Abstract
microRNAs (miRNAs) play a critical role in a variety of biological processes, including embryogenesis and the physiological functions of cells. Evolutionarily conserved microRNA-31 (miR-31) has been found to be involved in cancer, bone formation, and lymphatic development. We previously discovered that, in the sea urchin, miR-31 knockdown (KD) embryos have shortened dorsoventral connecting rods, mispatterned skeletogenic primary mesenchyme cells (PMCs) and shifted and expanded Vegf3 expression domain. Vegf3 itself does not contain miR-31 binding sites; however, we identified its upstream regulators Eve and Wnt1 to be directly suppressed by miR-31. Removal of miR-31’s suppression of Eve and Wnt1 resulted in skeletal and PMC patterning defects, similar to miR-31 KD phenotypes. Additionally, removal of miR-31’s suppression of Eve and Wnt1 results in an expansion and anterior shift in expression of Veg1 ectodermal genes, including Vegf3 in the blastulae. This indicates that miR-31 indirectly regulates Vegf3 expression through directly suppressing Eve and Wnt1. Furthermore, removing miR-31 suppression of Eve is sufficient to cause skeletogenic defects, revealing a novel regulatory role of Eve in skeletogenesis and PMC patterning. Overall, this study provides a proposed molecular mechanism of miR-31’s regulation of skeletogenesis and PMC patterning through its cross-regulation of a Wnt signaling ligand and a transcription factor of the endodermal and ectodermal gene regulatory network.
Keywords: MicroRNA-31, sea urchin, primary mesenchyme cells, miRNA target protector, post-transcriptional regulation, Even-skipped, Vegf signaling, Wnt
Summary statement:
This study demonstrates that miR-31 regulates skeletal development by directly suppressing Eve and Wnt1. Removing miR-31’s suppression of Eve and Wnt1 results in shorter skeletal spicules, PMC migration defects, and gene expression changes which recapitulate miR-31 knockdown phenotypes.
Introduction
miRNAs are small non-coding RNAs that play critical regulatory roles as fine-tuners of gene expression to regulate the physiological functions of cells (Bartel, 2009). miRNAs typically bind to the 3′ untranslated region (3’UTR) of target mRNAs, silencing translation and/or inducing target mRNA degradation (Bartel, 2009). miR-31, an evolutionarily conserved miRNA, has mostly been examined in the context of various cancers, and limited studies revealed its function in bone homeostasis, myogenesis, auto-immunity (Valastyan and Weinberg, 2010; Yu et al., 2018), lymphatic development (Pedrioli et al., 2010), and embryonic development (Stepicheva and Song, 2015).
Using the purple sea urchin (Strongylocentrotus purpuratus), we have previously identified miR-31 to regulate skeletogenesis and patterning of the skeletogenic primary mesenchyme cells (PMCs), by directly suppressing components of the PMC gene regulatory network (GRN) and indirectly regulating the Vegf signaling pathway (Stepicheva and Song, 2015). The molecular mechanism of how miR-31 regulates the Vegf signaling pathway to mediate directed migration of PMCs and skeletal formation remains unclear. The Vegf/VegfR signaling pathway is critical for the PMCs to form the skeletal rudiment, likely by providing differentiation and chemotactic cues from the ectoderm to the migrating PMCs, as well as activating the transcription of biomineralization genes in the PMCs (Duloquin et al., 2007; Ettensohn and McClay, 1986; McIntyre et al., 2014). VegfR10 is expressed specifically in the PMCs and is thought to respond to the Vegf3 ligand expressed in the ectoderm (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007). An overexpression of Vegf3 leads to supernumerary and abnormal branching of larval skeleton, and loss of Vegf3 or VegfR10 results in decreased expression levels of genes involved in biomineralization, such as p19, SM29, SM49, Msp130, and a lack of skeleton (Adomako-Ankomah and Ettensohn, 2014; Duloquin et al., 2007). Thus, Vegf signaling is critical for various aspects of sea urchin skeletogenesis. Additional signaling pathways, such as non-canonical Wnt (ncWnt), Nodal/BMP, MAPK and PI3 kinase pathways are also important in sea urchin skeletogenesis (Adomako-Ankomah and Ettensohn, 2013; Bradham et al., 2004; Croce et al., 2006; Duboc et al., 2004; Schlessinger, 2000).
Conservation of Vegf ligand and receptor recognition between sea urchins and humans has been demonstrated in Paracentrotus lividus (P. lividus), where Vegf3 knockdown resulted in complete loss of skeleton that was rescued by human VegfA mRNA (Morgulis et al., 2019). Vegf signaling in vertebrates is essential for blood vessel formation and this process has been proposed to be analogous to sea urchin skeletogenesis, since a common set of transcription factors (TFs) (Ets1/2, Erg, Hex, Tel, and FoxO) and signaling pathways (VEGFR, Notch, and Angiopoetin) important for vascularization are expressed and utilized in the sea urchin PMCs at the time of skeletal formation (Morgulis et al., 2019).
During the blastula stage, Vegf3 is likely to be indirectly regulated by Hox11/13b, since Hox11/13b KD resulted in a complete loss of Pax2/5/8 and Wnt5; and Wnt5 KD resulted in loss of Vegf3 expression (McIntyre et al., 2013). Hox11/13b activates Wnt5, which serves as a short-range signal specifying the border ectoderm (BE) which consists of a ring of ectodermal cells immediately bordering the endoderm anteriorly (McIntyre et al., 2013). In the sea urchin, Wnt5 binds to an unidentified Frizzled (Fzd) receptor that activates an unidentified activator (McIntyre et al., 2013; Nishita et al., 2010a; Nishita et al., 2010b). This unidentified factor, in turn, transcriptionally activates Pax2/5/8 in the BE (McIntyre et al., 2013). How Wnt5 signals remains unclear. Wnt5 is thought to signal primarily through the ncWnt signaling pathway (Nishita et al., 2010a; Nishita et al., 2010b). In other contexts, Wnt5 has also been reported to activate the canonical Wnt/β-catenin (cWnt/β-catenin) pathway (Mikels and Nusse, 2006). The dorsal ventral margin (DVM) is an area in the ectoderm restricted by Nodal signaling in the ventral ectoderm (VE) and BMP signaling in the dorsal ectoderm (DE) (McIntyre et al., 2014; McIntyre et al., 2013). Vegf3 is highly expressed at the intersection of Wnt5 and Nodal/BMP expression at the border ectoderm-dorsal ventral margin (BE-DVM) (McIntyre et al., 2014).Vegf3, along with several signaling molecules and TFs, such as Lim1, Nk1, and Pax2/5/8, are restricted to the BE-DVM to provide patterning inputs to the underlying mesenchyme where the PMC ventrolateral clusters reside to form the skeletal rudiments (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007; McIntyre et al., 2014; McIntyre et al., 2013; Rottinger et al., 2008). Additionally, the expression of Eve is important for the activation of Hox11/13b (Cui et al., 2014). In the sea urchin, Eve is a direct target of the cWnt/β-catenin signaling pathway (Peter and Davidson, 2010, 2011; Ransick et al., 2002). Knockdown of Eve in the Veg1 endoderm during the blastula stage resulted in decreased transcript levels of Hox11/13b, along with Wnt1, Wnt4, Wnt5, and Wnt16 (Cui et al., 2014).
Previously, we have shown that most miR-31 KD embryos have mislocalized PMCs that do not express VegfR10 (Stepicheva and Song, 2015). Interestingly, the Vegf3 expression domain observed in miR-31 KD blastulae is expanded and shifted anteriorly (Stepicheva and Song, 2015). We did not bioinformatically identify potential binding sites of miR-31 in Vegf3 or VegfR10, leading us to hypothesize that miR-31 regulates Vegf3 expression indirectly. The current study reveals that miR-31’s direct repression of Eve and Wnt1 indirectly regulates the expression of Vegf3. Further, Vegf3 expression domains correlate with the anterior migration distance of PMCs and the length of skeletal rods. Removal of miR-31’s direct suppression of both Eve and Wnt1 recapitulated miR-31 KD induced phenotypes. We also discovered a novel role of Eve in regulating skeletogenesis.
Materials and Methods
Animals
Adult Strongylocentrotus purpuratus (Sp) were obtained from Point Loma Marine Invertebrate Lab, Lakeside, California. Adult males and females were injected with 0.5 M KCl intracoelomically to obtain sperm and eggs. Filtered natural seawater (FSW) (collected from Indian River Inlet; University of Delaware) or artificial seawater (ASW) made from Instant Ocean© was used for embryo cultures incubated at 15°C.
Cloning
For generating Eve, Wnt1 and Wnt5 3’UTR luciferase reporter constructs, PCR primers of Eve, Wnt1 and Wnt5 3’UTR sequences were designed based on sequence information available from the sea urchin genome (echinobase.org) (Table S1). Amplified PCR products of Eve, Wnt1 and Wnt5 3’UTRs were first cloned into ZeroBlunt vector (Thermo Fisher Scientific, Waltham, MA) and then subcloned into the Renilla luciferase (Rluc) reporter construct. Mutations were generated within the miR-31 seed sequences using the QuikChange Lightning Kit (Agilent Technologies, Santa Clara, California). The two complete miR-31 seed sites within Eve 3’UTR were modified from 5’ TCTTGCC 3’ to 5’ TCCTACC 3’ at +28 and +652 positions. The position +1 is the first nucleotide after the stop codon. The truncated miR-31 seed site within Wnt1 3’UTR was modified from 5’ CTTGCC 3’ to 5’ CTCGAC 3’ at +2482 position. The truncated miR-31 seed site within Wnt5 3’UTR was modified from 5’ TCTTGC 3’ to 5’ TCCTAC 3’ at +3239 position to disrupt miR-31’s binding (Gregory et al., 2008; Stepicheva et al., 2015).
Each of the construct sequences was verified by DNA sequencing (Genewiz, South Plainfield, NJ). Firefly luciferase was used as a normalization control as previously described (Stepicheva et al., 2015). Luciferase constructs containing the Eve 3’UTR were linearized with EcoRI and luciferase constructs containing Wnt1 and Wnt5 3’UTRs were linearized with NotI. The constructs were in vitro transcribed using the mMessage machine kit with either T7 (for Eve, Wnt1 and Wnt5 RLuc mRNAs) or Sp6 (for Firefly luciferase mRNA) RNA polymerases (Ambion Inc, Austin, Texas). mRNAs were purified using Macherey-Nagel Nucleospin® RNA Clean-up kit (Macherey-Nagel, Bethlehem, PA) prior to injections.
To test if Eve and Wnt1 miRNA-31-TP phenotypes are due to an increase of translated protein, we cloned Eve and Wnt1 coding sequence (CDS) in ZeroBlunt vector (Thermo Fisher Scientific, Waltham, MA) (Table S1). Plasmids were linearized with BamHI and in vitro transcribed using T7 mMessage machine kit (Ambion Inc, Austin, Texas). CDS mRNA was injected into zygotes. Firefly mRNA was used as a control.
Dual luciferase quantification
The injection solutions for the dual-luciferase assay contained 20% sterile glycerol, 2 mg/ml 10,000 MW Texas Red lysine-charged dextran, 100–200 ng/μl Firefly mRNA and 100 ng/μl (Eve, Wnt1 and Wnt5 RLuc constructs) RLuc mRNA. 30–50 embryos were collected at the mesenchyme blastula stage (24 hpf). Dual luciferase assays were performed using the Promega Dual-Luciferase Reporter (DLR™) Assay Systems with the Promega GloMax 20/20 Luminometry System (Promega, Madision, WI). The RLuc values were normalized to the Firefly signal to account for microinjection volume differences. The data from the RLuc with mutated miR-31 seeds were normalized to the RLuc with wildtype (WT) 3’UTR construct.
Microinjections
Microinjections were performed as previously described (Cheers and Ettensohn, 2004; Stepicheva and Song, 2014) with modifications. All injection solutions were prepared in a 2.5μl solution consisting of 0.5 μl of 100% glycerol and 0.5μl of 2mg/ml 10,000 MW neutral non-fixable Texas Red dextran (Thermo Fisher Scientific, Waltham, MA). Approximately 1–2 picoliter (pl) was injected into each newly fertilized egg based on the size of the injection bolus at about one-fifth of the egg diameter. miR-31 inhibitor (Cel-miR-72) (miRCURY LNA power inhibitor, Qiagen, Germantown, MD) was used at a 30μM concentration (Stepicheva and Song, 2015). Texas Red dextran was used as an injection control (Thermo Fisher Scientific, Waltham, MA). For luciferase assays, 100–200ng of WT or mutated Rluc construct and 100–200ng of Firefly luciferase as injection control was used. miRNA target protector (TP) morpholinos were designed against validated miR-31 seed sites identified by dual-luciferase assays and their specific flanking sequences within the 3’UTRs of Eve and Wnt1 (GeneTools, LLC, Philomath, OR). The injection solution of Eve miR-31 TP contained 3μM or 300μM of the each of the two Eve miR-31 TPs and 6μM or 600μM of control TP, respectively. The control TP is against human β-globin and does not recognize sea urchin genes (as assessed by BLASTN against the sea urchin genome). The injection solution of control TP or Wnt1 miR-31 TP contained 30μM or 300μM of TP. The Eve+Wnt1 miR-31 TP cocktail solution contained 300μM of Wnt1 miR-31 TP and the two Eve miR-31 TPs. The control TP contained 900μM of TP.
To test for specificity of miR-31 TPs, Eve or Wnt1 transcripts were microinjected to examine their overexpression phenotypes. All injection solutions were prepared in a 2.5μl solution consisting of 0.5μl of 100% glycerol and 0.5μl of RNAse-free 2mg/ml 10,000 MW neutral non-fixable Texas Red dextran (Thermo Fisher Scientific, Waltham, MA) or 500ng of mCherry mRNA. Injection solution contained 3μg of Eve mRNA or 1.5μg of Wnt1 mRNA. Control mRNA consisted of 1.5–3μg of Firefly mRNA.
Immunofluorescence
Embryos were fixed and immunolabeled with 1D5 at 1:50, overnight to 2 days at 4°C as previously described (McClay et al., 1983; Sampilo et al., 2018) (gift from Dr. David McClay, Duke University). This was followed by goat anti-mouse Alexa488 (Thermo Fisher Scientific, Waltham, MA) conjugated secondary antibody at 1:300 for 1 hour at room temperature. Embryos were washed 3 times with PBS-Tween (0.05% Tween-20 in 1X PBS).
Whole mount in situ hybridization (WMISH)
Partial coding sequences of Eve, Hox11/13b, Wnt1, Wnt5, VegfR10, and Vegf3 were cloned into ZeroBlunt vector (Thermo Fisher Scientific, Waltham, MA) (Stepicheva et al., 2015; Stepicheva and Song, 2015). Pax2/5/8 CDS was generated by gBlock (Integrated DNA Technologies, Coralville, Iowa) and cloned into ZeroBlunt vector (Thermo Fisher Scientific, Waltham, MA). Wnt16 was cloned into pGEM-T Easy Vector (Martínez-Bartolomé and Range, 2019) (Gift from Dr. Ryan Range, Auburn University). Eve, Hox11/13b, Wnt1, Wnt5, Wnt16, Pax2/5/8, and Vegf3 were linearized with restriction enzymes EcoRI, BamHI, BamHI, NotI, BamHI, SpeI, and BamHI, respectively, using FastDigest™ (Thermo Fisher Scientific, Waltham, MA). Eve, Wnt5, Hox11/13b, Wnt1, and Pax2/5/8 were in vitro transcribed with Sp6 RNA polymerase, and Vegf3 and Wnt16 were in vitro transcribed with T7 RNA polymerase of the DIG RNA Labeling Kit (Millipore Sigma, St. Louis, MO).
