | 96.13 | 95.84 | 96.58 | 96.24 | 97.93 | 98.41 | 23.66 | 27.52 | 50.52 | 26.89 | 9.90 | 10.38 | |
| Min | 82.17 | 82.81 | 62.34 | 72.45 | 71.42 | 46.59 | 0 | 0.11 | 0.07 | 0.36 | 0.35 | 0.30 | |
Figure 5.
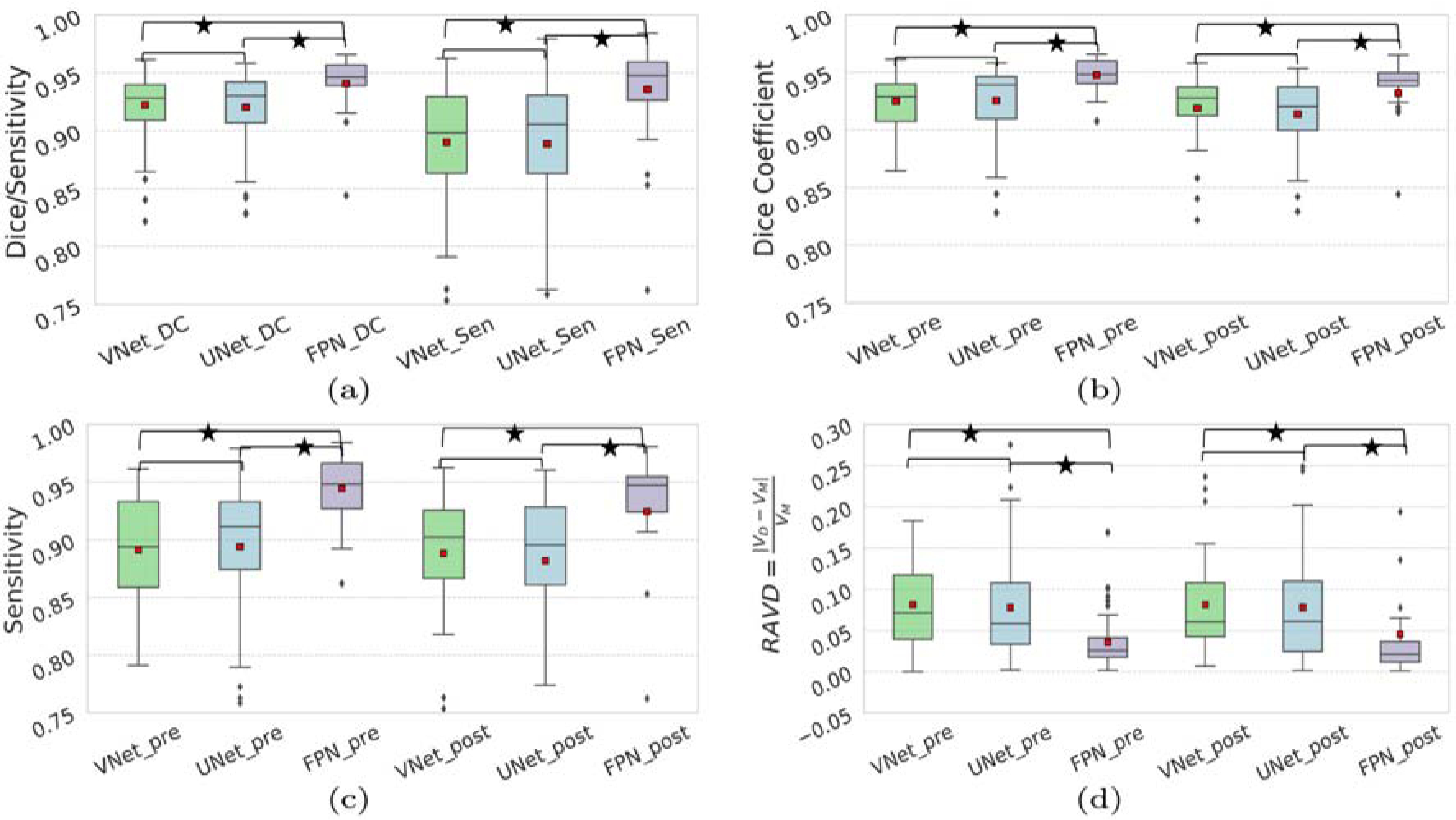
State-of-the-art comparison using evaluated score; (a) overall Dice coefficient and sensitivity, (b) Dice, (c) sensitivity for pre/post scans, and (d) RAVD. The small red squares show the average. The * corresponds to p < 0.05 according to the Wilcoxon signed-rank test. Comparisons without * corresponds to p ≥ 0.05.
