Abstract
Primary cilia are located at the dendritic tips of sensory neurons and house the molecular machinery necessary for detection and transduction of sensory stimuli. The mechanisms that coordinate dendrite extension with cilium position during sensory neuron development are not well understood. Here, we show that GRDN-1, the Caenorhabditis elegans ortholog of the highly conserved scaffold and signaling protein Girdin/GIV, regulates both cilium position and dendrite extension in the postembryonic AQR and PQR gas-sensing neurons. Mutations in grdn-1 disrupt dendrite outgrowth and mislocalize cilia to the soma or proximal axonal segments in AQR, and to a lesser extent, in PQR. GRDN-1 is localized to the basal body and regulates localization of HMR-1/Cadherin to the distal AQR dendrite. However, knockdown of HMR-1 and/or loss of SAX-7/LICAM, molecules previously implicated in sensory dendrite development in C. elegans, do not alter AQR dendrite morphology or cilium position. We find that GRDN-1 localization in AQR is regulated by UNC-116/Kinesin-1, and that correspondingly, unc-116 mutants exhibit severe AQR dendrite outgrowth and cilium positioning defects. In contrast, GRDN-1 and cilium localization in PQR is modulated by LIN-44/Wnt signaling. Together, these findings identify upstream regulators of GRDN-1, and describe new cell-specific roles for this multifunctional protein in sensory dendrite development.
Keywords: Cilia, GRDN-1, Girdin/GIV, Dendrite, Sensory neurons
INTRODUCTION
Neurons are highly polarized cells. Axons and dendrites house unique sets of structural and signal transduction proteins and exhibit distinct signaling and integration properties (Bentley and Banker, 2016; Takano et al., 2015). Sensory neurons represent a particularly intriguing example of neuronal polarization. The typically asynaptic dendrites in a subset of these neurons such as invertebrate chemo- and mechanosensory neurons and vertebrate photoreceptors and olfactory neurons contain specialized cilia at their distal ends (Keil, 1997; Menco, 1997; Rohlich, 1975; Ward et al., 1975). Cilia are evolutionarily conserved signaling organelles comprised of a microtubule-based axoneme core surrounded by a specialized membrane enriched in signaling molecules. Cilia are nucleated by a centriole-derived basal body that migrates toward the cell surface and docks at the plasma membrane; thus, the location of a cilium is necessarily dictated by the position of the basal body (Dawe et al., 2007; Elric and Etienne-Manneville, 2014; Tang and Marshall, 2012). How the location and anchoring of the basal body at the distal dendrite is coordinated with dendrite outgrowth during sensory neuron differentiation is not fully understood.
The C. elegans hermaphrodite contains 62 ciliated sensory neurons (Kazatskaya et al., 2020; Perkins et al., 1986; Ward et al., 1975). The majority of these neurons are born and differentiate in the embryo, with the exception of three types including the gassensing AQR and PQR neurons, which are generated postembryonically (Gray et al., 2004; Sulston and Horvitz, 1977; Sulston et al., 1983). Dendrites of sensory neurons extend anteriorly or posteriorly in the head and tail, respectively, and contain morphologically distinct cilia at their distal ends (Doroquez et al., 2014; Perkins et al., 1986; Ward et al., 1975). Sensory cilia are exposed either directly or indirectly to the external environment, or in the case of AQR and PQR, to the internal pseudocoelomic body fluid (Perkins et al., 1986; Ward et al., 1975; White et al., 1986). Since dendrite extension and ciliogenesis in the majority of ciliated sensory neurons occur during late embryonic development (Heiman and Shaham, 2009; Nechipurenko et al., 2017; Serwas et al., 2017), these processes are challenging to visualize in vivo. In contrast, AQR and PQR extend their dendrites during larval stages, allowing for investigations into the mechanisms underlying coordination of basal body localization and dendrite morphogenesis.
The multifunctional molecule GRDN-1/Girdin has recently been implicated in regulating basal body positioning and dendrite morphogenesis in C. elegans head amphid and URX/BAG sensory neurons, respectively (Cebul et al., 2020; Nechipurenko et al., 2016). The twelve pairs of sensory neurons in the bilateral amphid sense organs are born near the presumptive nose in the early embryo (Sulston et al., 1983). Following initial anterograde dendritic extension toward the developing nose (Fan et al., 2019), these dendrites ‘stretch’ from their attachment points at the nose as the cell bodies migrate posteriorly to their final positions (Heiman and Shaham, 2009; Low et al., 2019). Centrioles/basal bodies can be visualized in close proximity to the plasma membrane at the embryonic stages when amphid sensory dendrites begin to form (Nechipurenko et al., 2017; Serwas et al., 2017), indicating that in these neurons, basal bodies are likely anchored at the dendritic ends before the dendrites extend retrogradely. Similarly, the dendrites of the embryonically born URX and BAG sensory neurons extend retrogradely (Cebul et al., 2020), although localization of basal bodies has not been studied extensively in these neurons during development. In the amphid neurons, GRDN-1 acts cell autonomously to regulate basal body position and plays only a minor role in dendrite development (Nechipurenko et al., 2016). In contrast, in URX/BAG, GRDN-1 functions in the IL2so glia to regulate dendrite morphology; the role of this protein in URX/BAG ciliogenesis remains to be determined (Cebul et al., 2020) (M. Heiman, personal communication). These observations suggest that dendrite extension and basal body positioning are regulated by partly independent mechanisms in different neuron types, and that GRDN-1 contributes differentially to these processes in a cell type-specific manner.
In contrast to the retrograde dendrite extension exhibited by the embryonic amphid, URX, and BAG sensory neurons, the dendrites of the postembryonic AQR and PQR neurons extend anterogradely in a manner similar to mammalian olfactory neurons (Kirszenblat et al., 2011; Li et al., 2017; McEwen et al., 2008; White et al., 1986). The distal dendritic end of PQR is ensheathed by the process of the PHso2L glial cell, while the AQR distal dendrite does not appear to be associated with glia (Hall and Russell, 1991; White et al., 1986). LIN-44/Wnt signaling from posterior hypodermal sources acts via the LIN-17/Frizzled receptor in PQR to define the site of dendrite emergence from the PQR cell body (Kirszenblat et al., 2011). Moreover, translocation of the basal body to the PQR dendritic tip has been shown to require dynein (Li et al., 2017). Additional mechanisms for AQR/PQR (referred to henceforth as A/PQR) dendrite morphogenesis, basal body localization, or ciliogenesis are unknown.
Here we show that GRDN-1/Girdin acts via partly distinct pathways to regulate dendrite morphogenesis and basal body/cilium positioning in A/PQR. GRDN-1 is localized to the basal body in both neuron types, and tracks the dendritic growth cone as the AQR dendrite extends anterogradely during postembryonic development. Loss of grdn-1 results in mislocalization of basal bodies and cilia to ectopic subcellular locations in A/PQR. Moreover, A/PQR neurons exhibit a range of aberrant morphological phenotypes in grdn-1 mutants. Localization of HMR-1 cadherin to the AQR distal dendrite is disrupted in grdn-1 mutants; however, neuronal knockdown of HMR-1 alone, or together with loss of SAX-7/L1CAM function, does not affect AQR morphology or cilium position. We further demonstrate that UNC-116/Kinesin-1 and LIN-44/Wnt regulate GRDN-1 localization in AQR and PQR, respectively, and that both dendrite morphogenesis and cilium positioning in these neurons are compromised in the corresponding mutants. Taken together, our results describe a new cell type-specific role for GRDN-1 in modulating dendrite morphology and cilium positioning in two specialized sensory neuron types, and further highlight the importance of cellular context in the regulation of function of this versatile protein.
MATERIALS AND METHODS
C. elegans genetics
Animals were grown on nematode growth medium (NGM) at 20°C with E. coli OP50 bacteria as a food source. Transgenic animals carrying extrachromosomal arrays were generated by germline microinjection of plasmids at 10-50 ng/μl into wild-type animals. A transgenic line selected based on expression levels and transmission rate was subsequently crossed into the relevant mutant backgrounds for phenotypic analyses. For rescue experiments of grdn-1-dependent phenotypes, plasmids were injected into grdn-1(hmn1) mutant animals. At least two independent lines were examined for each extrachromosomal array generated in this work, and typically results from one representative line are reported. When necessary, unc-122p::gfp, unc-122p::mcherry, or srg-47p::gfp plasmids were used as co-injection markers (injected at 30-40 ng/μl). To knockout unc-116 specifically in neurons, tag-168p::Cre (nCre) plasmid (Flavell et al., 2013) was injected together with a co-injection marker in the unc-116(ce815) background containing a floxed unc-116 allele (Harterink et al., 2018). The presence of all mutations was confirmed by PCR and/or sequencing.
For experiments using larvae, eggs were collected either for 2 hrs at room temperature, or overnight at 15°C, on standard NGM plates seeded with E. coli OP50 bacteria and allowed to develop for approximately 24 or 48 hrs at 15°C prior to imaging. To obtain staged L1 animals, egg hatching was monitored continuously, and animals that hatched within a 30 min period were transferred to a separate plate and allowed to develop for 6-8 hrs at 25°C prior to imaging. All strains used in this study are listed in Table S1.
Molecular biology
The following promoters were used to drive expression in specific cells or tissues: gcy-32p (~0.7 kb, A/PQR and URX), gcy-36p (~1.0 kb, A/PQR and URX), rab-3p (~4.3 kb, gift from O. Hobert), and grdn-1p (~4.5 kb). cDNA sequences for dyf-19, arl-13, and grdn-1 (isoform a) (Nechipurenko et al., 2016) and xbx-1 (Kazatskaya et al., 2017) were subcloned under the relevant cell-specific promoters into a modified C. elegans expression plasmid pPD95.77 (a gift from A. Fire) or pMC10 (a gift from M. Colosimo) using standard cloning protocols. snb-1 cDNA was amplified from a mixed-stage N2 cDNA pool, verified by sequencing, and subcloned under the relevant cell-specific promoter. To generate GRDN-1ΔGCV, a fragment of grdn-1a cDNA lacking sequences corresponding to the GCV amino acids was amplified by PCR and subcloned into the relevant expression vector to replace the corresponding full-length grdn-1a fragment. The sequence of the mutated grdn-1 construct was verified by sequencing. All plasmids used in this work are listed in Table S2.
CRISPR/Cas9-mediated homologous recombination
The auxin-inducible degron (AID) sequence was inserted immediately upstream of the gfp sequence in the LP172 hmr-1(cp21[hmr-1::gfp + loxP]) strain (Marston et al., 2016) using CRISPR/Cas9-mediated homologous recombination as described (Dokshin et al., 2018). To induce homologous recombination, an injection mixture comprised of the following reagents was injected into LP172 animals: S. pyogenes Cas9 Nuclease V3 (0.25 μg/μl, IDT), CRISPR-Cas9 tracrRNA (1 μM, IDT), unc-122p::dsRed (40 ng/μl); donor template (0.11 μg/μl, megamer ssDNA fragment, IDT) with 120-bp homology arms, crRNA (0.02μg/μl, IDT): rCrArArUrArUrUrUrCrUrGrUrCrGrUrCrArCrGrUrGrUrUrUrUrArGrArGrCrUrArUrGrCrU (r denotes RNA nucleotides). AID insertion was verified by PCR and sequencing.
