Abstract
Background:
Although Clostridioides difficile infection (CDI) is known to involve the disruption of the gut microbiota, little is understood regarding how mucus-associated microbes interact with C. difficile. We hypothesized that select mucus-associated bacteria would promote C. difficile colonization and biofilm formation.
Methods/Results:
To create a model of the human intestinal mucus layer and gut microbiota, we used bioreactors inoculated with healthy human feces, treated with clindamycin and infected with C. difficile with the addition of human MUC2-coated coverslips. C. difficile was found to colonize and form biofilms on MUC2-coated coverslips and 16S rRNA sequencing revealed a unique biofilm profile with substantial co-colonization with Fusobacterium. Consistent with our bioreactor data, publicly available datasets and patient stool samples revealed that a subset of patients with C. difficile infection harbored high levels of Fusobacterium. We observed co-localization of C. difficile and F. nucleatum in an aggregation assay using adult patients and pediatric IBD patient stool and in CDI patient tissue sections. C. difficile strains were found to co-aggregate with F. nucleatum subspecies in vitro; an effect that was inhibited by blocking or mutating the adhesin RadD on Fusobacterium and removal of flagella on C. difficile. Aggregation was shown to be unique between F. nucleatum and C. difficile, as other gut commensals did not aggregate with C. difficile. Addition of F. nucleatum also enhanced C. difficile biofilm formation and extracellular polysaccharide production.
Conclusions:
Collectively, these data demonstrate a unique interaction of between pathogenic C. difficile and F. nucleatum in the intestinal mucus layer.
Keywords: Clostridium difficile, MUC2, mucus, biofilm
Graphical Abstract
LAY SUMMARY
Our work shows that the pathogen Clostridium difficile chemotaxes, aggregates and forms resilient biofilms with another pathogen Fusobacterium nucleatum in the intestinal mucus layer.
Introduction
Clostridioides difficile is the most common healthcare associated pathogen in U.S. hospitals, incurring billions of dollars in treatment costs each year1. Antibiotic disruption of the gut microbiota creates a favorable niche for C. difficile spore germination, proliferation, and ultimately toxin production and disease2. While several studies have investigated germination and toxin production, the colonization phase of C. difficile infection (CDI), which likely involves interaction with other gut microbes, remains relatively unknown. Comparisons of CDI and non-CDI stool have revealed alterations of the gut microbiota3, 4. CDI patients are frequently co-infected with other pathobionts, including Escherichia/Shigella, Salmonella, Staphylococcus, Klebsiella, and Fusobacterium3, 4. These findings imply that gut microbiome composition may influence C. difficile pathogenesis.
C. difficile localizes to the outer colonic MUC2 mucus layer with members of Bacteroidaceae and Enterobacteriaceae in murine models5. C. difficile is also present in the mucus layer of CDI patients6. However, the mechanism by which C. difficile associates with human MUC2 is uncharacterized. Moreover, little data exists regarding which members of the human gut microbiota interact with C. difficile. Despite the potential to influence the course of infection, the interactions between C. difficile and other mucus-associated bacteria remain poorly defined. To address these gaps in knowledge, we have focused on an entirely human-centered approach, employing human-derived MUC2, fecal bioreactors and patient samples. Our work points to the potential for mucus-associated microbes to interact with and modulate C. difficile.
Methods:
Bacterial Strain, Media, and Culture Conditions
Bacteria were grown in an Anaerobe Systems AS-580 chamber at 37°C in 10% CO2, 10% H2, and 80% N2 (supplemental methods). Bacteria were fluorescently-tagged with 10 μM CFDA-SE (Ex:Em: 428/528) or CMRA (Ex:Em: 548/576) (ThermoFisher) in PBS anaerobically at 37°C for 1 hr as described7. For growth curves, cultures of C. difficile R20291 and F. nucleatum subspecies nucleatum in BHIS were used to inoculate a chemically defined minimal medium (CDMM)8 at Optical density (OD)600nm =0.1. Growth was assessed by OD600nm and colony forming units (CFUs). For RNAseq, C. difficile R20291 was grown in CDMM supplemented with spent C. dificile R20291 CDMM medium (after 16 hrs incubation) or spent F. nucleatum subspecies polymorphum CDMM medium (supplemental methods).
Chemotaxis Assay:
For chemotaxis assays, C. difficile strains were grown in BHIS for 8 hrs, fluorescently-tagged with 10 μM CMRA, and suspended at OD600nm =2.0 in chemotaxis buffer (10 M potassium phosphate, 0.1 mM EDTA at pH 7.0) as previously described9. C. difficile was allocated to 96-well plates (200 μL) and capillary tubes filled with chemotaxis buffer with or without untagged F. nucleatum species (at OD600nm= 2.0) or 15 mM ethanolamine were added and incubated anaerobically at 37°C for 1 hr. Following incubation, capillary tube contents were assessed by a fluorescence plate reader and confirmed by microscopy (Nikon Eclipse 90i). To calculate the population of C. difficile that chemotaxed, the fluorescence from the capillary tubes (chemotaxed) was compared to the starting solution fluorescence (total) using the calculation chemotaxed/total x 100%.
Aggregation Assay:
Aggregation assays were performed as previously described for F. nucleatum10. Briefly, bacteria were washed with PBS and resuspended in anaerobic aggregation buffer (150 mM NaCl, 1 mM Tris/HCl, 0.1 mM CaCl2, and 0.1 mM MgCl2, pH 7) at OD600nm =1.5. Equal volumes (0.5 mL) of each bacteria were added to cuvettes, vortex-mixed for 10 sec, and read at OD600nm (time 0 hr). Samples were incubated for 1 hr at 37°C anaerobically and OD600nm was recorded (time 1 hr). Aggregation was calculated using the following equation: 100%- ((OD600nm at 0 hr –OD600nn at 1 hr) x 100%). For inhibition assays, F. nucleatum cells were pre-incubated with 50 mM L-arginine, D-galactose, or D-glucose for 5 min at 37°C, followed by addition of other bacterial strains. To assess aggregation of C. difficile and F. nucleatum within stool samples, we selected 8 stool samples from adult CDI patients and 4 stool samples from pediatric CDI patients with IBD (supplemental methods). The selected samples harbored high F. nucleatum levels by qPCR. Ten mg of each stool sample was suspended in 1.5 mL aggregation buffer and mixed thoroughly. To remove stool particulate, samples were centrifuged at 300 x g for 5 min. The supernatant was removed, 1 mL was transferred to microfuge tubes and incubated at 37°C for 1 hr. Following incubation, supernatants were removed and aggregates were transferred to microscope slides for FISH staining.
Mammalian Cell Culture
Mucus-producing human colon cells HT29-MTX (#12040401 Sigma), T84 (ATCC CCL-248), and LS174T (ATCC CL-188) were maintained in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) at 37°C, 5% CO2. For adhesion assays, 5 x 105 cells were seeded onto Corning Costar 12-well plates containing poly-lysine-coated coverslips and grown to confluence. MUC2 was purified from HT29-MTX and LS174T cells and human stool specimens as previously described7. Glycans were removed from MUC2 with 10 mM HCl heated in a boiling water bath (100°C) for 30 min and neutralized with 100 mM NaOH. MUC2 was ethanol-precipitated and treated with 100 mM NaOH for 20 hr at 23°C. The solution was neutralized with HCl and the denuded protein was ethanol-precipitated. Supernatants containing O-linked glycans were lyophilized and glycan concentrations were determined using the phenol sulfuric acid method.