Phenotyping
To measure the length of dorsoventral connecting rods (DVCs), ZEISS Observer Z1 microscope was used to take Z-stacks of differential interference contrast (DIC) and 1D5-immunolabeled images. N is the total number of embryos examined except where otherwise stated. ZEISS AxioCam105 color camera was used to take in situ images. AxioVision software was used to measure the length of DVCs, PMC migration distance, and in situ gene expressions. Zen 3.1 software (Carl Zeiss Microscopy, White Plains, NY) was used to determine the center of gastrulae in vegetal views to measure angles of Vegf3 expression domains, VE, and DE domains. Representative images were taken with Zeiss LSM 880 scanning confocal microscope using Zen software or ZEISS Observer Z1 using AxioVision software (Carl Zeiss Microscopy, LLC, White Plains, NY).
Real-Time quantitative PCR (qPCR)
To measure the transcriptional changes of genes within our model, and biomineralization genes, we used real-time quantitative PCR (qPCR). We injected 100 zygotes and collected them at blastulae (24 hpf) for injected control, miR-31 KD, Eve miR-31 TP, Wnt1 miR-31 TP, and Eve+Wnt1 miR-31 TP. Total RNA was extracted by using the Macherey-Nagel Nucleospin® RNA Isolation XS kit (Macherey-Nagel, Bethlehem, PA). cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). qPCR was performed using 2.5 embryo equivalents for each reaction with the Fast SYBR or PowerUp Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA) in the QuantStudio 6 Real-Time PCR cycler system (Thermo Fisher Scientific, Waltham, MA). Results were normalized to the mRNA expression of the housekeeping gene, ubiquitin, and shown as fold changes compared to injected control embryos using the ΔΔCt method as previously described (Stepicheva et al., 2015). Primer sequences were designed using the Primer 3 Program (Rozen and Skaletsky, 2000) and are listed in Table S1. 3–6 biological replicates were conducted.
Results
miR-31 indirectly regulates Vegf3 expression and skeletogenesis.
We previously found miR-31 KD embryos displayed expanded and anteriorly shifted Vegf3 expression domain compared to the injected control (Stepicheva and Song, 2015). miR-31’s regulation of Vegf3 is likely to be indirect, since it does not contain any predicted miR-31 binding sites (Stepicheva and Song, 2015). We propose a working model based on existing literature of how miR-31 indirectly regulates Vegf3 (Fig. 1A). In this model, Eve is the most upstream component of this pathway, since Eve controls the specification of the Veg1 lineage through its activation of Hox11/13b in the Veg1 endoderm at 24 hpf (Cui et al., 2014; Peter and Davidson, 2010). In addition to Eve, Wnt1 and Wnt16 also contribute to the activation of Hox11/13b in the Veg1 cells, and the positive feedback circuit of Hox11/13b, Wnt1, and Wnt16 occurs in Veg1 endodermal cells (Cui et al., 2014). The receptor and signaling pathways for Wnt1 and Wnt16 are Fzd5/8-ROCK/JNK and Fzd1/2/7-PKC, respectively; perturbation of either Fzd5/8, ROCK, Wnt16, Fzd1/2/7 or PKC leads to loss of skeleton (Croce et al., 2006; Martínez-Bartolomé and Range, 2019; Range et al., 2013). It has been implicated that Wnt16 acts downstream of Wnt1-Fzd5/8-JNK signaling (Martínez-Bartolomé and Range, 2019). Fzd1/2/7-PKC signaling then activates an unidentified activator of Pax2/5/8, which in turn, leads to the activation of Vegf3, as observed in L. variegatus (Lv) (McIntyre et al., 2013). LvWnt5 KD results in loss of Pax2/5/8 and Vegf3 expression in the BE-DVM, in addition to embryos lacking skeleton (McIntyre et al., 2013). This potentially indicates that LvWnt5 regulates Vegf3 and skeletogenesis. However, Wnt5 KD in S. purpuratus (SpWnt5) resulted in no change in expression levels of ectodermal genes nk1, sp5, unc4.1, hox7, IrxA, and msx, except for an increase in Wnt5 transcripts (Cui et al., 2014). This suggests that Wnt5 may not be the ligand that specifies the BE to activate Vegf3 in S. purpuratus, although this was not directly tested in this study (Cui et al., 2014). Unlike LvWnt5, SpWnt5 KD had no impact on Hox11/13b (Cui et al., 2014; McIntyre et al., 2013). Additionally, SpHox11/13b KD resulted in decreased Wnt1 and Wnt16 but did not alter Wnt5 levels (Cui et al., 2014). Thus, SpWnt1 and SpWnt16 are likely to function as the LvWnt5 in activating genes expressed in the border ectoderm in S. purpuratus (Cui et al., 2014).
Figure 1. miR-31 indirectly regulates Vegf3 expression domain and skeletogenesis by targeting Eve, Wnts, and Fzds.
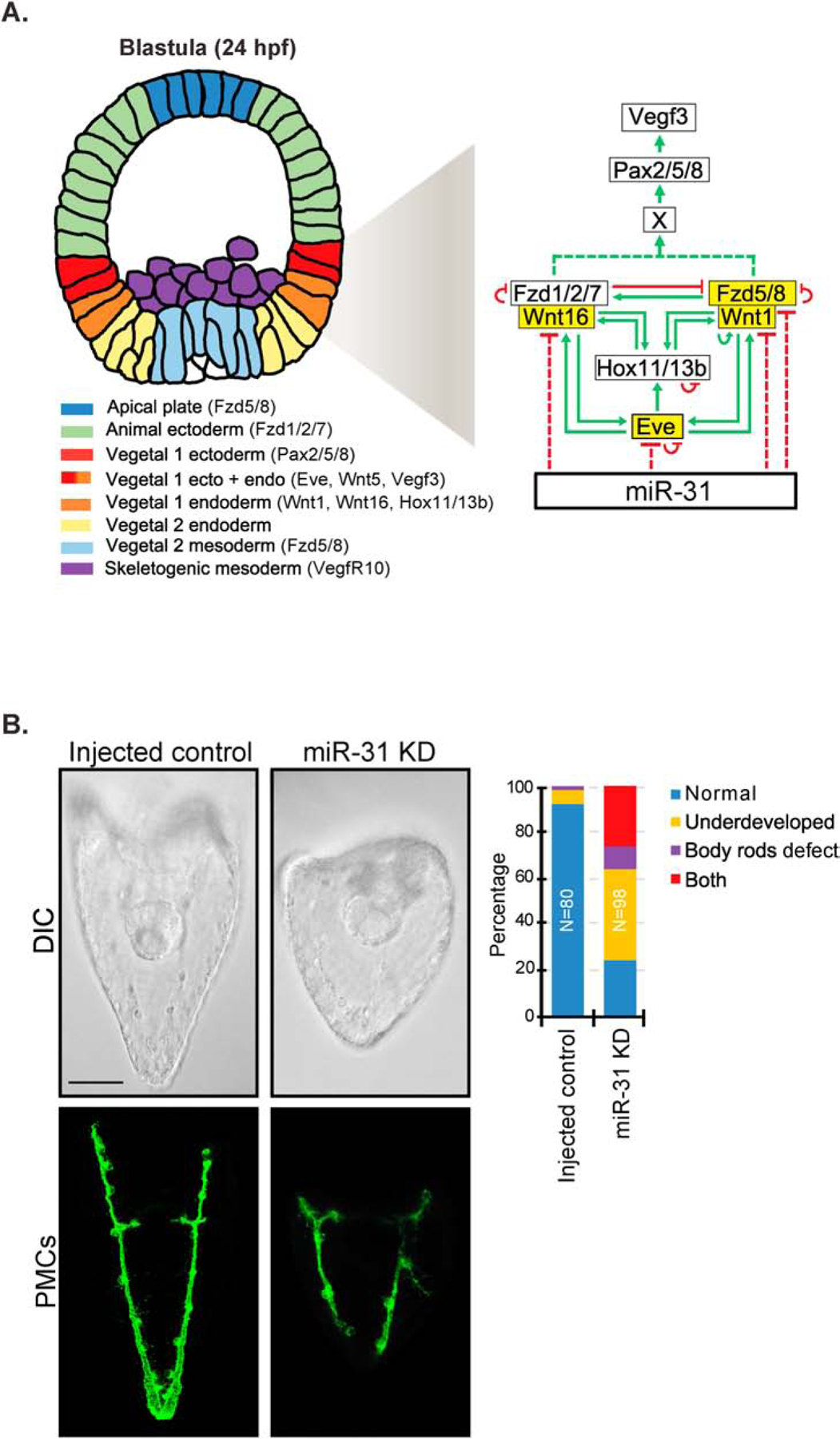
(A) A simplified proposed model of miR-31’s indirect regulation of Vegf3 and skeletogenesis through potential targets Eve, Wnt1, Wnt16, and Fzd5/8 during mesenchyme blastula (24 hpf). Genes highlighted in yellow are potential miR-31 targets. Eve is activated by canonical Wnt/β-catenin signaling and is expressed in both the Veg1 endoderm and ectoderm and activates Hox11/13b in the Veg1 endoderm (Cui et al., 2014; Peter and Davidson, 2011; Ransick et al., 2002). Wnt1 and Wnt16 also contribute to the activation of Hox11/13b in the Veg1 cells, and the positive feedback of Hox11/13b, Wnt1, and Wnt16 occurs in Veg1 endodermal cells (Cui et al., 2014). Wnt1 signals through Fzd5/8 receptor to activate the ncWnt/PCP-ROCK/JNK pathway, and Wnt16 signals through Fzd1/2/7 to activate the ncWnt/Ca2+-PKC pathway (Martínez-Bartolomé and Range, 2019; Range et al., 2013). Wnt1-Fzd5/8 and Wnt16-Fzd1/2/7 cross-regulate each other (Range et al., 2013). Wnt1-Fzd5/8 and/or Wnt16-Fzd1/2/7 signaling lead to the activation of an unidentified factor which activates transcription factor Pax2/5/8, leading to activation of Vegf3 expression (McIntyre et al., 2013; Rottinger et al., 2008). Eve, Hox11/13b, Fzd5/8 and Fzd1/2/7 are in part regulated through autorepression, while Wnt1 may be regulated through autoactivation (Cui et al., 2014; Cui et al., 2017; Range, 2018; Range et al., 2013). (B) The shorter DVC rods and aberrant PMC patterning defects observed in miR-31 KD gastrulae (Stepicheva and Song, 2015) persist into the larval stage (5dpf). A greater number of miR-31 KD larvae are underdeveloped and/or exhibit body rods that fail to meet at the posterior end of the embryo. Maximum intensity projection of Z-stack confocal images are shown. 2 biological replicates. N is the total number of larvae examined. Scale bar = 50μm.
Within this simplified model, we bioinformatically identified potential miR-31 binding sites within the 3’UTRs of Eve, Wnt1, Wnt5, Wnt16, and Fzd5/8 genes (Fig. 1A). The circuitries of Eve, Hox11/13b, Wnt1-Fzd5/8-JNK, and Wnt16-Fzd1/2/7-PKC are highly auto-regulated, as well as highly cross-regulated (Cui et al., 2014; Cui et al., 2017; Range, 2018; Range et al., 2013). Thus, perturbing one component would be likely to alter the expression of other genes in the pathway.
Previously, miR-31 KD induced defects in gastrulae, including shortened DVCs and PMC patterning defects (Stepicheva and Song, 2015). We examined the impact of miR-31 KD in larval development and found that injected control larvae (5 days post fertilization; dpf) have an elongated pyramidal body shape, whereas miR-31 KD larvae appear smaller and rounder (Fig. 1B). The control larvae have body rods that meet converge at the posterior end; however, miR-31 KD larvae have body rods that failed to meet at the posterior end, indicating that miR-31 KD defects are long-lasting and not recoverable (Fig. 1B).
Knockdown of miR-31 results in expanded and anteriorly shifted spatial expression of genes expressed in Veg1 cells.
To understand the mechanism of miR-31’s regulation of Vegf3, we tested the spatial expression of several genes expressed in the Veg1 ectodermal and endodermal cells in injected control and miR-31 KD mesenchyme blastulae at 24 hpf (Fig. 2). During this time, Eve and Wnt5 are expressed in both Veg1 endoderm and Veg1 ectoderm (same as Vegf3), whereas Hox11/13b, Wnt1, and Wnt16 are expressed in the Veg1 endoderm, and Pax2/5/8 is expressed in the Veg1 ectoderm (Cui et al., 2014). In miR-31 KD blastulae, all genes examined have a significant anterior shift in their expression domain, and all genes also have an expanded expression domain (except for Eve and Hox11/13b) (Fig. 2B), similar to the anterior shift and expansion of Vegf3 in the Veg1 endoderm and Veg1 ectoderm.
Figure 2. miR-31 KD results in expansion and anterior shift of Veg1 endodermal and Veg1 ectodermal gene expression domains.