Auxin-inducible degradation of HMR-1
Auxin-mediated HMR-1 degradation was achieved by adapting a previously published protocol (Zhang et al., 2015). Transgenic strains containing hmr-1::AID::gfp and either egl-17p::myr-mCherry or gcy-36p::myr-tagRFP to label AQR (see Table S1) were injected with gcy-32p::TIR1 plasmid and unc-122p::dsRed co-injection marker at 10 and 40 ng/μl respectively. L4 hermaphrodites containing hmr-1::AID::gfp, gcy-32p::TIR1 (selected on the basis of the unc-122p::dsRed co-injection marker), and egl-17p::myr-mCherry or gcy-36p::myr-tagRFP transgenes were placed on OP50-seeded NGM plates containing 2 mM auxin and allowed to age to young adults at 20°C overnight. Following a two-hour egg collection, adult animals were removed from auxin-containing plates, while eggs were allowed to develop at 15°C to L3 larvae or young adults. AQR neurons from animals carrying all transgenes were imaged on a spinning disk confocal microscope.
Microscopy
Larvae or one-day old adult hermaphrodites were immobilized in 10 mM tetramisole (Sigma) and mounted on 10% agarose pads set on microscope slides. Animals were imaged on an inverted spinning disk confocal microscope (Zeiss Axiovert with a Yokogawa CSU22 spinning disk confocal head). Complete z-stacks of A/PQR neurons were acquired at 0.27 μm intervals in Slidebook 6.0 (3i – Intelligent Imaging Innovations) using a Plan Apochromat 100X NA 1.4 or 63X NA 1.4 oil immersion objective. For experiments examining cilia position and/or GRDN-1 localization in the lin-44 and lin-17 mutant backgrounds, adult or larval animals were imaged on an upright THUNDER Imager 3D Tissue (Leica). Complete z-stacks of PQR were acquired at 0.22 μm intervals in Leica Application Suite X software using an HC Plan Apochromat 63X NA 1.40-0.60 oil immersion objective. Acquired optical sections were rendered into maximum intensity projections using Slidebook 6.0 or Image J (NIH, Bethesda, MD).
To assess microtubule polarity in AQR dendrites, 2D time-lapse images of wild-type and unc-116(e2310) mutant animals carrying a single copy of the integrated gcy-36p::ebp-2::gfp transgene (Harterink et al., 2018) (Table S1) were acquired on an inverted spinning disk confocal microscope (Zeiss Axiovert with a Yokogawa CSU22 spinning disk confocal head) with a Plan Apochromat 63X NA 1.4 oil immersion objective (300 ms exposure) or an upright THUNDER Imager 3D Tissue (Leica) using an HC Plan Apochromat 63X NA 1.40-0.60 oil immersion objective (200 ms exposure). 200-300 frames were recorded per animal. A/PQR morphology and localization of fluorophore-tagged proteins were quantified in Slidebook 6.0 or Image J. Kymographs were constructed using the MultipleKymograph plugin in Image J.
Statistical analysis
Prism 6 software (GraphPad, San Diego, CA) was used to carry out statistical analysis and generate all bar graphs and scatter plots.
RESULTS
GRDN-1 is localized to the basal body in AQR and PQR sensory neurons
Although the bipolar A/PQR neurons (Figure 1A) are known to be ciliated (Hall and Russell, 1991; White et al., 1986), cilium position and structure in these neurons have not been extensively characterized. In adult hermaphrodites, the cilia membrane-associated protein ARL-13::tagRFP (Caspary et al., 2007; Cevik et al., 2010; Li et al., 2010) was localized to the distal dendrites in both A/PQR (Figure 1B). Similar to observations in head sensory neurons, in which cilia emanate from the distal dendritic tips (Doroquez et al., 2014; Perkins et al., 1986; Ward et al., 1975), the cilium was present at the distal end of the PQR dendrite (Figure 1A–B) (Li et al., 2017). However, in AQR, the cilium was typically localized near, but not at, the distal dendritic tip (Figure 1A–B).
Figure 1.
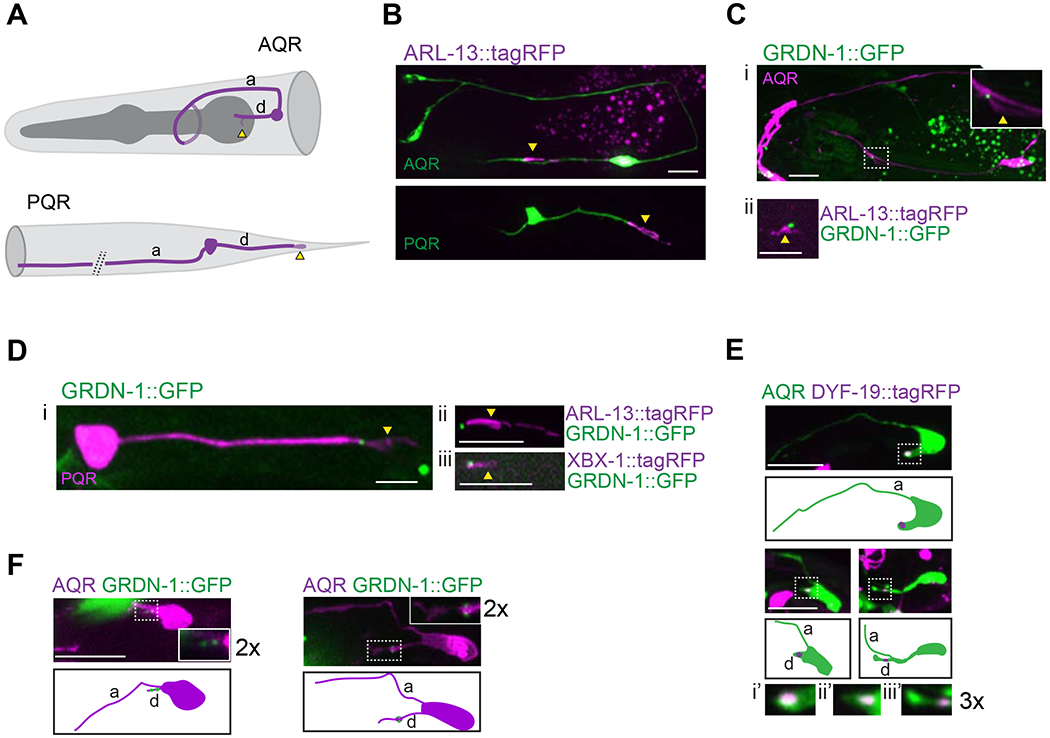
GRDN-1 is expressed in A/PQR and localizes to the cilia base in the distal dendrite.
(A) Diagram of AQR (top) and PQR (bottom) neurons in an adult hermaphrodite.
(B) Images of AQR (top) and PQR (bottom) in adults showing position of cilia labeled with gcy-32p::ARL-13::tagRFP. A/PQR were visualized via expression of gcy-37p::GFP.
(C-D) Images showing expression of GRDN-1::GFP in AQR (Ci) and PQR (Di), together with gcy-32p::ARL13::tagRFP in AQR (Cii) and PQR (Dii), and with gcy-32p::XBX-1::tagRFP in PQR (Diii). The cilium (dashed box) is enlarged (two-fold) in the inset in (Ci). A/PQR were visualized with gcy-32p::mCherry. GRDN-1::GFP was expressed under grdn-1 regulatory sequences in (Ci) and (Di) and gcy-36 regulatory sequences in (Cii) and (Dii-Diii). All images are of adult hermaphrodites.
(E) Images (Ei-Eiii, top) and cartoons (Ei-Eiii, bottom) showing localization of gcy-36p::DYF-19::tagRFP during the depicted stages of AQR dendrite outgrowth in L1 larvae. Dendritic tips (dashed boxes in Ei-Eiii, top) are enlarged three-fold in corresponding insets (Ei’-Eiii’). AQR was labeled with gcy-37p::GFP.
(F) Images (Fi-Fii, top) and cartoons (Fi-Fii, bottom) of L1 larval AQR neurons marked with egl-17p::myr-mCherry expressing gcy-36p::GRDN-1::GFP. Insets show two-fold magnification of distal dendrites (white dashed boxes).
Scale bars: 5 μm; anterior is at left. a - axon; d – dendrite; yellow arrowheads – cilium; white arrowheads – dendritic tips.
To investigate the expression and subcellular localization of GRDN-1 in A/PQR, we expressed the functional GFP-tagged full-length GRDN-1a isoform (Figure S1; henceforth referred to as GRDN-1::GFP) under its endogenous regulatory sequences (Nechipurenko et al., 2016). GRDN-1::GFP was expressed in A/PQR and localized to the cilia base in both neurons in adult hermaphrodites (Figure 1C–D). The dynein light intermediate chain protein XBX-1 localizes to the basal body and undergoes bidirectional movement along the ciliary axoneme (Schafer et al., 2003). The GRDN-1::GFP signal overlapped with the proximal-most region of the XBX-1::tagRFP signal in PQR (Figure 1Diii), suggesting that GRDN-1 localizes to the basal body in PQR.
AQR is born from the QR neuroblast in the L1 larval stage and completes migration to its final destination near the posterior bulb of the pharynx by ~9 hrs posthatching (Middelkoop and Korswagen, 2014; Sulston and Horvitz, 1977). Upon reaching its final location, AQR initiates neuritogenesis (Chai et al., 2012). In contrast to observations in PQR (Kirszenblat et al., 2011; Li et al., 2017), outgrowth of the axon precedes that of the dendrite in AQR (Figure 1E). We noted that when detectable, the basal body protein DYF-19::tagRFP was consistently associated with the distal segment of the extending AQR dendrite in L1 larvae, suggesting a coordination between dendrite outgrowth and basal body localization in AQR (Figure 1E). GRDN-1::GFP puncta were also found in proximity to the tip of the growing AQR dendrite, likely associated with both centrioles (Figure 1F) (Mao et al., 2012; Nechipurenko et al., 2016). The localization of GRDN-1::GFP and DYF-19::tagRFP in the extending AQR dendrite, together with the association of GRDN-1 with the basal body in PQR and amphid sensory neurons (Nechipurenko et al., 2016) (Figure 1Diii) suggests that GRDN-1 also associates with the basal body in the distal segment of the outgrowing AQR dendrite.
