Abstract
Introduction: Genomics and bioinformatics are useful methods for exploring unclear aspects of radiation effects on biological systems. Many radiation-induced alterations in irradiated samples are post-radiation time-dependent. This study aims to evaluate the post-irradiation effects of the gamma ray on human Jurkat cells.
Methods: Gene expression profiles of the samples harvested 6 and 24 hours after radiation to find the critical differential expressed genes and the related pathways. Samples are provided from Gene Expression Omnibus (GEO) and analyzed by ClueGO.
Results: Twnety-nine critical genes were determined as the important affected genes and 7 classes of related pathways were introduced. CCNE2, PSMD11, CDC25C, ANAPC1, PLK1, AURKA, and CCNB1 that were associated with more than 6 pathways were related to one of the determined pathway groups.
Conclusion: Cell protecting pathways were associated with the genes (HSPA5, HSPA8, HSP90B1, HMMR, CEBPB, RXRA, and PSMD11) which were related to the minimum numbers of pathways. The finding of this study corresponds to repair processes which depend on post-radiation time. It seems these sets of genes are suitable candidates for further investigation.
Keywords: Radiation, Bioinformatics, Gene expression, Pathway, Dysregulation
Introduction
It is reported that gamma radiation induces chromosomal aberration and DNA damages in the exposed samples.1,2 Genomics as a high-throughput method is applied to discover the widespread aspect of gamma irradiation effects on biological systems.3,4 Since the bioinformatics approach is tied to the high-throughput methods, bioinformatics plays a significant role in the interpretation of results of gamma irradiation effects on living organisms.5
Genomics as a method which studies the genome is applied to investigate the dysregulated genes after inducing alterations in internal or external conditions of living samples. In such a study, the gene expression profiles of samples are assessed to find the targeted genes. Finally, different types of dysregulated genes with different amounts of dysregulation are introduced.6,7 Like the other large scale methods, large numbers of dysregulated genes are determined in a genomics experiment. The study and evaluation of this gene set need powerful techniques such as bioinformatics and its various branches.8-10
The products of the genes play roles in the biochemical pathways to maintain life hemostasis and health condition. Alteration in the gene expression level leads to functional changes in the related pathways, which is reflected in the lifestyle of the treated sample.11,12 There are several databases including different biochemical pathways such as the KEGG database that is applied to analyze the dysregulated pathways by many researchers.13,14 Rezaei-Tavirani et al have introduced “Cytokine-Mediated Signaling Pathway” as distinctive dysregulated biological terms after skin laser therapy.15 Since the DNA repair process after irradiation is an important parameter and needs proper time to decrease damages,16 in the present study, the gene expression profiles of irradiated cells by gamma rays in 6 hours and 24 hours of “post-radiation time” are studied to find the critical dysregulated genes and biochemical pathways.
Materials and Methods
The gene profiles of GSM2792818-20 from GSE104222/GPL6480 in GEO, which were irradiated with 10 Gy γ-ray and harvested 6 hours after radiation, were selected as controls. The gene expression profiles of the samples after 24 hours of radiation were assigned to compare.
The feasibility of the comparison of samples was provided by the assessment of the studied gene expression profiles via boxplot analysis by the GEO2R program.
Among 250 differential expressed genes (DEGs) , 190 characterized individuals were selected to be analyzed. A P value < 0.001 and a fold change > 1.5 were considered statistically significant. If there was more than one isomer, the individual with the highest value of expression was selected. Therefore, 177 DEGs were selected to be analyzed in the next steps.
The related pathways for the 177 query DEGs were investigated in Wiki Pathways, KEGG, and REACTOME Pathways via ClueGO. The network specificity medium and P value < 0.05 were considered to find the related pathways. The pathways considering; term P value, term P value corrected with Bonferroni step down, group P value, and group P value corrected with Bonferroni step down were less than 0.05. Kappa score = 7 was resulted.