MUC2-coated coverslips and 96-well plates
Glass coverslips (18 mm) were coated with 1 mg/mL MUC2 using 3-aminopropyltriethoxysilane (APTS) as previously described11. High-binding 96-well plates (Costar) were also coated in 1 mg/mL mucin or O-linked glycans in Hank’s Salt Solution by overnight incubation at 4°C. For MUC2 adhesion, fluorescently-tagged bacteria at OD600nm =2 were incubated with coverslips or plates anaerobically at 37°C for 1 hr. After washing, adhesion was examined by plate reader and microscopy
Human Colonoid Monolayers
The colonoid (C104) line was obtained from Texas Medical Center Digestive Diseases Center Gastrointestinal Experimental Model Systems Core. All procedures used to generate organoid cultures were approved by the Baylor College of Medicine IRB committee. Differentiated human colonoid monolayers were generated as previously described12. Monolayers (passage 12) were stained with Hoechst for 10 min at room temperature and incubated for 1 hour with fluorescently-tagged C. difficile R20291 and/or F. nucleatum subspecies at 37°C, 5% CO2. Monolayers were thoroughly washed and fixed with 4% paraformaldehyde for 30 min at room temperature. Adhesion was examined by microscopy.
Biofilm Assays
Overnight cultures of C. difficile and F. nucleatum were diluted in BHIS to OD600nm =0.1 and 100 μL of each culture was added to the 96-well plates coated with 1 mg/mL MUC2. For C. difficile control wells, 200 μL of culture was added. Plates were sealed and anaerobically incubated at 37°C for 72 hours. Biofilms were washed with PBS, stained with 0.1% crystal violet and assessed by OD600nm on a plate reader. Biofilms were also stained with Ruby Red Biofilm Tracer (ThermoFisher) and imaged by microscopy. Biofilm were quantified with FIJI software (National Institutes of Health) by tabulating the mean pixel intensity. Polysaccharides were isolated from biofilms as described13 and examined by the phenol-sulfuric acid method. Proteins were extracted from biofilms using RIPA buffer complemented with Complete Protease Inhibitor Cocktail (Roche) and quantified by the BCA assay using bovine serum albumin standards (ThermoFisher cat#23209). Biofilms were also grown for 48 hrs anaerobically at 37°C, then treated with proteinase K (30 mA/mL), DNAse (50 kU/mL), EDTA (12.5 mM), and vancomycin (400 μg/mL) for an additional 24 hrs anaerobically at 37°C.
C. difficile growth on mucin-coated surfaces in human fecal bioreactors
Bioreactors were constructed from 100 mL media bottles fitted with 2-port Q-series GL45 bottle caps (Kinesis) using fecal samples, media and conditions previously optimized14 (supplemental methods). Bioreactors were inoculated with fecal samples, treated with clindamycin (250 μg/mL) to disrupt the community, and then HT29-MTX MUC2-coated or mock-PBS-coated coverslips were introduced into reactors. At the time of coverslip introduction, communities were also treated with 3 x 106 CFUs of C. difficile 2015 pre-grown in bioreactor medium14. After 3 days of co-incubation, MUC2-coated coverslips were removed for analysis.
Evaluation of bacterial RNA and gDNA
Bacterial RNA was isolated as previously described7. RNA sequencing was performed at the University of Houston Seq-N-Edit Core. Triplicates were pooled for each condition to limit the sample size. The NuGEN ovation complete prokaryotic RNAseq library preparation kit (Tecan Genomics) was used according to manufacturer’s recommendations. Library quality and quantification were assessed via a 2100 BioAnalyzer instrument (Agilent). cDNA libraries were sequenced on a NextSeq 500 system (Illumina) for 150 cycles according to manufacturer’s protocol. Analysis of the data was performed using PATRIC bacterial bioinformatics resource center15. FastQ files were used as input for the program Rockhopper to align, assemble, quantify reads and generate differential expression patterns.
gDNA from biofilms and stool was extracted using the Zymo gDNA isolation kit (Zymo) according to the manufacturer’s instructions with the addition of 2 rounds of bead homogenization. To assess Fusobacterium levels in patient stool samples, gDNA was examined by qPCR using FAST SYBR green master mix with primers (Fusobacterium forward: CAACCATTACTTTAACTCTACCATGTTCA; Reverse: GTTGACTTTACAGAAGGAGATTATGTAAAAATC) on a QuantStudio3 qPCR machine (Applied Biosystems). 16S rDNA amplicons from bioreactor biofilms were sequenced by MiSeq paired-end sequencing (supplemental methods). Additionally, 16S rDNA data (454 and Illumina; 16S regions of V3-V5 and V4) from published studies were downloaded from NCBI Sequence Read Archive database and sorted for relative Fusobacterium levels.
Patient information and human specimen collection
All patient specimens were collected under the appropriate IRBs at the University of Cincinnati Medical Center Hospital, Cincinnati, OH (Human Research Protection Program #2012-4147) and Vanderbilt University Medical Center, Nashville TN (Human Research Protections Program #170067) (supplemental methods).
Immunostaining, FISH and SEM
Cell monolayers were stained with Hoechst, incubated with fluorescently-tagged bacteria for 1 hr at 37°C, fixed with 4% paraformaldehyde and stained with rabbit anti-MUC2 antibody (Novus cat#NBP1-31231). FISH staining was performed on stool aggregates, bioreactor MUC2-coverslips, and patient slides. Slides were hybridized with the probe [6FAM]CATCCTGTACTGGCTCAC for C. difficile5, [Cy3]CTTGTAGTTCCGCCTACCTC for F. nucleatum16 or [Cy3]GCTGCCTCCCGTAGGAGT for total bacteria16 (Integrated DNA Technologies). For SEM, 104 C. difficile cells were incubated with LS174T monolayers or HT29-MTX-derived MUC2-coated coverslips for 1 hr at 37°C. For biofilms, cultures were incubated with HT29-MTX MUC2-coated coverslips or 96-well plates anaerobically for 72 hrs at 37°C. Following incubation, samples were washed with PBS, fixed in 2.5% glutaraldehyde, dehydrated in ethanol and coated with 20 nm of gold in a desktop sputtering system (Denton Desk II). Images were acquired on a scanning electron microscope (FEI XL-30FEG) at 12 kV.
Statistics
Comparisons between groups were made with either One-way or Two-way Analysis of Variance (ANOVA) with the Holm-Sidak post-hoc test via SigmaPlot (Systat Software, Inc., San Jose, CA). Graphs were generated using Graphpad (GraphPad Software, Inc. La Jolla, CA).