GRDN-1 regulates basal body and cilium position in AQR
We asked whether GRDN-1 is required for correct basal body positioning in A/PQR as is the case in amphid sensory neurons (Nechipurenko et al., 2016). While the putative null mutation in grdn-1 results in embryonic lethality (Nechipurenko et al., 2016), we had previously reported basal body positioning defects in amphid neurons in animals carrying viable grdn-1(hmnl) and grdn-1(ns303) hypomorphic alleles (Nechipurenko et al., 2016). The ns303 mutation is an early stop codon that is predicted to produce a single isoform lacking the N-terminal putative microtubule-binding Hook domain, whereas the hmn1 mutation is a late stop codon predicted to generate three truncated isoforms lacking a conserved PDZ binding motif and most of the predicted Gα-binding domain (Nechipurenko et al., 2016) (Figure S1). To quantify basal body localization in AQR, we scored DYF-19::tagRFP puncta as being present in the distal one-third of the dendrite, more proximal dendritic segments, or mislocalized to the cell body or axon. In wild-type AQR, DYF-19::tagRFP was found either within the distal one-third of the dendrite or in the dendritic regions slightly more proximal to the cell body in ~98% of neurons (Figure 2A–B). In contrast, DYF-19::tagRFP was either undetectable or mislocalized to the AQR soma or proximal axon segments in a fraction of examined grdn-1(ns303) and grdn-1(hmnl) mutants (Figure 2A–B). These results suggest that GRDN-1 likely regulates association of the basal body with the outgrowing AQR dendrite.
Figure 2.
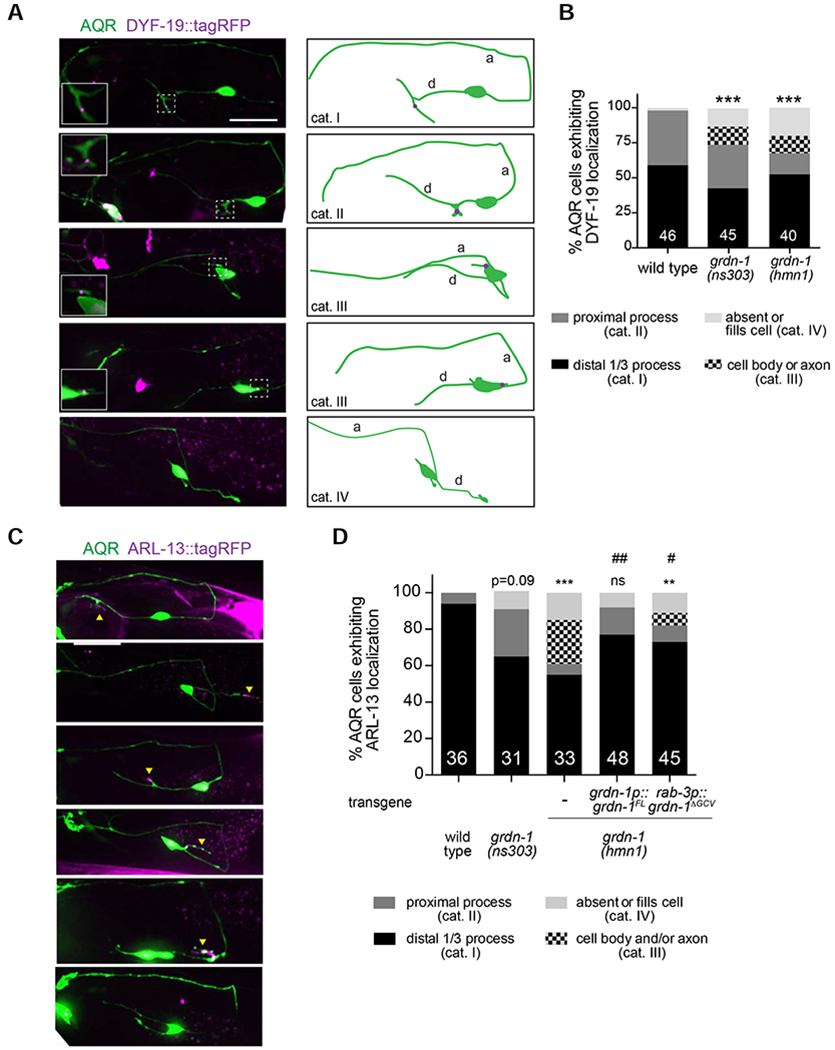
GRDN-1 regulates cilium position in AQR.
(A) Representative images (left) and cartoons (right) showing localization of the basal body marker gcy-36p::DYF-19::tagRFP in AQR neurons of adult wild-type and grdn-1 mutant animals. Images are classified into different phenotypic categories for quantification in (B). AQR was visualized with gcy-37p::GFP. Insets show two-fold magnification views of basal body regions (white dashed boxes).
(B) Quantification of DYF-19::tagRFP localization in adult AQR neurons in the indicated genetic backgrounds. *** indicates different from wild type at P<0.001 (Fisher’s exact test). Representative images for different phenotypic categories are indicated in (A).
(C-D) Representative images (C) and quantification (D) of gcy-32p::ARL-13::tagRFP localization in AQR neurons in animals of the indicated genotypes. Images are classified into different phenotypic categories for quantification in (D). AQR was visualized with gcy-37p::GFP. Yellow arrowheads and asterisks mark cilia and gut autofluorescence, respectively. ** and *** indicate different from wild type at P<0.01 and P<0.001 (or the shown P-value); ns – not significant; # and ## indicate different from grdn-1(hmn1) at P<0.05 and P<0.01, respectively (Fisher’s exact test).
In all image panels, a – axon; d – dendrite; anterior is at left. Scale bars: 10 μm. In all bar graphs, numbers indicate number of AQR neurons examined per genotype.
We noted that in grdn-1 mutants, mislocalized DYF-19::tagRFP was occasionally detected at the base of a short cellular process, suggesting that cilia were being assembled at ectopic cellular locations in AQR (Figure 2A). Consistent with this hypothesis, the ciliary membrane protein ARL-13::tagRFP labeled cellular protrusions on the soma and proximal axonal segments of AQR in grdn-1(hmnl) mutants (Figure 2C–D). Although we did not detect similar ectopic structures in the somas/axons in grdn-1(ns303) animals, cilia were located in proximal dendritic segments of AQR in a larger percentage of grdn-1(ns303) mutants as compared to wild-type animals (Figure 2D). These findings suggest that basal body migration to the cell surface is largely unaffected in grdn-1 mutants; instead, GRDN-1 may be required for correct basal body targeting to the distal sensory dendrite.
The ARL-13::tagRFP mislocalization phenotype in grdn-1(hmnl) animals was rescued upon expression of full-length wild-type grdn-1a cDNA under the endogenous grdn-1 promoter (Figure 2D). The C-terminus of GRDN-1 which is deleted in the grdn-1(hmnl) allele, contains a three amino acid putative PDZ-binding motif (GCV) (Figure S1) (Enomoto et al., 2006; Oshita et al., 2003). We previously showed that deletion of C-terminal sequences including but not limited to the GCV motif resulted in mislocalization of GRDN-1 in amphid neurons (Nechipurenko et al., 2016). However, neuronal expression of GRDN-1 lacking the GCV motif alone (grdn-1ΔGCV) under the panneuronal rab-3 promoter partially but significantly rescued the ARL-13::tagRFP localization defects in grdn-1(hmnl) mutants (Figure 2D). We infer that GRDN-1 acts in neurons to regulate cilia positioning in AQR, and that the PDZ-binding GCV motif is largely dispensable for this process.
GRDN-1 regulates AQR dendrite morphology
In the process of characterizing cilium position in adult hermaphrodites, we noted marked defects in AQR dendritic morphology in grdn-1 mutants. Phenotypes included misrouted dendrites which extended posteriorly, multipolar and unipolar rather than bipolar AQR morphology, dendrites and axons arising from a common process instead of opposites poles of the soma, as well as truncated dendrites (Figure 3A–B). While the penetrance of DYF-19::tagRFP localization defects were similar in both grdn-1 alleles, dendritic defects were more penetrant in grdn-1(hmnl) than grdn-1(ns303) mutants (Figure 3B). Gross axon morphology was not altered in either mutant background. The dendritic defects in grdn-1(hmnl) mutants were partly but significantly rescued upon expression of a wild-type grdn-1a cDNA under the endogenous grdn-1 or the panneuronal rab-3 promoter (Figure 3B–C). Although the C-terminal GCV motif in GRDN-1 was shown to be important for retrograde extension of dendrites in the gas-sensing URX neurons (Cebul et al., 2020), when expressed under the endogenous or pan-neuronal regulatory sequences, a GRDN-1 construct lacking this motif partly but significantly rescued AQR dendrite morphology in grdn-1(hmnl) mutants (Figure 3B). These results indicate that GRDN-1 acts neuronally to regulate dendritic morphogenesis in AQR, but unlike in URX, the GCV motif is not essential for GRDN-1 function in this neuron type.
Figure 3.
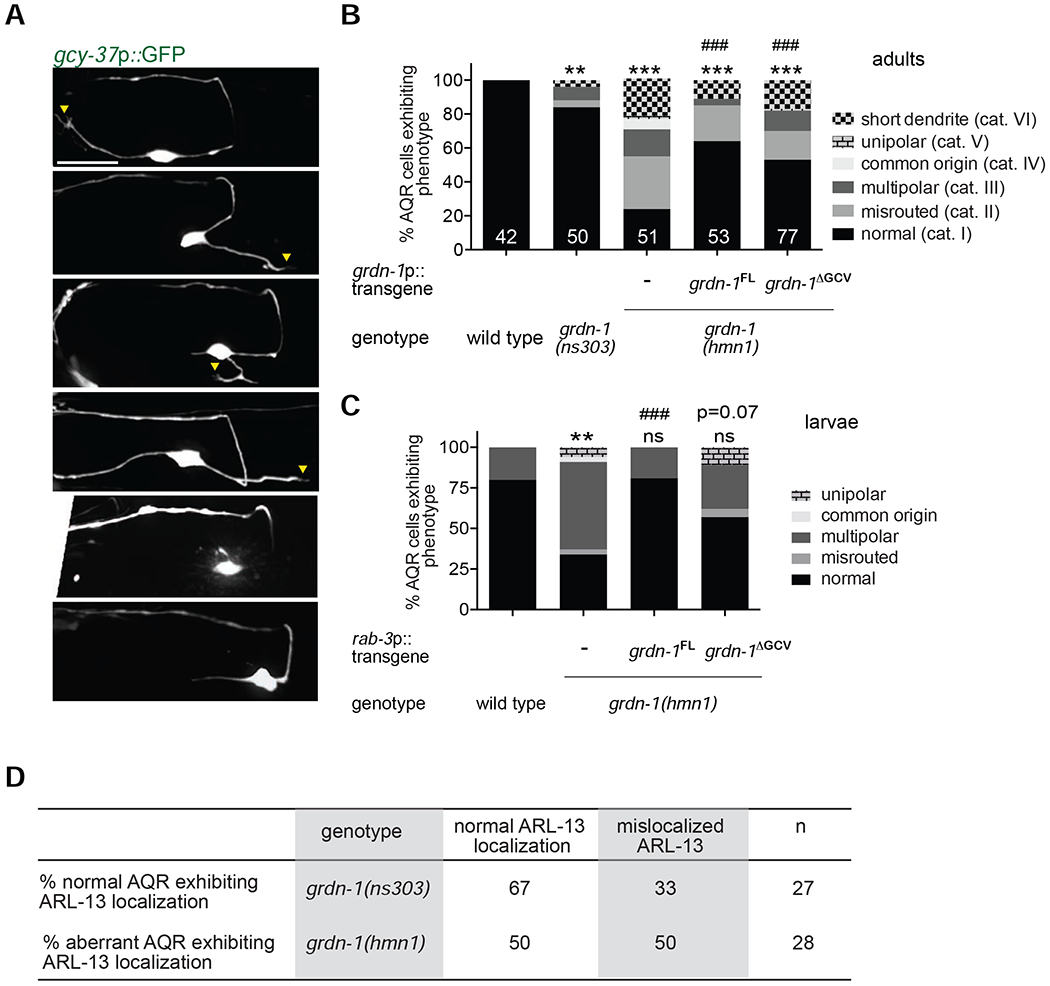
GRDN-1 is required for AQR dendrite development.
