Abstract
RNA-protein interaction networks govern many biological processes, but are difficult to examine comprehensively. We devised ribonucleoprotein networks analyzed by mutational profiling (RNP-MaP), a live-cell chemical probing strategy that maps cooperative interactions among multiple proteins bound to single RNA molecules at nucleotide resolution. RNP-MaP uses a heterobifunctional crosslinker to freeze interacting proteins in place on RNA, and then maps multiple bound proteins in single RNA strands by read-through reverse transcription and DNA sequencing. RNP-MaP revealed that RNase P and RMRP, two sequence-divergent but structurally related non-coding RNAs, share RNP networks and that network hubs define functional sites in these RNAs. RNP-MaP also identified protein interaction networks conserved between mouse and human XIST long non-coding RNAs and defined protein communities whose binding sites colocalize and form networks in functional regions of XIST. RNP-MaP enables discovery and efficient validation of functional protein interaction networks on long RNAs in living cells.
Ribonucleoproteins (RNPs), complexes made up of interacting RNA and protein, govern both mRNA regulation and the function of non-coding RNA (ncRNA)1,2. Understanding how RNPs assemble and function, often involving multi-component RNA-protein networks, is critical for characterizing biological mechanisms. Biochemical approaches have defined protein interactions required for a number of RNP assemblies1, and high-resolution structural approaches3-6 have transformed our understanding of small and large RNP architectures. Nonetheless, it remains challenging to characterize RNP assemblies and their interacting networks in living cells.
Current methods for characterizing RNPs in live cells suffer from several limitations. Crosslinking with ultraviolet light (UV), optionally aided by metabolic incorporation of photoactivatable nucleotides, captures RNP information in living cells7-11. Sites of protein crosslinking to RNA can be mapped transcriptome-wide, either without10,11 or with identification of binding sites for individual proteins (crosslinking and immunoprecipitation, CLIP and PAR-CLIP)7,8. However, UV-based crosslinking suffers from experimental biases and limited binding site resolution9, metabolic labeling probes a single substituted nucleobase at a time8, and CLIP strategies require a specific antibody or protein tag. Methods that focus on cataloging RNA-binding proteins, like mass spectrometry, do not readily locate protein-binding sites on RNA nor easily prioritize proteins in terms of function12. Major challenges unaddressed by current approaches are: (1) How do multiple proteins interact with an RNA to form networks, and (2) Which protein interaction networks drive function for an individual RNA?
Here we describe RNP-MaP, an experimentally concise strategy to locate protein interaction sites on RNA in live cells with nucleotide resolution and reveal multi-protein interaction networks integral to RNP function. After validation on RNPs with known structure, we used RNP-MaP to define functional protein interaction communities within the XIST long non-coding RNA (lncRNA), resulting in the discovery of an RNP network in the XIST E region that controls maintenance of the XIST particle. RNP-MaP will be widely useful for understanding RNP biology, particularly in defining functionally critical domains in large mRNAs and ncRNAs.
RESULTS
RNP-MaP Strategy
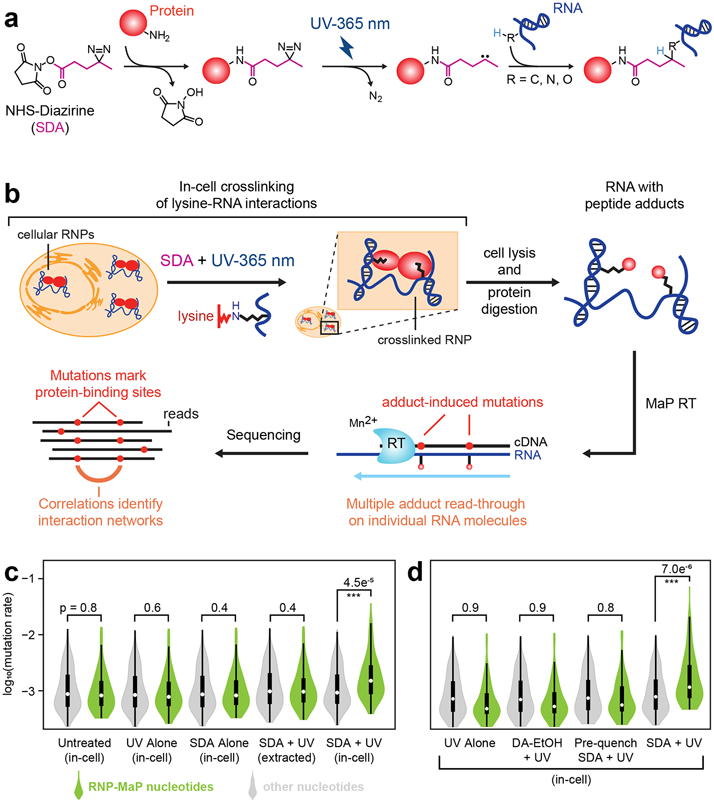
We identified a cell-permeable reagent, NHS-diazirine (SDA), that rapidly labels RNA nucleotides at sites of protein binding. SDA has two reactive moieties: a succinimidyl ester and a diazirine (Fig. 1a). Succinimidyl esters react to form amide bonds with amines, such that reaction occurs overwhelmingly with lysine side chains13. When activated with long-wavelength UV, diazirines form carbene or diazo intermediates14, which are broadly reactive toward nucleotide ribose and base moieties. Two-step reaction of SDA thus crosslinks protein residues with RNA with a distance governed by SDA linker length (4 Å) and side chain flexibility (~8 Å for lysine). Lysine is the second-most prevalent amino acid in RNA-binding domains (following arginine)15, and diazirine photo-intermediates are short-lived. SDA thus crosslinks short-range RNA-protein interactions relatively independently of local RNA structure or protein properties. Live cells are treated with SDA for ten minutes, excess reagent is quenched, and cells are exposed to UV light. SDA-treated cells are then lysed, and crosslinked proteins are digested to short peptide adducts.
Figure 1. RNP-MaP strategy for probing RNA-protein interaction networks in cells.
(a) Scheme for selective chemical crosslinking of proteins to RNA by SDA. (b) Workflow of the RNP-MaP experiment. (c) Nucleotide mutation rates after MaP reverse transcription, separated into RNP-MaP sites and non-sites (green and gray). Combinations of UV (365 nm, 3 J/cm2) and SDA (10 mM) were applied directly to cells (in-cell) or to protein-free RNA extracted from cells (extracted). The number of nucleotide mutation rates included in each distribution (n) are 77 RNP-MaP sites and 162 non-sites (d) RNA mutation rates for cells treated with UV and SDA, versus non-reactive controls (DA-EtOH or pre-quenched SDA). Representative data for Rmrp RNA from mouse embryonic stem cells (SM33) are shown. The n for distributions of RNP-MaP sites and non-sites is 82 and 157, respectively. For violin plots, circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values. P-values (Kolmogorov-Smirnov test, one-sided) are shown. If p-values are calculated comparing each condition to their untreated (in c) or UV alone (in d) counterparts, only reactivities of RNP-MaP nucleotides in the SDA and UV treated in-cell samples are significantly increased (p <0.05).
We detect SDA-mediated RNA-protein crosslinks using the MaP reverse transcription technology16 (Fig. 1b). With MaP, a relaxed fidelity reverse transcriptase reads through adduct-containing nucleotides and incorporates non-templated nucleotides into the product DNA at the site of RNA-protein crosslinks. Because reverse transcription reads through the adducts, RNP-MaP detects multiple protein crosslinks that co-occur on single RNA molecules (Fig. 1b). Sequencing the DNA product and locating sites of mutation thus reveals two key features of an RNA-protein complex: RNP-MaP adducts at individual nucleotides report locations of protein binding, and correlated crosslinking across multiple nucleotides reveals higher-order protein interaction networks. RNP-MaP thus detects both protein binding location and interaction network information with no requirement for pre-existing knowledge about the proteins involved. If desired, the involved proteins can subsequently be assigned by comparison to other information, such as CLIP datasets.
RNP-MaP Validation
The SDA reactivity at each nucleotide is the ratio of the MaP mutation rate for cells treated with SDA and UV compared to cells treated with UV only (Fig. S1). We derived universal normalization factors for each RNA nucleotide (U, A, C, and G) based on analysis of RNPs of known structure, enabling identification of protein-bound nucleotides (termed RNP-MaP sites) for arbitrary RNAs of interest (Fig. S1). RNP-MaP reactivities were reproducible (Fig. S2). Nucleotides with high reactivities are close to lysine amines in human U1, RNase P, and ribosome complexes3-6, whereas nucleotides distant from bound proteins rarely passed reactivity thresholds (Fig. S2). RNP-MaP sites occurred in both single-stranded and base-paired regions of RNA. Unpaired RNA regions showed higher reactivities for some RNAs (Fig. S2), which likely reflects binding preferences for single-stranded RNA. RNP-MaP sites occurred at all four nucleotides, with higher reactivities at uridine and adenosine (Fig. S2). RNP-MaP sites were not detected if UV or SDA were omitted, if RNA was first extracted from cells (removing protein) (Fig. 1c and Fig. S3), using a diazirine ethanol compound (DA-EtOH, no lysine-reactive group), or using pre-quenched SDA (Fig. 1d). RNP-MaP signals overlapped those of two orthogonal approaches, ΔSHAPE chemical probing17 and photo-lysine metabolic labeling18, which also identify protein-bound sites (Fig. S4). In sum, RNP-MaP identifies protein-proximal nucleotides, with good coverage across all four ribonucleotides in diverse structural contexts, in a manner strictly dependent on SDA and UV dosage and the presence of cellular proteins.
RNP-MaP defines protein interaction networks in the U1 snRNP
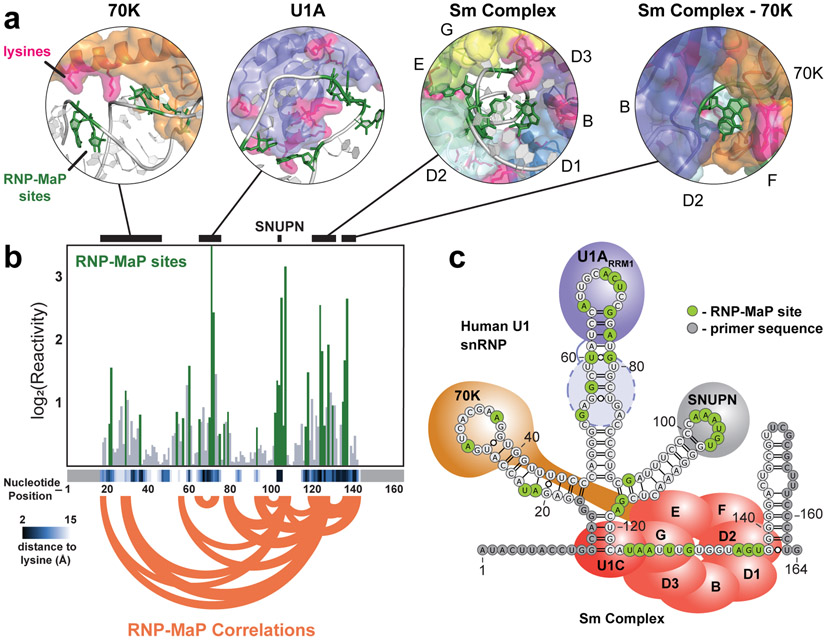
In human HEK293 cells, RNP-MaP sites clustered in regions of U1 RNA known to bind proteins5,19 (Fig. 2a, 2b), including at all four nucleotides and in both single-stranded and base-paired regions (Fig. 2c). The majority of RNP-MaP sites were within 9 Å of a lysine residue (Fig. 2b). We also identified RNP-MaP sites in U1 stem loop 2 (positions 48-91) that do not correspond to a known interaction site (Fig. 2c). These RNP-MaP sites presumably reflect binding by the second (currently unvisualized5) U1A RRM2 domain or an unidentified protein component of the U1 snRNP.
Figure 2. RNP-MaP defines protein interaction networks in the U1 snRNP.
(a) Structures surrounding lysine-RNA crosslinking sites (from 3CW14 and 4PKD5). (b) Bar graph of log2(average reactivity) from replicate experiments performed on U1 snRNA in HEK293 cells. RNP-MaP sites passing thresholds are shown in green. Nucleotide distance (Å) to nearest lysine amine are shown with heatmap (bottom). Black bars (top) indicate locations of structures represented in panel a. RNP-MaP correlations (top 10% in mutual information strength) are shown as orange arcs. Nucleotides that overlap amplification primers (light gray boxes) are not observable by RNP-MaP. (c) Secondary structure model of the human U1 RNP showing relative protein positions, RNP-MaP sites, and primer regions. Estimated location of U1A RRM2 binding (not visualized in structures) consistent with RNP-MaP signal is shown (dashed line oval).
RNP-MaP uniquely identifies correlated protein binding events that occur between sites on an RNA (Fig. 1b). Because the MaP reverse-transcription process reads through protein-RNA cross-links, multiple cross-link sites can be detected per single RNA molecule. We adapted a G-test framework20 to identify pairs of RNA nucleotides that are co-modified in a statistically significant manner, which we term RNP-MaP correlations (Fig. S1 and Fig. S5). RNP-MaP correlations are distinct from RNP-MaP sites, each providing an independent complementary measure of protein binding to RNA. Correlations require that a single RNA molecule form at least two crosslinks and arise from three scenarios: (i) a single protein that binds two locations on one RNA, (ii) two proteins that interact and bind two locations on one RNA, or (iii) two proteins are deposited at two locations on one RNA by a coordinated assembly process.
Networks of RNP-MaP correlations were consistent with the architecture of the U1 snRNP complex (Fig. 2b, 2c).The highest density and strength of correlations involved nucleotides bound by the Sm protein complex (Fig. 2b and Fig. S5), whose initial loading onto U1 is necessary for the maturation of the snRNP21. RNP-MaP correlations also detect long-distance interactions between 70K and the Sm complex important for U1 snRNP assembly. Correlations between the Sm core and the U1A protein were weak, consistent with independent binding by U1A and its expendability for splicing22. In sum, RNP-MaP reveals protein interaction networks, pinpoints the central hubs of these networks, and identifies interactions important for assembly and function in the U1 snRNP.
