Abstract
Correct reconstruction of macromolecular structure by cryo-electron microscopy (cryo-EM) relies on accurate determination of the orientation of single-particle images. For small (<100 kDa) DNA-binding proteins, obtaining particle images with sufficiently asymmetric features to correctly guide alignment is challenging. We apply DNA origami to construct molecular goniometers—instruments that precisely orient objects—and use them to dock a DNA-binding protein on a double-helix stage that has user-programmable tilt and rotation angles. We construct goniometers with fourteen different stage configurations to orient and visualize the protein just above the cryo-EM grid surface. Each goniometer has a distinct barcode pattern that we use during particle classification to assign angle priors to the bound protein. We use goniometers to obtain a 6.5 Å structure of BurrH, an 82-kDa DNA-binding protein whose helical pseudosymmetry prevents accurate image orientation using traditional cryo-EM. Our approach should be adaptable to other DNA-binding proteins as well as small proteins fused to DNA-binding domains.
Editor’s summary:
DNA origami orients single proteins on a surface to aid structure determination by cryo-electron microscopy.
Single-particle cryo-electron microscopy (cryo-EM) is a powerful technique for determining high-resolution structures of biological molecules1,2. A typical cryo-EM study involves adsorbing a specimen onto a grid, freezing it in a thin layer of vitreous ice, collecting cryo-EM images of individual particles, computationally sorting and aligning the particle images, and reconstructing a 3D structure from 2D particle images. In the final step, an algorithm refines orientations of the 2D particles and determines the 3D reconstruction via iterative expectation-maximization optimization. Refinements of the 3D reconstruction and 2D particle orientations typically converge to a correct solution when image data are sufficient in quantity, resolution, distribution of orientations, and conformational homogeneity. Even when these conditions are met, however, if particles lack obvious asymmetric features to guide their alignment—as is often the case for targets <100 kDa—the optimization is prone to getting trapped in a local minimum and thus yields a low-resolution electron density map. The alignment challenge has been addressed by docking the targets to protein scaffolds—such as antibody fragments3, bacterial ribosomes4, or engineered nanoparticles5—via a rigid attachment, and the structure of the full complex is determined together. However, these approaches fail when a rigid attachment is impractical.
In the 1980s, Ned Seeman envisioned programming DNA via self-assembly into custom nanostructures as a way to engineer positional control of proteins for crystallographic studies6. DNA ‘origami’ provides a means of building megadalton-sized nanostructures using a long single-strand DNA (ssDNA) scaffold to template the assembly of numerous short DNA oligonucleotide ‘staples’ into a custom shape7. Here we address the alignment challenge in cryo-EM by constructing nanoscale goniometers with DNA origami. Our study builds on previous work8, and includes several optimizations that help improve the resolution of 3D reconstructions.
We describe the design, characterization, and validation of molecular goniometers. Since the different orientations of small protein images are often too subtle to determine directly, we mapped unique angle configurations to large asymmetric features, or barcodes, on the DNA origami. We validated our approach by obtaining a 6.5 Å structure of BurrH9,10, an 82 kDa TALE-family DNA-binding protein whose crystal structure is known (PDB id: 4cja). We found that 3D structure maps built without the a priori information provided by the DNA origami (i.e., the tilt and rotation angle “priors”) did not fit as well to the known structure as the final map we constructed using the priors. We obtained structure maps using the same particle data, but without using the priors, at resolutions ranging from 7.4 Å to 10.8 Å (see Table 1).
Table 1 |. Resolution estimates of the BurrH 3D maps generated with and without angle priors.
FSC is calculated between the maps and BurrH crystal structure (PDB id: 4cj9) fitted to the maps. FSC 0.143 and FSC 0.5 criteria are used for the resolution estimates.
| FSC 0.143 (Å) | FSC 0.5 (Å) | |
|---|---|---|
| Relion with origami priors | 6.5 | 7.9 |
| Relion without priors | 7.4 | 9.1 |
| cisTEM without priors | 8.5 | 13.3 |
| cryoSPARC without priors | 10.8 | 27.4 |
Results
Our molecular goniometers enable user-controlled tilt and rotation of single proteins, coupled with large asymmetric features (barcode “bits”) to provide unambiguous identification of each protein orientation. We demonstrate the feasibility of determining the structure of a small protein attached to a host nanostructure by cropping out (or masking) the host particle and using only the subimages containing each protein together with its orientation information obtained from the host.
We show that a perfectly rigid link between the protein and the host nanostructure is not required, and may not be desired, for this style of structure determination. In fact, we found it useful to isolate and separately tune spatial and angular rigidity. Spatial rigidity is desirable when positioning the protein for well-isolated, well-centered extraction of low-background subimages, but care must be taken with the design as increasing separation between the protein and host can compromise rigidity, and vice versa. Angular rigidity between the protein and the goniometer is also needed, otherwise the angle priors do not have a meaningful relationship with the true orientation of the protein. On the other hand, excess angular rigidity may limit the particle-angle distribution and compromise reconstruction resolution. Therefore, our design allows for limited swiveling of the protein rotation angle, centered around a target angle. Finally, the goniometers must be designed to self-assemble with a sufficient yield to maintain precision and accuracy while satisfying these constraints.
Figure 1 summarizes the molecular goniometer design, which consists of a fixed chassis (gray), a programmable DNA stage (yellow) containing a protein-binding site (magenta), and barcode domains (teal) that uniquely identify each stage-angle configuration. The chassis resembles a tiny C-clamp that grasps the DNA stage, which is a 56-base-pair (bp) double helix anchored on both sides of the chassis aperture. The DNA stage orientation is set by the staple sequences in the anchoring regions of the chassis (dark gray) and by the 5’-to-3’ direction of the scaffold sequence within the origami.
Figure 1 |. Design and grid adsorption of molecular goniometers built from DNA origami.
a, A representative configuration is shown (+90° tilt, 0° rotation). DNA origami chassis (grey) grasps and controls the tilt and rotation of the 56-bp DNA stage (yellow) containing the 19-bp binding site for the BurrH protein (magenta). The TILT barcode bit (salmon) and ROTATION barcode domains (teal) identify each unique DNA stage configuration. b, 2D schematic of origami scaffold and staples. c, Chassis aspect ratio can promote the preferred orientation of goniometer adsorption onto the grid surface. d, The DNA stage is positioned with a 150 Å gap from the origami chassis. e, DNA stage tilt angle is set by the polarity of the origami scaffold route and the chassis attachment locations. f, Rotation angle is controlled by shifting the register of the DNA stage relative to the chassis. A 1-nt linear shift of the stage corresponds to a 34° angular rotation of the protein binding site. g, The DNA stage is flanked by unpaired 2-nt regions to provide rotational flexibility. An SD of 15° is used for the Gaussian rotation priors. h, A mixture containing BurrH and goniometers (at a 10:1 molar ratio) is deposited on a gold quantifoil grid with an amino graphene oxide support. A cropped region of 1 of 15,658 micrographs is shown. Circles indicate typical bound BurrH (yellow circle) and unbound BurrH (white circle). Scale bar: 100 nm.
The DNA stage (yellow) includes the 19-bp binding sequence of the BurrH DNA-binding protein (magenta). The outer edges of the goniometer chassis can be configured to display modular barcode domains, or “bits”, which form unique asymmetric patterns that enable particle classification. Our barcode design, which we expand upon below, includes seven bits (1–7) that specify the rotation angle of the DNA stage, and one bit (labeled ‘TILT’) that specifies the tilt angle of the DNA stage. The barcode pattern is set by including the corresponding ssDNA staples in the origami folding reaction. In Fig. 1a, the 4th bit of the rotation barcode is enabled, corresponding to a stage rotation angle of 0°. The remaining disabled rotation bits (1–3, 5–7) are shown as teal outlines. The location of the disabled tilt bit is outlined in salmon red.