(A-C) Representative images (A) and quantification (B, C) of morphological defects in adult (A, B) and L2-L3 larval (C) AQR neurons of the indicated genotypes. Images are classified into different phenotypic categories for quantification in (B). Qualitatively similar phenotypic categories in larval AQR visualized with egl-17p::myr-mCherry were used for quantification in (C). Yellow arrowheads mark presumptive cilia; a – axon; d – dendrite; anterior is at left. Scale bar: 10 μm. FL – full-length; ΔGCV – GCV motif deleted. ** and *** indicate different from wild type at P<0.01 and 0.001, respectively; ns – not significant; ### indicates different from grdn-1(hmn1) at P<0.001 or the indicated P-value; ns – not significant (Fisher’s exact test). Numbers indicate number of AQR neurons examined per genotype.
(D) Correlation between AQR dendrite morphology and cilia position in the indicated grdn-1 mutant strains.
The presence of multiple neurites and reversed direction of dendrite outgrowth in grdn-1 mutant AQR could arise from gross defects in neuronal polarity (Bentley and Banker, 2016; Yogev and Shen, 2017). To test this notion, we examined localization of the presynaptic protein SNB-1::tagRFP in AQR of wild-type and grdn-1(hmnl) mutants. We noted that regardless of neuronal morphology, SNB-1::tagRFP was restricted to the synaptic regions in AQR axons of both wild-type and grdn-1(hmnl) animals (Figure S2). We conclude that it is unlikely that defects in AQR morphology observed in grdn-1 mutants are due to global changes in neuronal polarity.
Since the basal body and cilium are associated with the distal end of the AQR dendrite, we asked whether cilium/basal body mislocalization and dendritic defects were correlated in grdn-1 mutants. We scored cilium position as ‘wild-type’ when ARL-13::tagRFP was localized to the distal segment of a presumed dendrite, regardless of the direction of dendrite outgrowth. We found that in grdn-1(hmnl) mutants, ARL-13: :tagRFP was present in the soma or proximal axonal segments in 50% of examined AQR neurons that exhibited altered morphology [grdn-1(ns303) animals were not examined due to the low penetrance of the AQR dendritic defect in this mutant background] (Figure 3D). Conversely, we observed ectopic ARL-13::tagRFP localization to the soma or axon in 33% of morphologically wild-type AQR neurons in grdn-1(ns303) mutants (Figure 3D). Together with the differential effects of grdn-1(hmnl) and grdn-1(ns303) mutations on basal body positioning and neuronal morphology, these results imply that GRDN-1 plays at least partly independent roles in basal body positioning and dendrite morphogenesis in AQR.
Mutations in grdn-1 affect cilia position and dendritic morphology in PQR
Although the dendrites of both AQR and PQR extend anterogradely in early larval stages, a key anatomical difference between AQR and PQR is that the distal end of the PQR, but not the AQR, dendrite is ensheathed by the PHso2L glial cell process (Hall and Russell, 1991; White et al., 1986). Recently, GRDN-1 was shown to act cell non-autonomously to regulate retrograde extension of glia-associated URX dendrites (Cebul et al., 2020). We asked whether GRDN-1 is required for anterograde extension of the glia-associated dendrite, and/or cilium position, in PQR.
While ARL-13::tagRFP was present at the distal tip of the PQR dendrite in 100% of wild-type animals, in grdn-1 mutants, ARL-13::tagRFP was mislocalized to the PQR cell body or proximal dendritic segments in a subset of examined animals (Figure 4A–B). As in AQR, the ARL-13::tagRFP mislocalization phenotype was more penetrant in grdn-1(hmn1) than grdn-1(ns303) mutants and was fully rescued upon GRDN-1ΔGCV expression under the pan-neuronal rab-3 promoter (Figure 4B). PQR also exhibited a range of dendritic morphological defects in grdn-1(hmnl) mutants that were qualitatively similar to, but less penetrant than, those observed in AQR (Figure 4C–D). We noted that PQR dendrites were significantly shorter in grdn-1(hmnl) mutants; this defect was also fully rescued by GRDN-1ΔGCV expression under the rab-3 promoter (Figure 4E). We conclude that GRDN-1 acts in neurons to regulate PQR cilium position and dendrite morphology, and the C-terminal GCV motif is not required for the examined phenotypes in this neuron type. Moreover, GRDN-1 appears to play a more prominent role in AQR than in PQR morphogenesis.
Figure 4.
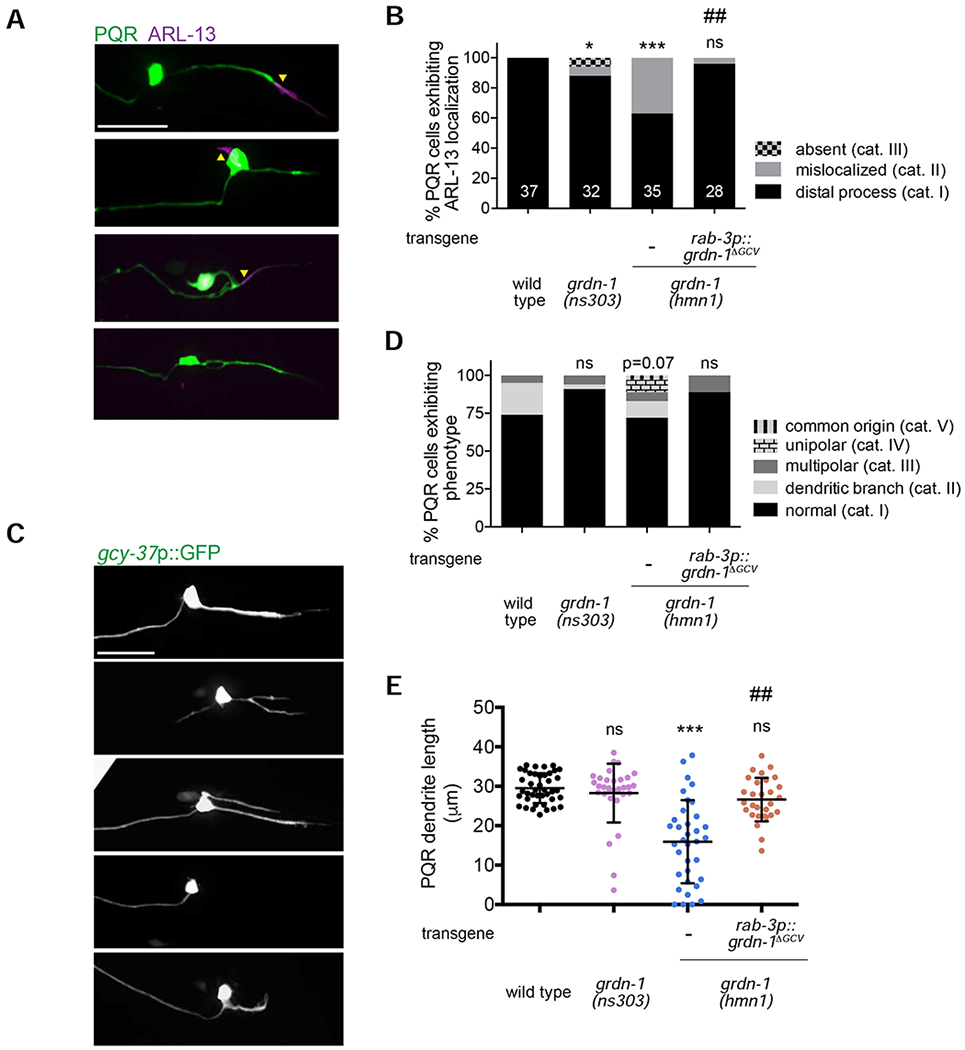
PQR morphology and cilium localization are differentially affected in grdn-1 mutants.
(A–B) Representative images (A) and quantification (B) of gcy-32p::ARL-13::tagRFP localization in PQR neurons of the indicated genotypes. Images are classified into different phenotypic categories for quantification in (B). PQR was visualized with gcy-37p::GFP. Yellow arrowheads mark cilia. * and *** indicate different from wild type at P<0.05 and P<0.001, respectively; ns – not significant; ## indicates different from grdn-1(hmn1) at P<0.01 (Fisher’s exact test).
(C–D) Representative images (C) and quantification (D) of morphological defects exhibited by PQR neurons of the listed genotypes. Images are classified into different phenotypic categories for quantification in (D). P-value is derived by Fisher’s exact test; ns – not significant (Fisher’s exact test).
(E) Quantification of PQR dendrite length in wild-type and grdn-1 mutant animals. *** indicates different from wild type at P<0.001; ns – not significant (Kruskal-Wallis test with Dunn’s test for multiple comparisons).
In all image panels, a – axon; d – dendrite; anterior is at left. Scale bars: 10 μm. In all bar graphs, numbers indicate number of PQR neurons examined per genotype.