Results:
C. difficile adheres to human intestinal MUC2 glycans
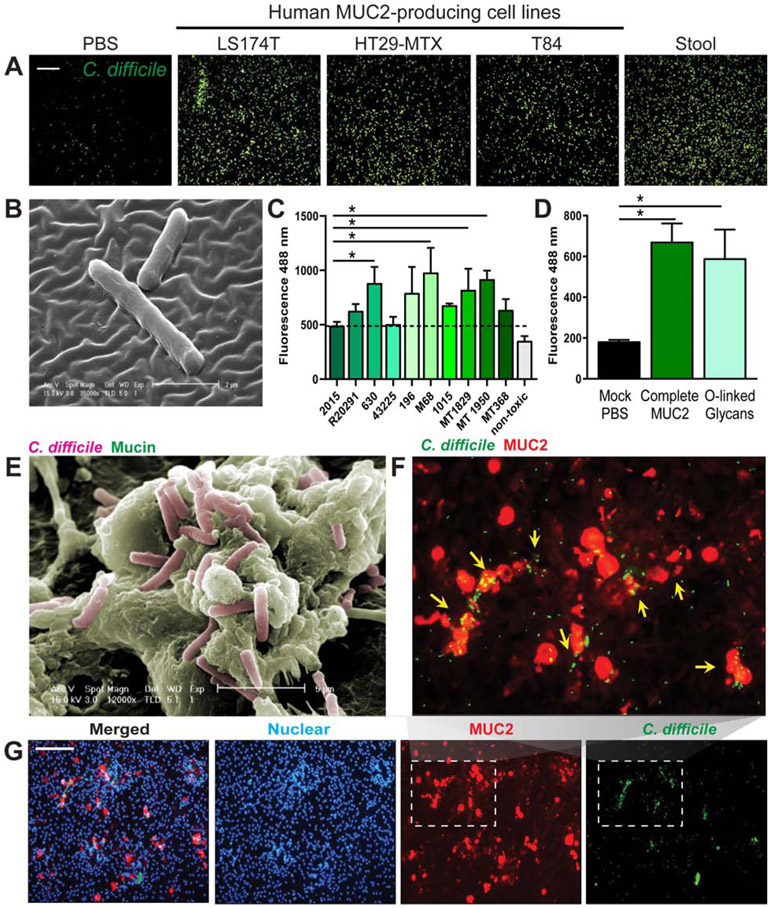
Adherence to mucus promotes colonization for numerous gastrointestinal pathogens. C. difficile has been identified in the colonic MUC2 mucus layer in a murine model of CDI5. However, little information exists on C. difficile adhesion to human MUC2. To examine the adhesive properties of C. difficile, human MUC2 was purified from several sources, including human mucin-producing LS174T, HT29-MTX and T84 cells and from human stool samples. Purified MUC2 was coated on glass coverslips and adhesion of fluorescently-tagged C. difficile to MUC2-coated coverslips was examined by microscopy after 1 hour incubation. We observed that C. difficile R20291 adhered to MUC2 isolated from all sources and was largely absent in control PBS-coated coverslips (Fig. 1A). To increase the resolution of the observed interaction between C. difficile and MUC2, we performed scanning electron microscopy (SEM) on HT29-MTX-MUC2 coverslips and observed C. difficile in the MUC2 layer (Fig. 1B). To examine if mucus binding is broadly conserved across multiple C. difficile lab strains and ribotypes (Fig. 1C, Supplemental Table 1), we examined adhesion of fluorescently-tagged C. difficile to MUC2 coated 96-well plates and found that all C. difficile strains could adhere to MUC2. We observed the greatest adhesion with C. difficile 630, M68, MT1850 and MT1950. These data indicate that MUC2 adhesion is functionally conserved across ribotypes. No adhesion was observed to methylcellulose, a polysaccharide commonly used to mimic the viscosity of a mucus environment (data not shown).
Figure 1: C. difficile adheres to purified human MUC2.
MUC2 was isolated from the supernatants of mucin-producing cell lines LS174T, HT29-MTX and T84, as well as from stool from healthy volunteers by a series of CsCl ultracentrifugation steps. MUC2 (1 mg/mL) was applied to coverslips linked via APTS. C. difficile R20291 was fluorescently-tagged with CFDA-SE and incubated with mucus-coated coverslips for 1 hour. A. Adhesion was visualized by microscopy (scale bar =50 μm) (representative image from n=4 replicates/repeated 3 independent times). B. Scanning electron microscopy of APTS-MUC2 coated coverslips demonstrating adhesion of C. difficile to HT29-MTX-derived MUC2 (representative image; n=3). C. Fluorescently-tagged C. difficile was added to 96-well plates coated with MUC2 purified from HT29-MTX and fluorescence values were obtained using a plate reader (n=8 replicates/repeated 3 independent times). D. Adhesion of C. difficile R20291 to mock (PBS), purified intact MUC2 or isolated MUC2-derived O-linked glycans (n=4 replicates/repeated 2 independent times). E. Co-localization of C. difficile and MUC2 in LS174T cells was also demonstrated by scanning electron microcopy (SEM) (representative image; n=3). F,G. CFDA-SE labeled C. difficile (green) was also found to co-localize with MUC2 (red) positive goblet cells in human colonoid monolayers by immunostaining (scale bar= 100 μM), which is clearly observed in the expanded inset above (F) (n=3 replicates, repeated 2 independent times). ANOVA, *p<0.05.
Mucin proteins are decorated with a wide range of glycans that serve as potential sites for adhesion by intestinal microbes. To identify C. difficile binding sites in human MUC2, O-linked glycans were removed by β-elimination and purified. We then repeated the adhesion assays with isolated O-linked glycans. Robust adhesion of C. difficile to extracted O-glycans was observed, and this pattern was similar to adhesion levels in the complete MUC2 protein (Fig. 1D). The data indicate that C. difficile adheres to the glycan component of the MUC2 mucin protein. We also sought to examine the interactions of C. difficile and MUC2 in live cells. Consistent with adhesion to purified MUC2, we observed that C. difficile R20291 co-localized with MUC2 in human mucin-producing LS174T cells by SEM after 1 hr incubation (Fig. 1E). Next, we examined C. difficile adhesion to MUC2 in a more physiologically relevant intestinal primary cell culture system. Colonoids derived from intestinal stem cells represent non-transformed epithelial cultures that harbor all the native intestinal cell types including MUC2 secreting goblet cells17. Similar to the observed patterns of adhesion in mucin-producing cell lines, C. difficile co-localized with MUC2 positive goblet cells in human colonoids (Fig. 1F, G); this can be seen more clearly in the expanded insert (Fig. 1F). These data are the first to demonstrate that C. difficile adheres to human MUC2, specifically binding to the O-linked glycans.
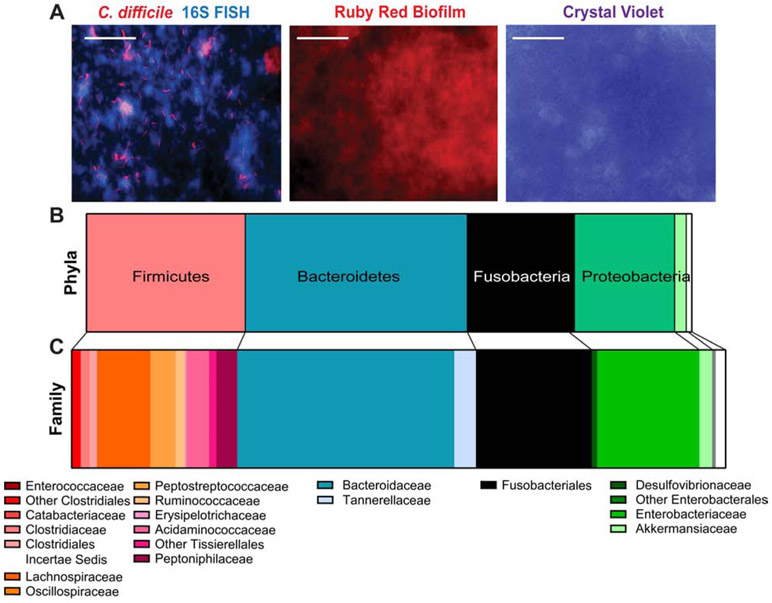
Microbes found in the mucus layer, including C. difficile, can be visualized growing as micro-colonies or biofilms. To identify bacterial members encountering C. difficile in a mucus-based microbial community, we turned to a modified bioreactor system. Fecal samples were cultivated and treated with clindamycin (250 μg/mL) to perturb microbial communities and mirror antibiotic-mediated microbial depletion during patient infection. Then, MUC2-coated coverslips were introduced into reactors and 3 x 106 CFU of C. difficile was added and incubated for 3 days to allow for mucus-associated microbial communities to form. FISH probes identified C. difficile and other microbial members marked by 16S FISH staining in the mucin-coated coverslips (Fig. 2A). Robust biofilms were observed, as denoted by ruby red biofilm tracer and crystal violet staining (Fig. 2A). By 16S rRNA gene sequencing, the dominant phyla of the mucus-associated microbiome included the phyla Firmicutes (26.3%), Bacteroidetes (36.7%), Proteobacteria (16.6%) and Verrucomicrobia (2.0%) (Fig. 2B). At the family level, within Firmicutes we observed high abundance Lachnospiraceae (8.3%), Ruminococcaceae (1.5%), Clostridiaceae (1.5%), and Clostridium XII sensu stricto (1.1%). Peptostreptococcaceae which contains C. difficile, compromised 4% of the sequences (Fig. 2C). In the Bacteroidetes, we observed primarily Bacteroidaceae (33.3%) and Tannerellaceae (3.4%). The majority of Proteobacteria were from the family Enterobacteriaceae (15.7%). Consistent with the presence of MUC2, we did observe the mucin degrader Akkermansia (1.9%). Surprisingly, 17.7% of sequences arose from the phylum Fusobacteria, family Fusobacteriaceae, a common oral microbiome constituent.