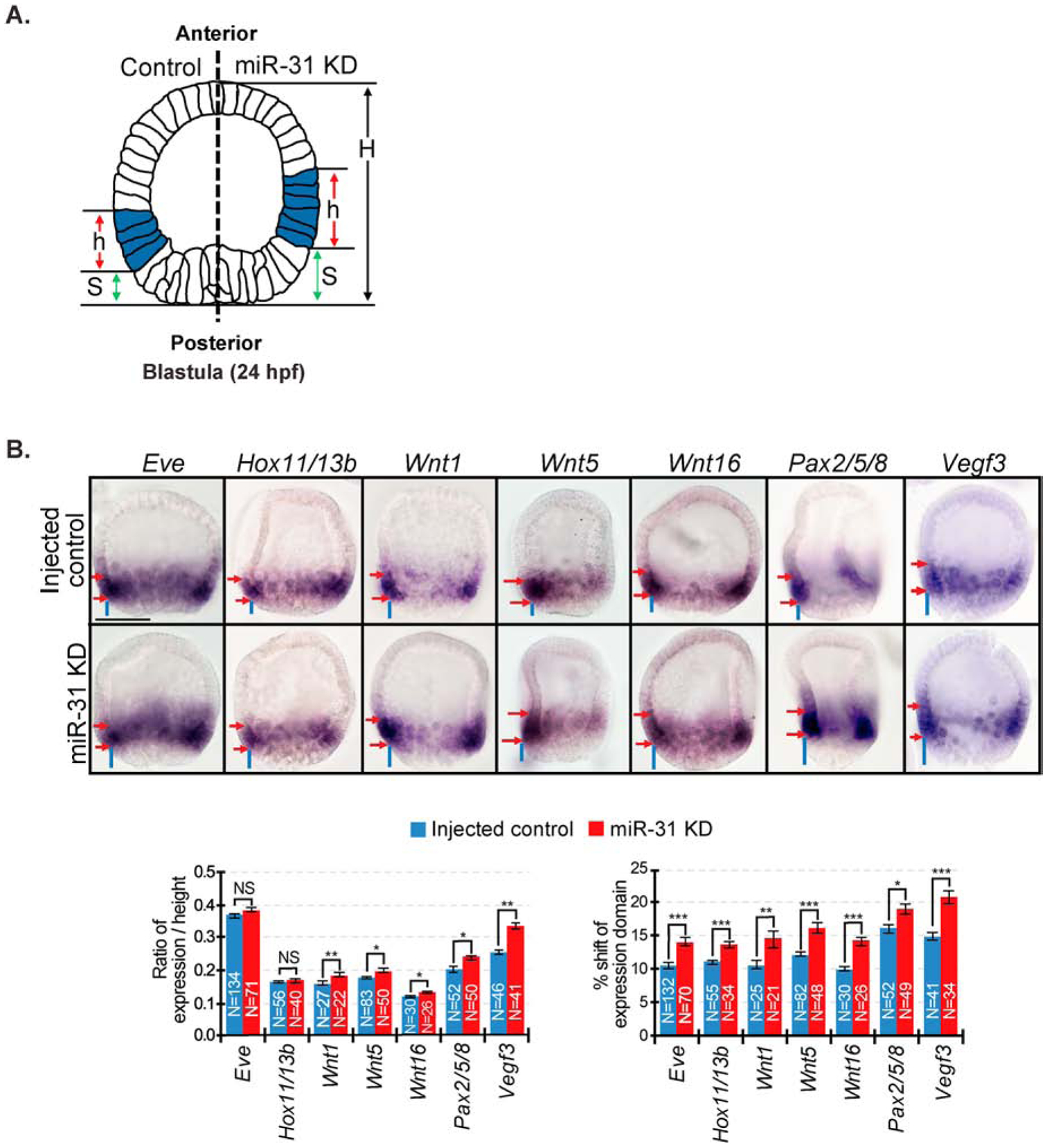
(A) The spatial expression domains (h) and anterior shift (S) were measured on both sides of each embryo. The average expression domain is calculated by taking the average of the ratio of h/H from each side. The anterior shift is measured by taking the average of the ratios of S/H from each side. (B) miR-31 KD mesenchyme blastulae (24 hpf) have an anterior shift in spatial expression domain of all genes expressed in the Veg1 endoderm and Veg1 ectoderm compared to control. miR-31 KD embryos have expanded expression domains of Wnt1, Wnt5, Wnt16, Pax2/5/8, and Vegf3 compared to control. Red arrows delineate expression domains. Blue lines indicate shift of expression domain. NS=Not significant, *p<0.05, **p<0.001, ***p<0.0001 using Student’s t-test. All error bars represent SEM. 2–3 biological replicates. Scale bar = 50μm.
miR-31 directly suppresses Eve and Wnt1.
Since Vegf3 does not contain a miR-31 binding site, we bioinformatically identified potential miR-31 targets within our proposed pathway (Fig. 1A). We found that Eve, Wnt1, Wnt16, and Fzd5/8 have predicted miR-31 regulatory sites within their 3’UTRs. We prioritized our analysis of Eve, Wnt1 and Wnt5 for the following reasons: Eve is the most upstream regulator in our model; Wnt1 (not Wnt16) has a significant effect on Veg1 ectodermal gene expression that controls PMC patterning (Cui et al., 2014); and LvWnt5 is critical for LvVegf3 expression (McIntyre et al., 2013). To test miR-31’s direct regulation of Eve, Wnt1, and Wnt5, their 3’UTRs were cloned downstream of the RLuc reporter construct. RLuc with WT or mutated miR-31 binding sites were co-injected with Firefly reporter construct as a normalization control into newly fertilized eggs. Mutated miR-31 binding sites will abolish target recognition by endogenous miR-31, preventing miR-31’s binding and regulation. Eve has two predicted miR-31 binding sites, and both Wnt1 and Wnt5 have predicted truncated miR-31 binding sites. Of the miR-31 seed sequence (TCTTGCC), Wnt1 has a mismatch of the first T nucleotide (nt) and Wnt5 contains a mismatch of the last C nucleotide. miRNA binding of 6 nts that is offset by a single nucleotide can also mediate detectable repression, but are less effective than a 7–8 nt perfect match (Bartel, 2018; Jan et al., 2011; Kim et al., 2016). Non-canonical sites (imperfect seed matches) have also been found to be effective in downregulating gene expression (Khorshid et al., 2013). Dual luciferase assays (24 hpf) indicate that miR-31 directly suppresses Eve and Wnt1. Wnt5 is not directly suppressed by miR-31, or experiences weak miR-31-mRNA binding affinity (Fig. 3). The relative change of Wnt1 luciferase read out was not as high as Eve luciferase readout, potentially indicating that miR-31’s binding to Wnt1 is not as strong as its binding to Eve.
Figure 3. miR-31 directly suppresses Eve and Wnt1.
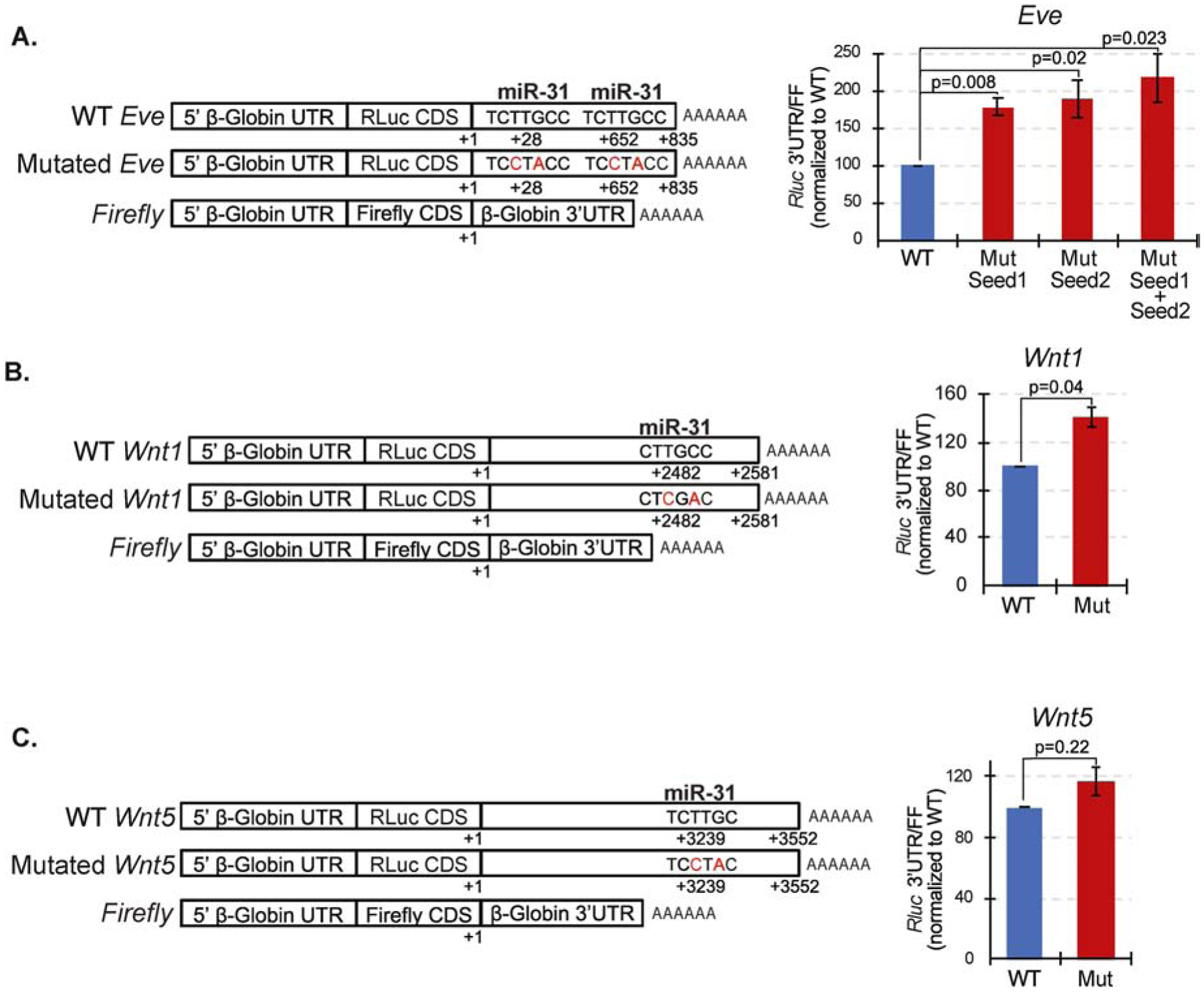
(A) Dual luciferase assays were conducted at mesenchyme blastula stage (24 hpf). The Rluc values were first normalized to the co-injected Firefly values. The ratios of normalized Rluc values with mutated miR-31 seed to the Rluc with wildtype (WT) miR-31 seed are presented. Luciferase readings of embryos injected with mutated (Mut) miR-31 3’UTR binding sites of Eve were increased significantly in comparison to embryos injected with the WT 3’ UTRs, indicating that miR-31 directly represses Eve. (B) Wnt1 has a mismatch of the first T nucleotide of the miR-31 seed sequence. Wnt1 is directly suppressed by miR-31. (C) Wnt5 contains a mismatch of the last C nucleotide of the miR-31 seed sequence. Wnt5 is not directly suppressed by miR-31 or experiences weak miRNA-mRNA binding affinity. 3–4 biological replicates. P-value was analyzed using Student’s t-test. All error bars represent SEM.
Removal of miR-31 suppression of Eve and overexpression of Eve result in skeletogenic and PMC patterning defects.
To test if the removal of miR-31 suppression of Eve had an impact in skeletogenesis, we designed miRNA target protector (TP) morpholinos against the validated miR-31 seed sites and their specific flanking sequences within Eve 3’UTRs (Remsburg et al., 2019; Staton and Giraldez, 2011; Stepicheva and Song, 2015). Newly fertilized eggs were microinjected with Eve miR-31 TP at two concentrations. TPs are synthetic morpholino oligonucleotides designed to bind to miR-31 regulatory seed sites of its target and unique nucleotides immediately flanking the seed, blocking miR-31’s regulatory function (Remsburg et al., 2019). We observed a dose-dependent 4% and 38% decrease in spicule length in 3 and 300μM of Eve miR-31 TP injected embryos compared to the control TP, respectively (Fig. 4A). Since we observed a change in spicule length, and PMCs are the only cells within the embryo that make the larval skeleton, we examined how PMC patterning is affected in Eve miR-31 TP injected embryos. Results indicate that Eve miR-31 TP gastrulae PMCs exhibited less anterior migration than the PMCs in control TP embryos (Fig. 4B). We also observed that Eve miR-31 TP injected larvae (5 dpf) appear smaller in size with body rods that often failed to converge at the posterior end compared to the control TP (Fig. 4C).
Figure 4. Removal of miR-31 suppression of Eve and Eve overexpression results in shortening of the DVCs and aberrant PMC patterning that persists into the larval stage.
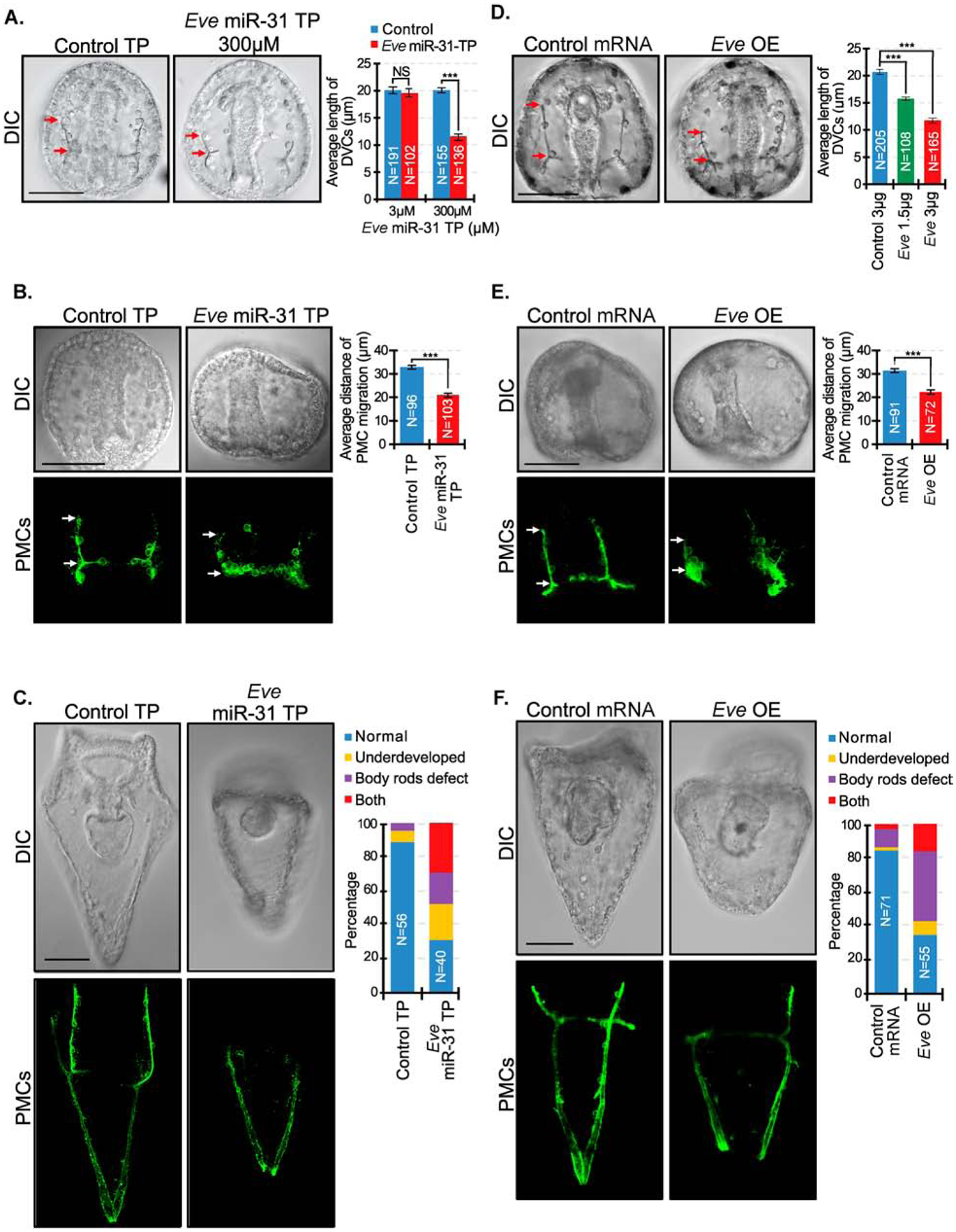
(A) Eve miR-31 TP injected gastrulae (48 hpf) had decreased DVC length in a dose-dependent manner compared to the control TP. Red arrows indicate the length of DVCs. P-value was analyzed using Student’s t-test. 3 biological replicates. NS=not significant. N is the total number of spicules examined. (B) Embryos were immunolabeled with PMC antibody, 1D5 (McClay et al., 1983). PMCs in Eve miR-31 TP injected embryos exhibit less anterior migration compared to the control injected embryos. P-value was analyzed using Student’s t-test. (C) Eve miR-31 TP injected larval stage (5 dpf) development displayed body rods that failed to meet at the posterior end and overall underdeveloped larvae compared to control. 3 biological replicates. (D) Overexpression of Eve (3 μg or 1.5 μg total mRNA in 2.5μl of injection stock solution) recapitulated shorter DVCs as in Eve miR-31 TP injected embryos in a dose-dependent manner. P-value was analyzed using Student’s t-test. N is the total number of spicules examined. (E) Eve overexpression resulted in similar PMC anterior migration defects in Eve miR-31 TP gastrulae. (F) Eve overexpression resulted in developmental delay and body rod defects, similar to Eve miR-31 TP larvae. P-value was analyzed using Student’s t-test. 3 biological replicates. ***p<0.0001. All error bars represent SEM. Scale bar = 50μm. N is the total number of embryos examined except where otherwise stated. Maximum intensity projection of Z-stack confocal images are presented for the PMC patterning.