Results
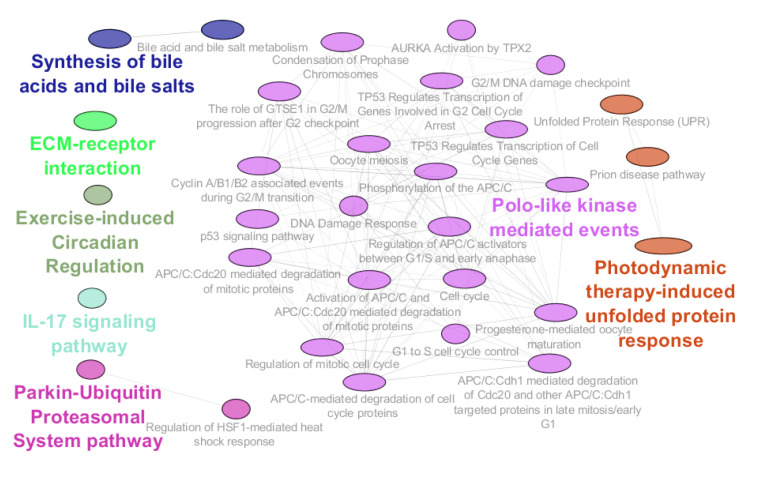
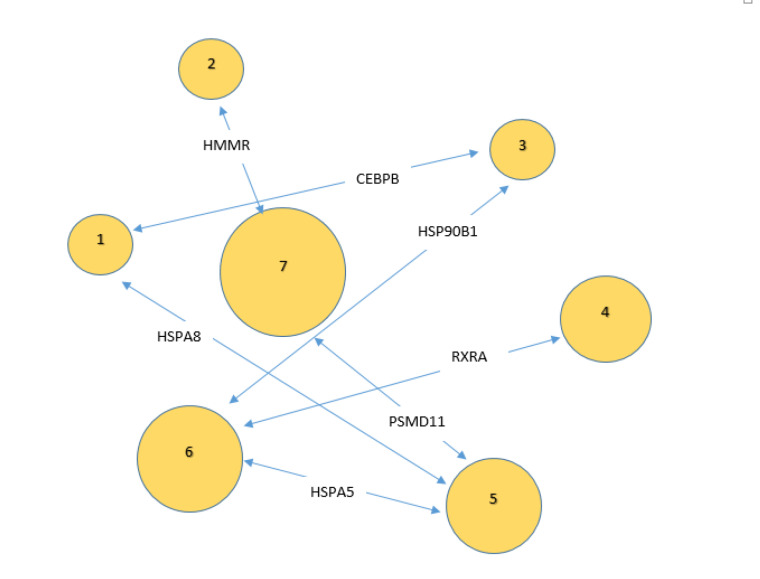
As it is shown in Figure 1, seven classes of pathways including 31 pathways were related to 29 DEGs among the 177 query genes. The introduced pathways and the related genes are shown in Table 1. As it is depicted in Table 1, about 16% of genes among 177 query DEGs are related to the identified pathways. The percentage of the associated genes for each pathway is presented in this table. The maximum percentage of associated genes (18.75%) is attributed to (Polo-like kinase mediated events). Based on the presence of an individual gene in several pathways, the frequency of the associated pathways for the query DEGs is calculated and presented in Table 2. CCNB1 is related to maximum pathways, while 11 DEGs are related to one pathway. Since the identified pathways are classified in the 7 groups of pathways, the associated pathway groups for the involved query DEG are determined and shown in Table 2. As shown in Table 2, there are several genes that are common between the introduced pathway groups. For better understanding, these genes and the related pathway groups are presented schematically in Figure 2. The pathways are split into 3 sets; individuals that are linked to 3 genes including groups 5 and 6, groups 1, 3, and 7 that are connected to two genes, and finally, pathway groups of 2 and 4 which are associated with one gene.
Figure 1.