4. Discussion
Infectious diseases are the second leading cause of death worldwide. In the preclinical studies of infectious diseases, animal models permit enhanced information through longitudinal studies for a better understanding of pathogenesis and drug development [23]. Specifically, animal models are important for Ebola, SARS, and MERS outbreaks to study the responses to vaccines and therapies in a manner that is often not possible with human subjects.
In automated liver segmentation, one of the most challenging aspects is to detect a clear boundary between liver and the nearby organs such as gallbladder, muscles, kidney, and stomach. Example images in Figure 4 suggests that FPN is capable of handling the diversity and has better boundary detection capability than VNet and UNet. The false negatives were also significantly reduced by FPN as seen in Figure 4. In this study of automated NHP liver segmentation, we obtained 94.77% overall Dice coefficient which is on par with the reported scores in the LiTS challenge [1]. Also, the proposed deep learning-based method FPN (Dice: mean 94.77%, Std. 1.43%, median 94.80%) significantly outperforms the atlas-based [8] (Dice: mean 89.0%, Std. 5.0%, median 91.0%) method.
In Table 2, we notice the overall scores for VNet, UNet, and FPN by our chosen metrics are comparable, however, a closer look at those metrics in Figure 5 indicates that the scores are compromised by a few outliers. The FPN significantly (p < 0.05) improves the Dice coefficients, indicating the effectiveness of the network. All three CNNs show better performance on the pre-scan images than the post-infection scans. Specifically, for the post-infection scans, the Dice scores are lower with more outliers (Figure 5.b). From the subject-wise outputs, we noticed that most of these outliers came from the scans with severely infected animals. Note that after exposure to the viruses, different animals have varying host responses to the viruses, causing variability in liver shape, size and density. In this study, only a few animals had shown severe changes in the post-exposure liver characteristics which were under-represented in our dataset. Well-represented training data might improve those scans’ outputs.
Regarding the computational time, the training was performed on a 16 core Intel Xeon E5 3.2 GHz CPU and three NVIDIA GeForce 12 GB TitanX GPUs. The average training time for one fold of the 10-fold cross-validation procedure took approximately 8 hours for any of the three CNNs. The inference time for a test scan was ~1.5 minutes for a 1 mm3 3D whole-body CT image.
An important part of these infectious disease studies is tracing the affected organs, such as liver, lung, spleen. Manual tracing of these organs’ boundary is a time-consuming task and suffers from intra- and inter-rater variability. Our proposed method provides effective results in this challenging segmentation task and establishes a baseline study for automated liver segmentation in NHPs. In a high consequence infectious disease, the liver is one of the most affected organs and has a spatially heterogeneous response where an automated segmentation method is highly beneficial to investigate viral pathogenesis as well as to test medical countermeasures. Effective countermeasure development and approval process would be enhanced by a rapid and reproducible method of quantification.