To test the role of miR-31’s direct post-transcriptional suppression of Eve, we examined if overexpression of Eve would result in similar phenotypes as Eve miR-31 TP, supporting the role of miR-31 in repressing Eve protein levels. We cloned and injected Eve CDS transcripts into newly fertilized eggs. Results indicate that overall Eve overexpression (OE) phenotypes mimicked that of Eve miR-31 TP injected embryos (Fig. 4D–F). Eve OE resulted in the dose-dependent shortening of DVCs and decreased PMC anterior migration compared to control (Fig. 4D, E). Eve OE also led to smaller larvae with body rods that failed to meet at the posterior end, similar to defects observed in Eve miR-31 TP, indicating that an increase in Eve impacts skeletogenesis and PMC patterning (Fig. 4).
Removal of miR-31 suppression of Wnt1 results in shorter DVCs, while Wnt1 overexpression results in supernumerary skeletal rudiments and exogastrulation.
Since we found miR-31 directly suppresses Wnt1 (Fig. 3), we examined how the removal of miR-31 suppression of Wnt1 impacts skeletogenesis and PMC patterning. We microinjected newly fertilized eggs with Wnt1 miR-31 TP to specifically block miR-31’s binding within the Wnt1 3’UTR. We observed a dose-dependent decrease in DVC length in Wnt1 miR-31 TP injected embryos compared to control TP embryos (Fig. 5A). PMC patterning in Wnt1 miR-31 TP (300μM) injected gastrulae at first glance displayed no apparent defects. However, measurements of the distance of their anterior migration revealed that the PMCs in Wnt1 miR-31 TP injected gastrulae did not migrate as far anteriorly, compared to the PMCs in control TP injected gastrulae (Fig. 5B). Wnt1 miR-31 TP injected larvae (5 dpf) exhibited body rods that failed to meet at the posterior end which may contribute to the rounded body appearance, as opposed to control TP larvae with body rods that meet at the posterior end, giving the larvae a pyramidal body shape (Fig. 5C). These are similar to defects observed in Eve miR-31 TP and Eve CDS injected larvae (Fig. 4C, F), suggesting that Eve and Wnt1 are likely to regulate a similar pathway that controls proper body rod formation.
Figure 5. Removal of miR-31 suppression of Wnt1 results in shorter DVC length while Wnt1 overexpression results in extra skeletal rudiments and exogastrulation.
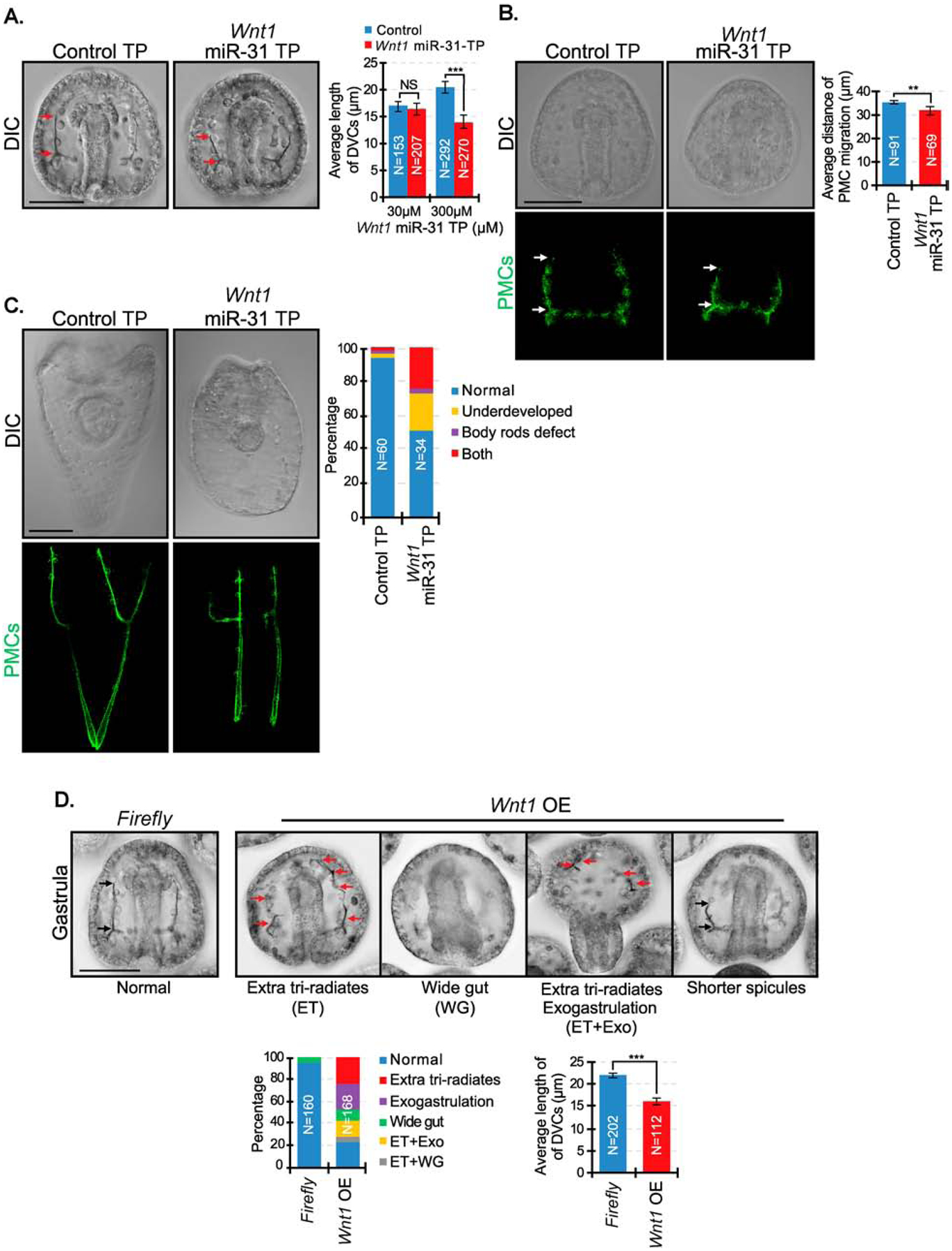
(A) Wnt1 miR-31 TP injected gastrulae (48 hpf) had decreased DVC length in a dose-dependent manner. Red arrows indicate the length of DVCs. P-value was analyzed using Student’s t-test. 2–3 biological replicates. NS=not significant. N is the total number of spicules examined. (B) Embryos were immunolabeled with PMC antibody, 1D5. PMC anterior migration is decreased in Wnt1 miR-31 TP injected embryos compared to the control injected embryos. P-value was analyzed using Student’s t-test. 2 biological replicates. White arrows indicate PMC migration distance. (C) Wnt1 miR-31 TP injected larvae (5dpf) appeared rounder with body rods that failed to meet at the posterior end compared to the control TP. 2 biological replicates. Maximum intensity projection of Z-stack confocal images are presented for the PMC patterning. (D) Overexpression of Wnt1 CDS resulted in multiple developmental defects. Red arrows indicate skeletal tri-radiates. Black arrows indicate the length of DVCs. P-value was analyzed using Student’s t-test. 4 biological replicates. N is the total number of spicules examined. Tri-radiates were counted through a series of Z-stack images. **p<0.001, ***p<0.0001. All error bars represent SEM. Scale bar = 50μm. N is the total number of embryos examined except where otherwise stated.
To mimic the effect of Wnt1 miR-31 TP where Wnt1 translation would be increased, we cloned and injected Wnt1 CDS transcripts (1.5μg/μl) into newly fertilized eggs. We observed that Wnt1 OE resulted in gastrulae with supernumerary skeletal tri-radiate rudiments (25%), exogastrulation (22%), widened gut (9%), or a combination of these phenotypes (19%) (Fig. 5D). In Wnt1 OE embryos with normal two spicule tri-radiates, the length of their DVCs was significantly shorter compared to control mRNA injected embryos. Wnt1 OE embryos did not survive well past the gastrula stage, with approximately 50% of the abnormal embryos undergoing exogastrulation (Fig. 5D). Higher concentrations (3μg/μl) resulted in embryonic lethality.
Removal of miR-31 suppression of both Eve and Wnt1 recapitulates skeletal and PMC patterning defects observed in miR-31 KD embryos.
To examine the impact of blocking miR-31’s suppression of both Eve and Wnt1, we microinjected a cocktail of Eve miR-31 TPs and Wnt1 miR-31 TP into newly fertilized eggs. We observed a significant decrease in the length of DVCs (Fig. 6A). Eve and Wnt1 miR-31 TP injected gastrulae also have more severe PMC patterning defects compared to the control TP, as well as compared to single Eve or Wnt1 miR-31 TP injections. Phenotypes observed in the Eve+Wnt1 miR-31 TP injected embryos were reminiscent of PMC anterior migration patterning defects observed in miR-31 KD embryos (Figs. 1B, 6B, 6C) (Stepicheva and Song, 2015). Eve+Wnt1 miR-31 TP injected larvae also exhibited body rods that failed to meet at the posterior end, similar to miR-31 KD larvae (Fig. 1B). These results suggest that miR-31’s regulation of Eve and Wnt1 contributes to the miR-31 KD induced defects.
Figure 6. Removal of miR-31 suppression of both Eve and Wnt1 results in similar skeletal and PMC patterning defects as miR-31 KD embryos.
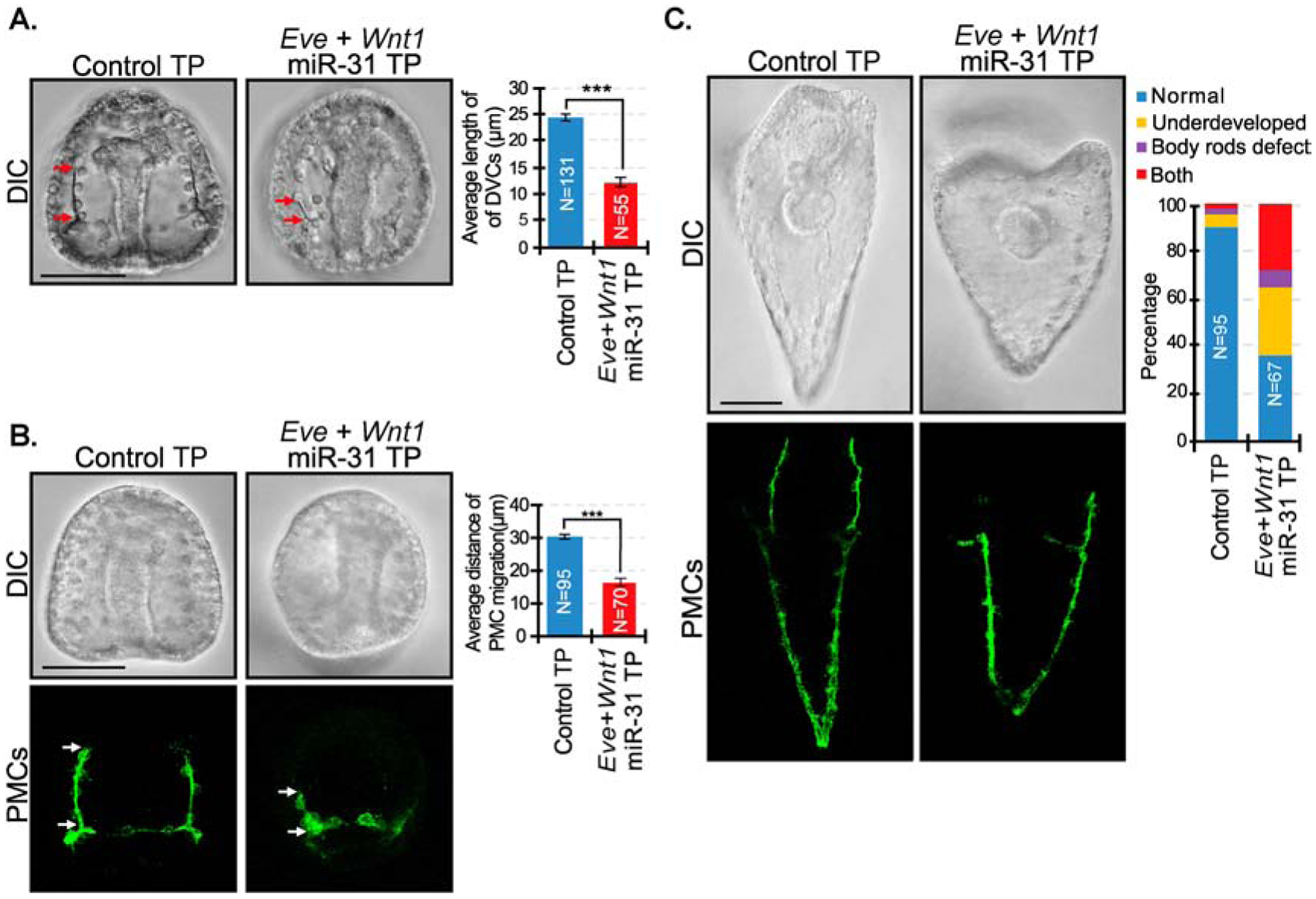
(A) A combination of Eve miR-31 TP and Wnt1 miR-31 TP resulted in a significant decrease in length of the DVCs compared to control TP gastrulae. Red arrows indicate the length of DVCs. P-value was analyzed using Student’s t-test. 2 biological replicates. N is the total number of spicules examined. (B) Eve+Wnt1 miR-31 TP injected embryos displayed severe anterior migration defects of PMCs compared to the control, reminiscent of defects observed in miR-31 KD gastrulae (Stepicheva and Song, 2015). P-value was analyzed using Student’s t-test. 3 biological replicates. (C) Compared to the control, Eve+Wnt1 miR-31 TP injected larvae (5 dpf) are smaller and have body rods that failed to meet at the posterior end, similar to miR-31 KD larvae. 3 biological replicates. **p<0.001, ***p<0.0001. All error bars represent SEM. Scale bar = 50μm. N is the total number of embryos examined except where otherwise stated. Maximum intensity projection of Z-stack confocal images are shown.
miR-31’s direct suppression of Eve regulates the increased spatial expression of Veg1 endodermal and ectodermal genes, and its direct suppression of Wnt1 regulates the anterior shift of gene expression.