Results of Pathway Analysis for the 177 Query DEGs. Each color refers to an individual class of pathways.
Table 1. Related Pathways for the 177 Query DEGs.
| Pathway | % AG | Associated Genes Found |
| 1Exercise-induced circadian regulation | 6.25 | [CEBPB, DNAJA1, HSPA8] |
| 2ECM-receptor interaction | 4.88 | [HMMR, SV2B, TNN, VWF] |
| 3IL-17 signaling pathway | 4.30 | [CEBPB, HSP90B1, MAPK15, SRSF1] |
| 4 Synthesis of bile acids and bile salts | 8.82 | [OSBPL1A, OSBPL6, RXRA] |
| 4Bile acid and bile salt metabolism | 6.98 | [OSBPL1A, OSBPL6, RXRA] |
| 5Regulation of HSF1-mediated heat shock response | 4.41 | [HSPA5, HSPA8, HSPH1] |
| 5 Parkin-Ubiquitin Proteasomal System pathway | 4.29 | [HSPA5, HSPA8, PSMD11] |
| 6Unfolded protein response (UPR) | 4.08 | [ATF3, CREBRF, HSP90B1, HSPA5] |
| 6 Photodynamic therapy-induced unfolded protein response | 14.81 | [ATF3, HSP90B1, HSPA5, TRIB3] |
| 6Prion disease pathway | 9.09 | [HSP90B1, HSPA5, RXRA] |
| 7Cell cycle | 4.03 | [ANAPC1, CCNB1, CCNE2, CDC25C, PLK1] |
| 7Oocyte meiosis | 4.84 | [ANAPC1, AURKA, CCNB1, CCNE2, CDC25C, PLK1] |
| 7p53 signaling pathway | 5.88 | [CCNB1, CCNE2, GTSE1, SESN2] |
| 7Progesterone-mediated oocyte maturation | 5.05 | [ANAPC1, AURKA, CCNB1, CDC25C, PLK1] |
| 7 Polo-like kinase mediated events | 18.75 | [CCNB1, CDC25C, PLK1] |
| 7APC/C-mediated degradation of cell cycle proteins | 5.81 | [ANAPC1, AURKA, CCNB1, PLK1, PSMD11] |
| 7APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 5.56 | [ANAPC1, AURKA, PLK1, PSMD11] |
| 7Regulation of APC/C activators between G1/S and early anaphase | 5.00 | [ANAPC1, CCNB1, PLK1, PSMD11] |
| 7APC/C:Cdc20 mediated degradation of mitotic proteins | 4.00 | [ANAPC1, CCNB1, PSMD11] |
| 7Phosphorylation of the APC/C | 15.79 | [ANAPC1, CCNB1, PLK1] |
| 7Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins | 5.26 | [ANAPC1, CCNB1, PLK1, PSMD11] |
| 7Condensation of Prophase Chromosomes | 4.05 | [CCNB1, HIST1H2BD, PLK1] |
| 7Regulation of mitotic cell cycle | 5.81 | [ANAPC1, AURKA, CCNB1, PLK1, PSMD11] |
| 7TP53 Regulates Transcription of Cell Cycle Genes | 8.16 | [AURKA, CCNB1, CCNE2, CDC25C] |
| 7TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest | 16.67 | [AURKA, CCNB1, CDC25C] |
| 7Cyclin A/B1/B2 associated events during G2/M transition | 12.00 | [CCNB1, CDC25C, PLK1] |
| 7G2/M DNA damage checkpoint | 4.21 | [CCNB1, CDC25C, HIST1H2BD, RNF168] |
| 7The role of GTSE1 in G2/M progression after G2 checkpoint | 5.13 | [CCNB1, GTSE1, PLK1, PSMD11] |
| 7AURKA Activation by TPX2 | 4.11 | [AURKA, HMMR, PLK1] |
| 7G1 to S cell cycle control | 4.41 | [CCNB1, CCNE2, PRIM1] |
| 7DNA damage response | 4.41 | [CCNB1, CCNE2, CDC25C] |
Note: Pathways are extracted from Wiki Pathways, KEGG, and REACTOME Pathways. Term P value, term P value corrected with Bonferroni step down, group P value, and group P value corrected with Bonferroni step-down were less than 0.05. %AG refers to %associated genes. The superscript numbers in column 1 is an indicator of group number.
