Abstract
While vaccines directed against the SARS-CoV-2 spike protein will have varying degrees of effectiveness in preventing SARS-CoV-2 infections, the severity of infection will be determined by multiple host factors including the ability of immune cells to lyse virus-infected cells. This review will discuss the complexity of both adaptive and innate immunomes and how a flow-based assay can detect up to 158 distinct cell subsets in the periphery. This assay has been employed to show the effect of age on differences in specific immune cell subsets, and the differences in the immunome between healthy donors and age-matched cancer patients. Also reviewed are the numerous soluble factors, in addition to cytokines, that may vary in the pathogenesis of SARS-CoV-2 infections and may also be employed to help define the effectiveness of a given vaccine or other antiviral agents. Various steroids have been employed in the management of autoimmune adverse events in cancer patients receiving immunotherapeutics and may be employed in the management of SARS-CoV-2 infections. The influence of steroids on multiple immune cells subsets will also be discussed.
Keywords: immunotherapy, adaptive immunity, immunity, cellular, T-lymphocytes, COVID-19
Introduction
The COVID-19 pandemic has affected every aspect of the population, including different age groups and patients with cancer. It is generally believed that preventive vaccines that will elicit antibody responses to the SARS-CoV-2 agent will greatly reduce the percentage of the population that becomes infected with the SARS-CoV-2 virus. In addition, antivirals, passive administration of anti-SARS-CoV-2 antibodies, and other agents such as steroids may lead to greater control of the disease. At this time, little is known about the effectiveness of the various anti-SARS-CoV-2 vaccine platforms being developed and administered, especially in terms of the induction of long-term immunity, reduction of severity of the disease, protection of SARS-CoV-2 variants that may arise, and toxicities.
One aspect of the control of the COVID-19 pandemic that has received limited attention is the role of the cellular immunome in apparently healthy individuals of different age groups, as well as in patients with cancer. It is quite possible that variations in the cellular immunome among individuals and among patients with different types and stages of cancer may well be a factor in (a) susceptibility to SARS-CoV-2 infection, (b) reduction of the severity of the infection, (c) determination of long-term immunity, (d) adverse events in the use of steroids, antivirals, or other agents to control the SARS-CoV-2 infection and (e) control of SARS-CoV-2 variants that may arise.
Most anti-SARS-CoV-2 vaccines being developed consist of the spike protein, or RNA or DNA, reflecting the sequence of that protein. Clinical trials are demonstrating the production of antibodies to SARS-CoV-2 and, in some cases, T-cell responses to the spike protein. While antibody responses to the virus will reduce or prevent infection, it is the cellular immunome response to the agent that is necessary to lyse viral-infected cells and thus limit the severity of the disease. Thus, in addition to the development of vaccines directed against the SARS-CoV-2 spike protein, vaccine platforms should also be developed consisting of proteins, RNA or DNA reflecting other structural components of the SARS-CoV-2 agent. The generation of T-cell responses to these components will thus enable the recognition of major histocompatibility complex (MHC)–peptide complexes of degradation and transport of these components to the surface of infected cells, resulting in lysis. If the SARS-CoV-2 agent is similar to other RNA viruses, in contrast to more stable internal structural proteins, most variants will be the reflection of changes in the spike or external protein, thus potentially limiting the effectiveness of vaccines directed only to that protein. The use of vaccines that elicit T-cell responses to multiple SARS-CoV-2 components may thus limit the severity of infections of such variants.