RNP-MaP reveals a protein interaction network conserved in RNase P and RMRP RNAs
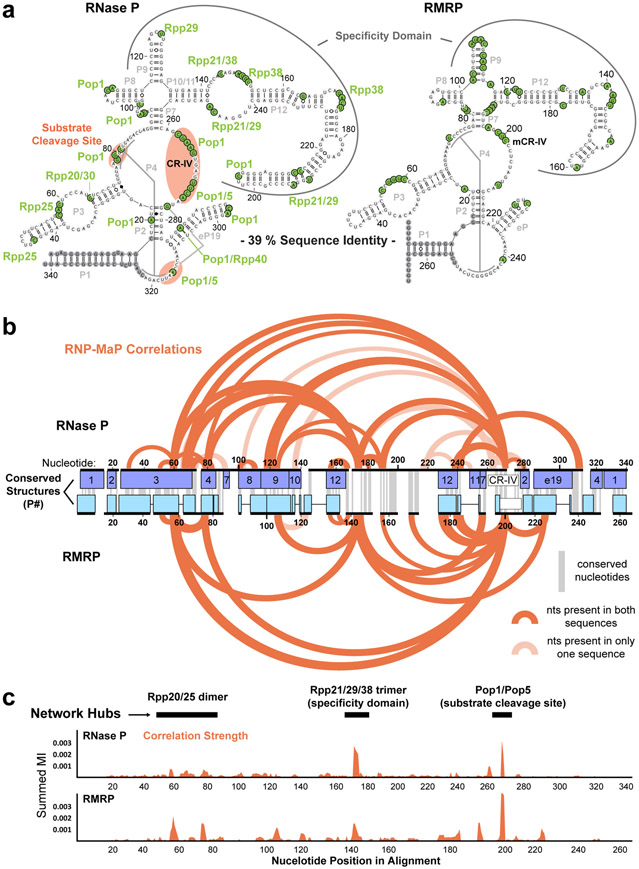
RNase P and RMRP are divergent, but structurally related, ncRNAs that bind intersecting sets of proteins to form RNP endonucleases that cleave distinct substrates6,20,23. Despite substantial differences in sequence, the two RNPs exhibit nearly identical RNP-MaP profiles (Fig. 3a). RNP-MaP sites identify 9 out of 10 known protein interactions with RNase P6. The matching patterns of RNP-MaP sites for RNase P and RMRP core domains suggest that most protein interaction sites are shared. Locations of RNP-MaP sites are also conserved between human and mouse homologs of RNase P and RMRP (Fig. S6).
Figure 3. RNP-MaP reveals conserved protein interaction networks in RNase P and RMRP RNAs.
(a) Secondary structures of human RNase P and RMRP RNAs annotated with RNP-MaP sites (green). Experiments performed with HEK293 cells. Proteins proximal to each site (based on nearest lysine) are labeled. Functional domains are indicated, and conserved base-paired structural regions (P#) are labeled (gray). (b) RNP-MaP correlations for human RNase P and RMRP, plotted on a structure-based sequence alignment. Correlations shown correspond to the top 10% of mutual information strength. Correlations that reflect linkages between nucleotides present in only one of the RNAs are shown in light shading. (c) Total strength of correlations (by mutual information, MI) at each nucleotide in the human RNase P and RMRP RNAs, plotted on the shared structure-based alignment. Protein interaction network hubs are labeled.
The patterns of through-space RNP-MaP interaction networks for the RNase P and RMRP RNAs are nearly identical after alignment by structural domains (Fig. 3b). The strongest correlations for RNase P define hubs involving the specificity domain, the substrate cleavage site, and the Rpp20/25 dimer (which links the specificity domain and cleavage site). These hubs are conserved in RMRP (Fig. 3c). Omission of proteins that comprise each hub suppresses or eliminates RNase P catalytic function24. Our RNase P and RMRP data thus identify shared protein interactions and functional interaction network hubs in sequence divergent RNAs and confirm conservation of RNP architecture in mice and humans.
RNP-MaP identifies conserved protein interaction networks in the XIST lncRNA
The 20-kb X-inactive specific transcript (RNA denoted Xist in mouse, XIST in human) controls X-chromosome dosage compensation in eutherian mammals25. The Xist/XIST sequence shows low conservation between mice and humans, despite accomplishing the same functions. We applied RNP-MaP to Xist/XIST protein interaction networks in mouse and human cells by enriching for Xist/XIST RNAs using an RNA antisense pull-down26 (Fig. S7). Data were obtained for 97% of nucleotides in Xist/XIST RNAs (17410 and 18708 nts); RNP-MaP sites (2139 and 3766, respectively) occurred at all four nucleobases and in both structured and unstructured regions (Fig. S8).
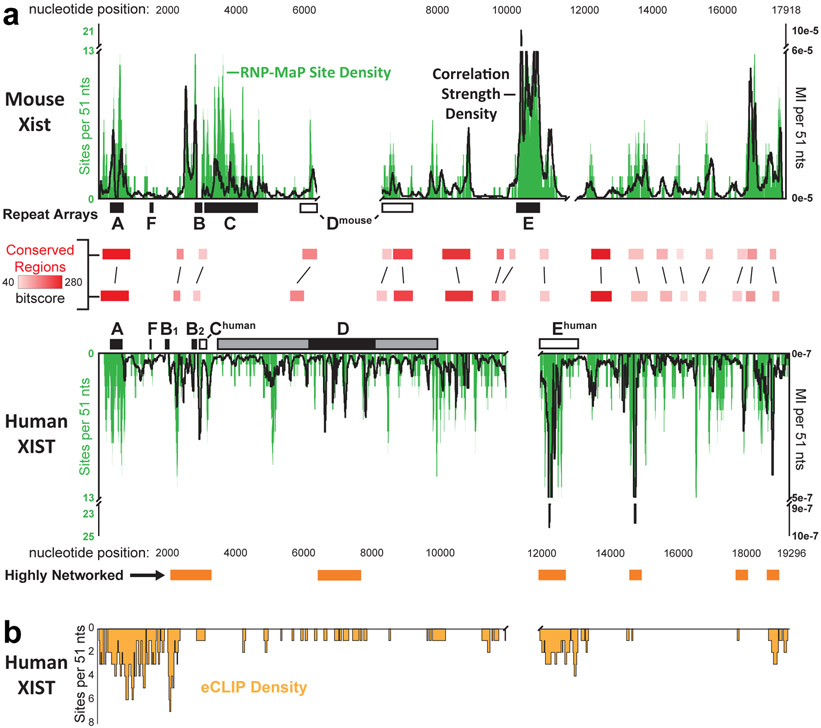
High RNP-MaP site density occurred in the Xist/XIST A, B, C, D, and E regions (Fig. 4a), which contain repetitive sequences important for Xist localization, assembly, and chromatin silencing27-32. Conservation of high RNP-MaP density in these regions occurred in both human and mouse RNAs despite changes in copy number (human XIST contains two copies of the B region) and in size, relative position, and sequence (C, D, and E regions differ extensively between humans and mice)33. We also discovered multiple additional regions of Xist/XIST that have not previously been defined as functional but which exhibit strong RNP-MaP signal density clearly conserved between mice and humans (Fig. 4a).
Figure 4. RNP-MaP identifies conserved protein interaction networks in the XIST lncRNA.
(a) Density of RNP-MaP sites (green, the total number of RNP-MaP sites per 51-nt window, left axis) across mouse Xist (top) and human XIST (bottom). Black lines indicate correlation strength densities (depth-normalized mutual information, MI, per 51-nt window, right axis). Human XIST regions that are highly networked (orange boxes, bottom) or have conserved local sequence alignments with mouse Xist (red boxes, middle) are emphasized. Rectangles labeled A-F show locations of tandem repeat arrays within each RNA: including well-defined repeat sequences (solid rectangles) and regions identified as homologous based on RNP-MaP similarity (open rectangles). The D region in human XIST contains a distinct repeat core (black) flanked by more degenerate repetitive elements (gray). Gaps were introduced to align RNAs by conservation and RNP-MaP similarity. Low sequencing depth prevented identification of correlations in human XIST A region; high repeat content prevented read alignment to Xist and XIST B regions. Experiments were performed in SM33 and HEK293 cells. (b) Density of eCLIP sites along the human XIST RNA, shown as the number of eCLIP sites within 51-nt windows. Based on 151 total sites from 30 proteins mapped reproducibly to XIST in K562 cells66.
We compared the RNP-MaP signal on human XIST to the most comprehensive set of enhanced CLIP (eCLIP) per-protein binding measurements available (from ENCODE34, obtained in K562 cells35). While there are differences between HEK293 and K562 cells, both female cell lines maintain silenced X chromosomes and XIST compartments, and eCLIP peaks are shared on genes of similar expression between cell lines35. We focused on proteins whose binding sites on XIST were reproducible between eCLIP replicates, yielding 30 proteins (from a total of 120 eCLIP experiments). Regions of XIST with more eCLIP sites had more RNP-MaP sites, especially over the XIST A and E regions (Fig. 4b). eCLIP site density is lower than RNP-MaP density across multiple regions, likely reflecting that only a subset of Xist-binding proteins have been mapped by eCLIP. Together, these data show that RNP-MaP identifies protein-binding sites in lncRNAs that are conserved between species and critical for function, in the absence of pre-existing knowledge about protein binding sites.
Protein-bound regions in the Xist/XIST RNAs form higher-order interaction networks with distinct levels of interactivity, which are features invisible to alternative strategies. Six highly networked regions occur in XIST, at least five of which are conserved in mouse (Fig. 4a). The E region of Xist/XIST represents an extreme example in which an extended region (spanning 1-1.5 kb) forms a cooperative protein interaction network, as evidenced by high correlation strength densities. In contrast, there also exist highly protein-bound regions, such as the C region of mouse Xist and portions of Xist/XIST D region, that do not show strong correlations and where proteins therefore bind relatively independently of one another (Fig. 4a). Thus, RNP-MaP reveals distinct local patterns of higher-order RNA-protein interaction networks, detected as low and high levels of network interactivity.
Communities of XIST-binding proteins
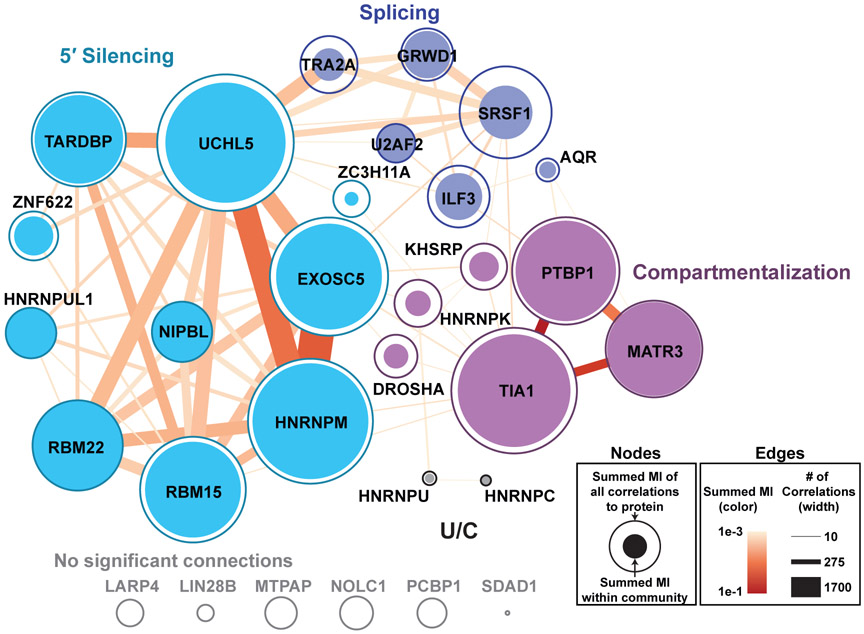
Using the same ENCODE34 eCLIP35 data, we assigned proteins to the interaction networks identified by RNP-MaP correlations. We performed a network analysis of high-confidence eCLIP sites that are linked by highly significant RNP-MaP correlations, revealing communities of proteins whose binding sites on XIST are networked together (Fig. 5, Table S1). We categorized these communities based on the functions of XIST sequences to which the proteins bind, yielding 5′ Silencing, Compartmentalization, Splicing, and U/C communities (Fig. 5 and Fig. S8). The communities are distinct: correlations between proteins from different communities occur significantly fewer times than expected based on the proximity of their binding sites (Table S2).
Figure 5. Communities of XIST-binding proteins.
Network graph of protein-bound sites (nodes) from eCLIP data on human XIST RNA linked by RNP-MaP correlations (edges). Communities were identified by maximizing modularity of the network68,71 while weighting by correlation strength (mutual information). Summed mutual information for all correlations linking each protein node to other proteins in its community (filled circles) or to all proteins (open circles) are represented as circles with proportional areas. Summed mutual information of all correlations in an edge (color) and total number of correlations in each edge (width) are shown. Low read depth in A and B repeat regions prevented correlation analysis of a portion of RBM15, SRSF1, UCHL5, and HNRNPK eCLIP sites at these locations, potentially underestimating their contributions to each network. Additionally, the known XIST-binding protein SPEN is not represented in eCLIP data.
Proteins in the 5′ Silencing community bind primarily to the 5′ region of XIST, including in the silencing-critical A region27. Community members include factors involved in XIST processing, XIST stability, and XIST-mediated silencing: UCHL536, EXOSC537, HNRNPUL138, and RBM1539, (Fig. 5). Silencing community members TARDBP and RBM22 are RNA-dependent regulators of transcription40. Binding sites for the 5′ Silencing community members show high interactivity consistent with forming a specific coordinated RNP on XIST.
The strongest inter-protein correlations occur between Compartmentalization community members PTBP1, MATR3, and TIA1, which bind in the XIST E region (Fig. 5 and Fig. S8). The XIST E region is critical for maintenance of the silenced X chromosome compartment30. PTBP1, MATR3, and TIA1 each undergo liquid-liquid phase-transitions to form RNA granules41-43, and PTBP1 and MATR3 interact on other RNAs44, features consistent with the formation of an XIST-mediated compartment.