Table 2. Frequency of Associated Pathways and Pathway Groups for Certain DEGs.
| R | Gene | Description | NAT | AGN |
| 1 | CEBPB | CCAAT/enhancer binding protein beta | 2 | 1, 3 |
| 2 | DNAJA1 | DnaJ heat shock protein family (Hsp40) member A1 | 1 | 1 |
| 3 | HSPH1 | heat shock protein family H (Hsp110) member 1 | 1 | 5 |
| 4 | HSPA5 | heat shock protein family A (Hsp70) member 5 | 4 | 5, 6 |
| 5 | HSPA8 | heat shock protein family A (Hsp70) member 8 | 3 | 1, 5 |
| 6 | HSP90B1 | heat shock protein 90 beta family member 1 | 4 | 3, 6 |
| 7 | HMMR | hyaluronan mediated motility receptor | 2 | 7, 2 |
| 8 | SV2B | synaptic vesicle glycoprotein 2B | 1 | 2 |
| 9 | TNN | tenascin N | 1 | 2 |
| 10 | VWF | von Willebrand factor | 1 | 2 |
| 11 | MAPK15 | mitogen-activated protein kinase 15 | 1 | 3 |
| 12 | SRSF1 | serine and arginine rich splicing factor 1 | 1 | 3 |
| 13 | OSBPL1A | oxysterol binding protein like 1A | 2 | 4 |
| 14 | OSBPL6 | oxysterol binding protein like 6 | 2 | 4 |
| 15 | RXRA | retinoid X receptor alpha | 3 | 4, 6 |
| 16 | PSMD11 | proteasome 26S subunit, non-ATPase 11 | 8 | 5, 7 |
| 17 | ATF3 | activating transcription factor 3 | 2 | 6 |
| 18 | CREBRF | CREB3 regulatory factor | 1 | 6 |
| 19 | ANAPC1 | anaphase promoting complex subunit 1 | 10 | 7 |
| 20 | CCNB1 | cyclin B1 | 19 | 7 |
| 21 | CCNE2 | cyclin E2 | 6 | 7 |
| 22 | CDC25C | cell division cycle 25C | 9 | 7 |
| 23 | PLK1 | polo like kinase 1 | 14 | 7 |
| 24 | AURKA | aurora kinase A | 8 | 7 |
| 25 | GTSE1 | G2 and S-phase expressed 1 | 2 | 7 |
| 26 | SESN2 | Sestrin 2 | 1 | 7 |
| 27 | HIST1H2BD | histone cluster 1, H2bd | 2 | 7 |
| 28 | PRIM1 | primase (DNA) subunit 1 | 1 | 7 |
| 29 | RNF168 | ring finger protein 168 | 1 | 7 |
Note:NAT and AGN refer to “number of the associated term” and “associated group number” respectively.
Figure 2.
The genes that are common between the introduced pathway groups and the related groups. Group size is proportional to the number of pathways which are included in the group. The groups of 5 and 6 are connected to 3 genes, while groups of 1, 3, and 7 are related to two genes and groups of 2 and 4 were linked by one DEG.