SARS-CoV-2 comprises four major structural elements: the transmembrane spike (S) glycoprotein that permits host cell entry by binding the human ACE2 receptor, a nucleocapsid (N) protein that facilitates viral replication, membrane (M) proteins, and small envelope (E) proteins.1 2 In individuals with active COVID-19 infection and in those in convalescent phases, CD4+ and CD8+ responses against SARS-CoV-2 proteins have been documented. In recovered patients, S-specific CD4+ T cells with an activated phenotype and CD8+ T cells expressing markers of cytotoxicity have been described.3 Predominantly, S-reactive CD4+ T cells are present in most patients with active and acute disease.4 5 Nearly all patients who have recovered from COVID-19 infection harbor CD4+ and CD8+ T cells that also recognize multiple regions of the N protein.6 In fact, very recent studies have shown that CD8+ T cells in particular largely recognize epitopes outside of the S protein, and T cells recognizing N protein are higher in frequency than T cells against S and other non-structural proteins after infection.7 8 In acute phase patients, it has been shown that specifically N-specific T cells skew towards a Th1 phenotype.9
Clinical data emerging thus far from trials with different vaccines have demonstrated differential induction of CD4+ and/or CD8+ responses against the receptor binding domain and other regions spanning the S protein; some of these immune responses have showed Th1 polarization with high IFNγ and TNFα production and limited IL-4 and IL-10 release.10–12 A vaccine platform targeting both the S and N proteins of the SARS-CoV-2 virus may be optimal. Recent preclinical work investigating an hAd5 vector encoding both S and N proteins in mice demonstrated that this vaccine construct elicited strong Th1 cellular and neutralizing humoral responses after prime boost, with CD4+ and CD8+ responses against both S and N epitopes.13
Immunotherapy is now playing a major role in the management of many cancer types. The mode of action of the vast majority of these immunotherapeutics is on multiple components of both the innate and adaptive cellular compartments, both in the periphery and in the tumor microenvironment. Thus, a concern will be the influence of the use of anticheckpoint monoclonal antibodies (MAbs), cytokines, anticancer vaccines, cellular therapies, or steroids to reduce autoimmune adverse events, on susceptibility to SARS-CoV-2 infection and/or severity of that infection.
As a consequence of the renaissance of cancer immunotherapy in recent years, there has been a much greater understanding of the human cellular immunome, especially in the complexities involving the interactions among and between different components of both the innate and adaptive arms of the immune system. To date, over 150 different phenotypes of human immune cells have been identified; these are traditionally characterized as the so called “parental” cell types: CD8+, CD4+, natural killer (NK), NK T cells, regulatory CD4+ T cells (Tregs), B cells, conventional (c) dendritic cells (cDCs), plasmacytoid (p) DCs, myeloid-derived suppressor cells (MDSCs), and monocytes. For each of these parental cell types, multiple refined subtypes have been identified, many of which have defined activation and/or suppressive functions (see table 1).
Table 1.
158 peripheral immune cell subsets analyzed by flow cytometry
| 1. Total CD4+ T cells | 2. Total CD8+ T cells | 4. Total B cells |
| PD-L1+ CD4 – activation/inhibition | PD-L1+ CD8 – activation/inhibition | PD-L1+ B cells – activation/inhibition |