Figure 2: C. difficile interacts with other mucus-associated microbes in bioreactor mucin-based biofilms.
Bioreactors were modified with a hanging insert containing MUC2-APTS-coated cover slips. Bioreactors were seeded with stool from healthy volunteers (2x donors, 3x fecal pools, n =4) and treated with clindamycin (250 μg/mL) followed by C. difficile (106 CFU). MUC2 coated inserts were examined for biofilm production, C. difficile localization and 16S rRNA sequencing. A. FISH immunostaining for C. difficile (red) and total 16S rRNA genes (all bacteria; blue). Biofilms on coverslips were stained with ruby red biofilm tracer and crystal violet staining (scale bar = 50 μm) (representative image; n=4/stain). The composition of mucin-coated inserts was examined by 16S sequencing which revealed a unique microbial community at the phylum (B) and family (C) level.
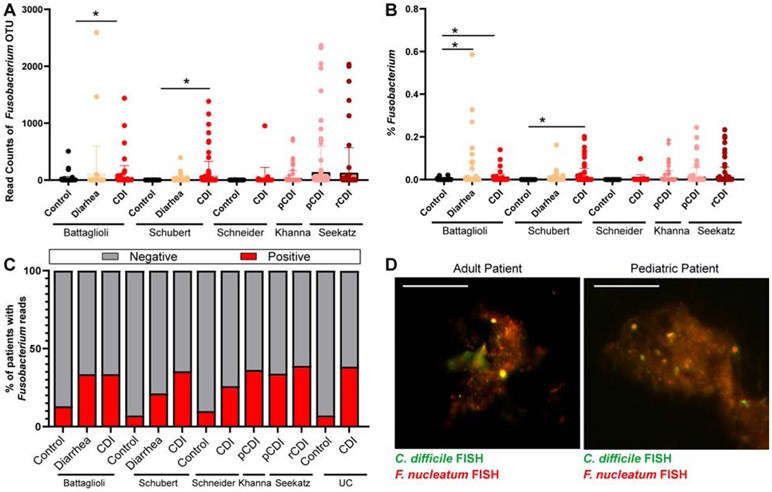
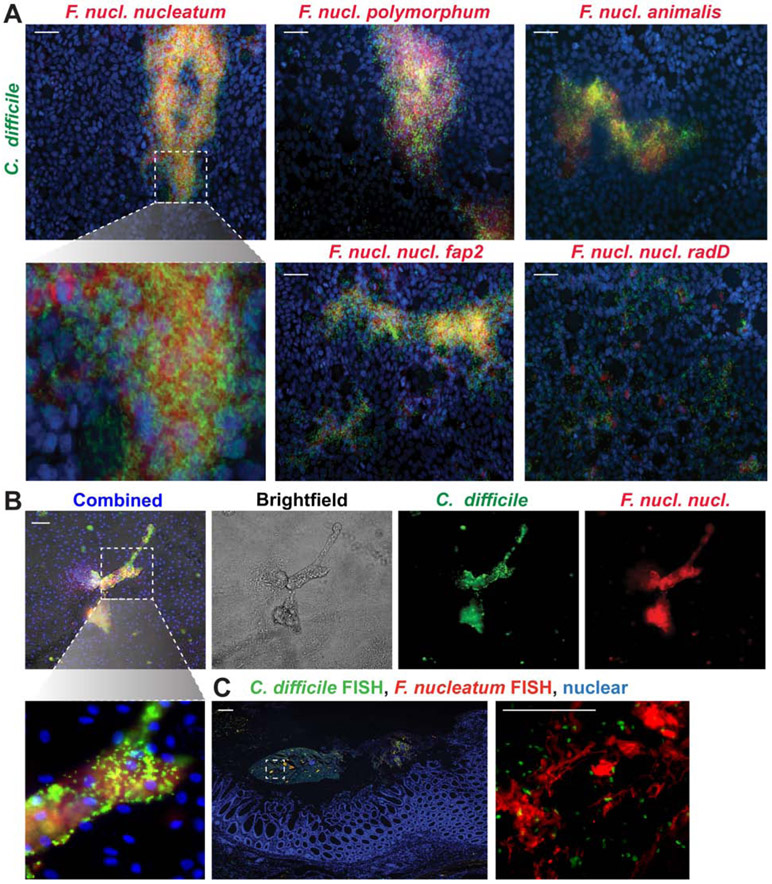
Fusobacterium plays a key role in multi-species biofilm formation in the oral cavity18, so we reasoned that members of the genus Fusobacterium may be an active participant in mucin-based intestinal biofilms with C. difficile. Three subspecies of F. nucleatum (nucleatum, polymorphum and animalis) were able to adhere to purified MUC2 and to the MUC2-producing cell line HT29-MTX (Supplemental Fig. 1A-C), suggesting that F. nucleatum can reside in the intestinal mucus layer. A previous report has pointed to the co-occurrence of F. nucleatum and C. difficile in colorectal cancer patients19. To determine whether F. nucleatum was present in non-cancer patients with CDI, we examined 16S sequencing data from publicly available datasets3, 4, 20-22. Data analysis from three published studies from Battaglioli21, Schubert4 and Schneider23 et al. revealed that patients with non-descriptive CDI harbored higher relative abundance of compared to the control population (Fig. 3A,B). Two additional studies from Khanna24 and Seekatz22 which only examined CDI patients revealed similar relative abundance levels of Fusobacterium OTUs for primary CDI24 and recurrent CDI22, 24 (Fig. 3A,B). In all studies, only subsets of patients had detectable Fusobacterium OTUs (Battaglioli21: Control=13.8%, CDI=31.1%; Schubert4.: Control=7.1%, CDI=35.5%, Schneider3: Control=7.4%; CDI=25.9%; Khanna24: pCDI=36.4%; Seekatz22: pCDI=33.9%; rCDI=38.9%)(Fig. 3C). Examination of another independent adult cohort of CDI patients from the University of Cincinnati revealed similar percentages of Fusobacterium positive samples by qPCR (38.4%)(Fig. 3C). Interestingly, we observed higher abundances of Fusobacterium in patients who experienced diarrhea, but were negative for CDI4, 21, indicating that Fusobacterium may be prevalent during times of microbial dysbiosis. Finally, we also examined stool from pediatric IBD patients, all of whom harbored detectable Fusobacterium by qPCR. Stool supernatants from adult and pediatric CDI patients who tested positive for Fusobacterium were collected, diluted in aggregation buffer and aggregates were examined by FISH for C. difficile and F. nucleatum (Fig 3D). We observed co-localization of both species in the patient stool samples, indicating that C. difficile may interact with Fusobacterium in a subset of patients.
Figure 3: Fusobacterium is present in a subset of patients with C. difficile infection (CDI).
16S rRNA genes in stool from published databases3, 4, 20-22 were examined for (A) number and (B) % abundance of Fusobacterium OTU reads. Samples included undefined CDI, recurrent CDI (rCDI) or primary CDI (pCDI) and age-matched controls (Battaglioli: control n=109, diarrhea n=114, CDI n=84; Schubert: control n=154, diarrhea n=89, CDI n=92; Schneider: control n=50, CDI n=29; Khanna pCDI n=88; Seekatz pCDI=148, rCDI n=82). C. Percentage analysis of patients who harbor Fusobacterium OTUs (positive, red) and patients without Fusobacterium OTUs (negative, grey) from previously published databases and a University of Cincinnati patient cohort (UC: control n=23, CDI n=59). Approximately 33.2% (averaged from all studies) of CDI patients had Fusobacterium reads. D. Aggregation of C. difficile and Fusobacterium in stool from adult patients with CDI (n=8) or pediatric patients with CDI and inflammatory bowel disease (IBD) (n=4) was examined by FISH using a probe for C. difficile (green) and Fusobacterium (red) (scale bar =50 μm). A representative image from an adult and pediatric patient are shown.