Discussion
Biological effects of gamma-ray radiation and human health care have been investigated for many years and useful information has been obtained but there are many aspects about the mechanism of resulted damages and also protective methods that are less clear by now.17-19 In the present study, the effect of post-radiation time on the gene expression profile of the exposure cells was investigated. Like other high-throughput studies,20,21 gamma radiation widely changed the gene expression profile of the irradiated cells so many different types of genes were dysregulated. Bioinformatics as a useful method is applied to resolve problems which are tied to the complex data of high-throughput investigations.22,23 Thus, among the large numbers of dysregulated genes 177 DEGs were candidates to be assessed. Pathway analysis is applied to screen and also elucidate the results of such experiments, and findings are published to introduce the limited numbers of important dysregulated genes and critical affected pathways.24-26 In Figure 1, various types of pathways which are related to the dysregulated genes are presented. Further investigation is needed to explain the details of Figure 2. As shown in Table 1, the determined pathways are classified into 7 pathway groups. The groups are different based on the kind of included pathways and also the number of pathways. “Polo-like kinase mediated events” is the main group which includes 21 pathways (about 72% of total pathways), while there are three groups (groups 1-3) that include only one pathway.
As it is depicted in Table 2, only 16% (29 DEGs) of the query genes are involved in the introduced pathways. Based on the attribution of DEGs in the pathways, the genes can be categorized as the following groups: first, based on the involvement of genes in the pathway groups, the genes that are presented merely in one group and the DEGs which are common between more than one groups; second, based on the number of related pathways for the genes, the genes with a high value of associated pathways and the genes with few relevant pathways.
As shown in Table 2, 11 genes (about 38% of the genes) are related to one pathway while 7 individuals are related to 2 pathways. There are 3 related pathways for two genes and also 4 pathways are attributed to 2 DEGs. The genes that are involved in more than 6 pathways including CCNE2, PSMD11, CDC25C, ANAPC1, PLK1, AURKA, and CCNB1 are the genes that were associated merely with class 7 of the pathways (Polo-like kinase mediated events).
It can be concluded that the main group of pathways is “Polo-like kinase mediated events” that includes 72% of pathways and the genes with a higher degree of participation in the pathway are related to this pathway class. The significant roles of cyclins such as CCNE2 and CCNB1 in the regulation of the cell cycle are highlighted in the many investigations.27-29 Roles of PSMD11, CDC25C, ANAPC1, and PLK1 in the cell cycle and response to DNA damages are highlighted by Torres-Ávila et al.30 Medina-Aguilar et al published a document about the role of AURKA (Aurora kinase A) in the regulation of the cell cycle.31 It seems that pathway group 7 is tied to the regulation of the cell cycle and cell proliferation. This finding corresponds with the nature of gamma exposure.
Considering Figure 2, seven important genes are introduced as a linker between the 7 pathway groups. HSPA5, HSPA8, HSP90B1, HMMR, CEBPB, RXRA, and PSMD11 are the genes that connect all the pathway groups. It can be concluded that all groups are connected directly or indirectly to each other. HSPA5, HSPA8, HSP90B1, RXRA, and PSMD11 are down-regulated while HMMR and CEBPB are up-regulated (the data are not shown). HMMR links groups 2 (ECM-receptor interaction) and 7. The up-regulation of genes which are involved in “ECM-receptor interaction” in adipocytes that are active mainly in energy storage is confirmed by Hyun-Jeong Lee et al.32 It seems the activation of this pathway is a protective response of cells against gamma exposure. “Exercise-induced Circadian Regulation” and “IL-17 signaling pathway” are connected by CEBPB. The up-regulation of CEBPB and the promotion of related functions increase the roles of these pathways in the protection of cells in the irradiated cells.
Conclusion
In conclusion, 29 critical genes were introduced as the dysregulated individuals by gamma-ray irradiation that is related to the lacks in the pathways which are mainly involved in cell proliferation and activation of protective pathways. It seems that the activation of protective pathways is supported in the cells with longer post-radiation time.
Ethical Considerations
Not applicable.
Conflict of Interests
The authors declare no conflict of interest.
Acknowledgments
Shahid Beheshti University of Medical Sciences supported this research.