5. Conclusion
In this work, we investigated the efficacy of CNN-based methods for automated liver segmentation in nonhuman primates using whole-body CT scans. Published research has focused on liver tumor segmentation in human subjects, whereas for animal models the liver segmentation task is limited likely due to the challenging nature of the effort. To the best of our knowledge, this is the first work that proposes a CNN-based automated segmentation method for liver in NHPs with active viral infections. Our study is aimed to find a suitable deep learning based method for NHPs’ liver segmentation. Future work will include data augmentation with spatial deformations [24], which may help to represent the scans from heavily infected animals well in the training. We also plan to apply cascaded CNNs [25] in auto-context mode [5] which have been shown effective in many segmentation tasks. In addition, we plan to apply our FPN-based method on human subjects to see if it is translatable to humans.
Acknowledgements
This project has been partially funded by the Center for Infectious Disease Imaging (CIDI), Clinical Center, National Institute of Health (NIH), and the Integrated Research Facility (IRF), National Institute of Allergy and Infectious Disease (NIAID), NIH. In addition, this project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. This work was supported in part through the prime contract of Laulima Government Solutions (LGS), LLC, with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under contract no. HHSN272201800013C and Battelle Memorial Institute (BMI)s, a former prime contract with NIAID under contract no. HHSN272200700016I. Ji Hyun Lee performed this work as a former employee of MedRelief as a subcontract of BMI, and a current employee of Tunnell Government Services as a subcontract of LGS. Marcelo Castro and Byeong-Yeul Lee performed this work as former employees of MedRelief as a subcontract of BMI and current employees of LGS. Richard S Bennett and Yu Cong performed this work as former employees of BMI and current employees of LGS. Authors would like to thank Philip Sayre, Christopher Bartos, and the comparative medicine team for acquiring imaging and animal care respectively. The authors thank Jiro Wada for his expertise in helping to prepare this manuscript. This work was also partially supported by NIAID Interagency Agreement NOR15003-001-0000. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, of the institutions and companies affiliated with the authors or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethical approval, human and animal rights
All animals used in this research project were cared for and handled humanely according to the following policies: the U.S. Public Health Service Policy on Humane Care and Use of Animals (2015); the Guide for the Care and Use of Laboratory Animals (2011); and the U.S. Government Principles for Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (1985). All animal work at NIAID was performed in a maximum containment BSL-4 facility [26] were accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
References
- [1].Christ P, LiTS Liver Tumor Segmentation Challenge (LiTS17). URL https://competitions.codalab.org/competitions/17094
- [2].Han X, Automatic liver lesion segmentation using a deep convolutional neural network method, CoRR abs/1704.07239. arXiv:1704.07239. URL http://arxiv.org/abs/1704.07239
- [3].Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A, H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes, IEEE transactions on medical imaging 37 (12) (2018) 2663–2674. [DOI] [PubMed] [Google Scholar]
- [4].Chlebus G, Schenk A, Moltz JH, van Ginneken B, Hahn HK, Meine H, Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing, Scientific reports 8 (1) (2018) 15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tu Z, Bai X, Auto-context and its application to high-level vision tasks and 3d brain image segmentation, IEEE transactions on pattern analysis and machine intelligence 32 (10) (2009) 1744–1757. [DOI] [PubMed] [Google Scholar]
- [6].Jin Q, Meng Z, Sun C, Wei L, Su R, RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans, arXiv preprint arXiv:1811.01328. [DOI] [PMC free article] [PubMed]