GRDN-1 regulates localization of HMR-1 cadherin, but not that of SAX-7 L1CAM, in AQR
Multiple cell adhesion molecules mediate dendrite morphogenesis across species including in C. elegans (Berger-Muller and Suzuki, 2011; Dong et al., 2013; Lefebvre, 2017; McLachlan and Heiman, 2013; Salzberg et al., 2013; Seong et al., 2015). The HMR-1 classical cadherin, SAX-7 L1 cell adhesion molecule, DLG-1 Discs large homolog 1, and DYF-7 Zona Pellucida domain proteins are expressed in the embryonically born amphid sensory neurons in C. elegans and act redundantly to mediate the initial stages of dendrite extension in these cells (Fan et al., 2019). DYF-7 is also critical for retrograde extension of amphid, but not URX/BAG, sensory dendrites (Cebul et al., 2020; Heiman and Shaham, 2009; Low et al., 2019). Recently, GRDN-1 and SAX-7 were shown to mediate anchoring of URX/BAG sensory dendrites at the nose of the developing embryo (Cebul et al., 2020). Girdin also physically interacts with components of the cadherin-catenin complex during Drosophila embryogenesis (Houssin et al., 2015) and in human cancer cells (Wang et al., 2018), and regulates cadherin localization in some cellular contexts (Ichimiya et al., 2015; Muramatsu et al., 2015; Weng et al., 2014). We asked whether GRDN-1 acts via one or more of these identified cell adhesion molecules to regulate AQR morphology.
We first examined whether candidate cell adhesion molecules are expressed in the developing AQR neuron. We found that endogenously tagged HMR-1::GFP protein localized to the distal segment of the emerging AQR dendrite in wild-type larvae (Figure 5A), similar to its localization pattern at the distal tips of dendrites in embryonic amphid sensory neurons (Fan et al., 2019). Like DYF-19 and GRDN-1, HMR-1::GFP remained associated with the distal dendritic segment, as the AQR dendrite extended anteriorly (Figure 5A–B). We also observed localization of endogenously tagged SAX-7::GFP along the AQR dendrite, as well as in the soma and axon, of wild-type larvae (Figure S3A). However, DYF-7::sfGFP expressed under its endogenous promoter (Low et al., 2019) was detected in the AQR cell bodies and/or dendrite in only four out of 13 examined animals, suggesting that this expression may be an artifact of transgene overexpression (Figure S3B). We did not observe expression of endogenously tagged DLG-1::mNG protein in larval AQR (n=30).
Figure 5.
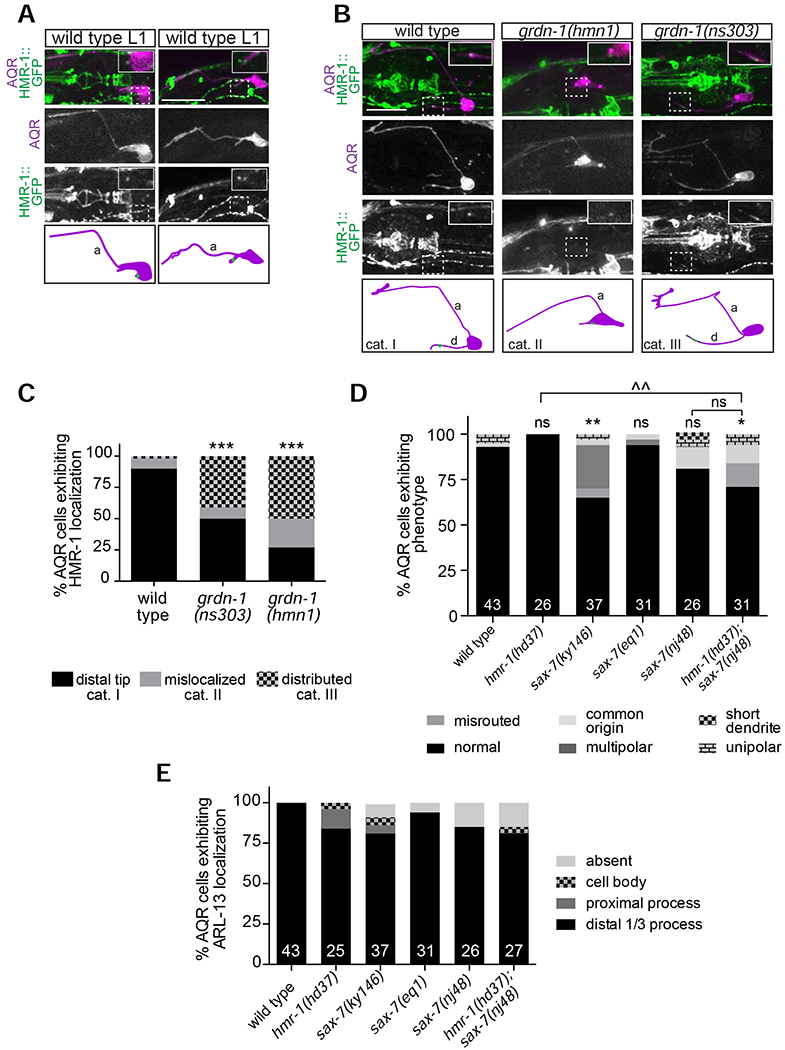
GRDN-1 regulates HMR-1 localization in AQR dendrites.
(A–B) Images (top panels) and cartoons (bottom panels) showing localization of endogenously tagged HMR-1::GFP in AQR at different stages of dendrite outgrowth in wild-type L1 larvae (A) and in L2-L3 larvae of the indicated genotypes (B). Images are classified into different phenotypic categories for quantification in (C). HMR-1::GFP localization in AQR (dashed boxes) is enlarged two-fold in all insets. AQR was visualized with egl-17p::myr-mCherry. White arrowheads mark HMR-1::GFP in AQR; a – axon; d – dendrite; anterior is at left. Scale bars: 5 μm.
(C) Quantification of HMR-1::GFP localization in L2-L3 larvae. Representative images for different phenotypic categories are indicated in (B).
(D–E) Quantification of AQR morphology (D), and gcy-32p::ARL-13::tagRFP localization (E) in adult animals of the indicated genotypes. In all bar graphs, numbers indicate number of AQR neurons examined per genotype. The phenotypes of the mutants indicated in (E) are not statistically different from wild type. *, ** and *** indicate different from wild type at P<0.05, P<0.01 and P<0.001, respectively; ^^ indicates different between indicated genotypes at P<0.01; ns – not significant (Fisher’s exact test).
We next asked whether GRDN-1 regulates AQR morphogenesis by modulating localization of HMR-1 and/or SAX-7. In both grdn-1(hmnl) and grdn-1(ns303) mutants, the HMR-1::GFP signal was no longer localized to a well-defined punctum at the distal dendrite but instead was more diffusely distributed, absent, or aberrantly localized to the AQR cell body (Figure 5B–C). HMR-1::GFP localization was similarly affected in both grdn-1 mutant backgrounds (Figure 5C). In contrast, localization of SAX-7::GFP in AQR was unaffected in grdn-1(hmnl) mutants, and conversely, GRDN-1::GFP localized normally to the distal AQR dendrite in sax-7(nj48) animals (Figure S3A and S3C). We conclude that GRDN-1 is necessary for HMR-1, but not SAX-7, localization in AQR.
Reduction of HMR-1 function and/or loss of SAX-7 is not sufficient to alter AQR morphology or cilium positioning
Since GRDN-1 regulates HMR-1 localization, we investigated whether AQR morphogenesis or cilium positioning is altered in hmr-1 mutants. Embryonic lethality of hmr-1 null and hmr-1(RNAi) animals precludes examination of postembryonically generated AQR neurons in these genetic backgrounds (Costa et al., 1998). However, the hmr-1(hd37) allele is predicted to specifically disrupt the neuronal hmr-1b isoform and is viable (Steimel et al., 2010) (www.wormbase.org). Despite altered HMR-1::GFP localization in AQR in grdn-1 mutants, we observed no or minor defects in AQR morphology or cilia position in hmr-1(hd37) animals (Figure 5D–E). Reasoning that this hypomorphic mutation may not result in sufficient reduction of hmr-1 function to cause significant defects in AQR, we depleted HMR-1 in a subset of sensory neurons including AQR using the auxin-inducible protein degradation (AID) system (Holland et al., 2012; Nishimura et al., 2009; Zhang et al., 2015). We introduced the degron sequence into the endogenous gfp-tagged hmr-1 locus and generated strains expressing this fusion protein together with the TIR1 F-box protein driven under the gcy-32 promoter that is transcriptionally active in A/PQR and URX (Yu et al., 1997). Levels of HMR-1::GFP fluorescence intensity in AQR dendrites of L3 larvae were significantly decreased in the presence of auxin (Figure S4A). AQR morphology was again largely unaffected in animals raised under these conditions (Figure S4B). Thus, although GRDN-1 is required for proper HMR-1 localization in AQR, neuron-specific reduction of HMR-1 function alone does not appear to be sufficient to alter AQR morphology or cilium position. However, we are unable to exclude the possibility that HMR-1 retains residual function in hmr-1(hd37) mutants or upon auxin-mediated depletion in AQR.
Since sax-7 is also expressed in AQR and is known to function redundantly with hmr-1 in several developmental contexts (Fan et al., 2019; Grana et al., 2010), we examined whether SAX-7 contributes to AQR morphogenesis. We observed weak but significant defects in AQR dendritic morphology in animals carrying the sax-7(ky146) but not the sax-7(nj48) strong loss-of-function or sax-7(eq1) null alleles (Figure 5D). Since the sax-7(ky146) allele has been reported to generate aberrant readthrough transcripts particularly in the nervous system (Wang et al., 2005), it is possible that the low penetrance AQR defects observed in this mutant background arise due to neomorphic SAX-7 function. AQR morphological defects in sax-7(nj48) mutants were not enhanced by additional loss of hmr-1 (Figure 5D). Similarly, we did not observe significant defects in AQR cilium positioning in any of the examined sax-7 alleles or in hmr-1(hd37); sax-7(nj48) double mutants (Figure 5E). Together, these results suggest that HMR-1 and SAX-7 may not be required for dendrite outgrowth and cilium positioning in AQR, or that these proteins act redundantly with additional molecules to regulate these aspects of AQR development.
UNC-116/Kinesin-1 regulates GRDN-1 localization in AQR
The mechanisms that regulate GRDN-1 function and localization in the context of sensory dendrite development or ciliogenesis are unknown. The KIF5a kinesin-1 motor was previously shown to regulate Girdin localization to the Golgi and cell adhesion contacts in mammalian cells (Muramatsu et al., 2015). We examined whether UNC-116/Kinesin-1 similarly regulates GRDN-1 localization in the AQR dendrite. Since null mutations in unc-116 result in embryonic lethality (Byrd et al., 2001), we examined animals carrying the hypomorphic unc-116(e2310) allele (Patel et al., 1993; Yan et al., 2013). We found that GRDN-1::GFP was localized to the soma in ~65% of examined unc-116(e2310) mutants, as compared to 12% of wild-type animals (Figure 6A–B). Consistent with a role for GRDN-1 in localizing HMR-1 to distal AQR dendrites, HMR-1::GFP puncta were also frequently mislocalized to the cell body in unc-116(e2310) animals (Figure 6C–D).
Figure 6.