The molecular goniometer is composed of a 9344 nucleotide (nt) scaffold and 233 staples (strand diagram, Fig. 1b). Colors of boxed scaffold regions and staples indicate the corresponding features from Fig. 1a. The DNA stage is composed of a cloned scaffold segment and a staple strand (Figure 2). The scaffold arrangement in the dark gray zone is modified, along with corresponding barcode regions, to change the angle settings.
Figure 2 |. Scaffold-integrated BurrH binding site and its relation to folded goniometers.
a, p9344-BurrH plasmid map. The custom scaffold was created by cloning an insert containing a BurrH binding region (yellow, magenta), an ampicillin resistance gene (green), and a plasmid origin of replication (blue) into M13mp18-RF. b, Representative goniometer scaffold path schematic (left) and cartoon 3D render (right), both annotated with the location of the cloned insert (orange). c, Sequence detail and 3D render of the DNA stage, which consists of the scaffold-integrated BurrH binding region (bottom) and a single complementary 56-nt staple strand (top). d, Sequence detail and 3D render of the BurrH binding site (magenta) and binding residues (blue), based on PDB id 4cja.
To ensure that the molecular goniometer chassis adsorbs onto the grid in a desired orientation relative to the electron beam direction, we designed the chassis cross-section to have a narrow aspect ratio (Fig. 1c). We found that a cross-sectional aspect ratio of w/h < 0.6 helped approximately 90% of the goniometers adsorb to the grid surface in a preferred orientation (face-up or face-down), positioning the DNA stage without obscuring the protein (Extended Data Figure 1a–b).
To minimize protein image artifacts due to delocalized signal from the origami, we positioned the DNA stage to ensure a large gap between the protein and chassis. The aperture is 230 Å wide along the helical axis of DNA stage, which is centered 170 Å from the inner chassis wall (Fig. 1d). Smaller than that, delocalized signal originating from the origami will overlap with the protein particle image, interfering with downstream refinement steps (Extended Data Figure 2c–d).
The tilt angle of the DNA stage is controlled by the anchor locations on the chassis, and by the polarity of the scaffold strand (Fig. 1e). Rerouting the scaffold onto the complementary strand within the origami design, in the opposite 5’-to-3’ direction relative to the chassis, flips the stage tilt angle by from +90° to –90°. Therefore, goniometer designs with the +90 and –90° tilt angles each require a separate set of staple strands.
The rotation angle of the protein-binding site is controlled by the register of the stage sequence relative to the goniometer chassis (Fig. 1f). Assuming 10.5 bp per helical turn, a 1-nt shift corresponds to a 3.4 Å translation along the stage axis, and a 34.3° rotation. To modify the stage rotation angle, we can modify the scaffold route in the surrounding anchoring region (dark gray, Figs. 1a & 1b), with corresponding updates to the complementary staple sequences (Fig. 1b, dark gray). We designed goniometers with seven possible rotation angles, and assigned the rotation angle 0° to the middle angle. Thus, the possible rotation angles, rounded to the nearest integer, are +103°, +69°, +34°, 0°, –34°, –69°, and –103°. Figure S3 explains how we define the goniometer tilt and rotation angles.
Each tilt and rotation angle configuration is paired with a unique barcode pattern that is similarly implemented via local modifications to the scaffold route and corresponding staple sequences. The 1-nt resolution of DNA stage registration corresponds to a rotation angle step size of ~34° which, despite some intrinsic torsional flexibility, may not provide sufficient angle coverage for cryo-EM structure determination. To ensure sufficient variance in the distribution of rotation angles, the DNA stage region is flanked on both sides by 2-nt ssDNA regions (blue), as shown in Fig. 1g.
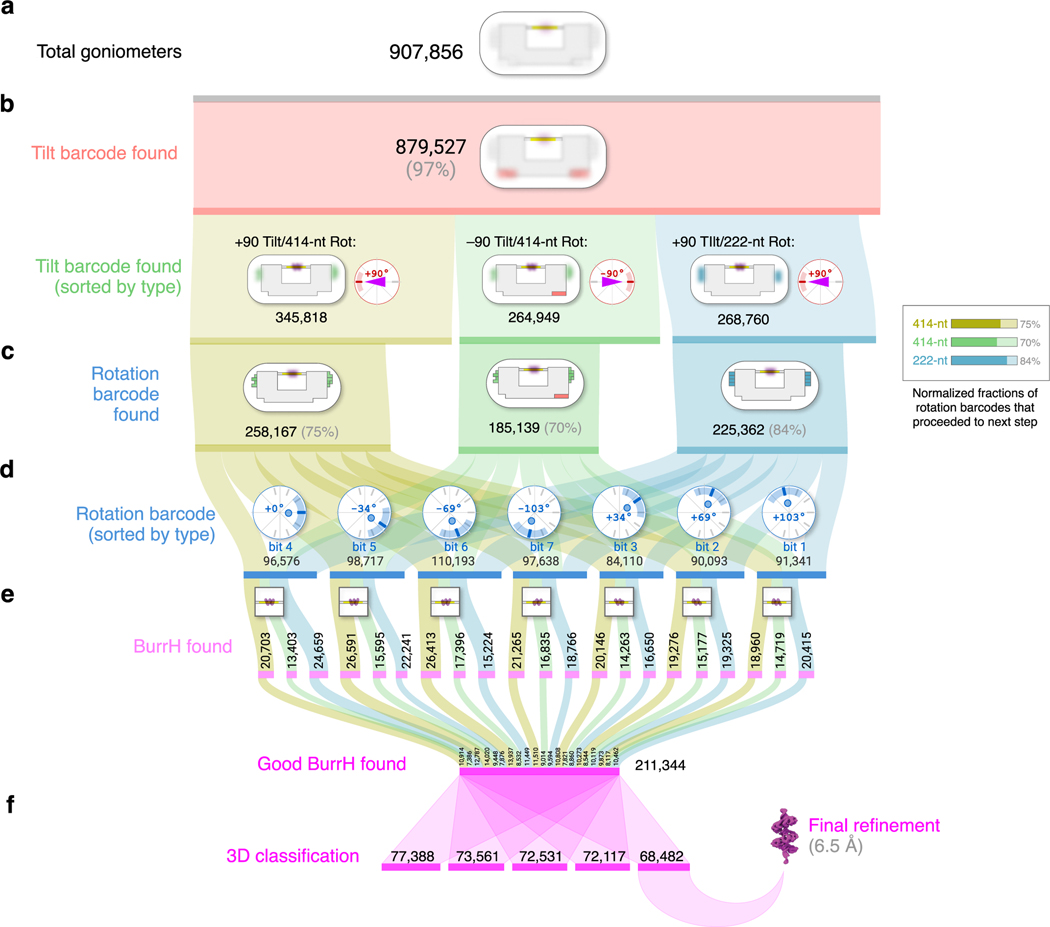
A pooled mixture with equal fractions of all goniometer designs are adsorbed onto holey gold grids with an amino-graphene oxide support11; Fig. 1h shows a representative micrograph region. We collected a total of 17,725 images. We were able to identify 908k goniometer particles. Of those, 669k (74%) had identifiable tilt and rotation barcodes, 398k (44%) had BurrH present. We selected 211k BurrH particles with protein present and low background signal. Thus, 23% of all goniometers had good BurrH particles present, and representing an average of 12 good BurrH particles per image.