Please cite this article as follows: Vafaee R, Nikzamir A, Razzaghi M, Rezaei Tavirani S, Ahmadzadeh A, Emamhadi MA. An investigation of post-radiation gene expression profiles: a System bology study. J Lasers Med Sci. 2020;11(suppl 1):S101-S106. doi:10.34172/jlms.2020.S16.
References
- 1.Kumar DS, Chakrabarty D, Verma AK, Banerji BK. Gamma ray induced chromosomal aberrations and enzyme related defense mechanism in Allium cepa L. Caryologia G CitolCitosistematicaCitogenet. 2011;64(4):388–97. [Google Scholar]
- 2.Teoule R. Radiation-induced DNA damage and its repair. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;51(4):573–89. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- 3.Kadhim MA, Moore SR, Goodwin EH. Interrelationships amongst radiation-induced genomic instability, bystander effects, and the adaptive response. Mutat Res. 2004;568(1):21–32. doi: 10.1016/j.mrfmmm.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 4.Matsui A, Kobayashi J, Kanno SI, Hashiguchi K, Miyaji M, Yoshikawa Y. et al. Oxidation resistance 1 prevents genome instability through maintenance of G2/M arrest in gamma-ray-irradiated cells. J Radiat Res. 2020;61(1):1–13. doi: 10.1093/jrr/rrz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgakilas AG, Pavlopoulou A, Louka M, Nikitaki Z, Vorgias CE, Bagos PG. et al. Emerging molecular networks common in ionizing radiation, immune and inflammatory responses by employing bioinformatics approaches. Cancer Lett. 2015;368(2):164–72. doi: 10.1016/j.canlet.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Taylor J, Mi X, North K, Binder M, Penson A, Lasho T. et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood. 2020;136(13):1477–86. doi: 10.1182/blood.2020006868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St-Cyr S, Aubin-Horth N. Integrative and genomics approaches to uncover the mechanistic bases of fish behavior and its diversity. Comp BiochemPhysiolA Mol Integr Physiol. 2009;152(1):9–21. doi: 10.1016/j.cbpa.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Safaei A, Rezaei-Tavirani M, Sobhi S, Akbari ME. Breast cancer biomarker discovery: Proteomics and genomics approaches. Iran J Cancer Prev. 2013;6(Suppl):45–53. [Google Scholar]
- 9.Zamanian Azodi M, Rezaei-Tavirani M, Rostami-Nejad M, Rezaei-Tavirani M. Comparative bioinformatics characteristic of bladder cancer stage 2 from stage 4 expression profile: a network-based study. Galen. 2018;7:e1279. doi: 10.22086/gmj.v0i0.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G. et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T. et al. Abscisic acid‐activated SNRK2 protein kinases function in the gene‐regulation pathway of ABA signal transduction by phosphorylating ABA response element‐binding factors. Plant J. 2005;44(6):939–49. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 12.Peukert K, Staller P, Schneider A, Carmichael G, Hänel F, Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16(18):5672–86. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. [PubMed] [Google Scholar]
- 14. Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2020. 10.1093/nar/gkaa970 [DOI] [PMC free article] [PubMed]
- 15.Rezaei-Tavirani M, Tavirani MR, Zamanian Azodi M, Moravvej Farshi H, Razzaghi M. Evaluation of skin response after Erbium: Yttrium–Aluminum–Garnet laser irradiation: a network analysis approach. J Lasers Med Sci. 2019;10(3):194–99. doi: 10.15171/jlms.2019.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banáth JP, MacPhail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64(19):7144–9. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 17.Graupner A, Eide DM, Instanes C, Andersen JM, Brede DA, Dertinger SD. et al. Gamma radiation at a human relevant low dose rate is genotoxic in mice. Sci Rep. 2016;6:32977. doi: 10.1038/srep32977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi NM, Nair CK. Protection of DNA and membrane from gamma radiation induced damage by gallic acid. Mol Cell Biochem. 2005;278(1-2):111–7. doi: 10.1007/s11010-005-6940-1. [DOI] [PubMed] [Google Scholar]