The Splicing community includes proteins that control splicing (U2AF2, SRSF1, TRA2A, AQR, and ILF3)45-47 and a chromatin modulator (GRWD1)48. All Splicing community proteins, except for TRA2A, bind to XIST at exon-exon junctions (Fig. S8), consistent with a function in splicing of XIST transcripts. The smallest community (U/C) includes two HNRNPs (U and C) that interact with one another49 but do not strongly interact with other communities and interact sparsely across XIST, suggesting that these proteins play more independent roles in XIST function or that methodological constraints precluded assignment to a single representative community. Together, network analysis reveals how RNP-MaP defines RNP communities with distinct levels of networking (low versus high), each associated with critical lncRNA functions.
RNP-MaP reveals interaction sites for Compartmentalization community proteins
The XIST E region, critical for maintaining the silenced X chromosome compartment29,30, is distinguished by strong inter-protein network connectivity (Fig. 4a), and includes binding sites for the proteins PTBP1, MATR3, and TIA1 in the Compartmentalization community (Fig. 5 and Fig. S8). PTBP1, MATR3 and TIA1 proteins are implicated in formation of RNA foci41-43, likely through multivalent RNA-protein interactions, consistent with the highly interactive protein network we observe in the XIST E region (Fig. 4a). We therefore investigated the role these protein interaction networks play in XIST particle formation.
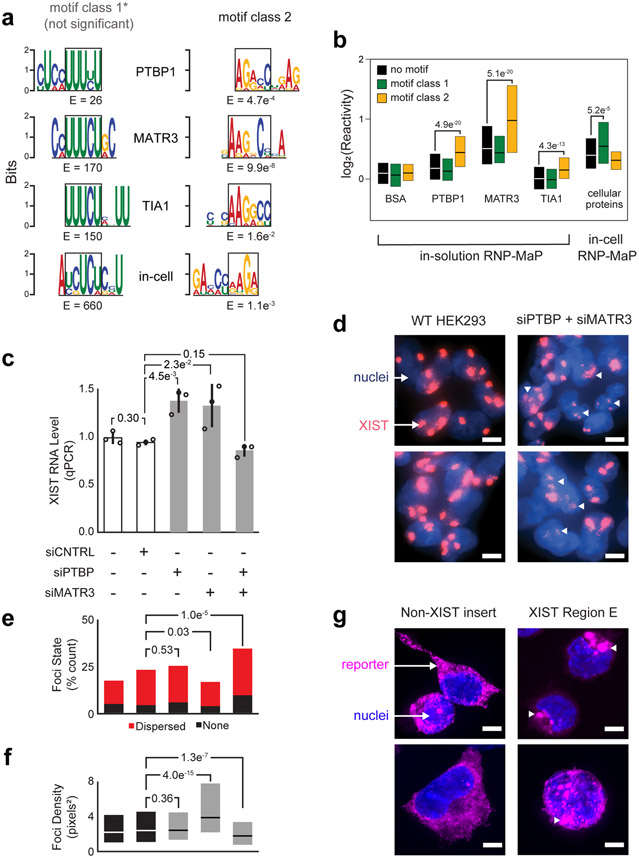
We examined binding by PTBP1, MATR3, and TIA1 in a simplified system using recombinant proteins and a synthetic RNA spanning the human XIST E region. PTBP1, MATR3, and TIA1 each bound the XIST E region RNA with similar RNP-MaP patterns (Fig. 6a and S9). Binding occurs at pyrimidine-rich sequences, similar to motifs previously defined in vitro50 and by CLIP methods51-53. However, binding to pyrimidine-rich motifs (termed class 1 here) was not significantly enriched (Fig. 6a) relative to the abundance of these motifs in the XIST E region. Instead, a purine-rich motif (called class 2) was significantly enriched in RNP-MaP sites for each protein, both in the reconstituted system and in cells (Fig. 6a). Class 1 and 2 motifs each align to reveal a 4-6 nt core sequence motif. Under our simplified conditions, PTBP1, MATR3, and TIA1 show higher RNP-MaP reactivity with class 2 than with class 1 motifs (Fig. 6b). In cells, due to the differing conditions and proteins present, strong RNP-MaP signals occur at both class 1 and 2 motifs and throughout the E region (Fig. 6b).
Figure 6. PTBP1 and MATR3 interactions with E region, and functional control of XIST particle formation.
(a) RNP-MaP enriched 9-mer sequence motifs (by MEME)69 using purified components for each indicated protein, and comparison with motifs observed in cell. Motifs are shown as position weighted matrices and E-values, aligned to a shared core sequence (black boxes) for class 1 and class 2 motifs. (b) Boxplots of RNP-MaP log2(reactivity) at class 1 and 2 motifs or other nucleotides (no motif) in the XIST E region, either for simplified conditions with synthetic RNA and indicated recombinant protein (in-solution) or with native XIST and cellular proteins in HEK293 cells (in-cell). The number of nucleotide reactivities included in each distribution (n), from left to right, were 631, 230, 293, 651, 235, 293, 651, 235, 293, 651, 235, 292, 668, 229, and 293. Box limits indicate first and third quartiles; central lines indicate medians. Significant increases in reactivity compared to no motif nucleotides are indicated, and – with the exception of TIA1 no motif and class 1 motif sites – reactivities in-cell and in-solution are all significantly increased compared to BSA (p <0.05, Kolmogorov-Smirnov test, one-sided). (c) Relative expression levels of XIST RNA in HEK293 cells, as a function of PTBP and MATR3 knockdown by short interfering (si)RNAs. Standard deviation (error bars), means (bar heights) of replicate measurements (n = 3, open circles), and P-values (Student’s t-test, two-sided) are shown. (d) Visualization of XIST foci for wildtype (WT) HEK293 cells and cells depleted of PTBP and MATR3 by siRNA treatment. Dispersed and non-punctate XIST foci are emphasized with white triangles. XIST in cells imaged by fluorescent in situ hybridization (FISH, red) using labeled antisense oligonucleotides; nuclei labeled with DAPI (blue). (e) Quantification of dispersed and absent XIST foci as a function of PTBP and MATR3 depletion (HEK293 cells). Conditions are ordered as per panel c. P-values (Chi-square goodness of fit test) are shown. The total number of cells counted (n) were 107, 396, 200, 286, and 304, respectively. (f) Effect of PTBP and MATR3 depletion on XIST foci density. Conditions are ordered as per panel c. P-values (Kolmogorov-Smirnov test, two-sided) are shown. The total number of XIST foci observed (n) were 311, 1000, 746, 533, and 863, respectively. Box limits indicate first and third quartiles; central lines indicate medians. (g) Localization of non-XIST control and XIST E region-containing RNA reporters (by FISH, magenta); nuclei labeled by DAPI (blue). Condensed foci formed by XIST E region reporters are emphasized by white triangles. The percentage of cells with granule phenotypes were 9% (1 of 11) and 89% (8 of 9), respectively. P-values (binomial test and chi-squared goodness of fit) were significant (7.8 e−8,, 1.13 e−13). Data from e and f were combined from two biological replicate experiments (performed and imaged on different days); d and g are representative images. Scale bars in d and g are 5 μm.
Despite their highly significant RNP-MaP signal, class 2 motifs have not been detected by CLIP for PTBP1, MATR3, or TIA1 (Fig. S9). This discrepancy is consistent with two models: (i) conditions and proteins in cells alter the intrinsic binding preferences of PTBP1, MATR3, and TIA1, or (ii) nucleotide biases of CLIP methods (for uridines9) mask binding to pyrimidine-poor class 2 motifs (Fig. S10). Overall, RNP-MaP identifies a larger set of protein-binding sites than CLIP: 93% of eCLIP sites in XIST (of 151 analyzed) contain 3 or more RNP-MaP sites (enrichment p-value = 0.019, compared to randomized eCLIP sites), but only 35% of RNP-MaP sites (of 3766) fall within an eCLIP site.
PTBP1 and MATR3 interaction with E region controls XIST particle formation
PTBP1 and MATR3, major components of the Compartmentalization community, bound to the E region RNA at lower concentrations and showed higher overall RNP-MaP reactivities than TIA1 (Fig. 6b, S9 and S10), and PTBP1 and MATR3 have more high-frequency eCLIP signals in the XIST E region (nts in the top 0.001%) than TIA1 (324 and 539, versus 73, respectively). We therefore focused further functional analysis on PTBP1 and MATR3 and depleted each protein in HEK293 cells by RNA interference, achieving 75% and 85% knockdown, respectively. Knockdown of either PTBP1 or MATR3 individually resulted in a ~40% increase in XIST RNA levels, whereas co-depletion of both PTBP1 and MATR3 returned XIST to normal levels (Fig. 6c). Compared to normal HEK293 cells (which have 2-5 silenced X chromosomes per cell), XIST foci in cells depleted of both PTBP1 and MATR3 were more dispersed or frequently absent (Fig. 6d and 6e) and remaining foci were significantly less dense (Fig. 6f). The dispersion observed upon PTBP1 and MATR3 depletion resembles the effects of CIZ1 depletion and E region deletion in mouse Xist29,30. These data suggest that PTBP1 and MATR3, whose binding in the E region forms an exceptionally interactive network, function to maintain the human XIST particle.
We inserted the highly protein-interactive E region into an RNA reporter and compared its expression and localization to reporters containing other highly protein-interactive XIST regions or a non-XIST sequence. The E region-containing reporter, but not other reporters, formed large foci in cells (Fig. 6g). These E region foci, while formed in the cytoplasm, are similar in size to native XIST particles observed in HEK293 cells (Fig. 6d). E region foci appear to trigger cellular deformations, and the E region-containing reporter is less stable than other tested sequences (Fig. S10). Highly interactive RNP networks, partially deleterious out of context, thus appear to intrinsically assemble on XIST E region RNA. E foci likely include granule-associated proteins like PTBP1, MATR3, and TIA1, and further work is necessary to identify components of these non-canonical granules. Still, these data support a role for the E region in organization of the XIST compartment29,30 and highlight the ability of RNP-MaP to discover and characterize novel motifs in ncRNAs, whose functions reflect interconnected RNA-protein networks.
DISCUSSION
RNP-MaP enables rapid and concise characterization of functionally important RNA-protein interaction networks. Protein binding sites are identified across an RNA with low sequence and structure biases, interaction networks are distinguishable by their correlation patterns, and functionally important hubs are revealed by their binding site density and interconnectivity.
RNP-MaP currently requires read depths of 103 for sites and 104 for correlations, and maximum correlation distance is governed by the length of reverse transcriptase products (currently ~500 nts). RNP-MaP detects RNA-protein interactions conserved between species and critical for function without pre-existing knowledge of the interacting proteins, and can be integrated with other information to reveal protein identity. Coupling RNP-MaP with complementary CLIP and mass spectrometry approaches will enable definition of cellular RNP networks in unprecedented detail.
RNP-MaP revealed insights into the assembly of small RNPs U1, RNase P, and RMRP. Each RNP has multiple interaction network hubs, and the strongest interaction hubs in each RNA correspond to regions central to RNP assembly and activity: the Sm complex assembly site in U1 and the substrate cleavage sites in RNase P and RMRP. The unique ability of RNP-MaP to distinguish interaction networks by their correlation strength and density will aid in discovery and prioritization of functional elements in large noncoding, messenger and viral RNAs.
Prior analyses of the mouse Xist RNA revealed that repeat-containing regions are structurally dynamic and accessible for protein binding, and these regions were proposed to function as “landing pads” for proteins17. Our RNP-MaP study now directly reveals that repeat sequences in Xist/XIST are extensively bound by highly networked proteins. Depleting the highly networked Compartmentalization community members PTBP1 and MATR3 induces dispersal of native XIST RNA foci in cells, and E region RNA is sufficient to promote foci formation in a heterologous reporter RNA in cells. These data suggest that protein interaction networks in the E region support formation of a phase-separated particle in vivo, which is consistent with recent orthogonal analyses of Xist particle shape and composition54 and a conserved role in this process for MATR3, PTBP1 and the Xist E region in mice55. The many other highly-connected protein interaction networks identified by RNP-MaP in Xist/XIST suggests additional biology that warrant future study.
Most noncoding RNAs and untranslated regions of mRNAs function by recruiting proteins and forming higher-order RNA-protein complexes. The ability of RNP-MaP to identify function-critical RNA regions and their interconnected protein networks will enable focused exploration of the thousands of ncRNAs and mRNA UTRs whose overall functions and specific internal functional elements are unexplored56. RNP-MaP can be further applied to reveal how protein interaction networks form and dissociate in both coding and non-coding RNAs, how networks differ between cell types, and how networks change in response to stimuli.
ONLINE METHODS
Cell culture
Adherent mammalian cells used in chemical probing experiments, either SM3326 or HEK293 cells, were grown to 80-90% confluency in either 6-well plates (for targeted priming) or 10-cm dishes (for RNA antisense pulldown). HEK293 cells were cultured in DMEM with 10% FBS. SM33 cells were cultured in embryonic stem cell media [DMEM high glucose with sodium pyruvate, 15 % FBS, 0.1 mM non-essential amino acids (Gibco), 2 mM L-glutamine, 0.1 mM β-mercaptoethanol,1000 U/mL leukemia inhibitory factor (ESGRO, Millipore Sigma)]. Cultures were grown with 100 U/mL penicillin and 100 μg/mL streptomycin. To induce expression of the Xist RNA, SM33 cells were supplemented with 2 μg/mL doxycycline 16 hours before treatment. For all experiments when performing biological replicates, chemical probing and sequencing library preparation were performed on distinct populations of cells on different days.
In-cell crosslinking with SDA
SDA (NHS-diazirine, succinimidyl 4,4’-azipentanoate, Thermo Fisher) was selected from a small screen of commercially available heterobifunctional reagents capable of crosslinking RNA and protein. For 6-well plates, cells were washed once in 1 mL PBS, then covered with 900 μL PBS. To these cells, 100 μL of 100 mM SDA in DMSO was added with concurrent manual mixing. For controls, 100 μL of neat DMSO was added. Cells were treated with SDA for 10 minutes in the dark at 37 °C, then excess SDA was quenched with a 1/9 × volume of 1 M Tris-HCl, pH 8.0 (111 μL). For SM33 cells, which remained adherent during treatment, quenching was performed for 5 minutes in the dark at 37 °C. For HEK293 cells, which detached upon treatment, cells were pelleted at 1000 ×g for 3 minutes immediately after addition of quencher. Cells were washed once with PBS (and pelleted again if not adherent) and then resuspended in 400 μL of PBS in a well of a 6-well plate. To crosslink labeled proteins to RNAs, SDA-treated and untreated cells were placed on ice and exposed to 3 J/cm2 of 365 nm wavelength ultraviolet light (about 9 minutes in a UVP CL1000 equipped with five 8-Watt F8T5 black lights) at a distance of 4 inches from the light source. When the amount of SDA used for treatment, the amount of UV light exposure, or the compound used for crosslinking was varied no other changes were made to the procedure. When performing crosslinking procedure on cells grown in 10 cm dishes, reagent volumes used were multiplied by a factor of five relative to the 6-well procedure.