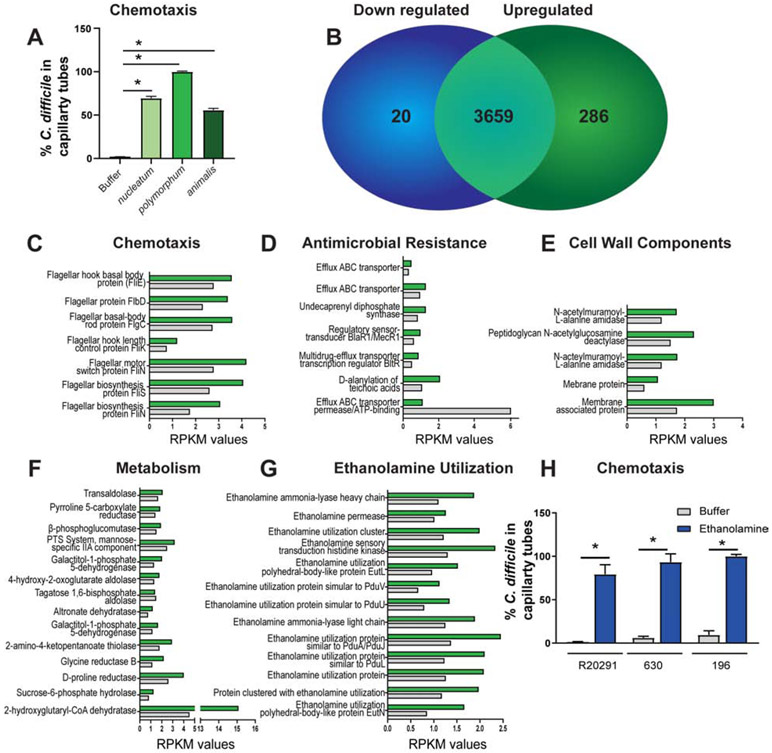
Based on our observation that C. difficile and F. nucleatum formed aggregates in patient stool samples, we reasoned that C. difficile may be chemoattracted to F. nucleatum. To address this hypothesis, we examined chemotaxis of fluorescently-tagged C. difficile R20219 towards F. nucleatum subspecies (nucleatum, polymorphum and animalis) using a standard capillary chemotaxis assay (Fig. 4A). After 1 hr incubation, C. difficile was found to enter capillary tubes containing F. nucleatum subspecies. To elucidate how F. nucleatum influences C. difficile, we performed a global transcriptional analysis of C. difficile R20291 incubated with F. nucleatum metabolites (Fig. 4B-G). Overall, 286 C. difficile genes were upregulated in response to F. nucleatum and 20 genes were downregulated (Fig. 4B). Among upregulated gene clusters in C. difficile, genes encoding chemotaxis, antimicrobial resistance, cell wall components, and metabolism, were observed (Fig. 4C-F). Consistent with our chemotaxis data, we found that 7 upregulated genes were related to bacterial chemotaxis (fliE, flbD, flgG, fliK, fliN, fliS)(Fig. 4C). We also found increased expression of 13 of 19 ethanolamine utilization genes in C. difficile in response to F. nucleatum metabolites (Fig. 4G). Ethanolamine has been previously reported to modulate C. difficile pathogenesis25 and is a chemotactic agent for other enteric pathogens. In a chemotaxis assay, C. difficile strains R20291, 630 and 196 were found to be attracted to 15 mM ethanolamine (Fig. 4H). These findings suggest that chemical signals may mediate interactions between C. difficile and F. nucleatum.
Figure 4: F. nucleatum metabolites influence C. difficile chemotaxis and gene expression.
A. Chemotaxis of fluorescently-tagged C. difficile R20291 into capillary tubes containing F. nucleatum subspecies nucleatum, polymorphum and animalis over 1 hr incubation (n=5 replicates, repeated 4 independent times). B-G. RNAseq analysis of pooled samples (n=3) of C. difficile after incubation with F. nucleatum subspecies polymorphum spent media (green bars) or C. difficile spent media (grey bars). B. 20 genes were downregulated and 286 genes were upregulated. C. difficile genes upregulated in response to F. nucleatum metabolites included (C) chemotaxis, (D) antimicrobial resistance, (E) cell wall components, (F) metabolism and (G) ethanolamine utilization. Data expressed as Reads Per Kilobase of transcript, per Million mapped reads (RPKM). H. Chemotaxis of fluorescently-tagged C. difficile strain R20291, 630 and 192 into capillary tubes containing 15 mM ethanolamine (n=5 replicates, repeated 3 independent times). One Way ANOVA, * p <0.05.
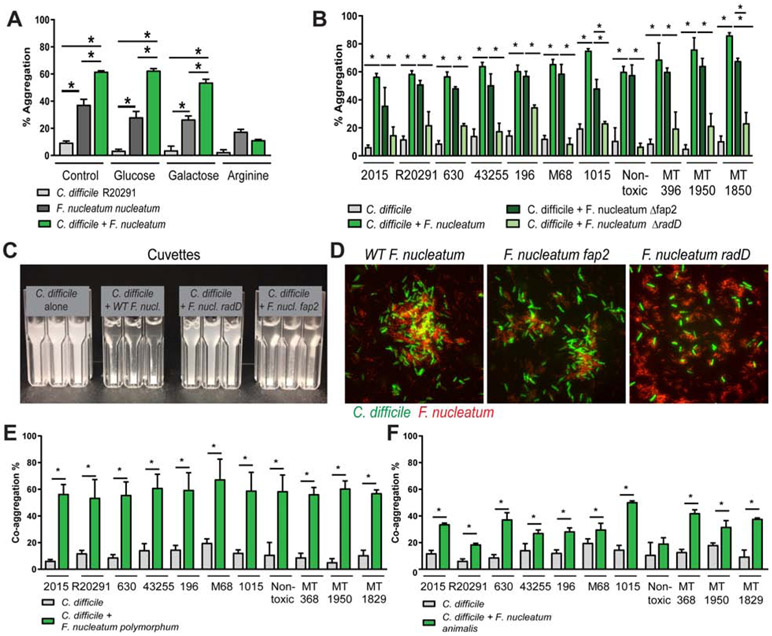
Bacterial surface adhesion and co-aggregation is essential for the development of multispecies biofilm communities18. To assess co-aggregation between C. difficile and F. nucleatum, strains were incubated in aggregation buffer for 1 hour and cell:cell aggregation was monitored by optical density (OD600nm). Aggregation of co-incubated strains compared to independently incubated controls was calculated based on the OD600 at time 0 and after 1 hr of anaerobic incubation at 37°C. C. difficile R20291 alone had minimal auto-aggregation (9.5 ± 1.2) compared to F. nucleatum subspecies nucleatum auto-aggregation (37.4 ± 4). However, co-incubation resulted in 61.8 ± 1% aggregated cells (Fig. 5A). F. nucleatum is known to express several proteins involved in adhesion, including lectin Fap2 which binds to Gal-GalNac26, and RadD which has been characterized as an arginine-dependent adhesin27. To elucidate the role of surface adhesion proteins in co-aggregation, F. nucleatum subspecies nucleatum was pre-incubated with either neutral glucose, Fap2-inhibiting galactose or RadD-inhibiting arginine27. While no impact on aggregation was observed in the presence of glucose or galactose (Fig. 5A), pre-incubation with arginine blocked co-aggregation, suggesting that RadD is a mediator of C. difficile-F. nucleatum adhesion. In a parallel approach, aggregation of C. difficile with F. nucleatum strains lacking either RadD or Fap2, were compared to aggregation of wild-type F. nucleatum (Fig. 5B). In agreement with our observation that pre-incubation with galactose did not effectively block co-aggregation, C. difficile strains incubated with F. nucleatum strains lacking Fap2 only exhibited a modest reduction in aggregation, while strains lacking RadD yielded reduced aggregation (Fig. 5B). This aggregation can be appreciated visually in cuvettes (Fig. 5C), where C. difficile alone remains un-aggregated and addition of wild-type F. nucleatum to C. difficile causes an accumulation of aggregates at the bottom of the cuvette. Similar to our aggregation calculations, the Fap2 mutant aggregates with C. difficile, while the RadD mutant has reduced aggregation with C. difficile. Microscopy of fluorescently-tagged bacteria also demonstrated the ability of wild-type F. nucleatum subspecies nucleatum to form aggregates with C. difficile, while F. nucleatum subspecies nucleatum ΔradD failed to exhibit the same degree of aggregation with C. difficile (Fig. 5D). Taken together, these data indicate that RadD is the primary mediator of C. difficile-F. nucleatum inter-microbial adhesion.