- [7].Jiang H, Shi T, Bai Z, Huang L, AHCNet: An application of attention mechanism and hybrid connection for liver tumor segmentation in CT volumes, IEEE Access 7 (2019) 24898–24909. [Google Scholar]
- [8].Solomon J, Aiosa N, Bradley D, Castro MA, Reza S, Bartos C, Sayre P, Lee JH, Sword J, Holbrook MR, et al. , Atlas-based liver segmentation for nonhuman primate research, International Journal of Computer Assisted Radiology and Surgery (2020) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Milletari F, Navab N, Ahmadi S-A, V-net: Fully convolutional neural networks for volumetric medical image segmentation, in: 2016 Fourth International Conference on 3D Vision (3DV), IEEE, 2016, pp. 565–571. [Google Scholar]
- [10].Ronneberger O, Fischer P, Brox T, U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer, 2015, pp. 234–241. [Google Scholar]
- [11].Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S, Feature pyramid networks for object detection, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 2117–2125. [Google Scholar]
- [12].Perona P, Malik J, Scale-space and edge detection using anisotropic diffusion, IEEE Transactions on pattern analysis and machine intelligence 12 (7) (1990) 629–639. [Google Scholar]
- [13].Negahdar M, Beymer D, Syeda-Mahmood T, Automated volumetric lung segmentation of thoracic ct images using fully convolutional neural network, in: Medical Imaging 2018: Computer-Aided Diagnosis, Vol. 10575, International Society for Optics and Photonics, 2018, p. 105751J. [Google Scholar]
- [14].Zhao Q, Sheng T, Wang Y, Tang Z, Chen Y, Cai L, Ling H, M2det: A single-shot object detector based on multi-level feature pyramid network, in: Proceedings of the AAAI Conference on Artificial Intelligence, Vol. 33, 2019, pp. 9259–9266. [Google Scholar]
- [15].Li H, Xiong P, An J, Wang L, Pyramid attention network for semantic segmentation, arXiv preprint arXiv:1805.10180.
- [16].Kirillov A, Girshick R, He K, Dollár P, Panoptic feature pyramid networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2019, pp. 6399–6408. [Google Scholar]
- [17].Seferbekov SS, Iglovikov V, Buslaev A, Shvets A, Feature pyramid network for multi-class land segmentation., in: CVPR Workshops, 2018, pp. 272–275. [Google Scholar]
- [18].Sun Y, KP PS, Shimamura J, Sagata A, Concatenated feature pyramid network for instance segmentation, in: 2019 IEEE Fifth International Conference on Multimedia Big Data (BigMM), IEEE, 2019, pp. 297–301. [Google Scholar]
- [19].Abdulla W, Mask R-CNN for object detection and instance segmentation on Keras and TensorFlow (2017). URL https://github.com/matterport/Mask_RCNN
- [20].He K, Gkioxari G, Dollár P, Girshick R, Mask r-cnn, in: Proceedings of the IEEE international conference on computer vision, 2017, pp. 2961–2969. [Google Scholar]
- [21].Lin T-Y, Goyal P, Girshick R, He K, Dollár P, Focal loss for dense object detection, in: Proceedings of the IEEE international conference on computer vision, 2017, pp. 2980–2988. [Google Scholar]
- [22].MedPy, Metrics for segmentation evaluation. URL https://loli.github.io/medpy/metric.html#image-metrics-medpy-metric-image
- [23].Jelicks L, Tanowitz H, Albanese C, Small animal imaging of human disease: From bench to bedside and back, American Journal of Pathology 182 (2) (2013) 294–295. doi: 10.1016/j.ajpath.2012.11.015. [DOI] [PubMed] [Google Scholar]
- [24].Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, 3D U-Net: learning dense volumetric segmentation from sparse annotation, in: International conference on medical image computing and computer-assisted intervention, Springer, 2016, pp. 424–432. [Google Scholar]
- [25].Reza SM, Roy S, Park DM, Pham DL, Butman JA, Cascaded convolutional neural networks for spine chordoma tumor segmentation from mri, in: Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, Vol. 10953, International Society for Optics and Photonics, 2019, p. 1095325. [Google Scholar]
- [26].Byrum R, Keith L, Bartos C, Claire MS, Lackemeyer MG, Holbrook MR, Janosko K, Barr J, Pusl D, Bollinger L, et al. , Safety precautions and operating procedures in an (A) BSL-4 laboratory: 4. medical imaging procedures, JoVE (Journal of Visualized Experiments) (116) (2016) e53601. [DOI] [PMC free article] [PubMed] [Google Scholar]