Cellular fractionation and proteinase K lysis of SDA-treated cells
Crosslinked cells were pelleted at 1500 ×g for 5 minutes at 4 °C, washed once in cold PBS and pelleted again, and resuspended in cytoplasmic lysis buffer [10 mM KCl, 1.5 mM MgCl2, 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 0.1% Triton X-100]. Cells were lysed for 10 minutes at 4 °C with agitation. Nuclei were pelleted at 1500 ×g for 5 minutes at 4 °C, and cytoplasmic lysates were separated into new tubes. Nuclei were washed once in low-salt solution [10 mM KCl, 1.5 mM MgCl2, 20 mM Tris-HCl (pH 8.0), 1 mM DTT], incubated with agitation at 4 °C for 2 minutes, pelleted again, and then resuspended in proteinase K lysis buffer [40 mM Tris-HCl (pH 8.0), 200 mM NaCl, 20 mM EDTA, 1.5% SDS, 0.5 mg/mL proteinase K]. Components were added to cytoplasmic lysates to adjust to proteinase K lysis buffer concentrations. For samples from 6-well plates, 500 μL of cytoplasmic lysis buffer and 500 μL of proteinase K lysis buffer were used; 2.5 mL of each were used for 10-cm dish samples. Nuclear and cytoplasmic fractions were incubated for 2 hours at 37 °C with intermittent mixing. Nucleic acid was recovered through two extractions with 1 volume of 25:24:1 phenol:chloroform:isoamyl alcohol (PCA) and two extractions with 1 volume of chloroform.
Control SDA treatment of protein-free RNA extracted from cells
SM33 cells in 10-cm dishes were washed once in 5 mL PBS, then lysed in 2.5 mL proteinase K lysis buffer at 23 °C for 45 minutes. Nucleic acid was recovered through two extractions with 1 volume of PCA and two extractions with 1 volume of chloroform, and the resulting solution was buffer exchanged into PBS (PD-10 columns, GE Healthcare). The nucleic acid solution was incubated at 37 °C for 20 minutes before splitting into two equal volume portions (1.75 mL each). To one portion, a 1/9 volume of 100 mM SDA in DMSO was added, and a 1/9 volume of neat DMSO was added to the other. Each sample was incubated at 37 °C for 10 minutes in the dark. Each sample was spread evenly over a new 10-cm dish, placed on ice, and exposed to 3 J/cm2 of 365 nm wavelength ultraviolet light.
In-cell treatment with 5NIA SHAPE reagent
SM33 mouse embryonic stem cells were grown in 6-well plates. In-cell 5NIA treatment proceeded as described57. Cells were washed once in PBS, then covered with 900 μL serum-free embryonic stem cell media. To these cells, 100 μL of 250 mM 5-nitroisatoic anhydride (5NIA, Astatech) in anhydrous DMSO was added with concurrent manual mixing. For controls, 100 μL of neat DMSO was added instead. Cells were treated with 5NIA for 10 minutes at 37 °C, cells were washed once with 1 mL of PBS, then RNA was harvested from cells with TRIzol (Invitrogen) according to manufacturer’s specifications.
5NIA treatment of cell-extracted RNA
SM33 cells on 10-cm dishes were washed once in ice-cold PBS and resuspended in 2.5 mL ice-cold lysis buffer [40 mM Tris-HCl (pH 8.0), 25 mM NaCl, 6 mM MgCl2, 1 mM CaCl2, 256 mM sucrose, 0.5% Triton X-100, 1000 Units/mL RNasin (Promega), 450 Units/mL DNase I (Roche)]. Cells were lysed for 5 minutes at 4 °C with agitation. Nuclei were pelleted at 1500 ×g for 5 minutes at 4 °C, resuspended in 2.5 mL of proteinase K digestion buffer, and incubated for 45 minutes at 23 °C with agitation. RNA was extracted twice with one volume of PCA that had been pre-equilibrated with 1.1× folding buffer [111 mM HEPES (pH 8.0), 111 mM NaCl, 5.55 mM MgCl2], followed by two extractions with one volume of chloroform. RNA was buffer exchanged into 1.1× folding buffer over a desalting column (PD-10, GE Healthcare). After incubating for 20 minutes at 37 °C, RNA solution was split into two equal portions: One was added to a 1/9 volume of 250 mM 5NIA in DMSO, and the other was added to a 1/9 volume of neat DMSO. Both portions were incubated for 10 minutes at 37 °C.
In-cell crosslinking with photo-lysine
HEK293 cells in 6-well plates at ~60% confluency were washed once with PBS and then cultured for 16 additional hours in media with either 2 mM natural lysine or 2 mM photo-lysine (Medchem Express). After 16 hours, cells were washed once with 1 mL of PBS and then coated with a thin layer of 400 μL PBS. Cells were then crosslinked on ice with 10 J/cm2 of 365 nm wavelength UV light. Cells were washed once in PBS, pelleted at 1500 ×g, and resuspended in proteinase K lysis buffer. Proteins were digested for 2 hours at 37 °C. Nucleic acid was recovered through two extractions with 1 volume of PCA and two extractions with 1 volume of chloroform.
RNA precipitation and DNase treatment
Nucleic acids, including those treated after extraction from cells and those collected by Trizol or PCA after in-cell treatments, were precipitated by addition of a 1/25 volume of 5 M NaCl and 1 volume of isopropanol, incubation for 10 minutes at 23 °C, and centrifugation at 10,000 ×g for 10 minutes. The precipitate was washed once in 75% ethanol and pelleted by centrifugation at 7500 ×g for 5 minutes. Pellets from 6-well plates were resuspended in 50 μL of 1× DNase buffer and incubated with 2 units of DNase (TURBO, Thermo Fisher) at 37 °C for 1 hour. After the first incubation, 2 more units of TURBO DNase were added, and samples were incubated at 37 °C for 1 hour. Volumes were doubled for samples derived from 10-cm dishes. RNA was purified with Mag-Bind TotalPure NGS SPRI beads (Omega Bio-tek): A 1.8× volume of beads was added to DNase reactions and incubated 23 °C for 5 minutes followed by magnetic separation for 2 minutes. The solution was discarded, and beads were washed three times with 70% ethanol. RNA was eluted into 30 μL of nuclease-free water.
Antisense-mediated purification of Xist and XIST
In 50 μL of nuclease-free water, 10 μg of total nuclear RNA (from SM33 or HEK293 cells) was heated at 70 °C for 5 minutes and then immediately placed on ice for 2 minutes. To the RNA, 100 μL of 1.5× hybridization buffer [15 mM Tris-HCl (pH 7.0), 7.5 mM EDTA, 750 mM LiCl, 0.15% Triton X-100, 6 M urea], prewarmed to 55 °C, was added. RNA was pre-cleared for 15 minutes at 55 °C with 15 μL of streptavidin-conjugated magnetic beads (Dynabeads MyOne Streptavidin C1, Thermo Fisher) that were pre-washed and resuspended in 1× hybridization buffer. After magnetic separation, the pre-cleared supernatant was retained. Biotinylated antisense RNA capture probes26 (Guttman laboratory Caltech), specific to either mouse Xist or human XIST, were heated at 70 °C for 5 minutes, cooled on ice for 2 minutes, then diluted in 1× hybridization buffer. Each pre-cleared RNA sample was mixed with 72 ng of capture probes, and mixtures were incubated at 55 °C for 80 minutes with shaking. After probe hybridization, 30 μL of streptavidin magnetic beads, pre-washed and resuspended in 1× hybridization buffer, were added to RNA-probe mixtures, and incubation was continued at 55 °C with shaking for 20 minutes. Beads were captured by magnetic separation and washed twice with 200 μL of 1× hybridization buffer for 5 minutes each at 55 °C. Beads were resuspended in 60 μL of NLS elution buffer [20 mM Tris-HCl, pH 8.0, 10 mM EDTA, 2% N-lauroylsarcosine, 10 mM TCEP]. RNA was eluted from beads with three heating-cooling cycles where the temperature was ramped down from 95 °C to 4 °C and up to 95 °C in 1.5-minutes cycles. Beads were captured and RNA eluates saved. The same beads were then resuspended in 40 μL of NLS elution buffer and the elution procedure was repeated; the 40 μL eluate was added to the original 60 μL eluate. Captured RNA was purified (RNeasy MinElute Cleanup Kits, Qiagen). To reduce non-target RNA in the sample, RNAs were enriched again via a second capture: the procedure was identical to first capture except omitted the pre-clear step.
In vitro SDA crosslinking of T7-transcribed XIST E region with recombinant proteins
The E region of human XIST RNA (nucleotides 11900-13100 of NCBI NR_001564.2) was transcribed from a DNA template using T7 RNA polymerase (MEGAscript, Thermo Fisher), treated with DNase I (TURBO, Thermo Fisher), and purified via denaturing polyacrylamide gel electrophoresis. Product RNA was eluted from gels in nuclease-free water for 2 hours at 23 °C and concentrated with centrifugal filters (Amicon Ultra 10K, Millipore Sigma). Before SDA crosslinking, RNA was heat denatured at 98 °C for 2 minutes, then cooled on ice for 2 minutes before being diluted to 10 nM in 200 μl of RNP crosslinking buffer [1× PBS (pH 7.4), 1 mM MgCl2, 1 mM DTT] containing varying concentrations of recombinant XIST-binding proteins PTBP1, MATR3, or TIA1 (HEK293 recombinant, Origene) or BSA control protein (Millipore Sigma). RNPs were allowed to assemble for 30 minutes at 23 °C. 196 μl of mixtures were added to 4 μl of 100 mM SDA (in DMSO) in wells of a 6-well plate and incubated in the dark for 15 minutes at 23 °C. RNPs were crosslinked with 3 J/cm2 of 365 nm wavelength UV light. To digest unbound and crosslinked proteins, reactions were adjusted to 1.5% SDS, 20 mM EDTA, and 0.5 mg/ml proteinase K and incubated for 2 hours at 37 °C. RNA was purified once with 1.8× SPRI magnetic beads, purified again over an RNeasy MinElute column (Qiagen), and eluted into 14 μl of nuclease-free water.
MaP reverse transcription
MaP reverse transcription was performed using a revised protocol as described57,58. For smaller RNA targets (U1, RNase P, RMRP), 2 pmol of gene-specific primers (Table S3) were mixed with 500 ng of total nuclear RNA (or unfractionated total RNA when indicated). For MaP reverse transcription of enriched Xist/XIST RNAs or in vitro crosslinked XIST E region, 7 μL of final RNA product was mixed with 200 ng of random nonamer DNA oligonucleotides. When performing MaP reverse transcription on ribosomal RNA, 3 μg of total cytoplasmic RNA was mixed with 200 ng of random nonamers. To RNA-primer mixes, 20 nmol of dNTPs (5 nmol each base) was added (10 μL total volume of RNA, primers, and dNTPs), heated to 70 °C for 5 minutes, and then immediately placed at 4 °C for 2 minutes. To this template solution, 9 μL of freshly-made 2.22× MaP buffer [111 mM Tris-HCl (pH 8.0), 167 mM KCl, 13.3 mM MnCl2, 22 mM DTT, 2.22 M betaine] was added, and the mixture was incubated at 25 °C for 2 minutes. After adding 200 units of SuperScript II reverse transcriptase (Thermo Fisher), reaction mixtures were incubated for 10 minutes at 25 °C, 90 minutes at 42 °C, cycled 10 times between 42 °C and 50 °C with each temperature incubation 2-minutes long, and then heated to 70 °C for 10 minutes to inactivate enzyme. Reverse transcription reactions were buffer exchanged into TE buffer [10 mM Tris-HCl (pH 8.0), 1mM EDTA] (Illustra G-50 microspin columns, GE Healthcare).
Two-step PCR of small RNA MaP libraries
Small RNA sequencing libraries were generated using a two-step PCR strategy as described57,58. Briefly, 3 μL of cDNA from the reverse transcription reaction was used as template for step 1 PCR, using 20 cycles of gene-specific PCR (Q5 hot-start polymerase, New England Biolabs): 30 s at 98 °C, 20 × [10 s at 98 °C, 30 s at gene-specific annealing temperature, 20 s at 72 °C], 2 min at 72 °C. Each set of step 1 primers contained the same added handles to prime step 2 PCR (Table S3), in which Illumina adapters and multiplex indexing sequences were appended to the libraries. Step 1 PCR products were purified (SPRI beads, Mag-Bind TotalPure NGS, Omega Bio-tek, at a 1× ratio), and 2 ng of product was used as template for step 2 PCR. Step 2 PCR involved 30 s at 98 °C, 10 × [10 s at 98 °C, 30 s at 66 °C, 20 s at 72 °C], and 2 min at 72 °C. Step 2 PCR products were purified with SPRI beads at a 0.8× ratio and eluted into 15 μL of nuclease-free water.