To examine the impact of blocking miR-31’s direct suppression of Eve on expressions of genes in the proposed regulatory pathway (Fig. 1A), we microinjected Eve miR-31 TPs and examined the spatial expression of Eve, Hox11/13b, Wnt1, Wnt5, Wnt16, Pax2/5/8, and Vegf3 (Fig. 7A). All their expression domains were significantly expanded, except for Wnt16. However, all the genes examined did not have an anterior shift in gene expression as observed in miR-31 KD embryos. Of note is that Eve OE had similar expansion of expression domain of Vegf3 but did not cause its anterior shift in expression. (Fig. S1). Thus, while the expansion of Vegf3 expression domain in miR-31 KD embryos may be regulated by miR-31’s direct suppression of Eve, the anterior shift in gene expression of Vegf3 observed in miR-31 KD is not due to this regulation.
Figure 7. miR-31’s direct suppression of Eve regulates the increased spatial expression of Veg1 endodermal and ectodermal genes, and its direct suppression of Wnt1 regulates the anterior shift of gene expression.
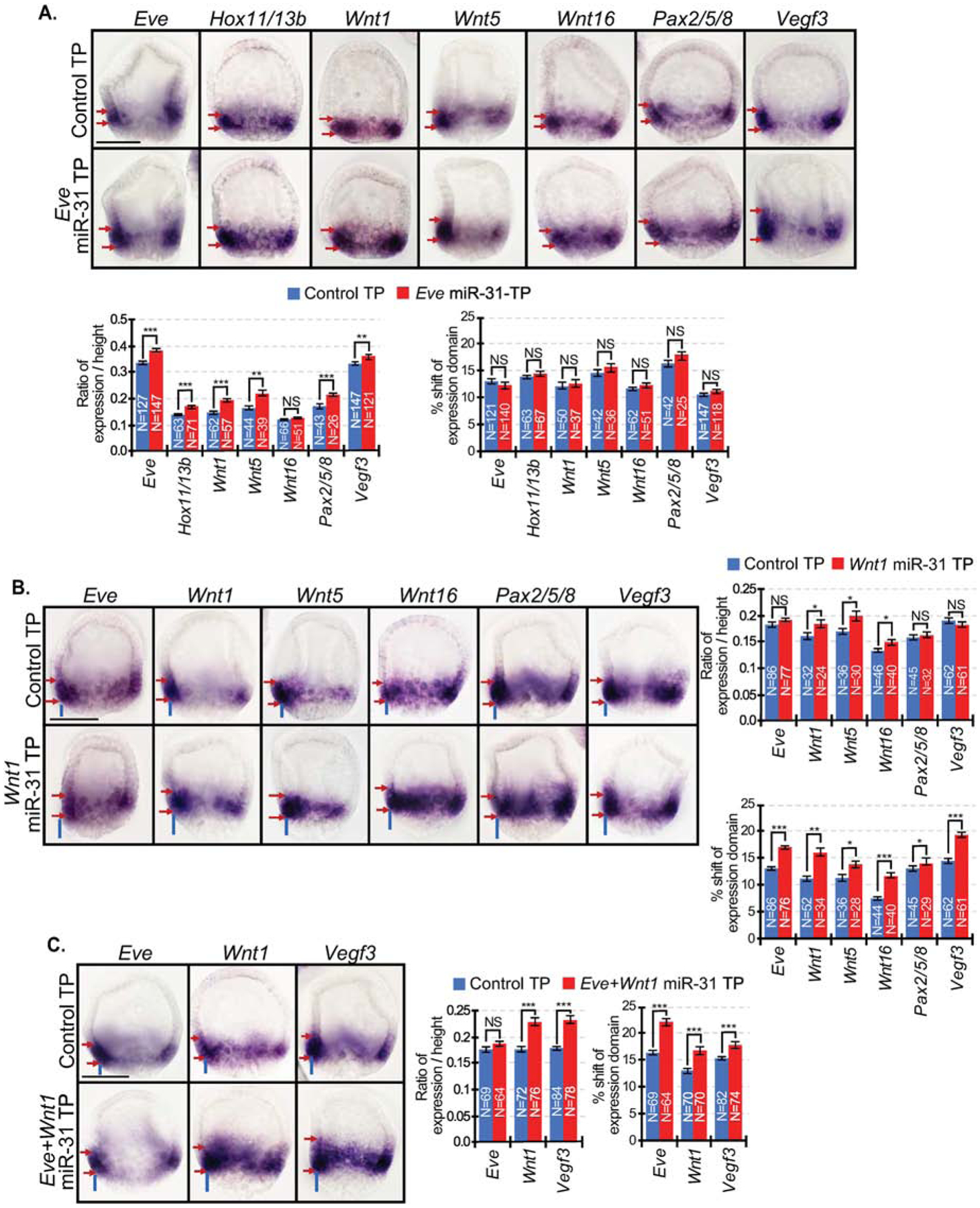
(A) In Eve miR-31 TP injected embryos, expression domains of all examined genes were significantly expanded with the exception of Wnt16. Presence of red arrows indicate an expansion of expression domain. (B) In Wnt1 miR-31 TP injected embryos, the expression domains of Wnt5 and Wnt16 were expanded compared to control TP. An anterior shift of gene expression domain was observed for all genes. Presence of blue lines indicates a shift of expression domain. (C) Eve+Wnt1 miR-31 TP injected embryos resulted in an anterior shift of the expression domains of Vegf3, Eve, and Wnt1. The expression domains of these genes also expanded, similar to that observed in miR-31 KD embryos. NS=not significant. *p<0.05, **p<0.001, ***p<0.0001 using Student’s t-test. All error bars represent SEM. 2–4 biological replicates. Scale bar = 50μm. N is the total number of embryos examined.
To test if miR-31’s direct regulation of Wnt1 results in the anterior shift in the Vegf3 expression domain in miR-31 KD embryos (Fig. 2B), we microinjected Wnt1 miR-31 TP to block miR-31’s specific binding within the Wnt1 3’UTR. We examined the spatial expression of Eve, Wnt1, Wnt5, Wnt16, Pax2/5/8 and Vegf3 in Wnt1 miR-31 TP injected embryos in the blastula stage at 24 hpf (Fig. 7B). The expression domains of Wnt1, Wnt5, and Wnt16 expanded compared to control TP. Importantly, an anterior shift in gene expression domain was observed for all genes expressed the Veg1 endoderm and Veg1 ectoderm, including Vegf3 (Fig. 7B).
To test the impact of removing miR-31’s direct regulation of both Eve and Wnt1, we microinjected both Eve and Wnt1 miR-31 TPs and tested for gene expression domains of Eve, Wnt1, and Vegf3. Results indicated that the spatial expression of Eve, Wnt1, and Vegf3 were all anteriorly shifted in their expression domains, but only Wnt1 and Vegf3 expression domains expanded (Fig. 7C), similar to that observed in miR-31 KD blastulae (Fig. 2).
Inhibition of miR-31 and removal of miR-31’s suppression of Eve and/or Wnt1 result in decreased levels of biomineralization transcripts compared to the control.
To identify the underlying molecular mechanism that led to skeletal defects, we examined the relative expression levels of biomineralization genes, such as P16, P19, p58A, SM29, SM37, SM49, SM50 (Adomako-Ankomah and Ettensohn, 2011; Cheers and Ettensohn, 2005; Livingston et al., 2006; Veis, 2011); PMC-specific cell surface protein, Msp130 (Leaf et al., 1987); and Otop2L, that may play a role in regulating cytosolic calcium levels in response to extracellular signals (Hughes et al., 2004; Hurle et al., 2003; Rafiq et al., 2014; Söllner et al., 2004). All these genes are highly expressed in the PMC ventrolateral clusters that give rise to the tri-radiate skeletal primordium, and in the PMCs at the tips of the larval body rods (with the exception of SM49) (McIntyre et al., 2014; Sun and Ettensohn, 2014). Results indicate that all perturbed embryos had decreased expression of biomineralization transcripts compared to the control TP (Fig. 8). No significant change was observed for P16 and Otop2L in any condition. miR-31 KD resulted in at least a 2-fold decrease in expression of P19, SM29, SM49, SM50, and Msp130. Msp130 is also decreased 2–3-fold in Wnt1 miR-31 TP and Eve+Wnt1 miR-31 TP injected embryos. All Eve miR-31 TP injected embryos had negligible average changes (less than 2-fold) compared to the controls, except for SM37. In contrast, Wnt1 miR-31- TP injected embryos had the strongest repressive effects on a number of transcripts, including p58A, SM29, SM37, SM49, SM50 and Msp130. Interestingly, the combination of Eve and Wnt1 miR-31 TPs seems to rescue the Wnt1 miR-31 TP effect on gene expression of several transcripts (p58A, SM29, SM50, and Msp130), suggesting that the effects of miR-31 on Eve and Wnt1 may have distinct and inverse impacts on the expression of these genes.
Figure 8. miR-31 KD, Eve miR-31 TP, Wnt1 miR-31 TP, and Eve+Wnt1 miR-31 TP injected embryos have decreased levels of biomineralization genes.
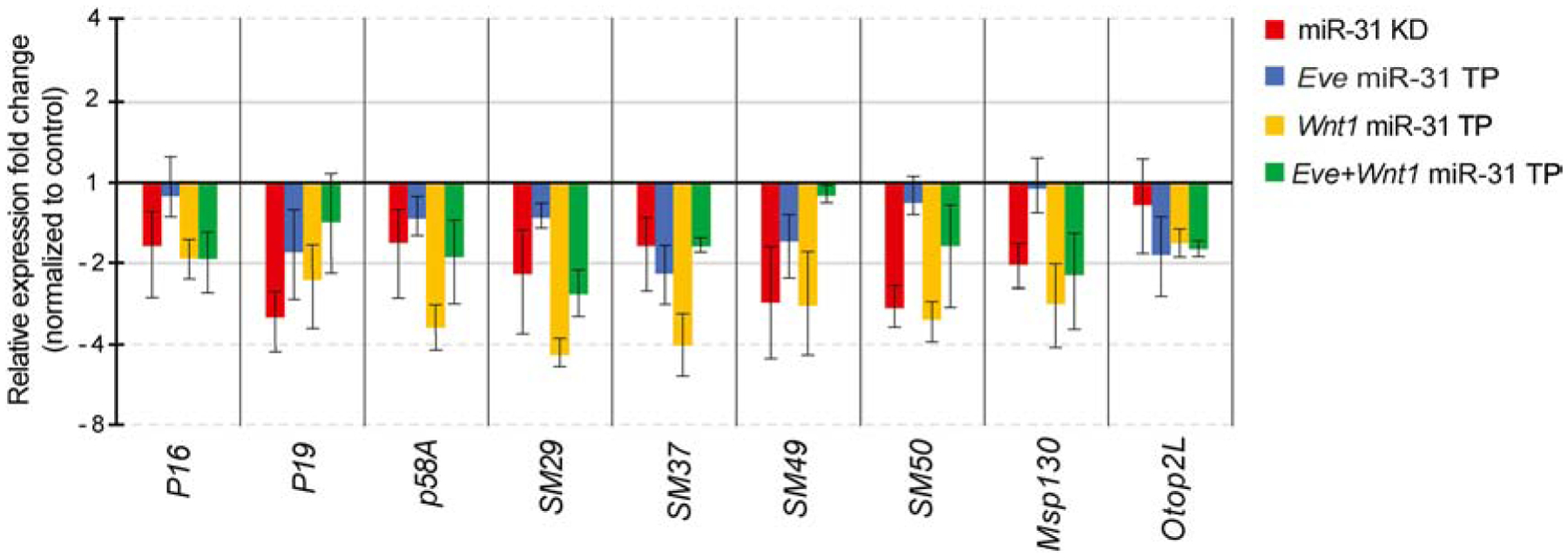
qPCR was used to measure the transcriptional changes of biomineralization and PMC genes P16, P19, p58A, SM29, SM37, SM49, SM50, Msp130 and Otop2L. All perturbed embryos have decreased expression of biomineralization transcripts compared to the control. miR-31 KD resulted in a ≥2-fold decrease in P19, SM29, SM49, SM50, and Msp130. All Eve miR-31 TP embryos have negligible average changes (<2-fold) compared to the controls, except for SM37. In contrast, Wnt1 miR-31 TP injected embryos have the strongest effects, resulting in more than a 2-fold decrease in p58A, SM29, SM37, SM49, SM50 and Msp130. Embryos were collected at mesenchyme blastula stage (24 hpf). All error bars represent SEM. 3–5 biological replicates.
miR-31 inhibitor injected embryos have delayed PMC ingression and an ectopic Vegf3 expression domain that correlates with PMC migration patterning defects.
To elucidate how miR-31 KD affects PMC ingression and migration patterning defects, we examined the expression of VegfR10 and Vegf3 and directed migration of PMCs during different developmental time points (Fig. 9). VegfR10 is the presumed receptor that responds to Vegf3 ligand and is expressed exclusively in all PMCs (Duloquin et al., 2007; Ettensohn and Adomako-Ankomah, 2019). We used the VegfR10 RNA probe to identify PMCs that underwent epithelial to mesenchymal transition (EMT) between 18 to 26 hpf (Fig. 9A) (Katow, 2015; Rizzo et al., 2006). Typically, while undergoing epithelial to mesenchymal transition (EMT), PMCs must loosen apical junctions to their neighbors, change shape, and breach the basal lamina to ingress into the blastocoel, giving these cells a ‘bottle-shape’; on the other hand, cells that have completed EMT have round cell shape (Anstrom, 1992; Ettensohn, 1999; Fink and McClay, 1985; Katow and Solursh, 1981; Lyons et al., 2012; Nakajima and Burke, 1996). This includes non-skeletogenic mesenchyme cells (NSMs) and PMCs. Using the cell shape as a criterion for EMT, we have documented the number of VegfR-10-expressing PMCs that are fully ingressed (round in cell shape) and PMCs that are undergoing ingression (bottle cell shaped) in control and miR-31 KD embryos in this series of time course experiment. In injected control blastulae, by 24 hpf, over 90% of PMCs have ingressed (Fig. 9A). In contrast, in miR-31 KD embryos at 24 hpf, only 50% of PMCs have ingressed. In fact, 30% of VegfR10-positive, bottled-shaped PMCs were still undergoing EMT at 26 hpf in miR-31 KD embryos, when nearly 100% of all PMCs in control embryos have fully ingressed. This delay in PMC ingression observed in miR-31 KD embryos was consistent throughout all times points examined and is unlikely due to transient delay caused by microinjections.
Figure 9. miR-31 KD embryos exhibit a delay in PMC ingression and express ectopic Vegf3 expression domain that correlates with PMC patterning defects.