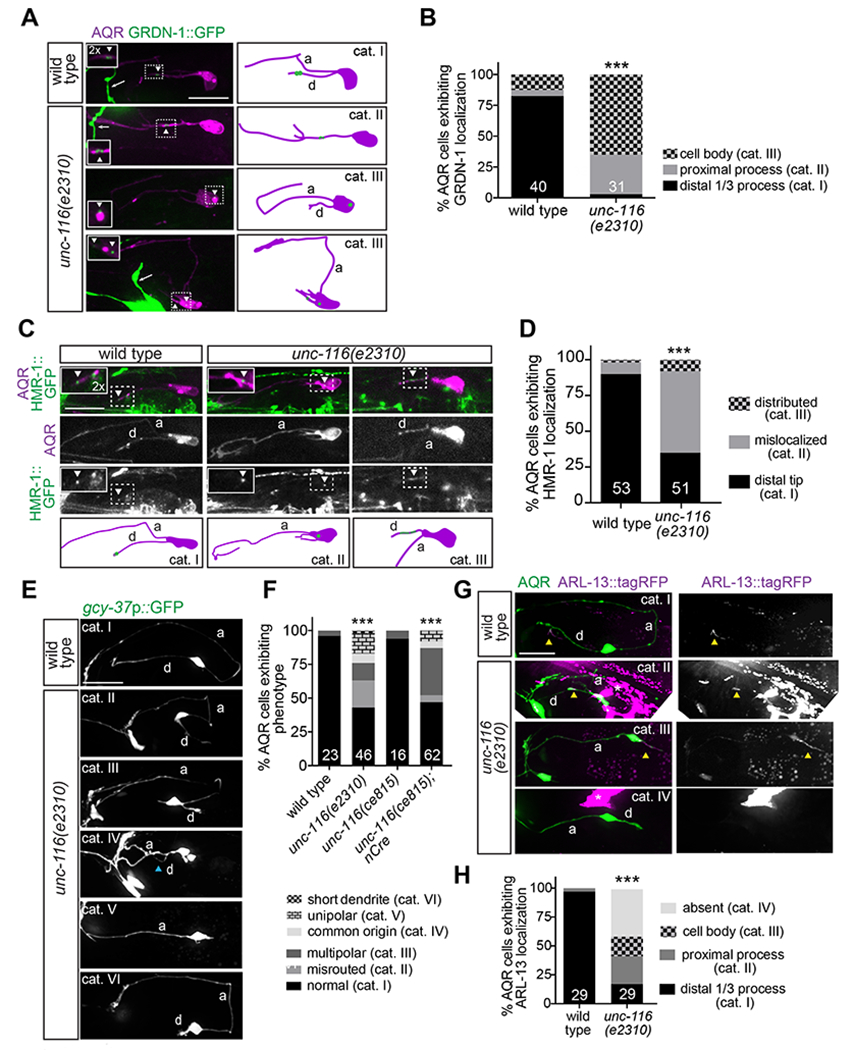
Kinesin-1/UNC-116 regulates GRDN-1 localization and AQR morphology.
(A) Representative images (left) and cartoons (right) of gcy-36p::GRDN-1::GFP localization in AQR of wild-type and unc-116 mutant larvae. Images are classified into different phenotypic categories for quantification in (B). AQR was labeled with egl-17p::myr-mCherry. GRDN-1::GFP signal in AQR (white dashed boxes) is magnified two-fold in all insets. White arrowheads mark GRDN-1::GFP; white arrows point to ASI sensory neuron labeled with srg-47p::GFP in the background.
(B) Quantification of GRDN-1::GFP localization in wild-type and unc-116 L2-L3 larvae. *** indicates different from wild type at P<0.001 (Fisher’s exact test). Representative images for different phenotypic categories are indicated in (A).
(C) Representative images (top panels) and cartoons (bottom panels) showing localization of the endogenously tagged HMR-1::GFP in wild-type and unc-116 mutant L2-L3 larvae. Images are classified into different phenotypic categories for quantification in (D). HMR-1::GFP signal in AQR (white dashed boxes) is magnified two-fold in all insets and marked with white arrowheads. AQR was labeled with egl-17p::myr-mCherry. (D) Quantification of HMR-1::GFP localization in wild-type and unc-116 L2-L3 larvae. Data for wild type are repeated from Figure 5C. Representative images for different phenotypic categories are indicated in (C). *** indicates different from wild type at P<0.001 (Fisher’s exact test).
(E–F) Representative images (E) and quantification (F) of AQR morphological phenotypes in the indicated genetic backgrounds. Images are classified into different phenotypic categories for quantification in (F). Blue arrowhead marks dendrite. *** indicates different from wild type at P<0.001 (Fisher’s exact test).
(G–H) Representative images (G) and quantification (H) of gcy-32p::ARL-13::tagRFP localization in AQR of adult wild-type and unc-116 mutant animals. Images are classified into different phenotypic categories for quantification in (H). Yellow arrowheads and white asterisk mark cilia and gut autofluorescence, respectively. *** indicates different from wild type at P<0.001 (Fisher’s exact test).
In all image panels, a – axon; d – dendrite; anterior is at left. Scale bars: 10 μm. In all bar graphs, numbers indicate number of AQR neurons examined per genotype.
Since UNC-116 regulates GRDN-1 localization in AQR, we asked whether unc-116 mutants display defects in AQR morphology or cilium positioning similar to those observed in grdn-1 mutant animals. As shown in Figure 6E, unc-116(e2310) mutants exhibited a range of AQR dendritic phenotypes that resembled those observed in grdn-1 mutants and included misrouted and truncated dendrites, as well as multipolar and unipolar neuronal morphologies. Moreover, animals in which unc-116 was knocked out in neurons via Cre-loxP-mediated recombination (Harterink et al., 2018), exhibited AQR morphological defects similar to those in unc-116(e2310) mutants (Figure 6F). In addition to altered morphology, unc-116(e2310) mutant AQR neurons displayed ectopic localization of ARL-13::tagRFP (Figure 6G–H), with a subset of examined cells lacking expression possibly due to cilia loss (Figure 6H). In contrast, PQR neurons in unc-116 mutants exhibited relatively weak dendritic and ciliary defects (Figure S5A–B and D–E). As in grdn-1 mutants, PQR dendrites in unc-116 animals were also significantly shorter compared to wild type (Figure S5C). We conclude that UNC-116 regulates GRDN-1 localization to the distal AQR dendrite, and is required for dendrite outgrowth and cilium positioning in AQR, and to a lesser extent, in PQR neurons.
UNC-116 has been shown to regulate dendritic microtubule polarity in a subset of C. elegans neurons such that the typical minus-end-out polarity of dendritic microtubules is reversed to plus-end-out orientation in unc-116 mutants (Yan et al., 2013). We asked whether altered dendritic microtubule polarity may in part account for the defects in GRDN-1 localization in unc-116 mutants in AQR. To address this issue, we compared the direction of movement of a GFP-tagged plus-end tracking protein EBP-2/EB1 in the AQR dendrites of wild-type and unc-116(e2310) mutants. 80-100% of EBP-2::GFP tracks were directed towards the soma in each examined wild-type AQR dendrite (Figure S5F–G), consistent with the minus-end-out orientation of dendritic microtubules in this neuron type. In contrast, the direction of EBP-2::GFP movement was highly variable in the AQR dendrites of unc-116 mutants. The fraction of EBP-2::GFP comets directed towards the AQR soma ranged from 100% in a subset of cells to 0% in others (Figure S5F–G), suggesting that dendritic microtubules may be oriented with their minus- or plus ends out or exhibit mixed polarity in unc-116 mutants. These observations suggest that dendritic microtubule orientation is altered in AQR in unc-116 mutants, and may in part underlie defects in GRDN-1 localization in this background.
LIN-44/Wnt and LIN-17/Frizzled regulate GRDN-1 localization in PQR
We noted that the PQR dendrite defects in grdn-1 mutants, although less penetrant, were qualitatively similar to those previously reported in Wnt signaling mutants (Kirszenblat et al., 2011). We hypothesized that GRDN-1 may mediate a subset of Wnt-dependent functions in PQR morphogenesis. LIN-44/Wnt and the LIN-17/Frizzled receptor were shown to be the primary regulators of dendrite outgrowth in PQR (Kirszenblat et al., 2011). We tested whether LIN-44 and LIN-17 also regulate GRDN-1 localization in PQR. While GRDN-1::GFP localized to the PQR distal dendrite in 97% of wild-type animals, only 20% of lin-44(n1792) and ~40% of lin-17(n671) mutants exhibited normal GRDN-1::GFP localization (Figure 7A–D). In the remaining fraction of mutant animals, GRDN-1::GFP localized to ectopic cellular compartments including the soma and dendritic base, more proximally in the posteriorly projecting or misrouted dendrite, or was undetectable (Figure 7A–D). These results indicate that Wnt signaling regulates GRDN-1::GFP localization in PQR.
Figure 7.
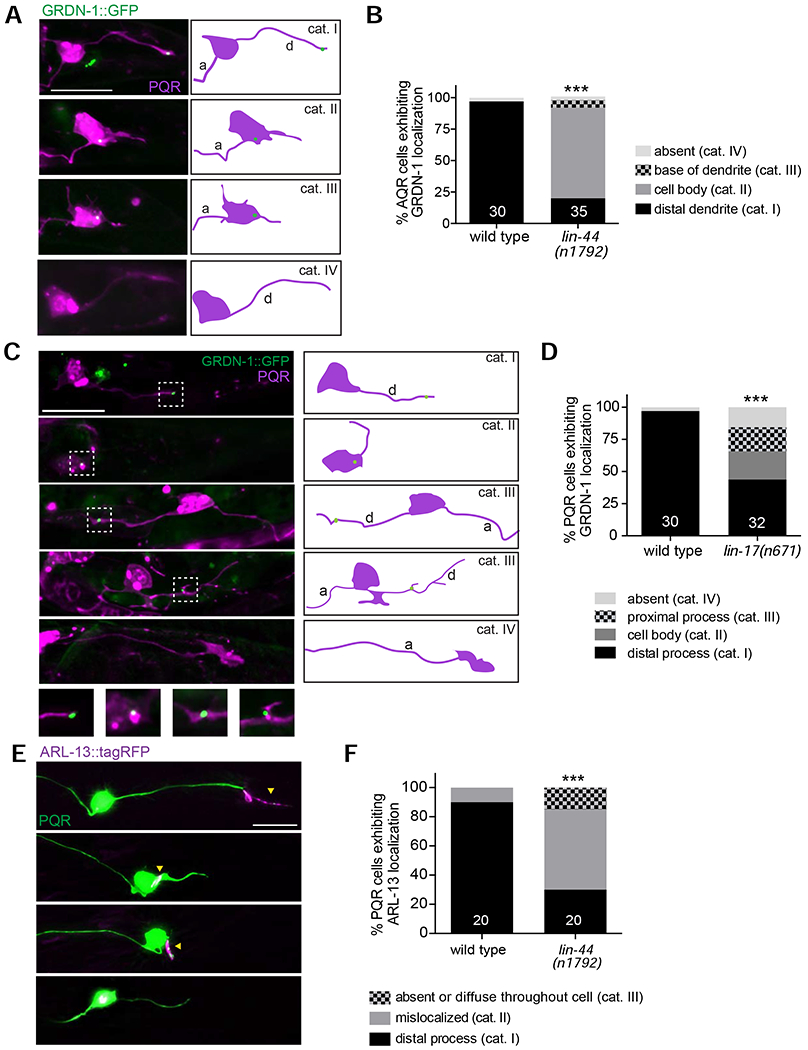
LIN-44/Wnt and LIN-17/Frizzled regulate GRDN-1 localization and cilium position in PQR.