Second-strand synthesis, fragmentation, and amplification of long RNA MaP libraries
For products of randomly primed MaP reverse transcription, buffer-exchanged cDNA was diluted to 68 μL with nuclease-free water. Each diluted cDNA was mixed with 8 μL of 10× Second Strand Synthesis Reaction Buffer and 4 μL Second Strand Synthesis Enzyme Mix (NEBNext, New England Biolabs), and reactions were incubated at 16 °C for 2.5 hours. The double-stranded DNA (dsDNA) products were purified with SPRI beads at a 0.8× ratio to favor longer products and exclude probe-templated products. Products were eluted into 15 μL of nuclease-free water. The dsDNA libraries were fragmented, multiplex indexed, and PCR amplified. To fragment libraries from total cytoplasmic RNA, 5 μL of 0.2 ng/μL dsDNA was combined with 10 μL of Tagment DNA Buffer and 5 μL of Amplicon Tagment Mix (Nextera XT DNA Library Prep Kits, Illumina). Mixtures were incubated at 55 °C for 5 min, then cooled to 10 °C. As soon as the temperature reached 10 °C, 5 μL of NT Buffer (Nextera XT DNA Library Prep Kits, Illumina) was added to neutralize the reaction, which was then incubated at 23 °C for 5 min. The entire reaction volume was used as a template for PCR with 15 μL of Nextera PCR Master Mix and 5 μL each of forward and reverse indexing primers (Nextera XT DNA Library Prep Kits, Illumina): 72 °C for 3 min, 95 °C for 30 s, 12 × [95 °C for 10 s, 55 °C for 30 s, 72 °C for 30 s], and 72 °C for 5 minutes. The final PCR products were purified with SPRI beads at a 0.65× ratio and eluted into 15 μL of nuclease-free water. For low concentration Xist and XIST capture libraries, 8 μL of capture product was fragmented with only 2 μL of Amplicon Tagment Mix, the concentration of index primers was halved during PCR, and PCR cycles were increased to 20.
Sequencing of MaP libraries
Size distributions and purities of amplicon and randomly primed libraries were verified (2100 Bioanalyzer, Agilent). Step 2 amplicon libraries (about 120 amol of each) were sequenced on a MiSeq instrument (Illumina) with 2 × 150 or 2 × 250 paired-end sequencing, depending on the length of the RNA target. Libraries derived from total cytoplasmic RNA were sequenced with 2 × 300 paired-end sequencing on a MiSeq instrument, combining reads from multiple runs until desired ribosomal RNA sequencing depth was achieved. Xist and XIST capture libraries were sequenced to desired depth via a combination of 2 × 300 paired-end runs on a MiSeq and 2 × 150 paired-end runs on a NextSeq 500 instrument.
Mutation counting and SHAPE profile generation with ShapeMapper 2 software
FASTQ files from sequencing runs, with the exception of capture libraries, were directly input into the ShapeMapper 2 software59 for read alignment and mutation counting. Crosslink-induced termination events are specifically omitted in this analysis as such stops do not contribute information beyond that measured in read-through events57. To ensure mutation rates were not affected by reduced fidelity at reverse transcription initiation sites, reads from capture libraries were trimmed by 14 nucleotides (primer length + 5 nts) after adapter sequences on each end. To accomplish this step for amplicon libraries, target FASTA files input to ShapeMapper 2 had primer-overlapping sequences and the first 5 nucleotides transcribed in RT set to lowercase, which eliminates these positions from analysis. To expedite analysis of long RNAs like Xist/XIST, corresponding FASTQs were split into ~10 subsets and run in multiple parallel ShapeMapper 2 instances before having their outputs recombined into single profiles. ShapeMapper 2 was run with --min-depth 5000 and --output-classified flags with all other values set to defaults. In an RNP-MaP experiment, the SDA+UV-treated samples are passed as the “modified” samples and UV-only treated samples as “unmodified” samples. The outputs “profile.txt”, “parsed.mut”, and “.map” files are required for RNP-MaP site, RNP-MaP correlation, and SHAPE analyses.
Identification of low SHAPE, low Shannon entropy regions of Xist and XIST using SuperFold
The SuperFold analysis software16 was used with in-cell and cell-extracted 5NIA experimental SHAPE data from mouse Xist and human XIST to inform RNA structure modeling by RNAStructure60. Default parameters were used to generate base-pairing probabilities for all nucleotides (with a max pairing distance of 600 nt), Shannon entropies for each nucleotide, and minimum free energy structure models.
ΔSHAPE of mouse RNase P and Rmrp
Normalized SHAPE reactivities for 5NIA-treated mouse RNase P and Rmrp RNAs were compared between in-cell treated samples and those treated after cell extraction using the ΔSHAPE program61. Default parameters were used, and the 5′ primer sequence, the 3′ primer sequence, and the first 5 nucleotides transcribed during the reverse transcription step were all masked to exclude them from analysis. Only nucleotides that passed the included Z-factor and standard score significance testing were mapped as ΔSHAPE sites.
Post-processing of mutation frequencies into RNP-MaP reactivities
Per-nucleotide mutation frequencies (number of mutation events/effective read depth) for both crosslinked (SDA+UV-treated) and uncrosslinked (UV-treated) samples were calculated from output ShapeMapper 2 profiles. RNP-MaP “Reactivity” was computed as the ratio of nucleotide crosslinked mutation frequency to uncrosslinked mutation frequency (SDA+UV rate/UV only rate). Exceptions are in Figure 1C and S1, where “Reactivity” refers to the ratio with a no treatment control as the denominator (treatment rate/no treatment rate). To be designated as RNP-MaP sites, nucleotide positions had to pass three quality filters: (1) sites were required to have at least 50 more mutation events in the SDA+UV-treated sample than the UV-treated sample; (2) site reactivities had to exceed the nucleotide-dependent empirical thresholds described in the next section; and (3) nucleotide reactivities were required to achieve a Z-factor greater than zero. .
Empirical derivation of RNP-MaP site nucleotide reactivity thresholds
Two biological replicates of RNP-MaP were performed on human U1 snRNA, RNase P RNA, and 18S and 28S rRNAs, each a part of RNA-protein complexes where atomic resolution structural data is available, enabling separation of nucleotides into two groups: those within 10 Å of protein (<10 Å) and those further than 10 Å from protein (>10 Å). For U1 snRNA, the binding site of the SNUPN protein has been mapped by crosslinking and mass spectrometry19, and distances between the three nucleotides surrounding the crosslink site and nearest amino acids were assumed to be less than 4 Å. For each RNA replicate, reactivities were further grouped by nucleotide identity (U, A, C, and G). The 90% reactivity value of nucleotides in a >10 Å group were set as background thresholds (BGX>10) and compared to the median (MEDall X) and standard deviations (SDall X) of reactivities for all nucleotides included in both <10 Å and >10 Å groups to create relative threshold factors (TX):
Relative threshold factors for each nucleotide from all eight replicates of the four RNAs were then averaged together, weighted by the number of nucleotides measured in each RNA, to obtain final empirically derived nucleotide relative threshold factors: 0.59 for U, 0.29 for A, 0.93 for C, and 0.78 for G. These factors represent the number of standard deviations from the median of nucleotide reactivity that must be achieved to be considered an RNP-MaP site. Factors can be applied to any RNA: to get exact nucleotide thresholds for an RNP-MaP experiment, the median reactivities for each nucleotide group (U, A, C, or G) are multiplied by their corresponding threshold factors. Factors were calculated from existing comprehensive datasets, however including more data from other RNPs or improving upon existing atomic resolution RNP structures could increase the precision of these threshold factors in the future.
Graphical display of RNP-MaP Reactivities and crosslinking sites
Violin plots representing distributions of RNP-MaP reactivities were generated using the vioplot package in R through the web tool BoxPlotR62. RNP-MaP crosslinking sites were superimposed onto atomic resolution structure models using PyMol63. Secondary structure projection images were generated using the (VARNA) visualization applet for RNA64.
RNP-MaP correlation analysis
Correlations between RNP-MaP sites were computed over 3-nucleotide windows using a previously described G-test framework (RingMapper)20. Windows were required to be separated by > 4 nucleotides, jointly covered by more than 10,000 sequencing reads, jointly co-mutated >50 times, and have background mutation rates below 6% (Fig. S2). Pairs of windows exhibiting G-test statistics > 20 (P < 10−5) in the SDA+UV treated sample and G < 10.83 (P > 0.001) in the UV-only sample were determined to be significantly correlated. Current technical limitations of MaP reverse transcription processivity (500-600 nucleotides) and sequencing instrument clustering (< 1000 nucleotides) limit distances of readily measured correlations to < 500 nucleotides.
RNase P and RMRP structural alignment
Corresponding helices, loops, and intervening regions in RNase P and RMRP RNAs were separated into structural domains (Fig. 3) and separately aligned with MUSCLE65. Region alignments were recombined to create the final alignment (Supplemental Data S1).
Calculation of Xist/XIST RNP-MaP site, correlation strength, and eCLIP site densities
To calculate the RNP-MaP site density for Xist and XIST (Fig. 4a and S8), nucleotides whose reactivities were in the top 5% of all reactivities (U, A, C, and G nucleotides evaluated separately) were identified. Site density was defined as the number of nucleotides in a centered 51-nucleotide window that were top 5% sites. Correlation strength density was defined as the sum of the mutual information of all nucleotides within the window normalized to (i.e., divided by) the read depth of the central nucleotide. We selected high confidence K562 cell eCLIP sites66 (Supplemental Data S2) within XIST that passed Irreproducible Discovery Rate thresholds (see www.encodeproject.org/eclip), meaning that sites were defined by signal peaks of similar amplitude in both eCLIP replicates. While other eCLIP experiments have been performed in more directly XIST-relevant cell lines (including human embryonic stem cells), the limited number of proteins tested and the lack of a shared systematic approach did not support inclusion in this analysis. The eCLIP site density (Fig. 4b) was defined as the total number of observed eCLIP sites within the 51-nucleotide window. For densities in Fig. 6, Fig. S9, and Fig. S10, all RNP-MaP sites were included (because signal only comes from a single protein) and centered nucleotide windows were shortened to 25 long (since the E region is only 1200 nts).
Identification of conserved sequence regions between mouse Xist and human XIST
To identify and rank areas of significant conservation between mouse Xist (NCBI NR_001463.3) and human XIST (NCBI NR_001564.2) RNA sequences, we performed a local alignment (BLASTn67) and retained all segments with E-values above 0 and with lengths > 100 nucleotides and ranked these segments by alignment bitscore.
Network analysis of XIST RNP-MaP correlation-linked eCLIP sites
To create a list of protein-linking correlations, we first counted the number of times nucleotides within our high confidence eCLIP sites were correlated in RNP-MaP data (links) and measured the summed total of mutual information within links. To ensure that links were not simply a product of eCLIP site number and proximity or the average length and density of RNP-MaP correlations, we randomly shuffled the location of RNP-MaP correlations and counted links achieved between protein pairs iteratively 2000 times, generating p-values for each protein pairing based on the number of links between them and the strength of those links (Table S1). Resulting links between protein pairs with p-values less than 0.05 were then used as edges connecting nodes (proteins) on a network map (Fig. 5). A maximum modularity of the network (0.419), weighted by the strengths of included links (mutual information) and without changing resolution, was calculated using Gephi68, and node sizes were adjusted manually to convey indicated relationships.
Identification of RNP-MaP enriched motifs in the XIST E region
MEME69 was used to identify motifs enriched by RNP-MaP in vitro. We first expanded each RNP-MaP nucleotide into 9-mer sites (extending by 4 on either side). Overlapping sites were combined iteratively until no overlapping sequences remained. Combined sites were used as MEME input in classic mode using a 0-order model of sequences, allowing for any number of motif repetitions in each sequence, and explicitly looking for 9-mer motifs. The top two motifs were retained for each in vitro experiment, and locations of all matching motifs in XIST E region were found using FIMO70 with a p-value threshold of 10-3. For the in-cell experiment, only sites from the E region were considered in MEME, and the first and fourth most significant motif (class 2 and class 1, respectively) were included in Fig. 6, S9, and S10. Use of background sets created from the XIST E region (nt 11900-13100) or increasing the order of the background model further strengthened the significance of class 2 motifs and weakened the significance of class 1 motifs.
XIST RNA reporter plasmid design
To create XIST region-containing reporters, we used inverse PCR and re-ligation to insert a multiple cloning site into the 3′ end of the pNL 3.2.CMV vector (Promega) between XbaI and FseI sites and to add the second intron from human HBA1 at nucleotide position 196 in the nanoluciferase coding region (native Xist/XIST is spliced in its central region). Plasmids with varying regions of XIST were subcloned into the XhoI and KpnI restriction sites, and a control plasmid was generated through inverse PCR (Table S3).
Plasmid transfection and purification of XIST reporter RNA for qPCR
HEK293 cells were plated at 100,000 cells/mL in 2-mL volumes per well of 6-well plates and then cultured for 24 hours at 37 °C. Each well was then transfected with a mixture of 0.6 μg of reporter plasmid and 1.8 μL of FuGENE 6 transfection reagent (Promega) in 200 μL of serum-free DMEM, and cells were cultured for an additional 48 hours at 37 °C. Transfected cells were pelleted at 1500 ×g for 5 minutes at 4 °C, washed once in cold PBS and pelleted again, and resuspended in 500 μL cytoplasmic lysis buffer. Cells were lysed for 10 minutes at 4 °C with agitation. Nuclei were pelleted at 1500 ×g for 5 minutes at 4 °C, and cytoplasmic lysates were separated into new tubes. Nuclei were washed once in low-salt solution, incubated with agitation at 4 °C for 2 minutes, pelleted again, and then resuspended in 500 μL proteinase K lysis buffer. NaCl, EDTA, SDS, and proteinase K were added to cytoplasmic lysates up to proteinase K lysis buffer concentrations. Nuclear and cytoplasmic fractions were incubated for 2 hours at 37 °C with intermittent mixing. Nucleic acid was recovered through two extractions with 1 volume of 25:24:1 PCA, two extractions with 1 volume of chloroform, and precipitation with 1/25 volume of NaCl and 1 volume of isopropanol.
siRNA transfection and purification of endogenous RNA for qPCR
HEK293 cells were plated at 20% confluence into wells of either 6-well (2 mL, for qPCR) or 12-well (1 mL, with each well containing a poly-L-lysine coated #1.5 coverslip [Neuvitro] plates for microscopy) and then cultured for 24 hours at 37 °C. Media was replaced with DMEM + 10% FBS lacking antibiotics, and each well was then transfected with a mixture of OPTIMEM (Gibco), siRNA, and Lipofectamine RNAiMAX (Thermo Fisher) according to manufacturer’s protocol. Final siRNA concentrations were 20 nM in transfections, and siRNAs included the MISSION siRNA Universal Negative Control #1 (Millipore Sigma), PTBP1 siRNA SASI_Hs01_00216644 (Millipore Sigma), PTBP2 siRNA SASI_Hs01_00201967 (Millipore Sigma), and MATR3 siRNA HSS114732 (Thermo Fisher). For siPTBP samples, siRNAs for both PTBP1 and PTBP2 were included. 24 hours after transfection, media was exchanged for new media including antibiotics. 72 hours after transfection, slides (using cells from 12-well plates) were prepared for microscopy, or cellular RNA was harvested (from 6-well plates; Trizol, Invitrogen) for qPCR.