Figure 5: F. nucleatum aggregates with C. difficile via the adhesin RadD.
A. C. difficile R20291 and F. nucleatum subspecies nucleatum were incubated together anaerobically in co-aggregation buffer. Aggregation was calculated using the equation: 100%- ((OD600nm at 0 hr –OD600nn at 1 hr) x 100%). For inhibition assays, F. nucleatum subspecies nucleatum was pre-incubated for 5 min with 50 mM L-arginine, D-galactose, or D-glucose anaerobically, followed by addition of C. difficile (n=3-4 replicates, repeated 4 independent times). Significance was determined by Multi-way ANOVA. B. Aggregation was examined with 11 C. difficile strains, wild-type F. nucleatum subspecies nucleatum, as well as F. nucleatum subspecies nucleatum Δfap2 and F. nucleatum subspecies nucleatum ΔradD. C. For visualization, cuvettes were examined for aggregation in C. difficile alone, C. difficile with wild-type F. nucleatum subspecies nucleatum, Δfap2 and ΔradD. D. Microscopy of CFDA-SE tagged C. difficile R20291 (green) and F. nucleatum subspecies nucleatum (wild-type (WT), Δfap2 or ΔradD) (red) (scale bar =50 μm) (n=3). Co-aggregation was also examined with C. difficile strains and (E) F. nucleatum subspecies polymorphum and (F) F. nucleatum subspecies animalis (n=4 replicates, repeated 3 independent times). One Way ANOVA, * p <0.05.
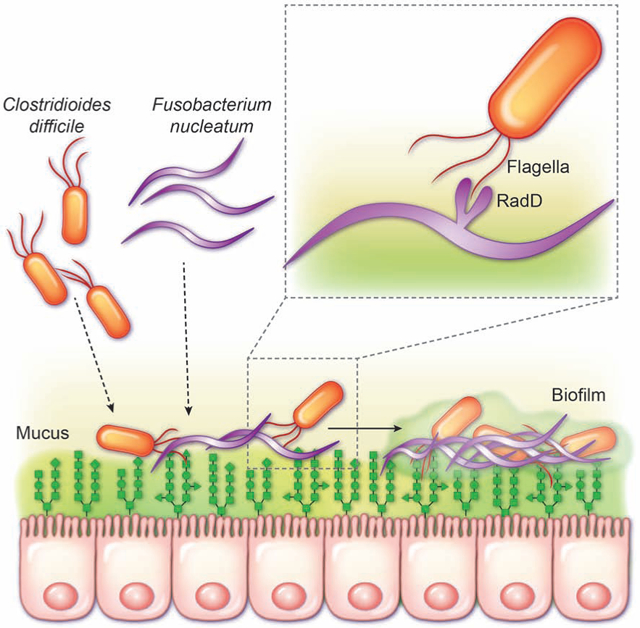
To investigate the conservation of co-aggregation of C. difficile to a broad range of F. nucleatum subspecies, aggregation assays were performed using F. nucleatum subspecies polymorphum and animalis (Fig. 5E,F). Consistent with our F. nucleatum subspecies nucleatum data, C. difficile strains also co-aggregated with F. nucleatum subspecies polymorphum and animalis (Fig. 5E,F). Of note, C. difficile exhibited the lowest aggregation with F. nucleatum subspecies animalis, which is the least auto-aggregative of the three species. Next, we examined the aggregation of C. difficile with other gut microbes to determine if aggregation was a unique feature of C. difficile and F. nucleatum. We analyzed 10 different intestinal microbes representing the phyla Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia (Supplemental Fig. 2A). No microbes were identified that had enhanced aggregation with C. difficile. Since flagella are important for host adhesion, we asked whether flagella could mediate the interaction between C. difficile and F. nucleatum RadD. To address this question, we sheared the flagella from C. difficile R20291 and examined aggregation of sheared flagella or flagella-free C. difficile with F. nucleatum subspecies nucleatum (Supplemental Fig. 2B). F. nucleatum was observed to aggregate to a similar degree with sheared flagella and intact C. difficile. Moreover, flagella-free C. difficile exhibited decreased aggregation with F. nucleatum compared to intact C. difficile. These data indicate that C. difficile flagella adhere to F. nucleatum RadD.
To determine if C. difficile and F. nucleatum would preferentially aggregate in the presence of the intestinal epithelium, fluorescently-tagged bacteria were incubated with mucin-producing HT29-MTX cells for 1 hour and then examined by microscopy (Fig. 6A). F. nucleatum subspecies nucleatum, polymorphum and animalis, as well as F. nucleatum subspecies nucleatum Δfap2 co-localized with C. difficile R20291 above the epithelium. In contrast, minimal aggregation was observed with C. difficile and F. nucleatum subspecies nucleatum ΔradD on HT29-MTX cells. We also confirmed this aggregation profile using human colonoids, where a similar pattern was observed (Fig. 6B). Finally, FISH analysis of surgical resections from patients with recurrent CDI revealed co-localization of C. difficile and Fusobacterium in the mucus layer above the epithelium (Fig. 6C). These data establish that C. difficile interacts with the F. nucleatum surface adhesin RadD to promote aggregation and that these two species aggregate in the mucus layer.
Figure 6. F. nucleatum strains aggregate with C. difficile R20291 on HT29-MTX cells and human colonoids.