| PD-1+ CD4 – activation/inhibition | PD-1+ CD8 – activation/inhibition | PD-1+ B cells- activation/inhibition |
| CTLA-4+ CD4 – inhibition | CTLA-4+ CD8 – inhibition | 5. Total NK |
| Tim-3+ CD4 – inhibition | Tim-3+ CD8 – inhibition | PD-L1+ NK – inhibition |
| 41bb+ CD4 – costimulation | 41bb+ CD8 – costimulation | PD-1+ NK – activation/inhibition |
| Ki67+ CD4 – proliferation | Ki67+ CD8 – proliferation | Tim-3+ NK – activation/inhibition |
| CD73+ CD4 – exhausted/suppressive | CD73+ CD8 – exhausted/suppressive | Ki67+ NK – proliferation |
| ICOS+ CD4 – activation | Naïve (CCR7+CD45RA+) CD8 | NKp30+ NK – activation |
| ICOS+ PD-L1+ CD4 – activation/inhibition | Central memory (CCR7+CD45RA-) CD8 | NKp46+ NK – activation |
| ICOS+ PD-1+ CD4 – activation/inhibition | PD-L1+ CM CD8 – activation/inhibition | NKG2D+ NK – activation |
| Naïve (CCR7+CD45RA+) CD4 | PD-1+ CM CD8 – activation/inhibition | CD226+ NK – adhesion/activation |
| Central memory (CCR7+ CD45RA-) CD4 | CTLA-4+ CM CD8 – inhibition | Mature (CD16+ CD56dim) NK – lytic |
| PD-L1+ CM CD4 – activation/inhibition | Tim-3+ CM CD8 – inhibition | PD-L1+ mature NK – inhibition |
| PD-1+ CM CD4 – activation/inhibition | 41bb+ CM CD8 – costimulation | PD-1+ mature NK – activation/inhibition |
| CTLA-4+ CM CD4 – inhibition | Ki67+ CM CD8 – proliferation | Tim-3+ mature NK – activation/inhibition |
| Tim-3+ CM CD4 – inhibition | CD73+ CM CD8 – exhausted/suppressive | Ki67+ mature NK – proliferation |
| 41bb+ CM CD4 – costimulation | Effector memory (CCR7- CD45RA-) CD8 | NKp30+ mature NK – activation |
| Ki67+ CM CD4 – proliferation | PD-L1+ EM CD8 – activation/inhibition | NKp46+ mature NK – activation |
| CD73+ CM CD4 – exhausted/suppressive | PD-1+ EM CD8 - activation/inhibition | NKG2D+ mature NK – activation |
| Effector memory (CCR7- CD45RA-) CD4 | CTLA-4+ EM CD8 – inhibition | CD226+ mature NK – adhesion/activation |
| PD-L1+ EM CD4 – activation/inhibition | Tim-3+ EM CD8 – inhibition | Functional intermediate (CD16+ CD56br) NK – lytic, cytokine production |
| PD-1+ EM CD4 – activation/inhibition | 41bb+ EM CD8 – costimulation | PD-L1+ functional intermediate NK – inhibition |
| CTLA-4+ EM CD4 – inhibition | Ki67+ EM CD8 – proliferation | PD-1+ functional intermediate NK – activation/inhibition |
| Tim-3+ EM CD4 – inhibition | CD73+ EM CD8 – exhausted/suppressive | Tim-3+ functional intermediate NK – activation/inhibition |
| 41bb+ EM CD4 – costimulation | EMRA (CCR7-CD45RA+) CD8 | Ki67+ functional intermediate NK – proliferation |
| Ki67+ EM CD4 – proliferation | PD-L1+ EMRA CD8 – activation/inhibition | NKp30+ functional intermediate NK – activation |
| CD73+ EM CD4 – exhausted/suppressive | PD-1+ EMRA CD8 – activation/inhibition | NKp46+ functional intermediate NK – activation |
| EMRA (CCR7-CD45RA+) CD4 | CTLA-4+ EMRA CD8 – inhibition | NKG2D+ functional intermediate NK – activation |
| PD-L1+ EMRA CD4 – activation/inhibition | Tim-3+ EMRA CD8 – inhibition | CD226+ functional intermediate NK – adhesion/activation |
| PD-1+ EMRA CD4 – activation/inhibition | 41bb+ EMRA CD8 – costimulation | Immature (CD16- CD56br) NK – cytokine production |
| CTLA-4+ EMRA CD4 – inhibition | Ki67+ EMRA CD8 – proliferation | PD-L1+ immature NK – inhibition |
| Tim-3+ EMRA CD4 – inhibition | CD73+ EMRA CD8 – exhausted/suppressive | PD-1+ immature NK – activation/inhibition |
| 41bb+ EMRA CD4 – costimulation | 3. Total Tregs | Tim-3+ immature NK – activation/inhibition |
| Ki67+ EMRA CD4 – proliferation | PD-L1+ Tregs – activation/inhibition | Ki67+ immature NK – proliferation |