Figure 3 describes how we classified the goniometers by their barcodes and then assigned tilt- and rotation-angle priors to each masked protein particle, which are used as inputs for building the initial 3D reconstruction. After cryo-EM micrograph data collection, particles are processed in a multi-step computational pipeline (Fig. 3a). Individual goniometer particles are picked and aligned into classes, which are then sorted into +90° and –90° tilt-angle classes according to the tilt-angle barcode domain (Fig. 3b).
Figure 3 |. Determination of protein classes with angle priors from goniometer image data.
a, Goniometer data processing pipeline. Top right: representative 2D class, prior to sorting by tilt barcode. b, Example +90° and –90° tilt classes, with barcode detail below. Only the –90° tilt class has the 582-nt barcode bit enabled. Magenta arrowhead indicates N-to-C orientation of BurrH. c, The 222-nt rotation barcode bit consists of 108-nt scaffold paired with 3 staples of 114-nt combined length. d, An alternate 414-nt bit design (186 nt scaffold + 228 nt combined staple length) was also used. e, 2D consensus classes derived from goniometers with stages configured at +90° and –90° tilt angles, and 7 rotation angles (0°, ±34°, ±69°, and ±103°). See Figure 5 for statistics and Extended Data Figure 4 for all 20 classes of each goniometer. Scale bars: (a–d) 20 nm, e, 10 nm.
The differences between each DNA stage configuration would be difficult to classify directly, so we rely on decorating the goniometers with machine-readable geometric patterns, or barcodes, that identify the DNA stage tilt and rotation configuration. Each barcode pattern consists of two bits that can be toggled on (or off) by routing (or not routing) the scaffold and staples to fold into small domains at the edges of the chassis. Figure S5 describes the process of barcode classification, in which image masks are used to perform focused classification of individual bits. Images are considered successfully classified when automatically assigned to a 2D class in which the barcode region matches one of the designed bit configurations. Images that give rise to classes that do not resemble one of the expected barcode configurations were discarded.
We used a single 582-nt bit (Fig. 3b) to identify frames with a –90° tilt angle. The tilt subclasses were further divided into groups by rotation-angle barcodes. We tested two rotation-angle barcode designs: a 222-nt bit (Fig. 3c), and a 414-nt bit (Fig. 3d). The 414-nt bit was used with both +90 and –90 tilt designs, while the 222-nt bit was used only for +90 tilt (Extended Data Figure 1), and data from all goniometer variants were combined for the final structure. Of the two barcode designs that we tried, the smaller rotation-angle barcode bit (Fig. 3c) seems to work best for classifying particles, perhaps because it projects farther from the edge of the origami: 84% of images containing 222-nt bits were successfully assigned into classes with an expected barcode configuration, versus 70–75% of images containing 414-nt bits.
After the goniometers are classified according to tilt and rotation barcodes, a mask is used to isolate the DNA stage and protein from the chassis. We generated 2D consensus classes for each angle configuration (Fig. 3e), in which different orientations of BurrH are clearly visible. Altogether, we tested 21 different goniometers (Extended Data Figure 3). Seven goniometers used a +90° tilt angle (with the tilt bit disabled), and each respective rotation angle (–103°, –69°, –34°, 0°, 34°, 69°, 103°) identified with a 222-nt bit. Seven goniometers also used a +90° tilt angle and same rotation angles, but with 414-nt rotation bit. Finally, seven goniometers used a –90° tilt angle (tilt bit enabled), and seven rotation angles identified with a 414-nt bit. All goniometers were pooled for the final 3D reconstruction (Figure 5).
Figure 5 |. Sankey diagram for particle selection, classification, prior assignment, and 3D reconstruction.
a, “Total goniometers” is the count of particles successfully identified as a DNA origami goniometer. b, “Tilt barcode found” means tilt barcode (Fig. 2b) was successfully classified. c, “Rotation barcode found” means both tilt barcode and rotation barcode (Figs. 3b–d) were successfully classified. d, Subclassification by rotation barcode for angle prior assignment. e, “BurrH found” describes goniometers from c, that contained a BurrH particle, and “Good” means the 2D BurrH subimage had a low background signal. f, Good BurrH particles were used as input for 3D classification, and the highest-resolution class was used for final refinement.
We used Gaussian priors to constrain the assignment of particle angle and position parameters within a window centered on a mean value. Prior to 3D reconstruction, we flipped the BurrH particle images derived from goniometers with –90° tilt so that all BurrH particles have the same +90° tilt angle. To constrain tilt angles for all particles, we used a Gaussian prior centered at +90° with a window size of ±30°. For the rotation angles, we used Gaussian priors centered at the rotation goal angles specific to each goniometer class, with a window size of ±45°. Figure S6 shows a sankey diagram of our pipeline, and Figure S7 includes twenty 2D class averages for each particle.
Figure 6 shows the final 3D reconstruction of BurrH using the a priori orientational information derived from our molecular goniometers (for detail, see Extended Data Figure 5), along with an assessment of the tilt and rotation angle accuracy, and a side-by-side comparison with 3D reconstructions built without and with the angle priors.
Figure 6 |. 3D refinement of BurrH structure using molecular goniometer-derived angle priors.
a, BurrH 2D class averages; 3 of 50 are shown, see Extended Data Figure 6. b, 3D reconstruction of BurrH using a priori tilt and rotation angles from molecular goniometers and FSC curve calculated from two half-maps of the final 3D reconstruction. FSC0.143 line crosses the curve at 6.5 Å resolution. c, BurrH consensus 2D class averages and 2D projections of the 3D reconstruction. A single 2D consensus was generated for each angle projection. Projections are low-pass filtered to a resolution of 30 Å. d, Euler angle distribution derived from 3D refinement, with blue-red color scale. e, Representative polar plots for stage tilt angles for +90° and –90° designs. The inner red tick mark represents the goal tilt angle, and the ±45° arc shows the bounds of the Gaussian tilt angle prior. The tilt angle distributions reported by Relion are plotted as histograms. Average tilt angles (magenta line) were +83° and –96°, respectively. f, Representative polar angle plots for seven stage rotation angles reported by Relion, derived from goniometers with +90° tilt and 414-nt barcode bits. The inner blue tick mark represents the goal rotation angle (integer value in blue text), and the ±45° arc shows the bounds of the Gaussian rotation angle prior. Average rotation angles are plotted as magenta lines (integer values in pink text). g, Comparison of 3D reconstructions obtained without angle priors (blue: cryoSPARC, red: cisTEM, yellow: Relion), and with priors (magenta: Relion with goniometer-derived angle priors), with BurrH crystal structure (cyan) fit to each density map. Areas where cyan is visible indicate a poor fit. Tilt-direction accuracy is compared to the true orientations derived from gonimeters. h, FSC curves showing goodness-of-fit of BurrH crystal structure into 3D cryo-EM density maps of BurrH. FSC curve is computed following a rigid body fit of BurrH crystal structure into each cryo-EM density map. FSC0.5 shown as a dashed line. Scale bars in a, c: 10 nm.
To build our 3D reconstruction of BurrH aided by goniometer-derived angle priors, we used the software Relion12 and custom Python code, with the following approach: With both tilt- and rotation-angle constraints enabled, we first generated an initial electron density map that became the starting structure for the 3D reconstruction. We then aligned all particles to the initial map with both tilt and rotation angle constraints enabled. Finally, during the subsequent 3D classification of particles and the refinement of the best 3D class, we removed the rotation-angle constraint. We kept the tilt-angle constraint enabled at every step of 3D reconstruction because we were highly confident about the direction of protein particles bound to the stage DNA.