(A-D) Representative images (A, C with cartoons at right) and quantification (B, D) of gcy-36p::GRDN-1::GFP localization in PQR of wild-type and lin-44(n1792) (A, B) and lin-17(n671) (C, D) larvae. Images are classified into different phenotypic categories for quantification in (B and D). Insets (Di’-Div’) show two-fold magnification of the corresponding PQR dendritic regions inside white dashed boxes. PQR was labeled with egl-17p::myr-mCherry. White arrowheads indicate GRDN-1::GFP. *** indicates different from wild type at P<0.001 (Fisher’s exact test). Data for wild type in (D) are repeated from (B).
(E-F) Representative images (E) and quantification (F) of gcy-32p::ARL-13::tagRFP localization in PQR of adult wild-type and lin-44 mutant animals. Images are classified into different phenotypic categories for quantification in (F). Data for wild type are repeated from Figure S5E.Yellow arrowheads mark cilia. *** indicates different from wild type at P<0.001 (Fisher’s exact test).
In all image panels, a – axon; d – dendrite; anterior is at left. Scale bars: 10 μm. In all bar graphs, numbers indicate number of PQR neurons examined per genotype.
A role for Wnt signaling in regulating cilium position in PQR has not been previously described. To test whether mislocalization of GRDN-1 in lin-44 mutants is correlated with altered cilium positioning in PQR, we also examined localization of ARL-13::tagRFP in this neuron type. We found that ARL-13::tagRFP localized normally to the PQR distal dendrite in only 30% of lin-44(n1792) mutants, and was present on the soma or diffusely distributed throughout the neuron in 70% of examined animals (Figure 7E–F). We conclude that Wnt signaling regulates both dendritic morphology and cilium positioning in PQR, possibly in part via regulation of GRDN-1 localization.
DISCUSSION
Here we report that GRDN-1 acts in neurons to regulate dendrite outgrowth and cilia position in the postembryonic AQR and PQR sensory neurons. In both neuron types, GRDN-1 is localized to the basal body. In AQR, the basal body is associated with the distal dendrite as it undergoes anterograde extension in early larvae. We find that in grdn-1 mutants, the cilium in AQR is frequently mispositioned to the soma or proximal axonal segments, and dendrites exhibit a range of outgrowth defects. Similar defects in cilium localization and qualitatively similar but less penetrant defects in dendrite morphogenesis were also observed in PQR suggesting that GRDN-1 plays a more critical role in AQR development. We also show that GRDN-1 regulates HMR-1 but not SAX-7 localization in AQR, although loss of sax-7 or reduction of hmr-1 function singly or together does not affect examined aspects of AQR development. GRDN-1 localization is regulated by UNC-116/Kinesin-1 in AQR, and unc-116 mutants in turn exhibit AQR dendritic and cilium positioning phenotypes resembling those of grdn-1 mutants. In contrast, GRDN-1 localization in PQR is regulated by LIN-44/LIN-17 Wnt/Frizzled signaling. Together, these results describe a new cell-specific role for GRDN-1 and identify its upstream regulators in two ciliated sensory neuron types in C. elegans.
GRDN-1 may act via distinct mechanisms to regulate dendrite morphology and basal body/cilium positioning in AQR. This conclusion is based on the observations: 1) that the grdn-1(ns303) and grdn-1(hmnl) hypomorphic mutations result in qualitatively and quantitatively similar defects in basal body positioning in AQR, but only grdn-1(hmn1) animals exhibit severe defects in AQR dendrite outgrowth, and 2) that the dendrite outgrowth and basal body positioning defects appear to be uncorrelated in AQR in grdn-1 mutants. GRDN-1 also acts in distinct pathways to regulate dendrite morphology and basal body positioning in other C. elegans sensory neuron types. In amphid sensory neurons, GRDN-1 regulates basal body position in part via CHE-10/Rootletin and the apical junction component AJM-1, but has a minor effect on dendrite anchoring (Nechipurenko et al., 2016). In URX/BAG, GRDN-1 acts in glia to regulate dendrite morphology, but it is unknown whether GRDN-1 also plays a role in regulating basal body/cilium position in these sensory neurons (Cebul et al., 2020). Although HMR-1::GFP was mislocalized in AQR in grdn-1 mutants, we detected only minor defects in AQR dendritic morphology or cilia positioning upon reduction of hmr-1 function alone or in combination with loss-of-function alleles of sax-7, suggesting that the effectors of GRDN-1 in regulating different aspects of AQR development remain to be identified. Mammalian Girdin has been shown to act as a hub for multiple signaling pathways in different cellular contexts (Aznar et al., 2016; Garcia-Marcos et al., 2015). Since GRDN-1 is expressed in many neuronal and non-neuronal cell types in C. elegans (Nechipurenko et al., 2016), it will be interesting to determine the extent to which GRDN-1 function is modulated by diverse signaling pathways in different cell types.
We find that UNC-116/Kinesin-1 is required to correctly localize GRDN-1 to distal dendrites in AQR. Reduction or loss of UNC-116 function results in aberrant neuronal morphology and cilium positioning in AQR. Interestingly, although a subset of ciliated sensory neurons in C. elegans is less reliant on UNC-116 to establish dendritic minus-end-out microtubules due to the presence of a distal microtubule organizing center at their dendritic tips (Harterink et al., 2018), AQR dendritic microtubule orientation appears to nevertheless be disrupted in unc-116 mutants. In PQR, the centriole/basal body is translocated to the dendritic tip via dynein (Li et al., 2017). While a role for dynein in basal body translocation in AQR has not been described, it is possible that defects in dendritic microtubule polarity and corresponding defects in motor-driven translocation of GRDN-1 may account in part for the defects in GRDN-1 localization in this neuron type in unc-116 mutants.
GRDN-1 localization in PQR is regulated in part via LIN-44/Wnt and LIN-17/Frizzled. Mutations in lin-44 lead to strong defects in PQR dendritic morphology (Kirszenblat et al., 2011), and we show that these mutants also exhibit strong defects in PQR cilium positioning. Dendrite outgrowth initiates in PQR well after the neuron has completed its migration towards the tail following birth from the QL neuroblast (Kirszenblat et al., 2011). Wnt signaling appears to act very early in PQR development prior to completion of cell migration to specify the site and direction of dendrite outgrowth (Kirszenblat et al., 2011). Consistent with this hypothesis, LIN-17::GFP expression is largely undetectable in PQR at the time of dendrite growth initiation (Kirszenblat et al., 2011) (I.V.N. and P.S., unpublished observations). We speculate that Wnt signaling recruits GRDN-1 directly or indirectly to the site of dendrite emergence; defects in GRDN-1 localization subsequently disrupt dendrite development and ciliogenesis in PQR.
While AQR phenotypes are quantitatively similar in unc-116 and grdn-1(hmnl) mutants, PQR phenotypes are more penetrant in lin-44 than in grdn-1 mutant animals. It is possible that this difference arises due to the hypomorphic nature of examined grdn-1 alleles. Alternatively, GRDN-1 may function in parallel with other molecule(s) downstream of Wnt signaling to regulate PQR morphogenesis. A feature shared by amphid and PQR, but not AQR, neurons is that their dendrites are associated with glia (Doroquez et al., 2014; Hall and Russell, 1991; Perkins et al., 1986; Ward et al., 1975; White et al., 1986). Glial cells of the amphid organ secrete the DEX-1 tectorin-like molecule which acts with DYF-7 to facilitate anchoring of amphid dendritic tips at the nose (Heiman and Shaham, 2009). We hypothesize that GRDN-1-independent, but possibly LIN-44-dependent, glial interactions may similarly maintain PQR dendrite morphology upon reduction of grdn-1 function. Indeed, mutations in lin-44 affect development of the PHso2L glia (Herman and Horvitz, 1994; Herman et al., 1995), and ablation of these glia result in dendritic extension defects, albeit relatively minor, in PQR (Kirszenblat et al., 2011). Structural interactions with surrounding epidermal and muscle cells via multiple protein complexes have also been shown to play a major role in regulating the complex dendritic arbors of the PVD nociceptive neurons (Inberg et al., 2019). The more penetrant dendritic phenotype of grdn-1 mutants in AQR suggests that in the absence of interaction with glia or other surrounding cells, GRDN-1-dependent pathways may play a more critical role in regulating dendrite outgrowth in this neuron type.
Girdin domain organization is highly conserved, and this protein is considered to be a hub for multiple signal transduction pathways in different cell types (Aznar et al., 2016; Garcia-Marcos et al., 2015). We speculate that in C. elegans, GRDN-1 may similarly integrate multiple extrinsic and intrinsic signaling cues and couple them to diverse downstream effectors in order to regulate distinct phenotypes in a cell type-specific manner. Results from this work may guide future investigations into the underlying molecular mechanisms, and provide additional insights into the diverse roles of this multifunctional protein in different cellular contexts.
Supplementary Material
GRDN-1 localizes to the basal body in the postembryonic ciliated AQR and PQR sensory neurons.
Basal body and cilium location is disrupted in grdn-1 mutants in AQR and PQR.
GRDN-1 also regulates dendrite outgrowth in AQR, and to a lesser extent in PQR.
UNC-116/Kinesin-1 regulated GRDN-1 localization in AQR.
GRDN-1 localization in PQR is regulated by LIN-44/Wnt and LIN-17/Frizzled.