| CD73+ EMRA CD4 – exhausted/suppressive | PD-1+ Tregs – suppression | NKp30+ immature NK – activation |
| CTLA-4+ Tregs – suppression | NKp46+ immature NK – activation | |
| ICOS+ Tregs – suppression | NKG2D+ immature NK – activation | |
| CD45RA+ Tregs – highly expandable in vitro | CD226+ immature NK – adhesion/activation | |
| CD49d- Tregs – suppression | Unconventional (CD16- CD56dim) NK – non-lytic, non-cytokine production | |
| Ki67+ Tregs – proliferation | ||
| CD38+ Tregs – suppression | ||
| HLA-DR+ Tregs – suppression | ||
| 6. Total NK-T | 9. Total MDSC | 10. Total Monocytes |
| PD-L1+ NK-T – inhibition | PD-L1+ MDSC – inhibition | PD-L1+ monocytes – inhibition |
| PD-1+ NK-T – activation/inhibition | PD-1+ MDSC – activation/inhibition | PD-1+ monocytes – activation/inhibition |
| Tim-3+ NK-T – inhibition | CD16+ MDSC – immature/suppression | Classical monocytes – phagocytic |
| Ki67+ NK-T – proliferation | Monocytic (CD14+ CD15-) MDSC | PD-L1+ classical monocytes – inhibition |
| 7. Total cDC | PD-L1+ mMDSC – inhibition | PD-1+ classical monocytes- activation/inhibition |
| PD-L1+ cDC – inhibition | PD-1+ mMDSC – activation/inhibition | Intermediate monocytes phagocytic/proinflammatory |
| PD-1+ cDC – activation/inhibition | CD16+ mMDSC – immature/suppression | PD-L1+ intermediate monocytes – inhibition |
| Tim-3+ cDC – inhibition | Granulocytic (CD14- CD15+) MDSC | PD-1+ intermediate monocytes- activation/inhibition |
| Ki67+ cDC – proliferation | PD-L1+ gMDSC – inhibition | Non-classical monocytes – proinflammatory |
| 8. Total pDC | PD-1+ gMDSC – activation/inhibition | PD-L1+ non-classical monocytes – inhibition |
| PD-L1+ pDC – inhibition | CD16+ gMDSC – immature/suppression | PD-1+ non-classical monocytes- activation/inhibition |
| PD-1+ pDC – activation/inhibition | ||
| Tim-3+ pDC – inhibition | ||
| Ki67+ pDC – proliferation |
Ten parental phenotypes are identified as well as refined subsets of each relating to maturation and function. Expected function based on expression of specific markers within each subset is indicated.
cDC, conventional dendritic cells; CM, central memory; CTLA-4, cytotoxic T lymphocyte-associated protein-4; EM, effector memory; EMRA, terminally differentiated effector memory; gMDSCs, granulocytic myeloid-derived suppressor cells; ICOS, inducible T-cell costimulator; mMDSCs, monocytic MDSCs; NK, natural killer; PD-1, programmed cell death-1; pDC, plasmacytoid DC; PD-L1, programmed cell death ligand-1; Tim-3, T-cell immunoglobulin and mucin domain-3; Tregs, regulatory T cells.
Assay of immune cell subsets
Multiple types of assays are employed to characterize human immune cells.14–16 Investigators at the National Cancer Institute have employed a flow cytometry-based assay that requires 5×106 peripheral blood mononuclear cells (PBMCs), usually obtained from one tube of blood. The assay interrogates the multiple components of both the innate and adaptive immune systems seen in table 1. After processing, cells are frozen and stored so that analyses of a given individual’s PBMCs obtained at different times can be assayed under identical conditions. The materials and methods used in this assay have been previously described in detail.17 18 Multiple prior studies have employed the 123-subset assay19–26 (online supplemental table S1). However, additional immune cell subsets have recently been identified as having biologic functions. The most recent assay being employed now includes analyses of 158 immune cell subsets (table 1).