A. Representative images of mucin producing HT29-MTX monolayers incubated with CFDA-SE fluorescently tagged C. difficile R20291 (green) and CMRA fluorescently-tagged F. nucleatum subspecies nucleatum, nucleatum Δfap2, nucleatum ΔradD, polymorphum or animalis (red) after 1 hr (scale bar =50 μm)(n=3 replicates, repeated 3 independent times). B. Representative images of human colonoids (organoids) incubated with C. difficile R20291 (green) and F. nucleatum subspecies nucleatum after 1 hr (scale bar =50 μm) (n=3 replicates, repeated 2 independent times). Higher magnification inset below highlights co-localization. C. Representative image of C. difficile and Fusobacterium FISH staining in surgical resections from patients with recurrent CDI (n=4 patients). Positive staining for C. difficile (green) and Fusobacterium (red) were observed at 40x (left) (scale bar = 50 μm) and 400x (inset right) (scale bar = 100 μm)
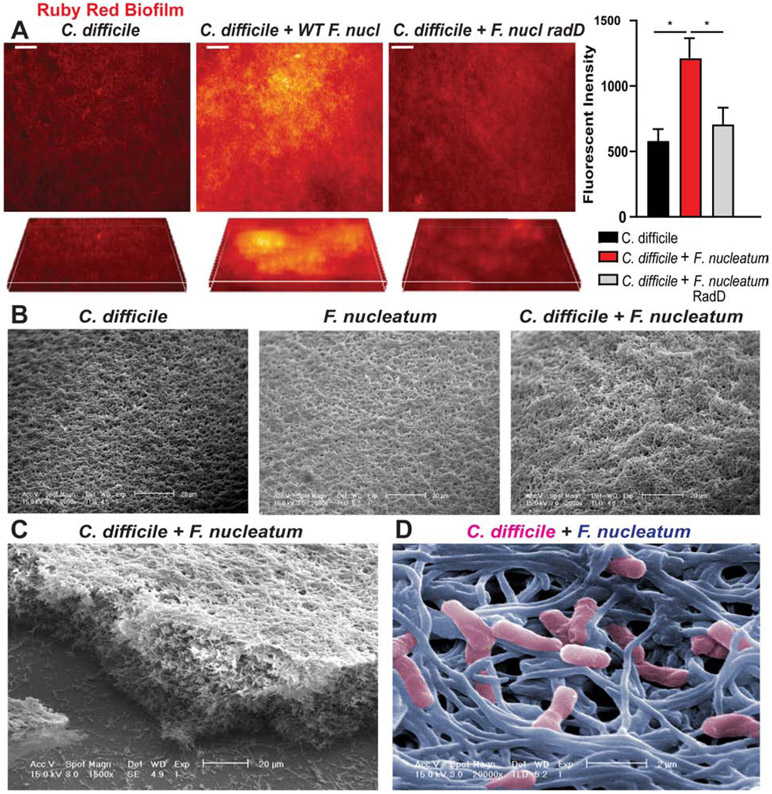
Aggregation is the initiating process in biofilm development, which is an important factor in colonization. Since we observed significant aggregation between C. difficile and F. nucleatum, we next sought to determine if F. nucleatum influenced C. difficile biofilm formation. Biofilm density present on MUC2-coated coverslips was visualized by ruby red biofilm tracer (Fig. 7A). Consistent with our aggregation data, wild-type F. nucleatum subspecies nucleatum enhanced C. difficile biofilm formation. The F. nucleatum subspecies nucleatum ΔradD derivative failed to enhance biofilm formation to the same degree as the wild-type strain (Fig 7A, Supplemental Fig. 3A). Examination of biofilms by scanning electron microscopy revealed the additive effect of F. nucleatum subspecies nucleatum on biofilm formation (Fig. 7B-D). Importantly, biofilms contain both C. difficile and F. nucleatum, as seen in the pseudo-colored SEM (Fig. 7D), indicating that F. nucleatum and C. difficile likely have a synergistic relationship. F. nucleatum subspecies polymorphum and animalis also enhanced biofilm for all C. difficile strains (Supplemental Fig. 3B,C), further supporting our hypothesis that these interactions are broadly conserved. To assess which components of the biofilm were enhanced by F. nucleatum, we examined the major components of biofilm: DNA, proteins and polysaccharides (Supplemental Fig. 4A-C). Co-cultures with C. difficile and F. nucleatum exhibited increased DNA, proteins and soluble extracellular polysaccharides (EPS) compared to either C. difficile alone or C. difficile-F. nucleatum subspecies ΔradD biofilms. No changes were observed in insoluble extracellular polysaccharides (IEPS) or intracellular polysaccharides (IPS) (Supplemental Fig. 4C). Finally, we sought to determine if F. nucleatum enhancement of C. difficile biofilms had an effect on biofilm stability. Biofilms harboring both C. difficile and F. nucleatum were more resistant to Proteinase K, DNAse, EDTA and the antibiotic vancomycin (Supplemental Fig. 4D). Collectively, this work points to F. nucleatum as an interacting member of the mucus-associated microbiome that promotes C. difficile aggregation and enhances biofilm formation.
Figure 7. F. nucleatum bolsters C. difficile biofilms.
A. Biofilms of C. difficile R20291 with or without wild-type F. nucleatum subspecies nucleatum or F. nucleatum subspecies nucleatum ΔradD were grown on HT29-MTX-derived MUC2 coverslips anaerobically for 72 hrs. Biofilm extracellular matrix was visualized with ruby red biofilm tracer and examined with 2D and z-stack analysis (scale bar =50 μm)(n=3 replicates, repeated 3 independent times). Quantification performed by FIJI for relative fluorescent intensities. B. Scanning electron microscopy (SEM) of mucin-based biofilms (scale bar =20 μm) (representative image; n=3 replicates). C. Robust biofilms were observed via SEM by lateral views of C. difficile and F. nucleatum (scale bar =20 μm) (left image). D. Representative falsely colored SEM image with C. difficile (pink) and F. nucleatum (blue) (scale bar = 2 μm).
Discussion:
Recent in-depth analysis of the gut microbiome has revealed that intestinal disorders are often polymicrobial in origin or manifestation28. Polymicrobial interactions are particularly important in the setting of infection and chronic disease, as microbes can interact synergistically to induce virulence traits, modify the environmental niche, or modulate the host immune response. In the setting of C. difficile infection, these interactions are incompletely understood. Here we show that F. nucleatum adheres to C. difficile, influences key genes in C. difficile for chemotaxis and niche development, and promotes aggregation and biofilm formation. These findings were replicated using eleven C. difficile strains and three F. nucleatum subspecies, indicating the shared abilities of these strains to interact with each other. This work also identified RadD and flagella as key adhesins involved in bacterial-bacterial interactions between C. difficile and F. nucleatum.
F. nucleatum is well-characterized in the oral cavity, where it forms highly structured, multispecies communities that can promote oral diseases like gingivitis/periodontitis18. F. nucleatum adheres to several oral Gram-positive microbes, so it is unlikely that the interaction between F. nucleatum and C. difficile is unique. However, one interesting feature of F. nucleatum and C. difficile is horizontal gene transfer. Genome analysis of F. nucleatum subspecies polymorphum ATCC 10953 found that 16 conserved gene clusters were from C. difficile29. Additionally, it appears that F. nucleatum subspecies polymorphum has obtained 5 composite ribozyme/transposons, similar to C. difficile’s CdISt-IStrons. These data support our hypothesis that these two species can act synergistically.
Although oral bacteria poorly colonize the healthy intestine, increased quantities of oral microbes, including Fusobacterium, have been reported in the gut microbiota of patients following antibiotic intake30-33. Consistent with these studies, our analysis of published 16S rRNA gene data sets4, 21 indicate that healthy subjects typically harbor lower Fusobacterium abundances. Fusobacterium has also been isolated from the intestinal mucus-associated microbiota of IBD patients, a population that experiences increased morbidity in terms of C. difficile infection34. Data from our pediatric cohort supports potential interactions between C. difficile and F. nucleatum in IBD patients. Although we do not have data on outcomes of patients harboring greater numbers of Fusobacterium OTUs, we speculate that F. nucleatum may be a confounding or disease-modifying factor in C. difficile infection since this species promotes robust biofilm formation.
Our RNAseq data revealed that C. difficile ethanolamine related genes in response to F. nucleatum metabolites. Ethanolamine is an end-product of phosphatidylethanolamine catabolism, an abundant phospholipid in both mammalian and bacterial membranes. Other GI pathogens, including enterohemorrhagic Escherichia coli (EHEC) and Salmonella, recognize ethanolamine as a signal to modulate virulence and colonization gene expression35, 36. Ethanolamine also impacts C. difficile pathogenesis25, 37. F. nucleatum encodes three glycerolphosphoryl diester phosphodiesterase proteins (FN1891, FN1908, and FN1954) that can generate ethanolamine38. While ethanolamine membrane diffusion in other microbial species is low, diffusion is amplified by F. nucleatum’s ethanolamine permease (FN0089)39. Similar to our findings, F. nucleatum upregulates ethanolamine utilization genes in the oral microbe S. gordonii38. As a result, we speculate that intestinal F. nucleatum-produced ethanolamine can influence C. difficile and perhaps modulate pathogenesis.