Figure 6a shows representative good 2D class averages (for additional classes, Extended Data Figure 6). The final 3D reconstruction of BurrH has all the repeats and the secondary structures resolved with an estimated resolution of 6.5 Å (Fig. 6b). The 2D consensus class averages and the corresponding 2D projections of the 3D map showed qualitative agreement (Fig. 6c). Consensus class averages were obtained by aligning cropped regions of goniometer class averages for each tilt-rotation angle pair, as in Fig. 2e, and weighting the intensity in proportion to the number of particles in each class. The Euler angle values reported by Relion in the final refinement are distributed in a ring around the “equator” of the angle map, which is expected given the tilt and rotation angles prescribed by the goniometers (Fig. 6d). The mean rotation-angle values reported by Relion were within ±45° of the goal angles for all goniometer designs (Fig. 6e,f). See Extended Data Figure 7 for the complete angle distributions of all designs.
To evaluate the utility of our overall approach, we performed three additional 3D reconstructions of BurrH without the use of a priori orientation information (i.e., the angle priors) provided by the molecular goniometers. We used cryoSPARC13, cisTEM14, Relion12 for these reconstructions, and then compared the results to our own approach, which used Relion along with the origami-derived priors.
For 3D reconstructions without angle priors, we used the same initial map and the particles from the final refinement step of the 3D reconstruction with the angle priors. In Fig. 6g, the 3D maps were each fit to the PDB structure, and the tilt-direction accuracy for each map was based on a comparison with the true angles derived from the origami particles. In all reconstructions that did not use angle priors, some fraction of particles ranging from 20–53% are misaligned with respect to the stage DNA axis (Fig. 6g). The BurrH crystal structure (Fig. 6g, cyan) does not fit well in the 3D volumes obtained from the reconstructions without angle priors and the correlation between the BurrH crystal structure and the 3D reconstruction volumes gets worse with the fraction of misaligned particles (Fig. 6h).
Discussion
The success of our approach depends on multiple key features of the goniometers, many of which we can evaluate in the context of a closely related work published in 2016 that described a DNA origami molecular support for protecting proteins from air-water interface exposure during cryo-EM sample preparation8. The device was used to obtain a 15 Å reconstruction of the transcription factor p53—a 160 kDa tetramer. We adopted several design and analysis concepts from this study, including sub-image masking to isolate the protein prior to reconstruction and the general aim of controlled protein orientation. The authors also identified many challenges that limited the initial success of the approach, including a low yield of intact particles, signal delocalization due to imaging defocus, and difficulty in controlling the protein orientation. In our design, we sought to maximize the yield of intact protein-bearing particles per micrograph, especially since the goniometer particles are massive (>6 MDa) and occupy a large footprint on the grid (>2000 nm2). We attempted using ssDNA staples to create the DNA stage, but the fraction of goniometers with bound protein was typically below 10%, similar to the previous study8. Cloning one strand of the DNA stage into the origami scaffold increased the occupancy to 60%, presumably due to enrichment of intact protein-binding substrates (Fig. 2). The unoccupied stages may be due to a missing staple oligo, or some strain in the BurrH binding region despite the 2-nt ssDNA gaps, or dissociation during sample prep. Adopting a chassis with a narrow aspect ratio and deposition onto amino graphene oxide grids boosted the fraction of particles in desirable orientations by 30% relative to a chassis design with a wider aspect ratio and deposition onto grids with a carbon film support (Extended Data Figure 1). To mitigate superposition of the signal from the support structure and the target protein, and to facilitate image cropping and analysis, we designed our chassis to position the DNA stage far from the chassis (Fig. 1a, d).
Contrary to what might be expected, our designs intentionally did not maximize the rigidity of the DNA stage with respect to the chassis. We wanted to obtain uniform sampling of rotation angles to avoid compromising the resolution of the 3D reconstruction15. Therefore, we included 2-nt gaps flanking the DNA stage to provide rotational flexibility and angle variance. If future studies require greater rotational angle precision, our designs could be modified to reduce one or both of the 2-nt gaps to 1-nt gaps or 0-nt nicks, or enzymatically ligated to further increase rigidity. Additional studies could establish design rules for finely tuning the accuracy and precision of rotation angles.
We tested goniometers designed with +90° and –90° tilt angles, but were unable to resolve any difference between the designs with the amount of data we collected. While we only tested two tilt angles, each with seven rotation angles, the goniometer allows for additional angles. The chassis accommodates tilt angles from 65° to 115° (or –65° to –115°), with a 7–10° step-size, based on rerouting the scaffold through alternate DNA stage anchor locations. The DNA stage can be set to 21 distinct rotation values, representing a 17° step size. Further testing will be needed to determine if and when different angle configurations are useful, for example for proteins that exhibit symmetry along a different axes compared to BurrH.
Our data analysis pipeline required careful bookkeeping of the relative and absolute orientations of the goniometers and the protein sub-particles after masking. To simplify the process, we used the goniometer chassis as a reference frame, and “flipped” the orientation of certain 2D images at two stages of our pipeline. Helpfully, the DNA stage serves as a built-in chirality standard, since DNA is known to be right-handed. First, the goniometers can adsorb onto the grid surface in face-up or face-down orientations. We therefore flipped the face-down goniometer images so all images matched the face-up orientation, reducing the number of goniometer image classes by 50% and increasing the image count in each remaining class. Second, after cropping out the DNA origami and prior to 3D reconstruction, we flipped the protein particle images derived from the –90° tilt goniometers to make the N-to-C polarity consistent for all protein particles, reducing 14 types of orientation classes (2 tilts x 7 rotations) to 7 (1 tilt x 7 rotations). Our software tracks the provenance of each image, allowing for the visualization of Relion-generated angle distributions for images derived from +90°-tilt and –90°-tilt goniometers (Fig. 6d).
An important consideration for our method is the relationship between the goal angles designed into each goniometer, the true orientation angles of each physical particle, and the estimated angles of each particle image reported by the refinement software. Discrepancies between the initial goal and final estimated angles may be caused by goniometer stage-angle inaccuracy, by software angle-estimation inaccuracy, or both. The tilt and rotation angle distributions plotted as polar histograms Figure 6 are estimated angles from the refinement. While most of the distributions have a clear peak at the goal angle, all rotation-angle distributions include angles that are a full 180° away from the goal angle. The distribution derived from the 0°-rotation goniometer lacks a clear peak at the goal angle, perhaps because the 2D projections of BurrH near that angle are highly symmetric and prone to angular drift during refinement, while asymmetric projections are more readily aligned. Follow-up studies should elucidate the underlying causes of these discrepancies and how to correct for them. In principle, the 0°-rotation goniometer variants may have a systematic error in the design or sample preparation, and should be further validated using a protein more asymmetric than BurrH. However, an angle-specific error would be surprising, given that all variants use identical DNA stage construction and source DNA (i.e. the p9344-BurrH scaffold and a single 56-nt complementary staple), and differ only in register by 1-bp increments, as described in Figure 1f. On the other hand, it is not straightforward to determine when the software-estimated angle differs from the true angle. If that were the case, any 3D refinement could be improved simply by discarding all misaligned particle images. We compared goniometer-drived 2D consensus classes of BurrH and 2D projections generated from the 3D reconstruction at the corresponding tilt and rotation angles in Figure 6c. Agreement between the 2D classes and projections indicates that the goniometers indeed control the BurrH orientation at the prescribed angles, including at the 0° rotation.