jitc-2020-002087supp001.pdf (118.8KB, pdf)
Age and immune subsets
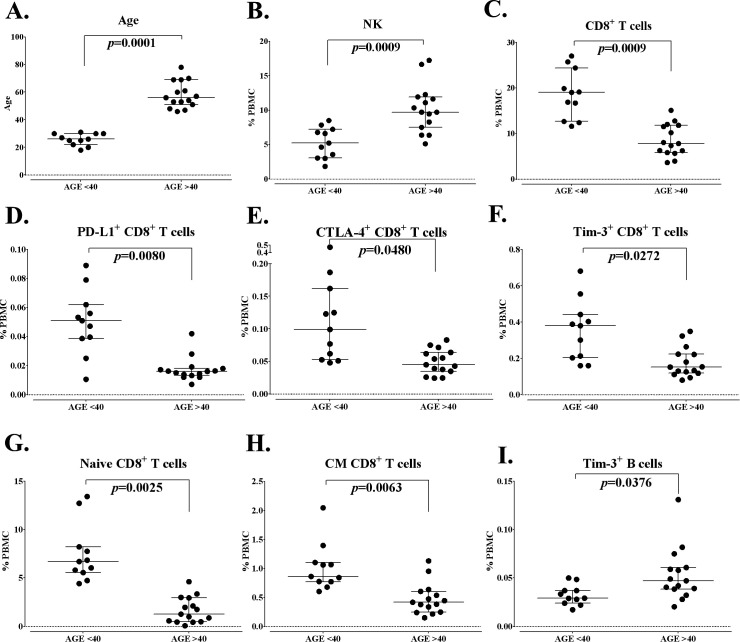
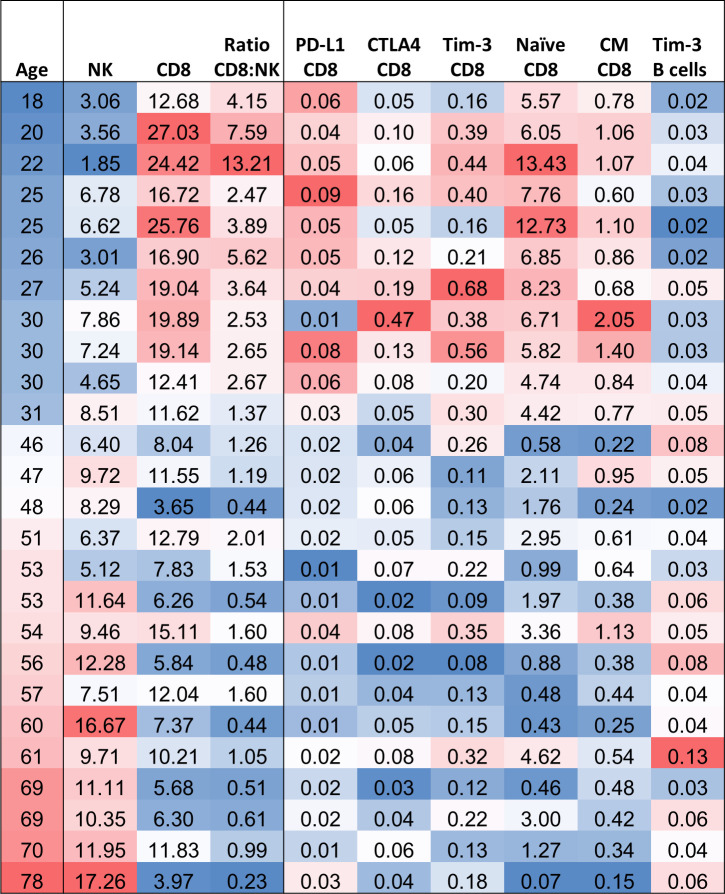
One observation that is emerging in the COVID-19 pandemic is differences in the severity of SARS-CoV-2 infection among different age groups. A prior study18 using the assay described defined differences in the immunome in apparently healthy individuals who are less than 40 years of age versus those older than 40 years; this arbitrary distinction was made because cancer incidence greatly increases in individuals over 40 years old. Figure 1A shows the ages of the two groups from which PBMCs were analyzed. This prior study18 showed higher levels of absolute lymphocyte counts (ALC) in individuals less than 40 years versus those older than 40 years. Figure 1B, C reveal that individuals older than 40 have higher levels of NK cells, but, in contrast, have lower levels of CD8+ T cells. This observation is extended when one analyzes more refined subsets, with examples of differences in individual subsets shown in figure 1D–I involving specific subtypes of CD8+ T cells and B cells. While most CD8+ refined subsets do not differ with age, it appears that apparently healthy individuals over 40 have lower levels of CD8+ T cells with potential suppressive properties (figure 1D–F). Figure 2 shows a heat map depicting the gradient in changes of immune cell subsets observed with age (18–78 years); as seen, NK cells increase with age, while CD8+ T cells decrease. This gradient is also seen with the different refined subsets of CD8+ T cells and B cells.