Plasmid transfection for XIST reporter microscopy
HEK293 cells (1 mL) were plated at a concentration of 10,000 cells/mL in wells of a 12-well plate, with each well containing a poly-L-lysine coated #1.5 coverslip (Neuvitro). After 24 hours of growth, 50 μl of transfection mix [20 μl of 15 ng/μl reporter plasmid, 0.9 μl of Fugene 6 (Promega), and 79 μl of DMEM] was added to cells.
qPCR of endogenous and reporter RNAs
Equal quantities of RNA (30 ng) from nuclear, cytoplasmic, or total Trizol RNA fractions were used generate first-strand cDNA in random hexamer-primed reverse transcription reactions (SuperScript II, Thermo Fisher). Triplicate mixtures of 2.5 μL of template cDNA, 2.5 μL of 2 μM primers, and 12.5 μL of Maxima SYBR Green qPCR Master Mix (Thermo Fisher) in 25 μL total reactions were prepared for each sample. Reaction mixtures from matched no-reverse transcriptase controls were also prepared. Primer sets (Table S3) included those specific to the reporter gene, an endogenous RNA, and 18S ribosomal RNA (as a normalization control). qPCR was performed on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher) with steps of 5 min at 95 °C, 40 cycles of [15 s at 95 °C, 30 s at 65 °C, 40 s at 72 °C], using a melting curve to confirm single major products. Fluorescence readings were taken at elongation steps (72 °C). All specific signals were observed at least 8 cycle thresholds earlier (256-fold more signal) than no-reverse transcriptase controls. Signals were averaged across triplicate qPCRs, normalized to 18S rRNA signal, then normalized to either control reporter or control siRNA signals, depending on the experiment. XIST reporters were expressed at 50 % the level of endogenous XIST, as assessed by RT-qPCR.
RNA FISH of XIST RNA and XIST RNA reporters
FISH probes labeled with Quasar 570 Dye and antisense to human XIST (SMF-2038-1) and the nanoluciferase mRNA (custom) were ordered from LGC Biosearch. Custom design parameters were masking level 5, oligo length 19, and minimum spacing length 2. At 72 (for siRNA) or 48 (for reporter expression) hours post-transfection, each well was washed once with 1 ml PBS, fixed with 1 ml of 3.7% formaldehyde in PBS for 10 minutes at room temperature, then washed twice with PBS. Cells were permeabilized with 1 ml of 70% ethanol overnight at 4 °C. After removing ethanol, cells were incubated in wash buffer 1 [20% Wash Buffer A (LGC Biosearch), 10% formamide] for 5 minutes at room temperature, and then coverslips were transferred to a humidified chamber with cells facing down onto 100 μl of hybridization buffer [90% Stellaris RNA FISH Hybridization Buffer (LGC Biosearch), 10% formamide, 125 nM antisense probes]. After overnight incubation in the dark at 37 °C, coverslips were transferred into 12-well dishes and incubated in 1 ml of wash buffer 1 for 30 minutes at 37 °C in the dark, then counterstained for 30 minutes at 37 °C with 5 ng/μl DAPI in 1 ml of wash buffer 1. Coverslips were washed a final time in 1 ml of Wash Buffer B (LGC Biosearch) before being mounted onto a microscope slide with 12 μl of Vectashield Mounting Medium (Vector Laboratories) and sealed. RNA FISH z-stack images of XIST reporters were captured using a 100×/1.3 oil objective on an Olympus IX81 Microscope and were deconvoluted using the AutoQuant X software. Z-stack images of native XIST particles were captured using a 100×/1.3 oil objective on a Nikon Eclipse Ti Microscope. Z-stacks were collapsed into maximum intensity projections and quantified using the Fiji software. XIST foci density was measured as the pixel area within XIST foci passing a fluorescence intensity background threshold (3680).
DATA AVAILABILITY
Raw and processed sequencing datasets analyzed in this report will be made available upon reasonable request and have been deposited in the Gene Expression Omnibus database (GSE152483).
CODE AVAILABILITY
ShapeMapper2, deltaSHAPE, SuperFold, and RingMapper software used for analysis are available here (http://weeks.chem.unc.edu/software.html) and here (https://github.com/Weeks-UNC). MEME, VARNA, PyMol, and Gephi are third party open source software.
Supplementary Material
Figure S1. RNP-MaP site and correlation determination. Scheme outlining determination of (a) RNP-MaP sites and (b) correlations. For (a), depth threshold and mutation count above background threshold were set to eliminate noise at signal margins. A Z-factor above 0 is achieved only when 99% confidence intervals of UV only and SDA + UV rates do not overlap. To limit false positives, nucleotide-specific thresholds were set at the level of highest reactivity among nucleotides distant from proteins in RNPs of known structure (U1, RNase P, 18S, and 28S), expressed as the number of standard deviations from median reactivities of all instances of each nucleotide (see Methods). For (b) 3 nt windows and 4 nt separations were set to accommodate the size of longer and more complex mutation and deletion events observed during MaP reverse transcription. Thresholds of 10,000 depth, 6% background mutation rate, and 50 co-mutation events limit noise and false positives.
Figure S2. RNP-MaP discriminates protein-bound nucleotides independent of local RNA structure and nucleotide identity. (a) Correlation (Pearson’s r) between log2(reactivity) of nucleotides from biological replicate RNP-MaP experiments in HEK293 cells on U1, RNase P, RMRP, 18S, and 28S RNAs. Correlations are shown for all nucleotides (blue) and RNP-MaP sites (green). The number of nucleotides measured for each RNA is indicated above each graph in parentheses. (b) Nucleotide log2(reactivity) from RNP-MaP experiments as a function of distance to protein lysine side chain amine groups. Data are from two biological replicates on human U1, RNase P, 18S, and 28S RNAs, obtained in HEK293 cells. Nucleotides analyzed (n) include 1260 RNP-MaP sites (green) and 11,184 non-sites (black). The significance of the difference between RNP-MaP site and non-site distance distributions is shown (Mann-Whitney U-test, one-sided). (c) RNP-MaP site and non-site nucleotide log2(reactivity) from experiments in HEK293 cells (18S, 28S, U1, RNase P, RMRP, XIST RNAs) or SM33 cells (Xist RNA), grouped as unpaired nucleotides (U) and paired nucleotides (P). Violin plots are shown. For the top row, from left to right, the number of nucleotide reactivities included in each distribution (n) was 62, 46, 126, 16, 176, 70, 326, 22, 138, 68, 232, and 22. For the bottom row, n was 1189, 272, 1991, 171, 2905, 375, 4039, 198, 8867, 1568, 6350, 558, 8695, 2900, 6247, and 866. RNP-MaP sites are significantly more reactive than non-sites (top row p = 1.8e−9, 2.4e−8, 1.2e−22, 4.2e−15, 6.4e−22, 1.2e−15; bottom row p = 4.5e−112, 9.1e−74, 5.4e−124, 3.8e−87, 0.0, 0.0, 0.0, 0.0; one-sided Kolmogorov-Smirnov test). Reactivities of single-stranded RNP-MaP sites are significantly higher than those of paired sites (top row p = 4.1e−4, 6.2e−3, and 0.01; bottom row p = 2.7e−3, 0.12, 0.26, and 2.5e−23; one-sided Kolmogorov-Smirnov) except for 28S (p = 0.12) and Xist (p = 0.26), but both single-stranded and base paired regions clearly show higher reactivity at RNP-MaP sites than non-sites. (d) RNP-MaP nucleotide log2(reactivity) grouped as protein-proximal (< 10 Å) and protein-distant (> 10 Å) nucleotides. Reactivities are shown for each nucleobase. Data obtained for U1, RNase P, 18S, and 28S RNAs; in HEK293 cells. Nucleobase-specific thresholds (black dotted lines) for RNP-MaP sites were obtained from the standard deviations of reactivity for all of each nucleobase within an experiment, multiplied by corresponding empirically-derived threshold factors (U – 0.59, A – 0.29, C – 0.93, G – 0.78); these values are added to the corresponding nucleobase reactivity medians to compute final thresholds. The total number of nucleotides analyzed in each distribution (n) are: U1 (28, 36, 28, 28, 20, 38, 40, and 32), RNase P (56, 46, 70, 42, 76, 98, 96, and 110), 18S (491, 279, 520, 293, 483, 490, 631, and 436), and 28S (750, 502, 843, 588, 1114, 1078, 1446, and 1196). For violin plots in panels c and d, white circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values.
Figure S3. RNP-MaP signal in living cells is dependent on SDA and UV. (a) Nucleotide mutation rates after MaP reverse transcription of U1 and RNase P RNA. Combinations of UV (3 J/cm2) and SDA (10 mM) were applied directly to SM33 cells (in-cell) or to (protein-free) RNA extracted from cells (extracted). Among extracted controls, slightly enhanced reactivity at RNP-MaP nucleotides was detected in the U1 RNA, likely a result of residual undigested Sm proteins. The number of nucleotide mutation rates included in each distribution (n) are 13 RNP-MaP sites and 118 non-sites in U1 RNA and 34 sites and 256 non-sites in RNase P RNA (b) MaP nucleotide mutation rates for Rmrp RNA in SM33 cells as a function of SDA and UV dose. When SDA concentration was varied, UV dose was 3 J/cm2. When UV energy was varied, SDA concentration was 10 mM. In the UV gradient, n included 185 non-sites and 54 sites, while in the SDA gradient n included 194 non-sites and 45 sites. For violin plots, white circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values. P-values (Kolmogorov-Smirnov test, one-sided) are indicated. If p-values are calculated comparing each condition to their untreated counterparts (in a), only reactivities of RNP-MaP nucleotides in the SDA and UV treated in-cell samples (and the U1 extracted sample) are significantly increased (p <0.05).
Figure S4. Orthogonal chemical probing and metabolic labeling approaches corroborate RNP-MaP. (a) Comparison with ∆SHAPE61. ΔSHAPE identifies changes in SHAPE reactivity between RNA in cells and in solution, identifying regions of RNA that interact with cellular components, including proteins. RNP-MaP and ΔSHAPE sites are superimposed on secondary structure diagrams for mouse Rmrp and RNase P RNAs: ΔSHAPE sites overlapped RNP-MaP sites in mouse Rmrp and RNase P RNAs. Of 65 RNP-MaP sites (green shading) in Rmrp and RNase P RNAs, 22 (34%) are ΔSHAPE sites (lines), and 52 (80%) are within five nucleotides of ΔSHAPE sites by contact distance (which counts distances across base pairs in the accepted secondary structure as a length of one nucleotide). In-cell SHAPE protections (green) and enhancements (purple) are shown as lines. Primer-binding sequences were excluded from analysis (gray spheres), and solid grey and dotted grey lines indicate pseudoknot helices and predicted tertiary base-pair (inferred from human RNase P structure), respectively. (b) Comparison with metabolic labeling by photo-lysine. Photo-lysine contains a diazirine group, which is photo-activated, like SDA, with 365 nm UV light. Photo-lysine metabolic labeling substitutes for the first NHS-ester reaction step in SDA treatment. UV activation of photo-lysine-labeled cells yielded reactivity patterns similar to RNP-MaP for human RMRP RNA, albeit at reduced rates. Photo-lysine is less reactive with RNA than is SDA (left), as it is likely incorporated into proteins with low efficiency and requires a shorter crosslinking distance. Growing cells with a 1:4 natural lysine to photo-lysine ratio before crosslinking eliminates mutation signal (data not shown), confirming that observed signal is from protein-incorporated photo-lysine and not free photo-lysine in solution. For violin plot, white circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values. The number of nucleotide mutation rates included in each distribution (n) is 230. Correlation (Pearson’s r) between SDA and photo-lysine reactivity (right) is shown. (c) RNP-MaP sites (green circles) called from reaction in-cell with SDA (left) or photo-lysine (right), superimposed on secondary structure of RMRP. All photolysine reactive regions are also reactive by SDA.
Figure S5. RNP-MaP interaction networks are highly reproducible. Top 10% of RNP-MaP correlations by mutual information (arcs) and the per-nucleotide summed mutual information for RNP-MaP correlations (purple peaks) from two biological replicates, plotted by nucleotide position (nt) for human U1, RNase P, and RMRP RNAs. Experiments performed in HEK293 cells.
Figure S6. RNP-MaP sites are conserved between mouse and human RNAs. RNP-MaP sites for mouse and human homologues of RNase P and RMRP, superimposed on secondary structure projections of each RNA. Structural domains are labeled in gray, the percent sequence identities between RNAs are indicated, and gray lines denote pseudoknot helices. Proteins in close proximity to each RNP-MaP site on RNase P (based on distance to lysine amine groups) are labeled (green lettering). Experiments performed in SM33 or HEK293 cells.
Figure S7. Gene-specific and random priming strategies for RNP-MaP library preparation. Schemes outlining generation of RNP-MaP sequencing libraries from (a) small RNA and gene- or region-specific targets and (b) long RNA targets. Gene-specific priming is recommended for RNAs or regions of an RNA < 500 nucleotides in length, and enrichment and random priming is suggested for longer RNAs.