Interactions between C. difficile and F. nucleatum are of key interest because of the potential impact each strain may have on the host and on each other. We speculate that interspecies interactions could be some of the earliest events that occur as C. difficile colonizes the host. As a result, we believe that an improved understanding of how C. difficile interacts with mucus-associated microbes will continue to clarify the mechanisms of CDI.
Supplementary Material
WHAT YOU NEED TO KNOW:
Background: Clostridium difficile is an enteric pathogen that causes diarrhea and life-threatening inflammation. C. difficile infection is accompanied by changes in the gut microbiota; however, it is unclear how these microbes influence C. difficile.
New Findings: Using bioreactors, patient stool, and human colonoids, we demonstrate that C. difficile aggregates with the opportunist pathogen Fusobacterium nucleatum. Moreover, these species form robust biofilms that depend on the Fusobacterium protein RadD.
Limitations: We do not have data on outcomes of patients harboring greater numbers of Fusobacterium OTUs, although we speculate that F. nucleatum may be a confounding factor in C. difficile infection.
Impact: This work indicates that F. nucleatum acts synergistically with C. difficile in the mucus layer. As a result, F. nucleatum should be considered when addressing C. difficile infection, especially in populations known to harbor higher Fusobacterium levels.
Acknowledgments
We would like to thank Dr. Xi-Lei Zeng for her expertise and preparation of the colonoid monolayers used in this study. We would also like to thank Dr. Chenggang Wu at UT Health for his intellectual input on the project.
Funding: This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (Grant P30-DK-56338 to Texas Medical Center Digestive Disease Center, Gastrointestinal Experimental Model Systems), National Institutes of Health (NIH) K01DK12319501 (MAE), KO1DK12186901 (ACE), T32DK07644 (WR), F32AI136404 (HAD), R01AI123278 (RAB), U01AI124290 (RAB, KWG, TS), and unrestricted research support from BioGaia AB (RAB, JV, Stockholm, Sweden).
Footnotes
Conflict of Interest:
J. Versalovic serves on the scientific advisory boards of Biomica, Plexus Worldwide and Seed Health, and J. Versalovic and R. Britton receive unrestricted research support from Biogaia, AB. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement :
The raw data supporting the conclusions of this manuscript can be found under the following BioSample accession numbers: SAMN15790567, SAMN15790568, SAMN15790569, SAMN15790570, SAMN15790571, SAMN15790572.
References:
- 1.Dubberke ER, Carling P, Carrico R, et al. Strategies to prevent Clostridium difficile infections in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol 2014;35 Suppl 2:S48–65. [DOI] [PubMed] [Google Scholar]
- 2.Leffler DA, Lamont JT. Clostridium difficile Infection. N Engl J Med 2015;373:287–8. [DOI] [PubMed] [Google Scholar]
- 3.Schneider D, Thurmer A, Gollnow K, et al. Gut bacterial communities of diarrheic patients with indications of Clostridioides difficile infection. Sci Data 2017;4:170152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubert AM, Rogers MA, Ring C, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio 2014;5:e01021–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semenyuk EG, Poroyko VA, Johnston PF, et al. Analysis of Bacterial Communities during Clostridium difficile Infection in the Mouse. Infect Immun 2015;83:4383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engevik MA, Yacyshyn MB, Engevik KA, et al. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 2015;308:G510–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engevik MA, Luk B, Chang-Graham AL, et al. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamakawa K, Kamiya S, Meng XQ, et al. Toxin production by Clostridium difficile in a defined medium with limited amino acids. J Med Microbiol 1994;41:319–23. [DOI] [PubMed] [Google Scholar]
- 9.Bainer R, Park H, Cluzel P. A high-throughput capillary assay for bacterial chemotaxis. J Microbiol Methods 2003;55:315–9. [DOI] [PubMed] [Google Scholar]
- 10.Park J, Shokeen B, Haake SK, et al. Characterization of Fusobacterium nucleatum ATCC 23726 adhesins involved in strain-specific attachment to Porphyromonas gingivalis. International Journal of Oral Science 2016;8:138–144. [Google Scholar]
- 11.Landry RM, An D, Hupp JT, et al. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol 2006;59:142–51. [DOI] [PubMed] [Google Scholar]
- 12.Chang-Graham AL, Danhof HA, Engevik MA, et al. Human Intestinal Enteroids With Inducible Neurogenin-3 Expression as a Novel Model of Gut Hormone Secretion. Cell Mol Gastroenterol Hepatol 2019;8:209–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aires CP, Del Bel Cury AA, Tenuta LM, et al. Effect of starch and sucrose on dental biofilm formation and on root dentine demineralization. Caries Res 2008;42:380–6. [DOI] [PubMed] [Google Scholar]
- 14.Robinson CD, Auchtung JM, Collins J, et al. Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates. Infect Immun 2014;82:2815–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wattam AR, Davis JJ, Assaf R, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 2017;45:D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karygianni L, Follo M, Hellwig E, et al. Microscope-based imaging platform for large-scale analysis of oral biofilms. Appl Environ Microbiol 2012;78:8703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011;141:1762–72. [DOI] [PubMed] [Google Scholar]
- 18.He X, Hu W, Kaplan CW, et al. Adherence to streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb Ecol 2012;63:532–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukugaiti MH, Ignacio A, Fernandes MR, et al. High occurrence of Fusobacterium nucleatum and Clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J Microbiol 2015;46:1135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna S, Montassier E, Schmidt B, et al. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment Pharmacol Ther 2016;44:715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battaglioli EJ, Hale VL, Chen J, et al. Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seekatz AM, Rao K, Santhosh K, et al. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med 2016;8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davido DJ, Richter F, Boxberger F, et al. Butyrate and propionate downregulate ERK phosphorylation in HT-29 colon carcinoma cells prior to differentiation. Eur J Cancer Prev 2001;10:313–21. [DOI] [PubMed] [Google Scholar]
- 24.Khanna R, Chande N, Nelson RL. Treatment of an initial infection with clostridium difficile in patients with inflammatory bowel disease. Inflamm Bowel Dis 2013;19:2223–6. [DOI] [PubMed] [Google Scholar]
- 25.Nawrocki KL, Wetzel D, Jones JB, et al. Ethanolamine is a valuable nutrient source that impacts Clostridium difficile pathogenesis. Environ Microbiol 2018;20:1419–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abed J, Emgard JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016;20:215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan CW, Lux R, Haake SK, et al. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 2009;71:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tay WH, Chong KK, Kline KA. Polymicrobial-Host Interactions during Infection. J Mol Biol 2016;428:3355–71. [DOI] [PubMed] [Google Scholar]
- 29.Karpathy SE, Qin X, Gioia J, et al. Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium. PLoS One 2007;2:e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol 2019;17:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 2011;17:1971–8. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y, Eun CS, Lee AR, et al. Fusobacterium Isolates Recovered From Colonic Biopsies of Inflammatory Bowel Disease Patients in Korea. Ann Lab Med 2016;36:387–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Meij TGJ, de Groot EFJ, Peeters CFW, et al. Variability of core microbiota in newly diagnosed treatment-naive paediatric inflammatory bowel disease patients. PLoS One 2018;13:e0197649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balram B, Battat R, Al-Khoury A, et al. Risk Factors Associated with Clostridium difficile Infection in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. J Crohns Colitis 2019;13:27–38. [DOI] [PubMed] [Google Scholar]
- 35.Gonyar LA, Kendall MM. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 2014;82:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kendall MM, Gruber CC, Parker CT, et al. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. MBio 2012;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janoir C, Deneve C, Bouttier S, et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 2013;81:3757–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrickson EL, Wang T, Beck DA, et al. Proteomics of Fusobacterium nucleatum within a model developing oral microbial community. Microbiologyopen 2014;3:729–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 2010;8:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript can be found under the following BioSample accession numbers: SAMN15790567, SAMN15790568, SAMN15790569, SAMN15790570, SAMN15790571, SAMN15790572.