Figure 1.
Differences in standard parental immune cell types and refined subsets in healthy donors under and over the age of 40. (A) Healthy donors included in this analysis were separated as younger (age less than 40 years, n=11) and older (age greater than 40 years, n=15). (B, C) Standard parental immune cell types that were different between healthy donors under and over age 40. (D–I) Representative graphs are shown for notable refined subsets related to activation and maturation, with differences between healthy donors under and over the age of 40 indicated. Graphs display median frequency as a percentage of PBMCs with 25–75 percentiles. Differences were defined by an adjusted p<0.05, the median of groups showing a >50% difference, and a frequency above 0.01% of PBMCs. P value was calculated using the Mann-Whitney test and with Holm adjustment made for multiple comparisons using the number of standard immune cell types with a frequency above 0.01% of PBMCs (n=9). For refined subsets, Holm adjustment was made using the number of subsets within each standard subset with a frequency above 0.01% of PBMCs (n=29 for CD4+ T cells, 25 for CD8+ T cells, 5 for regulatory T cells (Tregs), 14 for NK cells, 3 for NKT cells, 4 for B cells, 2 for conventional dendritic cells (cDCs), 3 for plasmacytoid DCs (pDCs) and 15 for MDSCs). Figure adapted from Lepone.18 CTLA-4, cytotoxic T lymphocyte-associated protein-4; MDSCs, myeloid-derived suppressor cells; NK, natural killer; PBMCs, peripheral blood mononuclear cells; PD-L1, programmed cell death ligand-1.
Figure 2.
The influence of age on the cellular immunome. Heatmap of age and standard and refined subsets that were different between healthy doors under and over the age of 40. Red: higher frequency, Blue: lower frequency. CTLA-4, cytotoxic T lymphocyte-associated protein-4; NK, natural killer; PD-L1, programmed cell death ligand-1.
Severe SARS-CoV-2 infection is characterized by lymphopenia, with reduced CD8+ T cells associated with higher levels of ICU admission and decreased survival.27 Remaining CD8+ cells express higher levels of exhaustion markers such as PD-1, Tim-3, and CTLA-4 with increasing disease severity.28 29 The lower levels of CD8+ T cells seen in older individuals along with the increased prevalence of suppressive markers echo the immune subset picture seen in severe COVID-19. This may begin to explain the older population’s higher risk of morbidity and mortality with this disease: the decreased number and effector function of CD8+ subsets at baseline would suggest impaired antiviral immunity and decreased capacity for viral clearance. As for the higher levels of NK cells in older age groups, one might expect that this would support a more robust antiviral response in COVID-19. However, NK cells are also negatively impacted by the lymphopenia seen with COVID-19, and preliminary data suggest that remaining cells either develop an exhausted phenotype or traffic to the lungs where they may contribute to inflammatory damage.30 The more severe illness seen in older patients, including the potential for greater inflammatory lung injury, may be due to one or more factors, including higher levels of NK cells, macrophages or a TH17-driven phenomenon. This will clearly be the focus of future investigations. There are too few subjects in the presented data sets to draw reliable conclusions or correlations from a racial subgroup analysis. However, this would be a critical area for future study in larger populations.