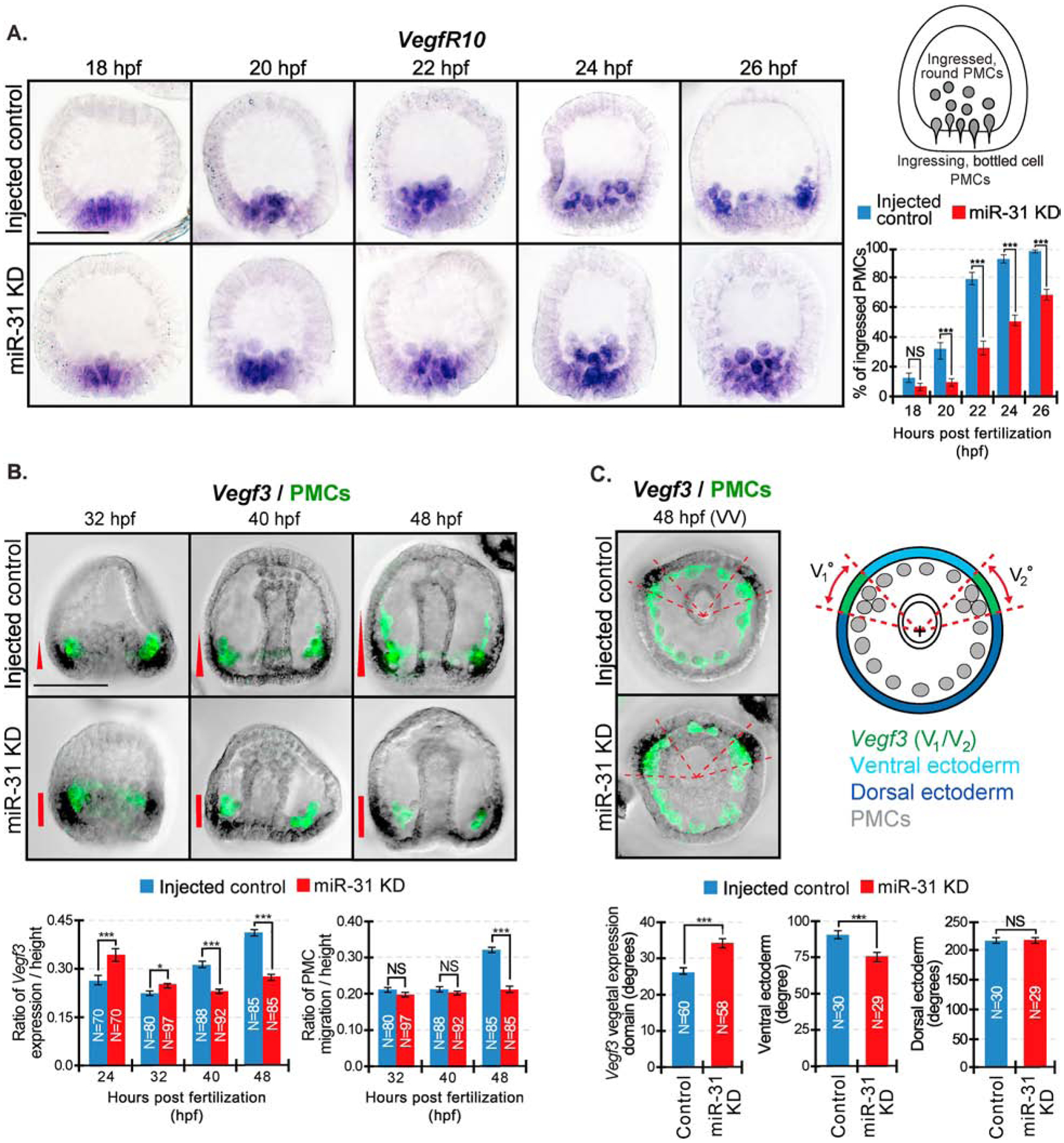
WMISH was performed on embryos to visualize Vegf3 or VegfR10 followed by PMC immunolabeling with the 1D5 antibody. (A) Control and miR-31 KD embryos were collected at various time points spanning PMC ingression. VegfR10-expressing PMCs were categorized as either “ingressed, round” PMCs or “ingressing, bottle cell” PMCs. VegfR10-positive PMCs were counted through a series of Z-stack images. The percentage of “ingressed, round” PMCs from each embryo were calculated from the total of VegfR10-expressing PMCs. Average percentage was taken from each time point. For all conditions, a total of 30 embryos were measured for 3 biological replicates. NS=not significant, ***p<0.0001 using Cochran-Mantel-Haenszel test. All error bars represent SEM. 3 biological replicates. Scale bar = 50μm. (B) Embryos undergoing gastrulation were collected between 32 to 48 hpf. Embryos were first hybridized with Vegf3 RNA probe and followed with immunolabeling against 1D5 antibody that recognizes the PMCs (McClay et al., 1983). In control gastrulae, PMCs have migrated anteriorly in parallel to the Vegf3 gradient at 48hpf. In miR-31 KD gastrulae at 48 hpf, PMCs are clustered next to the concentrated Vegf3 expression domain. The general trend of Vegf3 expression domain correlates with the anterior distance of PMC migration. (C) Vegf3 expression is indicated in red. Zen software was used to determine the center of gastrulae in the vegetal view and to determine angles of Vegf3 expression, VE, and DE domains. In the vegetal view (VV), the measured angle of Vegf3 (V1/V2) expression domain in miR-31 KD is expanded into the VE compared to control. N is the total number of Vegf3 expression domains measured. 3 biological replicates. NS=not significant. *p<0.05, ***p<0.0001 using Student’s t-test. All error bars represent SEM. Scale bar = 50μm. Representative images were taken with ZEISS Observer Z1 microscope.
To test if a change in Vegf3 expression could explain PMC patterning defects, we examined Vegf3 expression and anterior migration of PMCs during various stages of gastrulation (32 to 48 hpf). We observed an interesting trend of Vegf3 expression that correlates with anterior migration of PMCs. At 24 hpf, the anterior-posterior expression domain of Vegf3 is significantly expanded in the miR-31 KD blastulae compared to the control (Stepicheva and Song, 2015) (Figs. 2B, 9B). However, from 40 to 48 hpf, when PMCs undergo anterior migration (Duloquin et al., 2007; Ettensohn and McClay, 1986; McIntyre et al., 2014), the expression domain of Vegf3 in the AP region remains unchanged and is decreased in miR-31 KD gastrulae compared to the control. The decreased Vegf3 expression domain in the miR-31 KD gastrulae correlates with their decreased anterior migration distance of PMCs, compared to the control. Further, in control gastrulae, Vegf3 is expressed along the AP axis of the ectoderm in a gradient that likely guides the PMCs to migrate toward the anterior pole of the embryo (Fig. 9B) (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007). In contrast, miR-31 KD gastrulae express concentrated Vegf3 at the junction of the BE-DVM, but seem to lack a Vegf3 gradient along the AP axis (Fig. 9B). At the same time that the expression of Vegf3 along the AP axis ceases to expand and stays unchanged in the miR-31 KD embryos (40–48 hpf), the expression domain of Vegf3 expands in the DV axis (Fig. 9C). To determine if Vegf3’s expression domain expands into the dorsal or ventral ectoderm, we measured the angles of these domains in between the Vegf3 expressing BE-DVM areas. The measured angle of the ventral ectoderm is significantly narrower in miR-31 KD gastrulae compared to control gastrulae, while the angle of dorsal ectoderm remains unchanged (Fig. 9C). This result indicates that the DV expression domain of Vegf3 in miR-31 KD gastrulae likely expands into the ventral ectoderm.
Discussion
This study identifies that miR-31 directly post-transcriptionally represses the transcriptional factor Eve and the signaling ligand, Wnt1, in the endodermal and ectodermal gene regulatory network. Regulation mediated by miR-31 ensures proper skeletogenesis and PMC patterning through Vegf signaling. Blocking miR-31’s suppression of Eve and Wnt1 recapitulates miR-31 KD phenotypes, indicating that miR-31 regulates skeletogenesis and PMC patterning in part through its regulation of these two gene targets. Specifically, we found miR-31’s suppression of Eve is responsible for the anterior expansion of Veg1 endodermal and ectodermal genes; and its direct suppression of Wnt1 regulates the anterior shift in gene expression in these domains. We also found that miR-31’s indirect regulation of Vegf3 impacts the anterior migration of PMCs. Overall, this work provides a better understanding of the molecular mechanism of how miR-31 regulates Vegf3 expression to regulate skeletogenesis and PMC patterning.
Since we observed aberrant Vegf3 spatial expression in miR-31 KD embryos (Stepicheva and Song, 2015), we examined the molecular impact of miR-31 on genes expressed in the same cells as Vegf3 (Veg1 endoderm and Veg1 ectoderm), focusing on genes in our proposed pathway (Fig. 1A). Results indicate that all genes tested, except for Eve and Hox11/13b, had expanded expression domains at 24 hpf (Fig. 2B). Interestingly, when miR-31 inhibitor was injected at a lower concentration (20μM), Eve expression domain was significantly expanded (Fig. S2). Since Eve and Hox11/13b are autoregulated by negative feedback, an increase of Eve or Hox11/13b protein may result in repression of its own transcription (Cui et al., 2014; Cui et al., 2017). It is possible that a higher concentration of injected miR-31 inhibitor (30μM) led to sufficient increase of translated Eve that induces autorepression of its own transcription. However, increased Eve protein may still act as an activator for target genes before autorepression is induced.
We also observed that in miR-31 KD blastulae, Wnt1, Wnt5, Wnt16, and Pax2/5/8 expression domain expanded compared to the injected control (Fig. 2B). Eve and Wnt1 are both activators of Wnt5 (Cui et al., 2014). Our results identified that miR-31 directly suppresses Eve and Wnt1, but not Wnt5 (Fig. 3). Removing miR-31’s suppression of Eve and Wnt1 may activate transcription of Wnt5. miR-31 KD may lead to increased translated Wnt1, which may autoactivate its own transcription (Cui et al., 2014). This increase in Wnt1 may also explain the expanded expression of Wnt16, since Wnt1 activates Wnt16 (Cui et al., 2014). We speculate that the increased expression of Pax2/5/8 in miR-31 KD embryos may result from increased Wnt1 and Wnt16. The expanded expression of Pax2/5/8 in the BE may, in turn, lead to increased expression of Vegf3 (Rottinger et al., 2008). Many of the genes in this proposed pathway are autoregulated; for example, Hox11/13b, Eve, Fzd5/8 and Fzd1/2/7 are autorepressive, while Wnt1 can activate its own transcription (Cui et al., 2014; Cui et al., 2017; Range, 2018; Range et al., 2013). Additionally, each of the components in the pathway is also potentially cross-regulating each other (Fig. 1A).
We observed that removing miR-31’s direct suppression of Eve resulted in decreased spicule length and abnormally clustered PMCs (Fig. 4A, B). These defects persisted into the larval stage, where Eve miR-31 TP larvae appeared smaller in size with body rods that often failed to meet at the posterior end (Fig. 4C). These phenotypes are similar but less severe than those observed in miR-31 KD embryos (Stepicheva and Song, 2015). In Eve miR-31 TP embryos, we also observed an expanded Eve expression domain, indicating that the level of translated Eve may not be high enough to autorepress its own transcription (Figs. 7A). Additionally, Eve miR-31 TP resulted in an expansion of Vegf3 and all genes proposed to be upstream of Vegf3 and downstream of Eve within our model, with the exception of Wnt16 (Figs. 1A, 7A).
As a control for Eve miR-31 TP and to test that PMC defects observed are due to increased translated Eve, we injected Eve CDS transcripts into zygotes. Similar to miR-31 KD (Stepicheva and Song, 2015) and Eve miR-31 TP embryos (Fig. 4A–C), Eve OE embryos had PMC defects that persisted into the larval stage (5dpf), indicating that Eve regulates skeletogenesis and PMC patterning (Figs. 1B, 4C, 4F). Removing miR-31’s suppression of Eve is sufficient to cause increased translation of Eve, which increases Vegf3 expression. This is demonstrated by Eve OE blastulae, which exhibit an increased and expanded Vegf3 expression, similar to miR-31 KD and Eve miR-31 TP blastulae (Fig. S1). In embryos injected with the lower concentration of miR-31 KD (20μM) (Fig. S2), we observed an expansion of Eve expression domain, while in embryos injected with a higher miR-31 KD concentration (30μM), we observed no change in Eve expression (Fig. 2B). This indicates that the higher concentration of miR-31 inhibitor at 30μM, which was used throughout this paper, may trigger Eve’s autorepression (Fig. 2B). The exact mechanism of how Eve regulates Vegf3 remains unclear. We speculate that Vegf3 is likely activated before Eve is able to repress its own transcription, since Vegf3 expression domain expanded in miR-31 KD, Eve miR-31 TP, and Eve OE, independent of the expression domain of Eve. In general, blocking miR-31’s suppression of Eve resulted in expanded expression domains of most genes tested within our proposed model that impacts skeletogenesis (Figs. 1A, 7A). As these genes impact PMC patterning and biomineralization, our results reveal a novel role of Eve in sea urchin skeletogenesis.
Previously, it was reported that Wnt1 is likely to be the ligand for the Fzd5/8-JNK/ROCK signaling pathway (Range et al., 2013). Perturbation of Fzd5/8 or ROCK signaling leads to skeletal defects (Croce et al., 2006). Embryos injected with Wnt1 miR-31 TP exhibited phenotypes that are relatively less severe than that of miR-31 KD or Eve miR-31 TP embryos (Fig. 5A–C). Blocking miR-31’s suppression of Wnt1 resulted in shortened DVCs and body rods that failed to meet posteriorly. However, PMC patterning was mildly affected (Fig. 5B). This may be due to miR-31’s weak binding affinity to Wnt1 compared to Eve (Fig. 3). Blocking miR-31’s direct suppression of Wnt1 resulted in an anterior shift in gene expressions in the Veg1 cells (Fig. 7B). Interestingly, Wnt1 KD causes a vegetal (posterior) shift of expression in Emx, (animal ectoderm), Lim1 (Veg1 ectoderm), Eve (Veg1 endoderm and endoderm), Hox11/13b (Veg1 endoderm), and FoxA (Veg2 mesoderm) (Cui et al., 2014). This indicates that a change in Wnt1 levels has an impact on Veg1 ectoderm, Veg1 endoderm, and Veg2 mesoderm gene expression domains. While Wnt1 miR-31 TP embryos exhibit mild phenotypes, embryos overexpressing Wnt1 CDS resulted in much more severe phenotypes than Wnt1 miR-31 TP or miR-31 KD embryos, including having supernumerary skeletal rudiments and exogastrulation (Fig. 5D). These incongruous results may be due to Wnt1’s possible autoactivation, where overexpression of Wnt1 would result in even more production of Wnt1, resulting in a more severe phenotype. Additionally, overexpression of Wnt1 would ectopically activate Wnt signaling in all cells, whereas Wnt1 miR-31 TP only activates Wnt signaling in cells that normally express Wnt1. Previous literature indicates that Wnt1 KD has no effect on PMC patterning, but does result in larvae with a partial gut that lack mouth formation (Wei et al., 2012). Consistent with previous literature, we observed approximately 50% of all Wnt1 CDS injected abnormal embryos underwent exogastrulation (Fig. 5D). Overall, results indicate that the anterior shift of Vegf3 expression due to miR-31’s direct suppression of Wnt1 is not sufficient to fully cause the skeletal and PMC patterning defects observed in miR-31 KD. Nonetheless, proper levels of Wnt1 are critical for proper skeletal rudiment formation and gastrulation.
Removing the combinatorial suppression of miR-31 on Eve and Wnt1 resulted in skeletal defects and PMC mispatterning (Fig. 6), reminiscent of miR-31 KD induced phenotypes (Stepicheva and Song, 2015) (Fig. 1B). Further, the combination of Eve and Wnt1 miR-31 TPs recapitulated expression domain changes of Eve, Wnt1, and Vegf3 in miR-31 KD embryos (Figs. 2B, 7C). This indicates that the expansion in expression of Vegf3 regulated by Eve, along with the anterior shift in Vegf3 expression domain regulated by Wnt1, are potentially the cause of skeletogenic and PMC patterning defects in miR-31 KD embryos.