We compared how three popular cryo-EM software tools would handle determination of the 3D electron density map and orientations of the 2D particles. We offer this comparison as a practical guide for cases where angle priors are necessary (i.e., when particles lack asymmetric features to guide alignment). The tools cryoSPARC and cisTEM do not accept user-specified Gaussian angle priors as input, so we were unable to test the quality of map they might build with that information. Moreover, the comparison may not reflect how the tools would perform given particles that had not already been picked, aligned, and masked from the DNA origami goniometers, which effectively serve as fiducial markers in the first step of image analysis. However, starting with the centered and masked BurrH particles allows for a well-controlled comparison of how the tools handle angle assignment specifically. We assumed the goniometers provide 100% tilt-direction accuracy as a reference, since the DNA origami is highly asymmetric, unambiguous in its orientation, and the protein can only bind to its substrate in a known direction. Although Relion and cisTEM both utilize a maximum-likelihood algorithm, Relion achieves greater tilt-direction accuracy (80%) without priors compared to cisTEM (55%). CryoSPARC uses a conjugate-gradient optimization which was the fastest, but also the least accurate (47%). Relion performed better than random in determining the correct tilt angles without any priors, perhaps due to performing a more extensive search of the optimization space. However, Relion was incorrect for enough particles (20%) that it could not match the resolution compared to using the priors.
Our final reconstruction of BurrH achieved a resolution of 6.5 Å using 68,482 particles. Directional Fourier shell correlation (dFSC) analysis indicates a sphericity of 0.973 for the dFSC map (Extended Data Figure 8). We collected micrographs at 22,000× magnification, corresponding to a pixel size of 1.82 Å, and maximum resolution of 3.64 Å. We used a defocus range of 0.5–1.5 μm. We also attempted using a lower defocus (0.3–0.5 μm), but experienced difficulty applying automated motion correction to those images. Including particles with higher defocus values does not appear to compromise resolution (Extended Data Figure 9). Removing particles whose estimated rotation angles deviated from the goniometer goal angle did not improve resolution (Extended Data Figure 10).
BurrH was expected to be a challenging target due to its small size and helical pseudosymmetry, but additional factors may further complicate its structure determination with cryo-EM. If flexibility of the DNA stage-BurrH complex resulted in some conformational heterogeneity of our dataset, then additional images and classification may drive the resolution higher. It is also possible that the current generation of computational tools has difficulty distinguishing dynamic variability in small structures; further algorithmic optimizations may be beneficial. Because we can selectively modify several design parameters, our goniometers may be useful for systematic and well-controlled testing of the limits of experimental and computational methods in cryo-EM studies of small proteins. The goniometers may also help evaluate practical strategies to compensate for poor alignment accuracy, such as future custom-tailored software pipelines, and engineering asymmetry into the protein or its DNA substrate.
We obtained an average of 51 goniometers per micrograph, 24% of which had a “good” BurrH used in the initial reconstruction. The overall goniometer particle density on the grid surface might be increased by reducing the mass and footprint of the origami by using a more compact design, and promoting uniform orientation of chassis adsorption onto the support surface, for example, using a custom affinity grid and staples with anchoring ssDNAs that extend from one face of the goniometer. We can also improve the capacity for sample multiplexing on a single grid by using more complex barcodes.
Of the six degrees of freedom (x, y, z, tilt, rotation, psi) that specify the absolute location of a particle on a cryo-EM grid, our molecular goniometers provide limited user-defined control for three of them (tilt, rotation, and z translation from the grid surface). Our method enables simultaneous collection of all programmed particle orientations and thus avoids radiation damage incurred by serial electron dosing that may be required by methods that rely on physical specimen tilting16,17. We designed the DNA origami particles to be large, asymmetric, and easy to pick and align, and so in effect they do provide local x, y, and psi values of the DNA stage relative to the origami chassis. The capability of prescribing the distance of the target from the surface of the grid could be useful for certain targets, perhaps including targets with masses greater than 100 kDa with some modifications to the origami design. It should also be possible to gain better control over additional degrees of freedom, such as the absolute x, y, and psi values.
In cases where a tool or algorithm could achieve 100% tilt direction accuracy without the aid of priors derived from the origami, goniometers would not be necessary. However, currently this level of accuracy is not always possible. Thus, our results demonstrate that the a priori information provided by the molecular goniometers improve resolution for cryo-EM studies of small DNA-binding proteins such as BurrH. Furthermore, the paucity of small (<100 kDa) proteins in the Electron Microscopy Data Bank may suggest that there are many targets of unknown structure that may benefit from our approach, either by incorporating their binding sequence into the DNA stage, or for non-DNA-binding proteins, fusion with a DNA-binding domain. In principle, RNAs can bind to the DNA stage directly via hybridization, or by docking to RNA-binding moieties that attach to the DNA stage. In addition to studying natural targets, our goniometers may be useful for de novo design of small DNA-binding proteins to enable and accelerate the molecular design of hybrid architectures18, such as DNA origami structures decorated with proteins that carry out binding or catalysis.
Despite great technical advancements in cryo-EM in recent years, significant limitations to its use remain19,20. Using nanotechnology to control the positions and orientations of particles may bridge the gap between the practical and theoretical limits of the method.
Online Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment.
Protein Expression and Purification.
The BurrH open reading frame (ORF) with a flexible linker and C-terminal HIS TAG (-GSGHHHHHH) was subcloned into pet24d+ expression vector (Genscript, NJ, USA). For protein concentration quantification, we added single tryptophan residue upstream of the HIS TAG. The BurrH plasmid was chemically transformed into E.coli strain BL21(DE3)pLysS (Promega, WI, USA) as described in the product manual. BurrH was expressed and purified as described10. Briefly,
10 ml sterile LB media supplemented with 100 μg/ml Ampicillin was inoculated with a single colony picked from an LB-agarose plate. The bacterial culture was grown overnight at 37°C with moderate shaking.
The next day, 10 ml of the overnight culture was added into 1L sterile LB media supplemented with 100 μg/ml Ampicillin and the 1L culture was grown at 37°C with moderate shaking for 4–6 hours until OD600 reaches 0.4–0.6.
Once OD600 reaches 0.4–0.6, culture was brought to 1mM IPTG with a sterile 1 M IPTG stock. The culture was grown at 37°C with moderate shaking for an additional 4 hours.
Cells were harvested by centrifuging the culture at 6000× rcf for 10 minutes at 4°C.
For long-term storage and easy lysis of E.coli cells, the cell pellets were kept at −80°C.
Cell pellets were lysed using B-PER Bacterial Protein Extraction Reagent (ThermoFisher Scientific, MA, USA). 4 ml B-PER reagent was added per gram of cell pellet. In addition, the lysis reagent was supplemented with 100 μg/ml lysozyme and 3U DNase I. The lysate was incubated at room temperature with moderate stirring for 15 minutes.
Lysate was centrifuged at 15,000× rcf for 15 minutes and the pellet was discarded.
Clear lysate was filtered using a 0.45-μm syringe filter (Millipore Sigma, MA, USA).
Lysate was mixed with 2 ml nickel NTA beads (Qiagen, MD, USA) equilibrated with wash solution (10% Glycerol, 150 mM NaCl, 20 mM Imidazole, 50 mM HEPES-Cl at pH 8.0). The lysate-and-nickel-beads mix was incubated at 4°C with moderate mixing for 30 minutes.
Beads were washed twice using 25 ml wash solution each time.