Acknowledgements
We are grateful to Max Heiman, Guangshuo Ou, Kang Shen, and Oliver Hobert for sharing reagents and advice, the Caenorhabditis Genetics Center for strains, and Max Heiman for discussion and sharing data prior to publication. We thank Hayley McMahon for technical assistance, Anna Kazatskaya and Alison Philbrook for comments on the manuscript, and additional members of the Sengupta lab for advice. This work was funded in part by the National Institutes of Health (R35 GM122463 – P.S.); startup funds from the Worcester Polytechnic Institute (I.V.N.); National Science Foundation (Cell and Molecular Visualization REU, 1359172 – S.L.) and Brandeis University (Division of Science Summer Undergraduate Research Fellowship – S.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aznar N, Kalogriopoulos N, Midde KK, and Ghosh P (2016). Heterotrimeric G protein signaling via GIV/Girdin: Breaking the rules of engagement, space, and time. Bioessays 38, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M, and Banker G (2016). The cellular mechanisms that maintain neuronal polarity. Nat Rev Neurosci 17, 611–622. [DOI] [PubMed] [Google Scholar]
- Berger-Muller S, and Suzuki T (2011). Seven-pass transmembrane cadherins: roles and emerging mechanisms in axonal and dendritic patterning. Mol Neurobiol 44, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DT, Kawasaki M, Walcoff M, Hisamoto N, Matsumoto K, and Jin Y (2001). UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32, 787–800. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, and Anderson KV (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 12, 767–778. [DOI] [PubMed] [Google Scholar]
- Cebul ER, McLachlan IG, and Heiman MG (2020). Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Development 147, dev180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, Cottell D, Katada T, Kontani K, and Blacque OE (2010). Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol 188, 953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Li W, Feng G, Yang Y, Wang X, and Ou G (2012). Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat Protoc 7, 2090–2102. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, and Priess JR (1998). A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol 141, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Farr H, and Gull K (2007). Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci 120, 7–15. [DOI] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liu OW, Howell AS, and Shen K (2013). An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell 155, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez DB, Berciu C, Anderson JR, Sengupta P, and Nicastro D (2014). A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in C. elegans. eLife 3, e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elric J, and Etienne-Manneville S (2014). Centrosome positioning in polarized cells: common themes and variations. Exp Cell Res 328, 240–248. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Ping J, and Takahashi M (2006). Girdin, a novel actin-binding protein, and its family of proteins possess versatile functions in the Akt and Wnt signaling pathways. Ann NY Acad Sci 1086, 169–184. [DOI] [PubMed] [Google Scholar]
- Fan L, Kovacevic I, Heiman MG, and Bao Z (2019). A multicellular rosette-mediated collective dendrite extension. eLife 8, e38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, and Bargmann CI (2013). Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 154, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, and Farquhar MG (2015). GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J Biol Chem 290, 6697–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grana TM, Cox EA, Lynch AM, and Hardin J (2010). SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev Biol 344, 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marietta MA, and Bargmann CI (2004). Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430, 317–322. [DOI] [PubMed] [Google Scholar]
- Hall DH, and Russell RL (1991). The posterior nervous system of the nematode Caenorhabditis elegans: serial reconstruction of identified neurons and complete pattern of synaptic interactions. J Neurosci 11, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M, Edwards SL, de Haan B, Yau KW, van den Heuvel S, Kapitein LC, Miller KG, and Hoogenraad CC (2018). Local microtubule organization promotes cargo transport in C. elegans dendrites. J Cell Sci 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman MG, and Shaham S (2009). DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, and Horvitz HR (1994). The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120, 1035–1047. [DOI] [PubMed] [Google Scholar]
- Herman MA, Vassilieva LL, Horvitz HR, Shaw JE, and Herman RK (1995). The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83, 101–110. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, and Cleveland DW (2012). Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci USA 109, E3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssin E, Tepass U, and Laprise P (2015). Girdin-mediated interactions between cadherin and the actin cytoskeleton are required for epithelial morphogenesis in Drosophila. Development 142, 1777–1784. [DOI] [PubMed] [Google Scholar]
- Ichimiya H, Maeda K, Enomoto A, Weng L, Takahashi M, and Murohara T (2015). Girdin/GIV regulates transendothelial permeability by controlling VE-cadherin trafficking through the small GTPase, R-Ras. Biochem Biophys Res Commun 461, 260–267. [DOI] [PubMed] [Google Scholar]
- Inberg S, Meledin A, Kravtsov V, Iosilevskii Y, Oren-Suissa M, and Podbilewicz B (2019). Lessons from worm dendritic patterning. Annu Rev Neurosci 42, 365–383. [DOI] [PubMed] [Google Scholar]
- Kazatskaya A, Kuhns S, Lambacher NJ, Kennedy JE, Brear AG, McManus GJ, Sengupta P, and Blacque OE (2017). Primary cilium formation and ciliary protein trafficking is regulated by the atypical MAP kinase MAPK15 in Caenorhabditis elegans and human cells. Genetics 207, 1423–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazatskaya A, Yuan L, Amin-Wetzel N, Philbrook A, de Bono M, and Sengupta P (2020). The URX oxygen-sensing neurons in C. elegans are ciliated. MicroPubl Biol, 10.17912/micropub.biology.000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil TA (1997). Functional morphology of insect mechanoreceptors. Microsc Res Tech 39, 506–531. [DOI] [PubMed] [Google Scholar]
- Kirszenblat L, Pattabiraman D, and Hilliard MA (2011). LIN-44/Wnt directs dendrite outgrowth through LIN-17/Frizzled in C. elegans neurons. PLoS Biol 9, e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL (2017). Neuronal territory formation by the atypical cadherins and clustered protocadherins. Semin Cell Dev Biol 69, 111–121. [DOI] [PubMed] [Google Scholar]
- Li W, Yi P, Zhu Z, Zhang X, Li W, and Ou G (2017). Centriole translocation and degeneration during ciliogenesis in Caenorhabditis elegans neurons. EMBO J 36, 2553–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wei Q, Zhang Y, Ling K, and Hu J (2010). The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol 189, 1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low IIC, Williams CR, Chong MK, McLachlan IG, Wierbowski BM, Kolotuev I, and Heiman MG (2019). Morphogenesis of neurons and glia within an epithelium. Development 146, dev171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JZ, Jiang P, Cui SP, Ren YL, Zhao J, Yin XH, Enomoto A, Liu HJ, Hou L, Takahashi M, et al. (2012). Girdin locates in centrosome and midbody and plays an important role in cell division. Cancer Sci 103, 1780–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DJ, Higgins CD, Peters KA, Cupp TD, Dickinson DJ, Pani AM, Moore RP, Cox AH, Kiehart DP, and Goldstein B (2016). MRCK-1 drives apical constriction in C. elegans by linking developmental patterning to force generation. Curr Biol 26, 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen DP, Jenkins PM, and Martens JR (2008). Olfactory cilia: our direct neuronal connection to the external world. Curr Top Dev Biol 85, 333–370. [DOI] [PubMed] [Google Scholar]
- McLachlan IG, and Heiman MG (2013). Shaping dendrites with machinery borrowed from epithelia. Curr Opin Neurobiol 23, 1005–1010. [DOI] [PubMed] [Google Scholar]
- Menco BP (1997). Ultrastructural aspects of olfactory signaling. Chem Senses 22, 295–311. [DOI] [PubMed] [Google Scholar]
- Middelkoop TC, and Korswagen HC (2014). Development and migration of the C. elegans Q neuroblasts and their descendants. WormBook, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu A, Enomoto A, Kato T, Weng L, Kuroda K, Asai N, Asai M, Mii S, and Takahashi M (2015). Potential involvement of kinesin-1 in the regulation of subcellular localization of Girdin. Biochem Biophys Res Commun 463, 999–1005. [DOI] [PubMed] [Google Scholar]
- Nechipurenko IV, Berciu C, Sengupta P, and Nicastro D (2017). Centriolar remodeling underlies basal body maturation during ciliogenesis in Caenorhabditis elegans. eLife 6, e25686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko IV, Olivier-Mason A, Kazatskaya A, Kennedy J, McLachlan IG, Heiman MG, Blacque OE, and Sengupta P (2016). A conserved role for Girdin in basal body positioning and ciliogenesis. Dev Cell 38, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, and Kanemaki M (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6, 917–922. [DOI] [PubMed] [Google Scholar]
- Oshita A, Kishida S, Kobayashi H, Michiue T, Asahara T, Asashima M, and Kikuchi A (2003). Identification and characterization of a novel Dvl-binding protein that suppresses Wnt signalling pathway. Genes Cells 8, 1005–1017. [DOI] [PubMed] [Google Scholar]
- Patel N, Thierry-Mieg D, and Mancillas JR (1993). Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proc Natl Acad Sci USA 90, 9181–9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, and Culotti JG (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117, 456–487. [DOI] [PubMed] [Google Scholar]
- Rohlich P (1975). The sensory cilium of retinal rods is analogous to the transitional zone of motile cilia. Cell Tissue Res 161, 421–430. [DOI] [PubMed] [Google Scholar]
- Salzberg Y, Diaz-Balzac CA, Ramirez-Suarez NJ, Attreed M, Tecle E, Desbois M, Kaprielian Z, and Bulow HE (2013). Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell 155, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JC, Haycraft CJ, Thomas JH, Yoder BK, and Swoboda P (2003). XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol Biol Cell 14, 2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong E, Yuan L, and Arikkath J (2015). Cadherins and catenins in dendrite and synapse morphogenesis. Cell Adh Migr 9, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwas D, Su TY, Roessler M, Wang S, and Dammermann A (2017). Centrioles initiate cilia assembly but are dispensable for maturation and maintenance in C. elegans. J Cell Biol 216, 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimel A, Wong L, Najarro EH, Ackley BD, Garriga G, and Hutter H (2010). The Flamingo ortholog FMI-1 controls pioneer-dependent navigation of follower axons in C. elegans. Development 137, 3663–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Takano T, Xu C, Funahashi Y, Namba T, and Kaibuchi K (2015). Neuronal polarization. Development 142, 2088–2093. [DOI] [PubMed] [Google Scholar]
- Tang N, and Marshall WF (2012). Centrosome positioning in vertebrate development. J Cell Sci 125, 4951–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Enomoto A, Weng L, Mizutani Y, Abudureyimu S, Esaki N, Tsuyuki Y, Chen C, Mii S, Asai N, et al. (2018). Girdin/GIV regulates collective cancer cell migration by controlling cell adhesion and cytoskeletal organization. Cancer Sci 109, 3643–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kweon J, Larson S, and Chen L (2005). A role for the C. elegans L1CAM homologue lad-1/sax-7 in maintaining tissue attachment. Dev Biol 284, 273–291. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, and Brenner S (1975). Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol 160, 313–337. [DOI] [PubMed] [Google Scholar]
- Weng L, Enomoto A, Miyoshi H, Takahashi K, Asai N, Morone N, Jiang P, An J, Kato T, Kuroda K, et al. (2014). Regulation of cargo-selective endocytosis by dynamin 2 GTPase-activating protein girdin. EMBO J 33, 2098–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Transact R Soc Lond B 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Yan J, Chao DL, Toba S, Koyasako K, Yasunaga T, Hirotsune S, and Shen K (2013). Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. eLife 2, e00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev S, and Shen K (2017). Establishing neuronal polarity with environmental and intrinsic mechanisms. Neuron 96, 638–650. [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, and Garbers DA (1997). Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci USA 94, 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, and Dernburg AF (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