Consistent with shortened spicules in miR-31 KD embryos, blocking miR-31’s suppression of Eve and/or Wnt1 resulted in the downregulation of several PMC-specific biomineralization transcripts (Fig. 8). P19, p58a, SM29, SM37, SM49, and SM50 biomineralization transcripts are expressed by the PMCs, present in the biomineral, and may be important for the skeletal mineralization process (Knapp et al., 2012; Mann et al., 2008; Sun and Ettensohn, 2014). P19 is highly expressed in the PMCs during all stages of embryogenesis (Veis, 2011). During the larval stage, P19, p58A, and SM50 are expressed in the PMCs that give rise to the larval body rods (Adomako-Ankomah and Ettensohn, 2011; Cheers and Ettensohn, 2005; Peled-Kamar et al., 2002; Sun and Ettensohn, 2014). Results indicate that only Wnt1 miR-31 TP injected embryos resulted in more than a 3-fold decrease of p58A, while both miR-31 KD and Wnt1 miR-31 TP injected embryos have about a 3-fold decrease of SM50 compared to the control. Since p58A is likely to be involved in depositing the calcite-based biomineral for skeletal synthesis (Adomako-Ankomah and Ettensohn, 2011), and accumulation of SM50 (LSM34) is required for initiation of spicule formation and subsequent morphogenesis (Peled-Kamar et al., 2002), their decreased expression may contribute to skeletal shortening and the failure of the tips of the body rods to converge in the miR-31 KD and Wnt1 miR-31 TP injected embryos. In Eve miR-31 TP treated embryos, the only transcript decreased more than 2-fold is SM37, which has the same cis-regulatory elements as SM50 and is regulated coordinately with SM50 throughout development (Lee et al., 1999). However, the function of SM37 is not known. Similar to SM37, P19, SM29, SM49, MSP130 and Otop2L transcripts are expressed in the PMCs, but their exact function in biomineralization is unknown (Leaf et al., 1987; Lee et al., 1999; Livingston et al., 2006; Sun and Ettensohn, 2014; Veis, 2011). In miR-31 KD embryos, the composite ≥2-fold decrease of P19, SM29, SM49, SM50 and Msp130 may all contribute to skeletal defects we observed, since they are expressed in the PMC ventrolateral clusters and in the PMCs at the tips of the larval rods (Sun and Ettensohn, 2014). Wnt1 miR-31 TP resulted in greater than 2-fold decrease in all genes examined, except for Otop2L, suggesting that this perturbation has a broad repressive effect on biomineralization and PMC transcripts. Interestingly, it appears that miR-31 may regulate Eve and Wnt1 differently, since the Wnt1 miR-31 TP has the strongest repressive effects, while the combination of both Wnt1 and Eve miR-31 TPs appears to rescue the Wnt1 miR-31 TP repressive effect for some biomineralization transcripts (p58A, SM29, SM50, and Msp130). These results suggest a complex and undefined regulatory relationship between Eve and Wnt1. Overall, the decreased trend of these biomineralization and PMC transcripts may partially explain the skeletal defects of shortened DVCs and the larval body rods failing to meet at the posterior end in all the perturbations examined. In addition, the body rods extend from the tri-radiate spicule, and the convergences of the body rods is dependent on its initial orientation (Brandhorst and Davenport, 2001; Gustafson and Wolpert, 1961; McClay et al., 1992). The shortened spicules we observed in miR-31 KD, Eve miR-31 TP, Wnt1 miR-31 TP, and Eve+Wnt1 miR-31 TP could also contribute to the failure of body rod convergence.
Removal of miR-31’s suppression of Eve and Wnt1 resulted in an expansion and anterior shift in Vegf3’s expression domain which correlated with PMC migration defects. Vegf3 plays an especially critical role in PMC patterning. Previous research indicates that Vegf3 KD embryos have increased Vegf3 expression and mispatterned PMCs in the posterior end of the embryo with a loss of skeleton (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007). This PMC patterning defect is similar in miR-31 KD embryos, where expanded Vegf3 expression potentially contributes to the PMC positioning defect (Stepicheva and Song, 2015). In the current study, we observed that VegfR10-expressing PMCs in miR-31 KD blastulae had delayed PMC ingression (Fig. 9A). Normally, PMCs are fully ingressed into the blastocoel by the mesenchyme blastula stage (24 hpf) (Bradham et al., 2004; Sun and Ettensohn, 2014; Wu et al., 2007). In miR-31 KD embryos, only 67% of the VegfR10-expressing PMCs have fully ingressed by 26 hpf. This consistent delay in ingression may contribute to the PMC positioning defects and shortened skeletal spicules, but exactly how miR-31 regulates ingression is not known. Additionally, Vegf3 is expressed in a gradient in the normal sea urchin gastrulae (Adomako-Ankomah and Ettensohn, 2013; Duloquin et al., 2007), similar to VEGF-A in vertebrates (Chauvet et al., 2013). We observed PMC anterior migration correlates with Vegf3 expression (Fig. 9B). Interestingly, in miR-31 KD blastulae (24 hpf), Vegf3 expression domain was expanded in the AP axis compared to control TP; however, by 40 hpf, Vegf3 expanded expression shifted from the AP to the DV axis (Fig. 9B, C). Since the expression of BE genes, such as Vegf3, is restricted by expression of Nodal and BMP in the DVM (McIntyre et al., 2013), the expansion of Vegf3 expression in the DV axis may be explained by the regulatory relationship between Wnt1, Nodal, and BMP signaling. During early cleavage stages (60-cell), Wnt1 indirectly activates Nodal by suppressing FoxQ2 to specify the AP axis (Range et al., 2013; Yaguchi et al., 2008). In the gastrula stage, Nodal activates BMP signaling to restrict Wnt1 to the posterior-ventral side (Duboc et al., 2010; Lapraz et al., 2009; Wei et al., 2012) (Fig. 10B). Later in the gastrula stage, Wnt1 prevents Nodal expression in the posterior region to establish the ciliary band in the DV axis (Wei et al., 2012). In turn, Nodal activates Not1, which represses Vegf3 in the ventral ectoderm and restricts Vegf3’s expression to the two lateral ectodermal domains and posterior ventral corners (BE-DVM) (Layous, 2020; Li et al., 2012; McIntyre et al., 2013). In miR-31 KD blastulae, we observed Wnt1 is expanded in the AP axis and shifted anteriorly compared to injected control embryos (Figs. 2, 9B). During gastrulation, this miR-31 KD-induced ectopic expression of Wnt1 may restrict Nodal and Not1 in the ventral ectoderm, resulting in expanded Vegf3 expression in the DV axis (Figs. 9C, 10B). The fine-tuning nature of miRNAs would not likely result in a dramatic loss of BE-DVM restriction, but rather a slight expansion of Vegf3. Currently, we do not know the exact mechanism of how miR-31 mediates the shift inVegf3 expression expansion from AP to DV axis but speculate that this may involve cross-regulation of Wnt1, Nodal and BMP signaling.
Figure 10. miR-31 directly represses components within the PMC GRN and transcripts upstream of Vegf3.
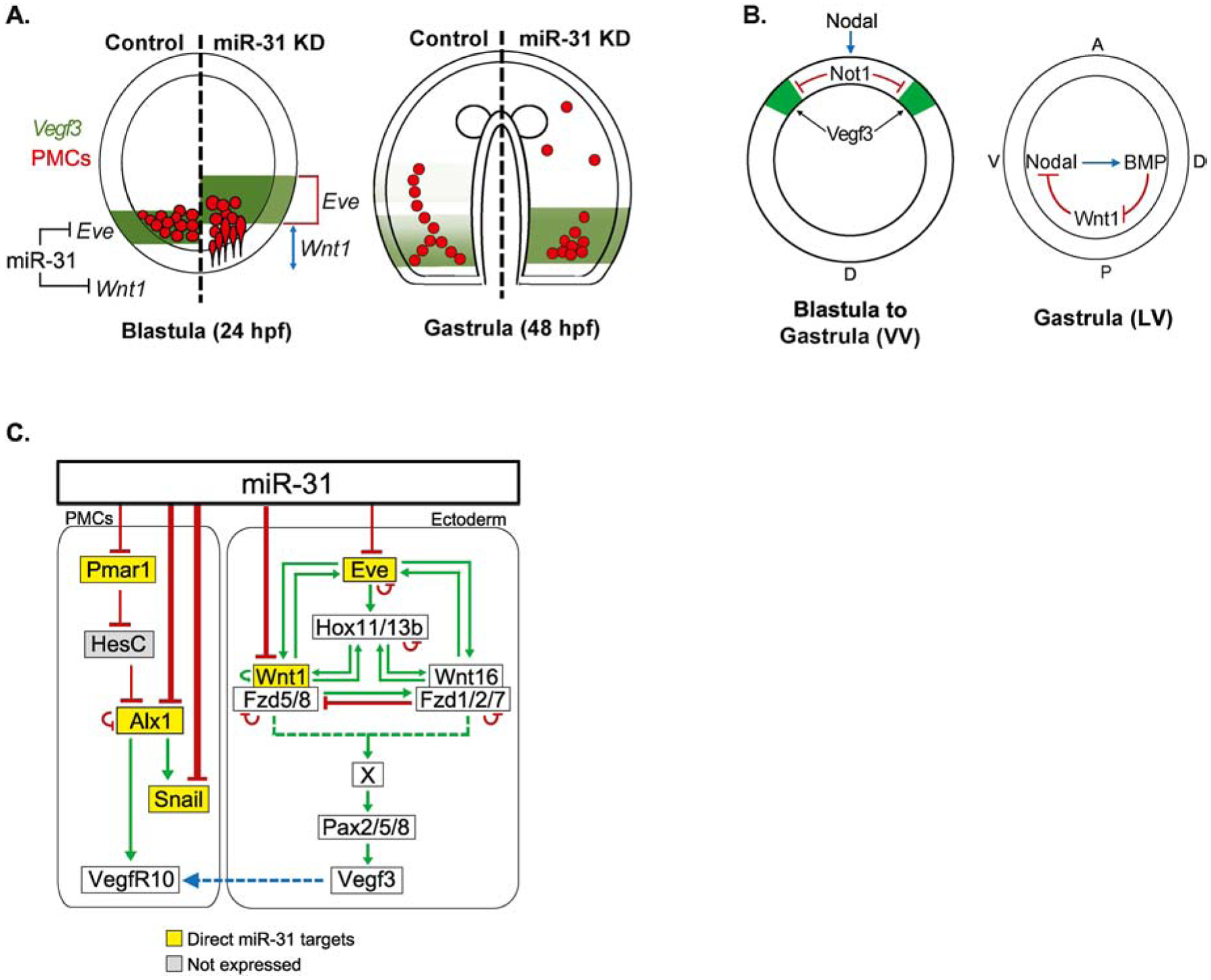
(A) miR-31’s regulation of Eve and Wnt1 indirectly impacts Vegf3 expression in the blastula stage. Removing miR-31’s direction suppression of Eve results in an expansion of Vegf3 expression domain, while removing miR-31’s direct suppression of Wnt1 results in an anterior shift of Vegf3 expression. (B) The expansion of Vegf3 in the BE-DVM in miR-31 KD embryos could be due to increased Wnt1, thus increasing Wnt1’s restriction of Nodal from the posterior-ventral region during gastrula stage. Decreased Nodal would result in less Not1 expression, leading to less Vegf3 restriction in the BE-DVM. VV = Ventral view; LV = Lateral view; A = anterior; P = posterior; V = ventral; D = dorsal. (C) We previously identified miR-31 directly suppresses Pmar1, Alx1 and Snail, which are expressed in the PMCs (Stepicheva and Song, 2015). cWnt/β-catenin triggers specification of PMCs by activating the transcriptional repressor Pmar1 expressed exclusively in the micromeres. Pmar1 inhibits transcriptional repressor HesC (Guss and Ettensohn, 1997; Oliveri et al., 2003; Revilla-i-Domingo et al., 2007). This leads to the activation of Alx1, which in turn, activates Snail and VegfR10 (Ettensohn et al., 2003; Rafiq et al., 2012; Sharma and Ettensohn, 2011). VegfR10 is expressed on the PMCs and is thought to respond to the Vegf3 ligand secreted by the ectoderm to guide patterning of PMCs. In this study, we identified Eve and Wnt1 as direct miR-31 targets upstream of Vegf3 activation.
Overall, this study identifies miR-31’s direct suppression of Eve and Wnt1 to be a potential underlying molecular mechanism of miR-31’s regulation of Vegf3 expression and skeletogenesis (Fig. 10A). miR-31’s suppression of Eve and Wnt1 defines the expression domain of Vegf3, which in turn, regulates skeletogenesis and PMC patterning. miR-31’s direct repression of Wnt1 may indirectly affect the expansion of Vegf3 in the DV axis, potentially via Wnt1’s regulation of Nodal which activates Not1 to restrict Vegf3 expression in the ventral ectoderm (Fig. 10B). Previously, we identified miR-31 to directly repress major TFs within the PMC GRN, such as Pmar1, Alx1, and Snail, in addition to a not well understood Vegf receptor, VegfR7 (Fig. 10C). Upon β-catenin activation in the micromeres, Pmar1 is activated in the PMC GRN. It represses a ubiquitously expressed repressor HesC (Guss and Ettensohn, 1997; Oliveri et al., 2003; Revilla-i-Domingo et al., 2007). Hence, HesC in the micromeres is repressed, allowing transcriptional activation of Alx1. Alx1 plays a central role in the PMC GRN by activating multiple TFs such as Snail, which is important for PMC EMT, and VegfR10 (Ettensohn et al., 2003; Khor et al., 2019; Rafiq et al., 2012; Sharma and Ettensohn, 2011). Removal of miR-31’s direct suppression of Alx1 and/or VegfR7 resulted in shorter skeletal spicules and mispatterned PMCs (Stepicheva and Song, 2015). Thus, our previous and current finding indicate that miR-31 suppresses the PMC GRN components, Eve, and Wnt1 to ensure proper skeletogenesis and PMC patterning.
Supplementary Material
Figure S1 Eve overexpression results in expanded expression domain of Vegf3. Overexpression of Eve (3 μg total mRNA in 2.5μl of injection stock solution) resulted in expansion of Vegf3 expression domain compared to control mRNA. This is similar to miR-31 KD and Eve miR-31 TP. NS=not significant, **p<0.001 using Student’s t-test. All error bars represent SEM. 3 biological replicates. Scale bar = 50μm. N is the total number of embryos examined.
Figure S2 Lower concentration of miR-31 KD results in expanded expression domain of Eve. miR-31 inhibitor injected at 20μM resulted in expansion and anterior shift of Eve expression domain. At a lower concentration (20μM), Eve may not sufficiently increase to autorepress itself. *p<0.05, ***p<0.0001 using Student’s t-test. All error bars represent SEM. 2 biological replicates. Scale bar = 50μm. N is the total number of embryos examined.
Highlights:
microRNA-31’s direct targeting of Eve and Wnt1 regulates skeletogenesis
Removal of miR-31’s suppression of Eve and Wnt1 mimics miR-31 KD ectopic Vegf3 expression
Eve plays a role in skeletogenesis, upstream of Vegf3 activation
Acknowledgements
The authors would like to thank David McClay (Duke University) for his kind gift of 1D5 antibody. We also thank Dr. Ryan Range (Auburn University) for the Wnt16 plasmid. This work is funded by University of Delaware Graduate Fellowship to NFS. NSF CAREER (IOS 1553338) to JLS and NIH NIGMS P20GM103446. We thank members of the Song lab in careful reading of the manuscript. The authors thank the anonymous reviewers for their invaluable comments.