BurrH was eluted using 10 ml elution solution (10% Glycerol, 150 mM NaCl, 250 mM Imidazole, 50 mM HEPES-Cl at pH 8.0) and passed through a 10-ml Pierce disposable column (ThermoFisher Scientific, MA, USA) to remove the beads from solution.
The final elution was dialyzed against BurrH solution (10 mM MgCl2, 150 mM NaCl, 50 mM HEPES-Cl at pH 8.0).
To remove any impurities, the dialyzed sample was passed through a “Superdex 200 Increase” size-exclusion column (GE HealthCare, PA, USA) equilibrated with the BurrH solution. The size-exclusion run was performed using the BurrH solution and the absorbance at 280 nm was used to track BurrH.
Construction of p9344-BurrH.
p9344-BurrH (GenBank MT081208) was created in two steps. First, an insert that included the Ampicillin resistance gene and plasmid origin was amplified by polymerase chain reaction from the pUC18 vector using primers:
5’-AAAAAAGAATTCCTTCCGCTTCCTCGCTCACTGACTC-3’
5’-AAAAAAGTCGACCTGCTCCCGGCATCCGCTTACAGAC-3’
The PCR product was cleaved by EcoRI and SalI, and ligated into the M13mp18 RF plasmid that was also cleaved by EcoRI and SalI. Next, the BurrH binding site was inserted into this construct. The 54-bp insert that comprises the BurrH binding site was ordered from IDT and inserted into the modified M13mp18 RF plasmid via SalI and HindIII restriction sites (Figure 2).
p9344-BurrH ssDNA Preparation.
XL1-Blue MRF’ Kan was chemically transformed with p9344-BurrH plasmid. Next, the transformed XL1-Blue MRF’ Kan p9344-BurrH cells were mixed with molten LB top agar and spread onto an LB Agarose plate that was supplemented with 30 μg/ml Kanamycin. The plate was stored at 37°C overnight. The next morning, a plaque was picked and inoculated into 10 ml 2XYT media supplemented with 30 μg/ml Kanamycin, 100 μg/ml Ampicillin and 10 mM MgCl2. The inoculated culture was incubated overnight at 37°C with moderate shaking. On the next day, uninfected XL1-Blue MRF’ Kan culture in 100 ml 2xYT was supplemented with 30 μg/ml Kanamycin and 10 mM MgCl2 was grown at 37°C until the OD600 reached 0.4–0.6, and then 100 ml of that culture was inoculated with the overnight-grown XL1-Blue MRF’ culture producing p9344-BurrH phage. The 100-ml inoculated culture was incubated at 37°C with moderate shaking for 5 hours. p9344-BurrH phage was recovered and p9344-BurrH ssDNA was purified as described previously22.
DNA Origami Designs and Preparation.
p9344-BurrH scaffold routings and initial Goniometer designs were made using Cadnano2.223 Optimization of staples (i.e. staple auto-breaking) and generation of pipetting instructions for the staple stocks were performed using a custom software toolkit (manuscript in preparation) developed in the Douglas Lab. Staple stocks for the goniometers were prepared using Labcyte Echo 525 (Labcyte, CA, USA). Folding reactions were performed using Bio-Rad MJ Research PTC-240 Tetrad thermal cycler. The temperature annealing ramp for the DNA Origami Goniometers was:
Incubate at 65°C for 00:10:00
- Incubate at 60°C for 01:00:00
- Decrease by 1.0°C every cycle
Cycle to step 2 for 20 more times
Folding was performed in 1XFOB20 (5 mM Tris-Base, 1 mM EDTA, 5 mM NaCl, 20 mM MgCl2 at pH 8.0). In the folding reaction, p9344-BurrH scaffold concentration was 20 nM and each staple concentration was 200 nM. The Goniometers were purified via PEG precipitation as described previously24. Briefly, 15% PEG-8000 solution (15% w/v PEG-8000, 5 mM Tris-Base, 1 mM EDTA, 500 mM NaCl, 20 mM MgCl2 at pH 8.0) were mixed with the folding reaction at a 1:1 ratio. The mix was centrifuged at 16,000× rcf for 25 minutes at room temperature. The supernatant was discarded, and the pellet resuspended in 1XFOB20. PEG precipitation was repeated one more time and the final pellet was resuspended in 1XFOB20 and stored at 4°C.
Negative-stain TEM Experiments.
Negative stain TEM experiments were performed on grids with different support surfaces for DNA-origami goniometer deposition. Prior to sample deposition and negative staining, ultrathin carbon coated quantifoil grids (Pacific grid tech, CA) were treated with a Pelco glow discharge unit (30 seconds hold/30 seconds glow discharge). For negative-stain grid preparation, 5 μl of 80 ng/ul DNA origami goniometer sample was deposited onto a glow-discharged ultra thin-carbon coated, graphene-oxide or amine-functionalized graphene-oxide grids (prepared in-house, as described11). The sample was incubated on the grids for 1 minute, then excess sample was wicked away using filter paper. Next, 10 μl of freshly prepared 2% uranyl formate (Electron Microscopy Sciences, PA) was applied to the grids and immediately wicked away using filter paper. A second round of 10 μl 2% uranyl formate was then applied onto the grids for 3 minutes before excess stain was wicked away and the grid left to dry. Micrographs of the negatively stained grids were collected on Tecnai T12 (FEI, OR) at 30,000× magnification.
Cryo-EM Sample Preparation and Data Acquisition,
For cryo-EM sample preparation, we used amine functionalized graphene-oxide grids prepared in-house as described previously11. Samples for cryo-EM were prepared by mixing 9 μl of 80 ng/μl DNA origami goniometer with 1μl of 2.5 μM BurrH in 1XFOB20 buffer supplemented with 1mM TCEP. Grids were prepared using Vitrobot (Thermo Fisher Scientific, MA) at 20°C. Prior to grid freezing in liquid ethane, the sample was incubated on the grid for 30 seconds and then blotted with a filter paper for 4 to 7 seconds. Micrographs were collected on a Talos Arctica microscope (Thermo Fisher Scientific, MA) operating at 200 kV with a K3 camera (Gatan, CA), at a nominal magnification of 22,000× corresponding to a physical pixel size of 1.82 Å. Total dose rate for imaging each hole is kept at 56 e/Å2. Samples containing goniometers with +90°-tilt/414-nt-bits and –90°-tilt/414-nt-bits were pooled together for the initial round of data collection. A sample of +90°-tilt/222-nt-bit goniometers were imaged in a separate round of data collection. All image data were combined for analysis, see Extended Data Figure 5.
Cryo-EM Image Processing and Protein 3D reconstruction.
Drift correction of the movie stacks was performed using MotionCor225. The CTF parameter estimation of the drift corrected micrographs was performed using GCTF26. Micrographs with ice contamination and with estimated resolution lower than 8Å were discarded from further analysis. Our data analysis pipeline for DNA origami goniometer classification included the following steps:
Goniometer particles were picked using Relion’s Laplacian of Gaussian Picking function18.
Particles were classified into 2D classes for particle clean-up using cryoSPARC13. Bad classes were removed from further analysis (Fig. 5b).
Good classes were recentered and aligned to a single reference (Fig. 5c–d).
All regions of the goniometer except the bottom (tilt) barcode were subtracted and filled with gaussian noise centered at zero with a standard deviation of one (Fig. 5e).