Figure S8. RNP-MaP captures protein interaction information across very long non-coding RNAs. (a) Read depth thresholds observed mouse Xist (orange) and human XIST (blue), shown on per-nucleotide basis. Read depths exceeded 10,000 for most nucleotides. Tandem repeat sequence arrays A-F (white boxes with black letters), regions homologous to repeat regions (white letters), the D region core (white box with thick black outline), and exon-exon junctions (thick vertical black lines) are shown for Xist and XIST. (b) Distribution of RNP-MaP sites as a function of nucleotide identify. RNP-MaP signals are observed at all four nucleotides, with an enrichment at adenosine and uridine. (c) RNP-MaP signal as a function of extent of local RNA structure. RNP-MaP shows relatively low biases, as shown by the percentages (bar graphs) of RNP-MaP site nucleotides that also have low SHAPE reactivity (Lo-SHAPE), have low SHAPE reactivity and low modeled Shannon entropy (Lo-SS), or are base paired in minimum free-energy models (paired). (d) Positions of 151 high confidence eCLIP sites for 30 XIST-binding proteins analyzed in this work, shown with RNP-MaP site density (top 5% reactive sites per 51 nucleotides) aligned to the XIST sequence. Of note, the eCLIP sites for human HNRNPK protein, whose mouse homologue mediates recruitment of Polycomb through the B and C regions of mouse Xist28,72, closely mirror RNP-MaP site density across the human XIST D region and overlap human XIST B and C regions, supporting a model wherein the B, C, and D regions of Xist/XIST have analogous functions73. Protein eCLIP sites are organized on density plots by identified community and ranked (from top to bottom) by decreasing summed mutual information of intra-community correlations.
Figure S9. Detection of PTBP1, MATR3, and TIA1 sites in the XIST E region in cells and in vitro. Detection of RNP-MaP sites (represented as 25 nt window densities) in the XIST region E RNA based on experiments in-cell (orange) or under simplified conditions with synthetic RNA and the indicated recombinant protein. In vitro RNP-MaP densities (gray shading with black and colored lines) are shown as a function of increasing amounts of PTBP1, MATR3, TIA1, and (negative control) BSA proteins (only highest BSA control concentration is shown in black, no effect seen). Locations of called eCLIP sites (thick colored rectangles) and average eCLIP replicate read depth (matching colored lines) are shown above each density plot, and class 1 and 2 motifs (small gray and black boxes) for each protein are shown below each density plot. The average transcriptome-wide eCLIP replicate read depth value (99.99 percentile) is indicated by a horizontal tick at right side of eCLIP intensity plots. RNP-MaP sites rely on read-through of modifications during RT while eCLIP reads terminate at modifications: thus, as expected, peaks of RNP-MaP density for PTBP1, MATR3, and TIA1 lie at the 5’ ends of associated eCLIP read peaks. Locations of eCLIP sites for other mapped proteins are shown at bottom. In-cell and 1000 nM in vitro data are from same experiments as represented in Figure 6a and 6b.
Figure S10. Extended data in support of Figure 6. (a) Correlation of log10(mutation rates) of T7-transcribed XIST E region RNA (protein-free); comparing RNA treated with SDA and UV versus RNA treated with UV alone. MaP RT-induced mutation rates for n = 1177 nucleotides are identical (Pearson’s r = 0.994). (b) log2(reactivity) of RNP-MaP site nucleotides (red) as a function of indicated increasing protein concentration, compared to non-RNP-MaP nucleotides (black). Reactivities shown are averages for all nucleotides of each indicated nucleobase (right); the number of nucleotides present in each group in each experiment (n) is indicated. (c) Occurrence of PTBP1, MATR3, and TIA1 eCLIP sites in the Xist E region relative to uridine density. eCLIP sites are only found within the uridine-rich region (containing class 1 motifs) but are absent in the uridine-poor region where RNP-MaP-identified class 2 motifs predominate. (d) Relative expression levels of reporter RNAs containing non-XIST control, XIST E region, and other highly interactive XIST (by RNP-MaP) sequences, shown for both cytoplasmic and nuclear fractions. E region insert includes nts 11900-13100 of XIST. Nucleotide positions in XIST for other inserted protein-interactive sequences are indicated. qPCR signal from XIST reporters were (first) normalized to co-transfected control reporter RNA signal (firefly luciferase) in each fraction, and then each reporter signal was normalized to the non-XIST insert control. Error bars represent standard error of the mean, and means (bar heights) of replicate measurements are shown (n=3, open circles). All differences between E region reporter and other constructs are significant (cytoplasmic fraction P = 7.0e−4, 2.8e−3, 2.2e−3, and 1.2e−3; nuclear fraction P = 0.28, 2.0e−4, 9.0e−4, and 1.7e−2; Student’s t-test, two-sided), except for the difference between XIST E and Non-XIST nuclear fractions (P = 0.28). Reporter RNA containing the XIST E region was expressed at significantly lower levels than RNA reporters containing other highly interactive XIST sequences, yet E region reporter was the only construct that directed foci-formation (Fig. 6b).
Table S1. Totals and summed mutual information (MI) of correlations linking pairwise combinations of XIST-binding protein eCLIP sites. P-values for both attributes were generated from 2000 randomizations (without replacement) of measured correlation locations along XIST (for nucleotides with sufficient read depth only). All measured correlations are included.
Table S2. Pairs of protein eCLIP sites significantly excluded (P-values > 0.95) from RNP-MaP data based on both total number of correlations and summed mutual information (MI) strength. Associated network communities, total numbers of each community pairing, and the percent of pairings that were significantly excluded are also listed.
Table S3. DNA sequences of MaP library preparation primers, plasmid construction, and qPCR primers. Sequences inserted during plasmid construction are separated from flanking plasmid sequences by dashes (−).
Supplemental Data S1. Structure-based sequence alignment of human RNase P and RMRP RNAs.
Supplemental Data S2. List of high confidence eCLIP sites on XIST organized by protein.
ACKNOWLEDGEMENTS
The work was supported by grants from the US National Science Foundation (MCB-1121024) and National Institutes of Health (R35 GM122532) to K.M. Weeks. C.A.Weidmann is a postdoctoral fellow of the American Cancer Society (ACS 130845-RSG-17-114-01-RMC). J.M.Calabrese was supported by NIH grant R01 GM121806. Xist and XIST antisense probes were provided by the M. Guttman laboratory (CalTech), and we thank M. Blanco (CalTech) for his initial support in their application. XIST eCLIP data from published work were provided upon request by the G.W. Yeo laboratory (UCSD), and we thank G.W. Yeo (UCSD), M. Corley (UCSD), and D. Sprague (UNC) for support in formatting these data for integration into this work and for helpful comments on the project.
Footnotes
COMPETING INTERESTS
A.M.M. is an advisor to and K.M.W. is an advisor to and holds equity in Ribometrix, to which mutational profiling technologies have been licensed.
REFERENCES
- 1.Gehring NH, Wahle E & Fischer U Deciphering the mRNP Code: RNA-Bound Determinants of Post-Transcriptional Gene Regulation. Trends in Biochemical Sciences vol. 42 369–382 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Guttman M & Rinn JL Modular regulatory principles of large non-coding RNAs. Nature vol. 482 339–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anger AM et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Pomeranz Krummel DA, Oubridge C, Leung AKW, Li J & Nagai K Crystal structure of human spliceosomal U1 snRNP at 5.5 resolution. Nature 458, 475–480 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo Y, Oubridge C, van Roon AMM & Nagai K Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5’ splice site recognition. Elife 4, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J et al. Cryo-EM Structure of the Human Ribonuclease P Holoenzyme. Cell 175, 1393–1404.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Ule J, Hwang HW & Darnell RB The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb. Perspect. Biol 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garzia A, Meyer C, Morozov P, Sajek M & Tuschl T Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods vols 118–119 24–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler EC, Van Nostrand EL & Yeo GW Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip. Rev. RNA 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeberg MA et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 14, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schueler M et al. Differential protein occupancy profiling of the mRNA transcriptome. Genome Biol. 15, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramanathan M, Porter DF & Khavari PA Methods to study RNA–protein interactions. Nature Methods vol. 16 225–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mädler S, Bich C, Touboul D & Zenobi R Chemical cross-linking with NHS esters: A systematic study on amino acid reactivities. J. Mass Spectrom 44, 694–706 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Das J Aliphatic diazirines as photoaffinity probes for proteins: Recent developments. Chemical Reviews vol. 111 4405–4417 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Krüger DM, Neubacher S & Grossmann TN Protein–RNA interactions: Structural characteristics and hotspot amino acids. RNA 24, 1457–1465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smola MJ, Rice GM, Busan S, Siegfried NA & Weeks KM Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc 10, 1643–1669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smola MJ et al. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U. S. A 113, 10322–10327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang T, Li XM, Bao X, Fung YME & Li XD Photo-lysine captures proteins that bind lysine post-translational modifications. Nat. Chem. Biol 12, 70–72 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kühn-Hölsken E et al. Mapping the binding site of snurportin 1 on native u1 snRNP by cross-linking and mass spectrometry. Nucleic Acids Res. 38, 5581–5593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustoe AM, Lama NN, Irving PS, Olson SW & Weeks KM RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci 116, 24574–24582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.So BR et al. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol 23, 225–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Will C In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 24, 4614–4623 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esakova O & Krasilnikov AS Of proteins and RNA: The RNase P/MRP family. RNA vol. 16 1725–1747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perederina A, Berezin I & Krasilnikov AS In vitro reconstitution and analysis of eukaryotic RNase P RNPs. Nucleic Acids Res. 46, 6857–6868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahakyan A, Yang Y & Plath K The Role of Xist in X-Chromosome Dosage Compensation. Trends in Cell Biology vol. 28 999–1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engreitz JM et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science (80-. ). 341, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wutz A, Rasmussen TP & Jaenisch R Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet 30, 167–174 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Colognori D, Sunwoo H, Kriz AJ, Wang CY & Lee JT Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol. Cell 74, 101–117.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridings-Figueroa R et al. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 31, 876–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunwoo H, Colognori D, Froberg JE, Jeon Y & Lee JT Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. U. S. A 114, 10654–10659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HJ et al. En bloc and segmental deletions of human XIST reveal X chromosome inactivation-involving RNA elements. Nucleic Acids Res. 47, 3875–3887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nesterova TB et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brockdorff N Local tandem repeat expansion in Xist RNA as a model for the functionalisation of ncRNA. Non-coding RNA 4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis CA et al. The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Nostrand EL et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moindrot B et al. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 12, 562–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciaudo C et al. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2, 0874–0882 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakaguchi T et al. Control of Chromosomal Localization of Xist by hnRNP U Family Molecules. Developmental Cell vol. 39 11–12 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Patil DP et al. M6 A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao R et al. Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap K et al. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 72, 525–540.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rayman JB, Karl KA & Kandel ER TIA-1 Self-Multimerization, Phase Separation, and Recruitment into Stress Granules Are Dynamically Regulated by Zn 2+. Cell Rep. 22, 59–71 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Gallego-Iradi MC et al. N-terminal sequences in matrin 3 mediate phase separation into droplet-like structures that recruit TDP43 variants lacking RNA binding elements. Lab. Investig. 99, 1030–1040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Attig J et al. Heteromeric RNP Assembly at LINEs Controls Lineage-Specific RNA Processing. Cell 174, 1067–1081.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long JC & Caceres JF The SR protein family of splicing factors: Master regulators of gene expression. Biochemical Journal vol. 417 15–27 (2009). [DOI] [PubMed] [Google Scholar]
- 46.De I et al. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat. Struct. Mol. Biol 22, 138–144 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Rigo F et al. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nature Chemical Biology vol. 8 555–561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimoto N et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res. 43, 5898–5911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hein MY et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Dominguez D et al. Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Mol. Cell 70, 854–867.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue Y et al. Genome-wide Analysis of PTB-RNA Interactions Reveals a Strategy Used by the General Splicing Repressor to Modulate Exon Inclusion or Skipping. Mol. Cell 36, 996–1006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uemura Y et al. Matrin3 binds directly to intronic pyrimidine-rich sequences and controls alternative splicing. Genes to Cells 22, 785–798 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Meyer C et al. The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. Mol. Cell 69, 622–635.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerase A et al. Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol 26, 331–334 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Pandya-Jones A et al. An Xist-dependent protein assembly mediates Xist localization and gene silencing. bioRxiv (2020) doi: 10.1101/2020.03.09.979369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R & Johnson R Towards a complete map of the human long non-coding RNA transcriptome. Nature Reviews Genetics vol. 19 535–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
SUPPORTING REFERENCES
- 57.Busan S, Weidmann CA, Sengupta A & Weeks KM Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies. Biochemistry 58, 2655–2664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sengupta A, Rice GM & Weeks KM Single-molecule correlated chemical probing reveals large-scale structural communication in the ribosome and the mechanism of the antibiotic spectinomycin in living cells. PLoS Biol. 17, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Busan S & Weeks KM Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reuter JS & Mathews DH RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smola MJ, Calabrese JM & Weeks KM Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry 54, 6867–6875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Development Core Team, R. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; vol. 1 (2011). [Google Scholar]
- 63.DeLano W . . Pymol: An open-source molecular graphics tool. Newsletter On Protein Crystallography (2002). [Google Scholar]
- 64.Darty K, Denise A & Ponty Y VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Nostrand EL et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J Mol Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 68.Bastian M, Heymann S & Jacomy M Gephi : An Open Source Software for Exploring and Manipulating Networks Visualization and Exploration of Large Graphs. Int. AAAI Conf. Weblogs Soc. Media 361–362 (2009) doi: 10.13140/2.1.1341.1520. [DOI] [Google Scholar]
- 69.Bailey TL & Elkan C Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Second Int. Conf. Intell. Syst. Mol. Biol 2, 28–36 (1994). [PubMed] [Google Scholar]
- 70.Grant CE, Bailey TL & Noble WS FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blondel VD, Guillaume JL, Lambiotte R & Lefebvre E Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp 2008, (2008). [Google Scholar]
- 72.Pintacuda G et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 68, 955–969.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sprague D et al. Nonlinear sequence similarity between the Xist and Rsx long noncoding RNAs suggests shared functions of tandem repeat domains. RNA 25, 1004–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RNP-MaP site and correlation determination. Scheme outlining determination of (a) RNP-MaP sites and (b) correlations. For (a), depth threshold and mutation count above background threshold were set to eliminate noise at signal margins. A Z-factor above 0 is achieved only when 99% confidence intervals of UV only and SDA + UV rates do not overlap. To limit false positives, nucleotide-specific thresholds were set at the level of highest reactivity among nucleotides distant from proteins in RNPs of known structure (U1, RNase P, 18S, and 28S), expressed as the number of standard deviations from median reactivities of all instances of each nucleotide (see Methods). For (b) 3 nt windows and 4 nt separations were set to accommodate the size of longer and more complex mutation and deletion events observed during MaP reverse transcription. Thresholds of 10,000 depth, 6% background mutation rate, and 50 co-mutation events limit noise and false positives.