Abbreviations:
- PMCs
primary mesenchyme cells
- GRN
Gene regulatory network
- miRNA-TP
microRNA target protector morpholino
- Eve
Even-skipped
- Wnt
Wingless-related integration site
- Vegf
Vascular endothelial growth factor
- DVC
Dorsoventral connecting rods
- KD
Knockdown
- OE
Overexpression
- TF
Transcription factor
- cWnt
canonical-Wnt
- ncWnt
non-canonical Wnt
- EMT
Epithelial to mesenchymal transition
- VE
Ventral ectoderm
- DE
Dorsal ectoderm
- BE-DVM
Border ectoderm-dorsal ventral margin
- AP
Anterior-Posterior
- DV
Dorsal-Ventral
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adomako-Ankomah A, Ettensohn CA, 2011. P58-A and P58-B: novel proteins that mediate skeletogenesis in the sea urchin embryo. Dev Biol 353, 81–93. [DOI] [PubMed] [Google Scholar]
- Adomako-Ankomah A, Ettensohn CA, 2013. Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development 140, 4214–4225. [DOI] [PubMed] [Google Scholar]
- Adomako-Ankomah A, Ettensohn CA, 2014. Growth factors and early mesoderm morphogenesis: insights from the sea urchin embryo. Genesis 52, 158–172. [DOI] [PubMed] [Google Scholar]
- Anstrom JA, 1992. Microfilaments, cell shape changes, and the formation of primary mesenchyme in sea urchin embryos. J Exp Zool 264, 312–322. [DOI] [PubMed] [Google Scholar]
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2018. Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradham CA, Miranda EL, McClay DR, 2004. PI3K inhibitors block skeletogenesis but not patterning in sea urchin embryos. Dev Dyn 229, 713–721. [DOI] [PubMed] [Google Scholar]
- Brandhorst BP, Davenport R, 2001. Skeletogenesis in sea urchin interordinal hybrid embryos. Cell Tissue Res 305, 159–167. [DOI] [PubMed] [Google Scholar]
- Chauvet S, Burk K, Mann F, 2013. Navigation rules for vessels and neurons: cooperative signaling between VEGF and neural guidance cues. Cell Mol Life Sci 70, 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA, 2004. Rapid microinjection of fertilized eggs. Methods Cell Biol 74, 287–310. [DOI] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA, 2005. P16 is an essential regulator of skeletogenesis in the sea urchin embryo. Dev Biol 283, 384–396. [DOI] [PubMed] [Google Scholar]
- Croce J, Duloquin L, Lhomond G, McClay DR, Gache C, 2006. Frizzled5/8 is required in secondary mesenchyme cells to initiate archenteron invagination during sea urchin development. Development 133, 547–557. [DOI] [PubMed] [Google Scholar]
- Cui M, Siriwon N, Li E, Davidson EH, Peter IS, 2014. Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo. Proc Natl Acad Sci U S A 111, E5029–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Vielmas E, Davidson EH, Peter IS, 2017. Sequential Response to Multiple Developmental Network Circuits Encoded in an Intronic cis-Regulatory Module of Sea Urchin hox11/13b. Cell Rep 19, 364–374. [DOI] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Saudemont A, Bessodes N, Mekpoh F, Haillot E, Quirin M, Lepage T, 2010. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development 137, 223–235. [DOI] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Besnardeau L, Lepage T, 2004. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell 6, 397–410. [DOI] [PubMed] [Google Scholar]
- Duloquin L, Lhomond G, Gache C, 2007. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134, 2293–2302. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, 1999. Cell movements in the sea urchin embryo. Curr Opin Genet Dev 9, 461–465. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Adomako-Ankomah A, 2019. The evolution of a new cell type was associated with competition for a signaling ligand. PLoS Biol 17, e3000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettensohn CA, Illies MR, Oliveri P, De Jong DL, 2003. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development 130, 2917–2928. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, McClay DR, 1986. The regulation of primary mesenchyme cell migration in the sea urchin embryo: transplantations of cells and latex beads. Dev Biol 117, 380–391. [DOI] [PubMed] [Google Scholar]
- Fink RD, McClay DR, 1985. Three cell recognition changes accompany the ingression of sea urchin primary mesenchyme cells. Dev Biol 107, 66–74. [DOI] [PubMed] [Google Scholar]
- Guss KA, Ettensohn CA, 1997. Skeletal morphogenesis in the sea urchin embryo: regulation of primary mesenchyme gene expression and skeletal rod growth by ectoderm-derived cues. Development 124, 1899–1908. [DOI] [PubMed] [Google Scholar]
- Gustafson T, Wolpert L, 1961. Studies on the cellular basis of morphogenesis in the sea urchin embryo. Directed movements of primary mesenchvme cells in normal and vegetalized larvae. Exp Cell Res 24, 64–79. [DOI] [PubMed] [Google Scholar]
- Hughes I, Blasiole B, Huss D, Warchol ME, Rath NP, Hurle B, Ignatova E, Dickman JD, Thalmann R, Levenson R, Ornitz DM, 2004. Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev Biol 276, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurle B, Ignatova E, Massironi SM, Mashimo T, Rios X, Thalmann I, Thalmann R, Ornitz DM, 2003. Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum Mol Genet 12, 777–789. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP, 2011. Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature 469, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katow H, 2015. Mechanisms of the epithelial-to-mesenchymal transition in sea urchin embryos. Tissue Barriers 3, e1059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katow H, Solursh M, 1981. Ultrastructural and time-lapse studies of primary mesenchyme cell behavior in normal and sulfate-deprived sea urchin embryos. Exp Cell Res 136, 233–245. [DOI] [PubMed] [Google Scholar]
- Khor JM, Guerrero-Santoro J, Ettensohn CA, 2019. Genome-wide identification of binding sites and gene targets of Alx1, a pivotal regulator of echinoderm skeletogenesis. Development. [DOI] [PubMed] [Google Scholar]
- Khorshid M, Hausser J, Zavolan M, van Nimwegen E, 2013. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods 10, 253–255. [DOI] [PubMed] [Google Scholar]
- Kim J, Siverly AN, Chen D, Wang M, Yuan Y, Wang Y, Lee H, Zhang J, Muller WJ, Liang H, Gan B, Yang X, Sun Y, You MJ, Ma L, 2016. Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways. Cancer Res 76, 6424–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp RT, Wu CH, Mobilia KC, Joester D, 2012. Recombinant sea urchin vascular endothelial growth factor directs single-crystal growth and branching in vitro. J Am Chem Soc 134, 17908–17911. [DOI] [PubMed] [Google Scholar]
- Lapraz F, Besnardeau L, Lepage T, 2009. Patterning of the dorsal-ventral axis in echinoderms: insights into the evolution of the BMP-chordin signaling network. PLoS Biol 7, e1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layous M, 2020. The tolerance to hypoxia is defined by a time-sensitive response of the gene regulatory network in sea urchin embryos, in: Khalaily L (Ed.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaf DS, Anstrom JA, Chin JE, Harkey MA, Showman RM, Raff RA, 1987. Antibodies to a fusion protein identify a cDNA clone encoding msp130, a primary mesenchyme-specific cell surface protein of the sea urchin embryo. Dev Biol 121, 29–40. [DOI] [PubMed] [Google Scholar]
- Lee YH, Britten RJ, Davidson EH, 1999. SM37, a skeletogenic gene of the sea urchin embryo linked to the SM50 gene. Dev Growth Differ 41, 303–312. [DOI] [PubMed] [Google Scholar]
- Li E, Materna SC, Davidson EH, 2012. Direct and indirect control of oral ectoderm regulatory gene expression by Nodal signaling in the sea urchin embryo. Dev Biol 369, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston BT, Killian CE, Wilt F, Cameron A, Landrum MJ, Ermolaeva O, Sapojnikov V, Maglott DR, Buchanan AM, Ettensohn CA, 2006. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev Biol 300, 335–348. [DOI] [PubMed] [Google Scholar]
- Lyons DC, Kaltenbach SL, McClay DR, 2012. Morphogenesis in sea urchin embryos: linking cellular events to gene regulatory network states. Wiley Interdiscip Rev Dev Biol 1, 231–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M, 2008. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Proteome Sci 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Bartolomé M, Range RC, 2019. A biphasic role of non-canonical Wnt16 signaling during early anterior-posterior patterning and morphogenesis of the sea urchin embryo. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay DR, Armstrong NA, Hardin J, 1992. Pattern formation during gastrulation in the sea urchin embryo. Dev Suppl, 33–41. [PubMed] [Google Scholar]
- McClay DR, Cannon GW, Wessel GM, Fink RD, Marchase RB, 1983. Patterns of Antigenic Expression in Early Sea Urchin Development., in: Raff W.R.J.a.R.A. (Ed.), Time, Space, and Pattern in Embryonic Development. A.R. Liss, New York, pp. 157–169. [Google Scholar]
- McIntyre DC, Lyons DC, Martik M, McClay DR, 2014. Branching out: origins of the sea urchin larval skeleton in development and evolution. Genesis 52, 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre DC, Seay NW, Croce JC, McClay DR, 2013. Short-range Wnt5 signaling initiates specification of sea urchin posterior ectoderm. Development 140, 4881–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R, 2006. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol 4, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis M, Gildor T, Roopin M, Sher N, Malik A, Lalzar M, Dines M, Ben-Tabou de-Leon S, Khalaily L, 2019. Possible cooption of a VEGF-driven tubulogenesis program for biomineralization in echinoderms. Proc Natl Acad Sci U S A 116, 12353–12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Burke RD, 1996. The initial phase of gastrulation in sea urchins is accompanied by the formation of bottle cells. Dev Biol 179, 436–446. [DOI] [PubMed] [Google Scholar]
- Nishita M, Enomoto M, Yamagata K, Minami Y, 2010a. Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol 20, 346–354. [DOI] [PubMed] [Google Scholar]
- Nishita M, Itsukushima S, Nomachi A, Endo M, Wang Z, Inaba D, Qiao S, Takada S, Kikuchi A, Minami Y, 2010b. Ror2/Frizzled complex mediates Wnt5a-induced AP-1 activation by regulating Dishevelled polymerization. Mol Cell Biol 30, 3610–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, McClay DR, 2003. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol 258, 32–43. [DOI] [PubMed] [Google Scholar]
- Pedrioli DM, Karpanen T, Dabouras V, Jurisic G, van de Hoek G, Shin JW, Marino D, Kälin RE, Leidel S, Cinelli P, Schulte-Merker S, Brändli AW, Detmar M, 2010. miR-31 functions as a negative regulator of lymphatic vascular lineage-specific differentiation in vitro and vascular development in vivo. Mol Cell Biol 30, 3620–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled-Kamar M, Hamilton P, Wilt FH, 2002. Spicule matrix protein LSM34 is essential for biomineralization of the sea urchin spicule. Exp Cell Res 272, 56–61. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH, 2010. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol 340, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH, 2011. A gene regulatory network controlling the embryonic specification of endoderm. Nature 474, 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiq K, Cheers MS, Ettensohn CA, 2012. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development 139, 579–590. [DOI] [PubMed] [Google Scholar]
- Rafiq K, Shashikant T, McManus CJ, Ettensohn CA, 2014. Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins. Development 141, 950–961. [DOI] [PubMed] [Google Scholar]
- Range RC, 2018. Canonical and non-canonical Wnt signaling pathways define the expression domains of Frizzled 5/8 and Frizzled 1/2/7 along the early anterior-posterior axis in sea urchin embryos. Dev Biol 444, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Range RC, Angerer RC, Angerer LM, 2013. Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol 11, e1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH, 2002. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol 246, 132–147. [DOI] [PubMed] [Google Scholar]
- Remsburg C, Konrad K, Sampilo NF, Song JL, 2019. Analysis of microRNA functions. Methods Cell Biol 151, 323–334. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, Davidson EH, 2007. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci U S A 104, 12383–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI, 2006. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus). Dev Biol 300, 35–48. [DOI] [PubMed] [Google Scholar]
- Rottinger E, Saudemont A, Duboc V, Besnardeau L, McClay D, Lepage T, 2008. FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development 135, 353–365. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Sampilo NF, Stepicheva NA, Zaidi SAM, Wang L, Wu W, Wikramanayake A, Song JL, 2018. Inhibition of microRNA suppression of. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, 2000. Cell signaling by receptor tyrosine kinases. Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- Sharma T, Ettensohn CA, 2011. Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development 138, 2581–2590. [DOI] [PubMed] [Google Scholar]
- Staton AA, Giraldez AJ, 2011. Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nat Protoc 6, 2035–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepicheva N, Nigam PA, Siddam AD, Peng CF, Song JL, 2015. microRNAs regulate beta-catenin of the Wnt signaling pathway in early sea urchin development. Dev Biol 402, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepicheva NA, Song JL, 2014. High throughput microinjections of sea urchin zygotes. J Vis Exp, e50841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepicheva NA, Song JL, 2015. microRNA-31 modulates skeletal patterning in the sea urchin embryo. Development 142, 3769–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Ettensohn CA, 2014. Signal-dependent regulation of the sea urchin skeletogenic gene regulatory network. Gene Expr Patterns 16, 93–103. [DOI] [PubMed] [Google Scholar]
- Söllner C, Schwarz H, Geisler R, Nicolson T, 2004. Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev Genes Evol 214, 582–590. [DOI] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA, 2010. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle 9, 2124–2129. [DOI] [PubMed] [Google Scholar]
- Veis A, 2011. Organic matrix-related mineralization of sea urchin spicules, spines, test and teeth. Front Biosci (Landmark Ed) 16, 2540–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Range R, Angerer R, Angerer L, 2012. Axial patterning interactions in the sea urchin embryo: suppression of nodal by Wnt1 signaling. Development 139, 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Ferkowicz M, McClay DR, 2007. Ingression of primary mesenchyme cells of the sea urchin embryo: a precisely timed epithelial mesenchymal transition. Birth Defects Res C Embryo Today 81, 241–252. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, 2008. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell 14, 97–107. [DOI] [PubMed] [Google Scholar]
- Yu T, Ma P, Wu D, Shu Y, Gao W, 2018. Functions and mechanisms of microRNA-31 in human cancers. Biomed Pharmacother 108, 1162–1169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Eve overexpression results in expanded expression domain of Vegf3. Overexpression of Eve (3 μg total mRNA in 2.5μl of injection stock solution) resulted in expansion of Vegf3 expression domain compared to control mRNA. This is similar to miR-31 KD and Eve miR-31 TP. NS=not significant, **p<0.001 using Student’s t-test. All error bars represent SEM. 3 biological replicates. Scale bar = 50μm. N is the total number of embryos examined.
Figure S2 Lower concentration of miR-31 KD results in expanded expression domain of Eve. miR-31 inhibitor injected at 20μM resulted in expansion and anterior shift of Eve expression domain. At a lower concentration (20μM), Eve may not sufficiently increase to autorepress itself. *p<0.05, ***p<0.0001 using Student’s t-test. All error bars represent SEM. 2 biological replicates. Scale bar = 50μm. N is the total number of embryos examined.