Classifications of the subtracted goniometers that focused on the left and right portions of the bottom barcode region were performed using Relion’s 2D classification function18. For the focused masked classifications, masks were created using EMAN2’s 2D mask drawing tool33. Left and right bottom barcode classifications were performed successively until the classification results converged. Finally, classes that could not be recognized based on the barcode information were discarded (Fig. 5f–h).
Images with particles that were oriented with a flipped orientation relative to a chosen reference are flipped so that side (rotation) barcodes could be classified in a single classification run. The particles were aligned to a common reference and all regions of the goniometer except the side (rotation) barcodes were subtracted and filled with gaussian noise centered at zero with a standard deviation of one. Left and right side barcode classifications were performed successively until the classification results converged. Finally, classes that could not be recognized based on the barcode information were discarded. Side barcode classification was performed separately for each goniometer class (Fig. 5i–l).
After goniometers were separated based on the barcode bits, we moved onto data analysis steps that only involve the target protein:
Goniometers were aligned to a common reference so that target protein was at the center (Extended Data Figure 5a).
Protein particles were picked with box size as large as possible avoiding any signal from the goniometer (Extended Data Figure 5b).
Protein particles were classified using Relion’s 2D classification tool with the psi angle restrained to maintain protein orientations fixed with respect to Goniometers12. Bad classes were removed from further processing (Extended Data Figure 5c).
Good classes were recentered keeping protein orientations fixed (Extended Data Figure 5d).
The initial 3D model was built using Relion’s 3D Initial Model function with additional arguments “--sigma_tilt 10 --sigma_psi 10 --sigma_rot 15” to keep the euler angles restrained around the a priori values12, Extended Data Figure 5e).
All particles were aligned to the initial 3D model using Relion’s 3D Classification utility for single class classification with regularization parameter (T) set to 6 for stronger particle alignment and with additional arguments “--sigma_tilt 10 --sigma_psi 10 --sigma_rot 15” to keep the euler angles restrained around the a priori values18, Extended Data Figure 5f).
Particles were classified into five 3D classes with regularization parameter set to 2 with additional arguments “--sigma_tilt 10 --sigma_psi 10 ” to keep the particle orientation restrained while allowing rotation angles to float (Extended Data Figure 5h).
3D class with the best resolution, orientational, and positional accuracy, were picked and refined using Relion’s 3D refinement utility with additional arguments “--sigma_tilt 10 --sigma_psi 10 ” to keep the particle orientation restrained allowing rotation angles float18, Extended Data Figure 5h).
In our data analysis pipeline, conversions between Relion12, cryoSPARC13, and cisTEM14 data formats, data editing, particle subtractions and alignments, global FSC, and dFSC calculations were performed using custom Python scripts (https://github.com/douglaslab/cryoorigami). Cryo-EM data analysis was performed on AWS GPU instances p2.16xlarge and g3.16xlarge (Amazon, WA).
Consensus Alignment of BurrH 2D Class Averages and BurrH Map 2D Projections.
Goniometer class averages were aligned to a single class using Relion’s 2D classification utility12. To focus the alignment on the BurrH density, a tight circular mask (140 Å) was used and the psi alignment angle was constrained to within ±9° of the goniometer alignment angle from the previous step (--sigma_psi 3). Finally, the aligned class averages were weighted in proportion to the number of particles in each goniometer class. 2D projections of BurrH map at Goniometer prescribed tilt and rot angles are generated using Relion’s relion_project utility12.
Model Fitting into Cryo-EM Maps, FSC, and dFSC Calculations.
The crystal structure of BurrH bound to DNA (PDB id: 4cja) was fitted into 3D maps using UCSF Chimera’s rigid body fitting utility27. FSC curves between the fitted 4cja maps at 3.64Å resolution and the 3D refinement maps were calculated using the in EMAN2 e2proc3d.py script28. Renders of the maps were generated using UCSF Chimera27. Directional FSCs (dFSCs) between the two half maps were calculated as described previously29,30. Briefly, dFSCs were calculated for 500 conical shells with an apex angle of 20°. Conical shell centers were evenly distributed on a Fibonacci sphere. The global FSC was calculated from an average of all directional FSCs, and the dFSC volume was generated from the sums of individual conical dFSCs. The dFSC values in overlapping conical regions were averaged, and the dFSC map was filtered using a 3×3×3 median filter for 3 iterations to remove noise. Resolutions in each direction were determined by FSC0.143 criteria. The sphericity of the dFSC volume is determined using 3DFSC web server (https://3dfsc.salk.edu/).
Extended Data
Extended Data Fig. 1. Comparison of grid adsorption orientation and signal delocalization for two chassis designs.
a, Representative negative-stain micrographs of chassis with 0.67 aspect ratio. b, Representative cryo-EM micrograph of chassis with 0.59 aspect ratio. Desired orientations assessed by manual counting. c, Top: Representative 2D class average of 0°-rotation goniometer from a. The chassis aperture exhibits a shadow that overlaps the DNA stage and BurrH protein. Bottom: Representative 2D class averages of BurrH, exhibiting artifacts that we hypothesized are due to delocalized origami signal in the 0.5–2.0 μm defocus range used for image acquisition. d, Top: 2D class of 0°-rotation goniometer from b designed to reduce shadow overlap. Bottom: Representative BurrH 2D class averages. Artifacts observed in c are significantly reduced. Scale bars: a, b: 100 nm, c, d: 25 nm (top), 10 nm (bottom).
Extended Data Fig. 2. How we defined “tilt” and “rotation” angles for the molecular goniometers.
Independent 2D views of a 3D object can be derived using only two orthogonal rotational transformations. In cryo-EM, the two orthogonal rotations can be referred to as tilt and rotation angles, respectively12,21. The reference coordinate system for the rotation operations can be chosen arbitrarily, and here we define the goniometer tilt angle as the angle between the stage DNA and the normal vector perpendicular to the goniometer face (the normal vector is parallel to the electron beam (Fig. 1a, orange line labeled e–) when the goniometer adsorbs in the desired face-up orientation. We define the goniometer rotation angle as the rotation angle with respect to the axis parallel to the DNA stage helical axis.
Extended Data Fig. 3. Cadnano design strand diagram schematics.
a, Bit and rotation angles for each column. b, +90 tilt, 414-nt rotation bit designs. TILT bit is inactive (red outline in bottom right corner of each schematic). c, –90 tilt, 414-nt rotation bit designs. TILT bit is active. d, +90 tilt, 222-nt rotation bit designs. TILT bit is inactive.
Extended Data Fig. 4. 2D cryo-EM class averages goniometers with BurrH.
a, +90 tilt, 414-nt bit b, –90 tilt, 414-nt bit (flipped), c, +90 tilt, 222-nt bit. Scale bars: 100 nm.
Extended Data Fig. 5. Cryo-EM data analysis workflow for BurrH 3D reconstruction and refinement.
a, Centering goniometer classes at tilt DNA. b, Picking protein particles from the center of goniometers. c, 2D classification of BurrH particles and removing “bad” classes. d, Recentering “good” classes. e, Building an initial model 3D model with tilt and rot angle constraints. f, Aligning BurrH particles to initial model with tilt and rot angle constraints and regularization parameter, T, set to 6 for better alignment. g, Classifying particles into five 3D classes with tilt angle constraint on and regularization parameter, T, set to 2 to minimize overfitting during classification. Best 3D class (magenta) is at 7.3 Å resolution. h, Refining the best 3D class with only the tilt angle constraint. Estimated resolution of the refined map is 6.5 Å.
Extended Data Fig. 6. 2D classification of goniometer stage DNA with BurrH.