Figure S2. RNP-MaP discriminates protein-bound nucleotides independent of local RNA structure and nucleotide identity. (a) Correlation (Pearson’s r) between log2(reactivity) of nucleotides from biological replicate RNP-MaP experiments in HEK293 cells on U1, RNase P, RMRP, 18S, and 28S RNAs. Correlations are shown for all nucleotides (blue) and RNP-MaP sites (green). The number of nucleotides measured for each RNA is indicated above each graph in parentheses. (b) Nucleotide log2(reactivity) from RNP-MaP experiments as a function of distance to protein lysine side chain amine groups. Data are from two biological replicates on human U1, RNase P, 18S, and 28S RNAs, obtained in HEK293 cells. Nucleotides analyzed (n) include 1260 RNP-MaP sites (green) and 11,184 non-sites (black). The significance of the difference between RNP-MaP site and non-site distance distributions is shown (Mann-Whitney U-test, one-sided). (c) RNP-MaP site and non-site nucleotide log2(reactivity) from experiments in HEK293 cells (18S, 28S, U1, RNase P, RMRP, XIST RNAs) or SM33 cells (Xist RNA), grouped as unpaired nucleotides (U) and paired nucleotides (P). Violin plots are shown. For the top row, from left to right, the number of nucleotide reactivities included in each distribution (n) was 62, 46, 126, 16, 176, 70, 326, 22, 138, 68, 232, and 22. For the bottom row, n was 1189, 272, 1991, 171, 2905, 375, 4039, 198, 8867, 1568, 6350, 558, 8695, 2900, 6247, and 866. RNP-MaP sites are significantly more reactive than non-sites (top row p = 1.8e−9, 2.4e−8, 1.2e−22, 4.2e−15, 6.4e−22, 1.2e−15; bottom row p = 4.5e−112, 9.1e−74, 5.4e−124, 3.8e−87, 0.0, 0.0, 0.0, 0.0; one-sided Kolmogorov-Smirnov test). Reactivities of single-stranded RNP-MaP sites are significantly higher than those of paired sites (top row p = 4.1e−4, 6.2e−3, and 0.01; bottom row p = 2.7e−3, 0.12, 0.26, and 2.5e−23; one-sided Kolmogorov-Smirnov) except for 28S (p = 0.12) and Xist (p = 0.26), but both single-stranded and base paired regions clearly show higher reactivity at RNP-MaP sites than non-sites. (d) RNP-MaP nucleotide log2(reactivity) grouped as protein-proximal (< 10 Å) and protein-distant (> 10 Å) nucleotides. Reactivities are shown for each nucleobase. Data obtained for U1, RNase P, 18S, and 28S RNAs; in HEK293 cells. Nucleobase-specific thresholds (black dotted lines) for RNP-MaP sites were obtained from the standard deviations of reactivity for all of each nucleobase within an experiment, multiplied by corresponding empirically-derived threshold factors (U – 0.59, A – 0.29, C – 0.93, G – 0.78); these values are added to the corresponding nucleobase reactivity medians to compute final thresholds. The total number of nucleotides analyzed in each distribution (n) are: U1 (28, 36, 28, 28, 20, 38, 40, and 32), RNase P (56, 46, 70, 42, 76, 98, 96, and 110), 18S (491, 279, 520, 293, 483, 490, 631, and 436), and 28S (750, 502, 843, 588, 1114, 1078, 1446, and 1196). For violin plots in panels c and d, white circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values.
Figure S3. RNP-MaP signal in living cells is dependent on SDA and UV. (a) Nucleotide mutation rates after MaP reverse transcription of U1 and RNase P RNA. Combinations of UV (3 J/cm2) and SDA (10 mM) were applied directly to SM33 cells (in-cell) or to (protein-free) RNA extracted from cells (extracted). Among extracted controls, slightly enhanced reactivity at RNP-MaP nucleotides was detected in the U1 RNA, likely a result of residual undigested Sm proteins. The number of nucleotide mutation rates included in each distribution (n) are 13 RNP-MaP sites and 118 non-sites in U1 RNA and 34 sites and 256 non-sites in RNase P RNA (b) MaP nucleotide mutation rates for Rmrp RNA in SM33 cells as a function of SDA and UV dose. When SDA concentration was varied, UV dose was 3 J/cm2. When UV energy was varied, SDA concentration was 10 mM. In the UV gradient, n included 185 non-sites and 54 sites, while in the SDA gradient n included 194 non-sites and 45 sites. For violin plots, white circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values. P-values (Kolmogorov-Smirnov test, one-sided) are indicated. If p-values are calculated comparing each condition to their untreated counterparts (in a), only reactivities of RNP-MaP nucleotides in the SDA and UV treated in-cell samples (and the U1 extracted sample) are significantly increased (p <0.05).
Figure S4. Orthogonal chemical probing and metabolic labeling approaches corroborate RNP-MaP. (a) Comparison with ∆SHAPE61. ΔSHAPE identifies changes in SHAPE reactivity between RNA in cells and in solution, identifying regions of RNA that interact with cellular components, including proteins. RNP-MaP and ΔSHAPE sites are superimposed on secondary structure diagrams for mouse Rmrp and RNase P RNAs: ΔSHAPE sites overlapped RNP-MaP sites in mouse Rmrp and RNase P RNAs. Of 65 RNP-MaP sites (green shading) in Rmrp and RNase P RNAs, 22 (34%) are ΔSHAPE sites (lines), and 52 (80%) are within five nucleotides of ΔSHAPE sites by contact distance (which counts distances across base pairs in the accepted secondary structure as a length of one nucleotide). In-cell SHAPE protections (green) and enhancements (purple) are shown as lines. Primer-binding sequences were excluded from analysis (gray spheres), and solid grey and dotted grey lines indicate pseudoknot helices and predicted tertiary base-pair (inferred from human RNase P structure), respectively. (b) Comparison with metabolic labeling by photo-lysine. Photo-lysine contains a diazirine group, which is photo-activated, like SDA, with 365 nm UV light. Photo-lysine metabolic labeling substitutes for the first NHS-ester reaction step in SDA treatment. UV activation of photo-lysine-labeled cells yielded reactivity patterns similar to RNP-MaP for human RMRP RNA, albeit at reduced rates. Photo-lysine is less reactive with RNA than is SDA (left), as it is likely incorporated into proteins with low efficiency and requires a shorter crosslinking distance. Growing cells with a 1:4 natural lysine to photo-lysine ratio before crosslinking eliminates mutation signal (data not shown), confirming that observed signal is from protein-incorporated photo-lysine and not free photo-lysine in solution. For violin plot, white circles indicate medians, box limits indicate the first and third quartiles, whiskers extend 1.5 times the interquartile range, and smoothed polygons show data density estimates and extend to extreme values. The number of nucleotide mutation rates included in each distribution (n) is 230. Correlation (Pearson’s r) between SDA and photo-lysine reactivity (right) is shown. (c) RNP-MaP sites (green circles) called from reaction in-cell with SDA (left) or photo-lysine (right), superimposed on secondary structure of RMRP. All photolysine reactive regions are also reactive by SDA.
Figure S5. RNP-MaP interaction networks are highly reproducible. Top 10% of RNP-MaP correlations by mutual information (arcs) and the per-nucleotide summed mutual information for RNP-MaP correlations (purple peaks) from two biological replicates, plotted by nucleotide position (nt) for human U1, RNase P, and RMRP RNAs. Experiments performed in HEK293 cells.
Figure S6. RNP-MaP sites are conserved between mouse and human RNAs. RNP-MaP sites for mouse and human homologues of RNase P and RMRP, superimposed on secondary structure projections of each RNA. Structural domains are labeled in gray, the percent sequence identities between RNAs are indicated, and gray lines denote pseudoknot helices. Proteins in close proximity to each RNP-MaP site on RNase P (based on distance to lysine amine groups) are labeled (green lettering). Experiments performed in SM33 or HEK293 cells.
Figure S7. Gene-specific and random priming strategies for RNP-MaP library preparation. Schemes outlining generation of RNP-MaP sequencing libraries from (a) small RNA and gene- or region-specific targets and (b) long RNA targets. Gene-specific priming is recommended for RNAs or regions of an RNA < 500 nucleotides in length, and enrichment and random priming is suggested for longer RNAs.
Figure S8. RNP-MaP captures protein interaction information across very long non-coding RNAs. (a) Read depth thresholds observed mouse Xist (orange) and human XIST (blue), shown on per-nucleotide basis. Read depths exceeded 10,000 for most nucleotides. Tandem repeat sequence arrays A-F (white boxes with black letters), regions homologous to repeat regions (white letters), the D region core (white box with thick black outline), and exon-exon junctions (thick vertical black lines) are shown for Xist and XIST. (b) Distribution of RNP-MaP sites as a function of nucleotide identify. RNP-MaP signals are observed at all four nucleotides, with an enrichment at adenosine and uridine. (c) RNP-MaP signal as a function of extent of local RNA structure. RNP-MaP shows relatively low biases, as shown by the percentages (bar graphs) of RNP-MaP site nucleotides that also have low SHAPE reactivity (Lo-SHAPE), have low SHAPE reactivity and low modeled Shannon entropy (Lo-SS), or are base paired in minimum free-energy models (paired). (d) Positions of 151 high confidence eCLIP sites for 30 XIST-binding proteins analyzed in this work, shown with RNP-MaP site density (top 5% reactive sites per 51 nucleotides) aligned to the XIST sequence. Of note, the eCLIP sites for human HNRNPK protein, whose mouse homologue mediates recruitment of Polycomb through the B and C regions of mouse Xist28,72, closely mirror RNP-MaP site density across the human XIST D region and overlap human XIST B and C regions, supporting a model wherein the B, C, and D regions of Xist/XIST have analogous functions73. Protein eCLIP sites are organized on density plots by identified community and ranked (from top to bottom) by decreasing summed mutual information of intra-community correlations.
Figure S9. Detection of PTBP1, MATR3, and TIA1 sites in the XIST E region in cells and in vitro. Detection of RNP-MaP sites (represented as 25 nt window densities) in the XIST region E RNA based on experiments in-cell (orange) or under simplified conditions with synthetic RNA and the indicated recombinant protein. In vitro RNP-MaP densities (gray shading with black and colored lines) are shown as a function of increasing amounts of PTBP1, MATR3, TIA1, and (negative control) BSA proteins (only highest BSA control concentration is shown in black, no effect seen). Locations of called eCLIP sites (thick colored rectangles) and average eCLIP replicate read depth (matching colored lines) are shown above each density plot, and class 1 and 2 motifs (small gray and black boxes) for each protein are shown below each density plot. The average transcriptome-wide eCLIP replicate read depth value (99.99 percentile) is indicated by a horizontal tick at right side of eCLIP intensity plots. RNP-MaP sites rely on read-through of modifications during RT while eCLIP reads terminate at modifications: thus, as expected, peaks of RNP-MaP density for PTBP1, MATR3, and TIA1 lie at the 5’ ends of associated eCLIP read peaks. Locations of eCLIP sites for other mapped proteins are shown at bottom. In-cell and 1000 nM in vitro data are from same experiments as represented in Figure 6a and 6b.
Figure S10. Extended data in support of Figure 6. (a) Correlation of log10(mutation rates) of T7-transcribed XIST E region RNA (protein-free); comparing RNA treated with SDA and UV versus RNA treated with UV alone. MaP RT-induced mutation rates for n = 1177 nucleotides are identical (Pearson’s r = 0.994). (b) log2(reactivity) of RNP-MaP site nucleotides (red) as a function of indicated increasing protein concentration, compared to non-RNP-MaP nucleotides (black). Reactivities shown are averages for all nucleotides of each indicated nucleobase (right); the number of nucleotides present in each group in each experiment (n) is indicated. (c) Occurrence of PTBP1, MATR3, and TIA1 eCLIP sites in the Xist E region relative to uridine density. eCLIP sites are only found within the uridine-rich region (containing class 1 motifs) but are absent in the uridine-poor region where RNP-MaP-identified class 2 motifs predominate. (d) Relative expression levels of reporter RNAs containing non-XIST control, XIST E region, and other highly interactive XIST (by RNP-MaP) sequences, shown for both cytoplasmic and nuclear fractions. E region insert includes nts 11900-13100 of XIST. Nucleotide positions in XIST for other inserted protein-interactive sequences are indicated. qPCR signal from XIST reporters were (first) normalized to co-transfected control reporter RNA signal (firefly luciferase) in each fraction, and then each reporter signal was normalized to the non-XIST insert control. Error bars represent standard error of the mean, and means (bar heights) of replicate measurements are shown (n=3, open circles). All differences between E region reporter and other constructs are significant (cytoplasmic fraction P = 7.0e−4, 2.8e−3, 2.2e−3, and 1.2e−3; nuclear fraction P = 0.28, 2.0e−4, 9.0e−4, and 1.7e−2; Student’s t-test, two-sided), except for the difference between XIST E and Non-XIST nuclear fractions (P = 0.28). Reporter RNA containing the XIST E region was expressed at significantly lower levels than RNA reporters containing other highly interactive XIST sequences, yet E region reporter was the only construct that directed foci-formation (Fig. 6b).
Table S1. Totals and summed mutual information (MI) of correlations linking pairwise combinations of XIST-binding protein eCLIP sites. P-values for both attributes were generated from 2000 randomizations (without replacement) of measured correlation locations along XIST (for nucleotides with sufficient read depth only). All measured correlations are included.
Table S2. Pairs of protein eCLIP sites significantly excluded (P-values > 0.95) from RNP-MaP data based on both total number of correlations and summed mutual information (MI) strength. Associated network communities, total numbers of each community pairing, and the percent of pairings that were significantly excluded are also listed.
Table S3. DNA sequences of MaP library preparation primers, plasmid construction, and qPCR primers. Sequences inserted during plasmid construction are separated from flanking plasmid sequences by dashes (−).
Supplemental Data S1. Structure-based sequence alignment of human RNase P and RMRP RNAs.
Supplemental Data S2. List of high confidence eCLIP sites on XIST organized by protein.
Data Availability Statement
Raw and processed sequencing datasets analyzed in this report will be made available upon reasonable request and have been deposited in the Gene Expression Omnibus database (GSE152483).