- 19.Aygün B, Alaylar B, Turhan K, Şakar E, Karadayı M, Al-Sayyed MIA. et al. Investigation of neutron and gamma radiation protective characteristics of synthesized quinoline derivatives. Int J Radiat Biol. 2020;96(11):1423–34. doi: 10.1080/09553002.2020.1811421. [DOI] [PubMed] [Google Scholar]
- 20.Carulli JP, Artinger M, Swain PM, Root CD, Chee L, Tulig C. et al. High throughput analysis of differential gene expression. J Cell Biochem. 1998;72(S30‒31):286–96. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<286::AIDJCB35>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Kim HK, Lee S, Kim Y, Park J, Min S, Choi JW. et al. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat Biomed Eng. 2020;4(1):111–24. doi: 10.1038/s41551-019-0505-1. [DOI] [PubMed] [Google Scholar]
- 22.Heidari MH, Razzaghi M, Akbarzadeh Baghban A, Rostami-Nejad M, Rezaei-Tavirani M, Zamanian Azodi M. et al. Assessment of the microbiome role in skin protection against UV irradiation via network analysis. J Lasers Med Sci. 2020;11(3):238–42. doi: 10.34172/jlms.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amiri-Dashatan N, Koushki M, Jalilian A, Ahmadi NA, Rezaei-Tavirani M. Integrated bioinformatics analysis of mRNAs and miRNAs identified potential biomarkers of oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2020;21(6):1841–8. doi: 10.31557/APJCP.2020.21.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rezaei Tavirani M, Rezaei Tavirani S, Rostami FT. Biochemical pathway analysis of gastric atrophy. Gastroenterol Hepatol Bed Bench. 2018;11(2):118–24. [PMC free article] [PubMed] [Google Scholar]
- 25.Rostami-Nejad M, Rezaei-Tavirani M, Zadeh-Esmaeel MM, RezaeiTavirani S, Akbari Z, Esmaeili S. et al. Assessment of cytokine-mediated signaling pathway dysregulation in arm skin after CO2 laser therapy. J Lasers Med Sci. 2019;10(4):257–63. doi: 10.15171/jlms.2019.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdue MP, Song L, Scélo G, Houlston RS, Wu X, Sakoda LC. et al. Pathway analysis of renal cell carcinoma genome-wide association studies identifies novel associations. Cancer Epidemiol Biomarkers Prev. 2020;29(10):2065–9. doi: 10.1158/1055-9965.EPI-20-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson D, Walker C. Cyclins and cell cycle checkpoints. Annu Rev PharmacolToxicol. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- 28.Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116(2):221–34. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 29.Ding L, Cao J, Lin W, Chen H, Xiong X, Ao H. et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci. 2020;21(6):1960. doi: 10.3390/ijms21061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres-Ávila JF, Espitia-Pérez L, Bonatto D, da Silva FR, de Oliveira IM, Silva LF. et al. Systems chemo-biology analysis of DNA damage response and cell cycle effects induced by coal exposure. Genet Mol Biol. 2020;43(3):e20190134. doi: 10.1590/1678-4685-GMB-2019-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina-Aguilar R, Marchat LA, Arechaga Ocampo E, Gariglio P, García Mena J, Villegas Sepúlveda N. et al. Resveratrol inhibits cell cycle progression by targeting Aurora kinase A and Polo-like kinase 1 in breast cancer cells. Oncol Rep. 2016;35(6):3696–704. doi: 10.3892/or.2016.4728. [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ, Jang M, Kim H, Kwak W, Park W, Hwang JY. et al. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction is involved in the depot-specific adipogenesis in cattle. PLoS One. 2013;8(6):e66267. doi: 10.1371/journal.pone.0066267. [DOI] [PMC free article] [PubMed] [Google Scholar]