Representative 2D classes of stage DNA with BurrH after one round of 2D classification. Classes with high background signal, with empty tilt DNA and DNA origami signal are removed before further 2D classification of BurrH bound to stage DNA. b, “Good” 2D classes retained for 3D reconstruction and refinement of BurrH. Scale bar is 20 nm.
Extended Data Fig. 7. Measured tilt, rotation, and psi angle distributions for all goniometer designs.
Polar plots and histograms follow the convention from Figure 6. The inner tick mark represents the goal angle, and the arc shows the bounds of the Gaussian angle prior. Average rotation angles are plotted as magenta lines for tilt and rotation plots.
Extended Data Fig. 8. Directional FSC Plots for BurrH.
a, Individual directional FSCs (gray lines), global FSC value (blue line) and ±1 standard deviation from global FSC (magenta dashed lines). b, Directional FSC map and its 2D projections. The minimum resolution (blue) and maximum resolution (magenta) are indicated for each 2D projection.
Extended Data Fig. 9. Comparison of particle count and defocus effects on final BurrH resolution.
a, ResLog plots showing FSC vs. resolution for 3D refinement of BurrH using 1,000 to 68,482 particles. b, Resolution estimates from FSC0.143 vs. particles count. The resolution plateaus at 40k particles, suggesting our resolution is not limited by particle count. c, BurrH resolution vs. maximum micrograph defocus used in 3D reconstruction. d, Resolution estimates from FSC0.143 vs. maximum defocus. The resolution reaches a plateau at 1.5 μm and the addition of higher defocus particles doesn’t comprise the resolution. Inset shows the distribution of BurrH particle defocus in μm.
Extended Data Fig. 10. Filtering particles by estimated rotation angle.
FSC and ResLog plots for BurrH 3D reconstructions in which particles are filtered when their estimated angles from the unfiltered 3D refinement differ from the goniometer goal angle. a, We compared three conditions: No filter (i.e., identical to Fig. 6), and two conditions in which particles with assigned rotation angles that differ by >45°, and >90° from the goal angle. b, FSC curves comparing the 3D reconstruction for the three filtering conditions. c, Plot of particle count versus resolution estimate. Filtering reduces the particle count available for the 3D reconstruction, which in turn reduces the final resolution (see Extended Data Figure 8b). Compared to the unfiltered reconstructions, removing particles in the indicated angle ranges does not improve the final BurrH resolution.
Supplementary Material
Supplementary Table 1 Staple sequences for goniometers with +90° tilt and 414-nt barcode bit.
Supplementary Table 2 Staple sequences for goniometers with −90° tilt and 414-nt barcode bit.
Supplementary Table 3 Staple sequences for goniometers with +90° tilt and 222-nt barcode bit.
Figure 4 |. Cryo-EM data analysis workflow for DNA origami goniometer barcode classification.
a, Picking goniometers. b, Removing bad goniometer classes (purple outlines). c, Preparing well-centered classes. d, Aligning goniometer classes to a reference. e, Subtracting top region of goniometers for bottom (tilt) barcode classification. f, Focused classification on the left-bottom tilt barcode. g, Focused classification on the right-bottom tilt barcode. h, Separating +90° tilt and –90° tilt goniometers based on the barcode classifications. i, Flipping –90° gonimeters to align BurrH N-to-C direction. j, Subtracting the central region of goniometers for rotation-barcode classification. Focused classification on k, left-side and l, right-side rotation barcodes, with representative “bad” classes. m, Separating goniometers into 7 sub-classes based on the rotation-barcode classifications. Scale bars: a: 100 nm, b–m: 20 nm.
Acknowledgments
We thank F. Wang for furnishing amino graphene oxide grids and protocols for early tests. We thank D. Bulkley and A. Myasnikov for help with cryo-EM data collection. We thank J. Brown and C. Gingold for help with visualization. TA was supported by Ruth L. Kirschstein NRSA Postdoctoral Fellowship F32GM119322. SMD is supported by the UCSF Program for Breakthrough Biomedical Research, Pew-Stewart Scholars Program, NSF CAREER Award 1453847, and NIH R35GM125027. YC is supported by NIH R01GM098672, R01HL134183, S10OD021741, and S10OD020054.
Footnotes
Competing interests
Authors declare no competing interests.
Data and Materials Availability
Source code is available at https://github.com/douglaslab/cryoorigami.The cryo-EM map of BurrH has been deposited in the Electron Microscopy Data Bank (EMD-21443). The cryo-EM movie files have been deposited at the Electron Microscopy Public Image Archive (EMPIAR-10373). The p9344-BurrH scaffold sequence has been submitted to GenBank (MT081208), and the plasmid deposited with AddGene (#140326).
References
- 1.Bai X-C, McMullan G & Scheres SHW How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci 40, 49–57 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Cheng Y Single-particle cryo-EM-How did it get here and where will it go. Science 361, 876–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S et al. Fabs enable single particle cryoEM studies of small proteins. Structure 20, 582–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C et al. Analysis of discrete local variability and structural covariance in macromolecular assemblies using Cryo-EM and focused classification. Ultramicroscopy 203, 170–180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Gonen S, Gonen T & Yeates TO Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system. Proc. Natl. Acad. Sci. U. S. A 115, 3362–3367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seeman NC Nucleic acid junctions and lattices. J. Theor. Biol 99, 237–247 (1982). [DOI] [PubMed] [Google Scholar]
- 7.Rothemund PWK Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Martin TG et al. Design of a molecular support for cryo-EM structure determination. Proc. Natl. Acad. Sci. U. S. A 113, E7456–E7463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juillerat A et al. BurrH: a new modular DNA binding protein for genome engineering. Sci. Rep 4, 3831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stella S et al. BuD, a helix-loop-helix DNA-binding domain for genome modification. Acta Crystallogr. D Biol. Crystallogr 70, 2042–2052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F et al. Amino and PEG-amino graphene oxide grids enrich and protect samples for high-resolution single particle cryo-electron microscopy. J. Struct. Biol 107437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Grant T, Rohou A & Grigorieff N cisTEM, user-friendly software for single-particle image processing. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldwin PR & Lyumkis D Non-uniformity of projection distributions attenuates resolution in Cryo-EM. Prog. Biophys. Mol. Biol (2019) doi: 10.1016/j.pbiomolbio.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Meents A, Gutmann S, Wagner A & Schulze-Briese C Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures. Proc. Natl. Acad. Sci. U. S. A 107, 1094–1099 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou K et al. Toward Precise Manipulation of DNA–Protein Hybrid Nanoarchitectures. Small 15, 1804044 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Glaeser RM How good can cryo-EM become? Nat. Methods 13, 28–32 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Lyumkis D Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem 294, 5181–5197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods References
- 21.Heymann JB, Chagoyen M & Belnap DM Common conventions for interchange and archiving of three-dimensional electron microscopy information in structural biology. J. Struct. Biol 151, 196–207 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Nafisi PM, Aksel T & Douglas SM Construction of a novel phagemid to produce custom DNA origami scaffolds. Synth. Biol 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas SM et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagenbauer KF et al. How We Make DNA Origami. Chembiochem 18, 1873–1885 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD & Huang CC UCSF Chimera—a visualization system for exploratory research and analysis. Journal of (2004). [DOI] [PubMed] [Google Scholar]
- 28.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Dang S et al. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan YZ et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Staple sequences for goniometers with +90° tilt and 414-nt barcode bit.
Supplementary Table 2 Staple sequences for goniometers with −90° tilt and 414-nt barcode bit.
Supplementary Table 3 Staple sequences for goniometers with +90° tilt and 222-nt barcode bit.