Immunome differences between cancer patients and apparently healthy donors
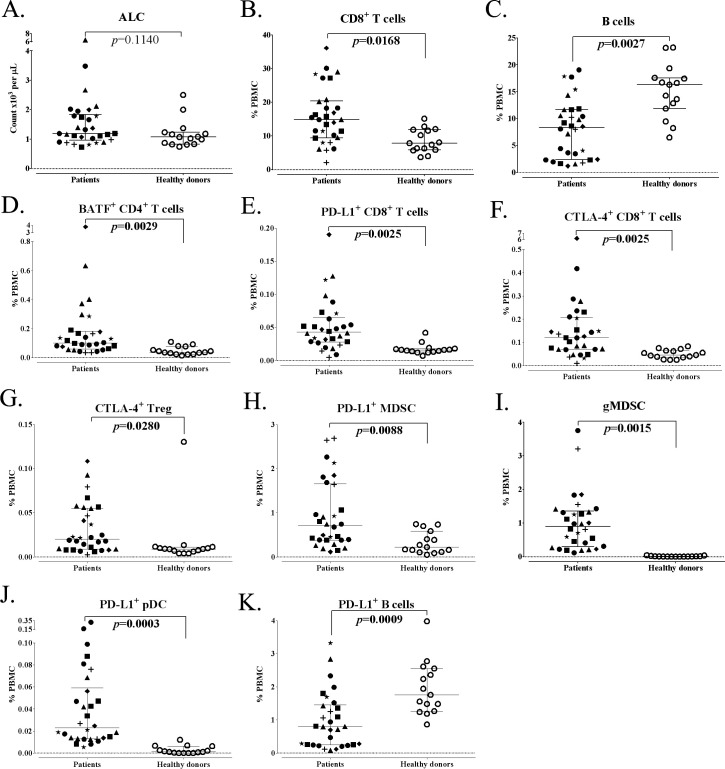
Studies have also been conducted18 to see if differences exist between apparently healthy donors and age-matched patients with a range of different cancers (figure 3A; online supplemental tables S2 and S3). While there was no difference in ALC between the two age-matched groups, clear differences were seen in CD8+ T cells and B cells (figure 3B, C), but not in the other major parental subsets. More profound differences were seen with the analyses of some, but not all, refined CD8+ subsets; in particular, age-matched cancer patients had much higher levels of CD8+ T cells with a suppressive phenotype (figure 3E, F). Notably, age-matched cancer patients had much higher levels of Tregs with a suppressive phenotype (figure 3G), and higher levels of specific MDSCs (figure 3H, I). Changes were also noted in specific, but not all, refined B cell subsets; B cells expressing PD-L1 were lower in age-matched cancer patients than in healthy donors (figure 3K). The assays described here could thus be employed to study any adverse effects that chemotherapy, steroids, or other agents may have on cancer patients being exposed to SARS-CoV-2. This assay has also been used to study the effects of various immune modulators and other so-called ‘non-immune-based therapeutics’ on the immune system of cancer patients in clinical trials. It should be pointed out that analyses of specific immune cells in the blood compartment may not reflect their presence in different tissue compartments such as lymph nodes and sites of viral infection, since immune responses can shift from one compartment to another. The timing of such shifts should also be considered. In this case, analyses of the peripheral immunome, with the ability to obtain blood samples at multiple time points during infection and convalescence, may help in the understanding of the pathogenesis of SARS-CoV-2. It is thus suggested that this and similar assays be employed in clinical studies of patients with COVID-19 to help determine (a) which individuals or groups are most susceptible to SARS-CoV-2 infection, (b) which individuals will have inapparent or mild versus severe disease, (c) the effects of vaccines or therapeutic agents on the immune system of individuals/patients, (d) the durability of response to vaccination and to whether there is a need for revaccination, (e) the relationship of antibody response versus cellular immune response post-SARS-CoV-2 vaccination and/or convalescence postinfection, and (f) the mechanisms involved in SARS-CoV-2 viral clearance and of convalescence of severe infections.
Figure 3.
Differences in standard parental immune cell types and refined subsets in age-matched advanced cancer patients and healthy donors. Patients with advanced cancer (n=30) and healthy donors (n=15) included in this analysis were age-matched above age 40. (A) ALC of cancer patients and healthy donors. (B, C) Standard parental immune cell types that were different between cancer patients and healthy donors. (D–K) Representative graphs are shown for notable refined subsets with differences between cancer patients and healthy donors. Graphs display median ALC or median frequency as a percentage of PBMCs with 25–75 percentiles. Cancer type is indicated by shape (square: GI (anal, colon, esophageal); n=6; triangle: pancreatic, n=6; star: breast, n=3; plus sign: mesothelioma, n=3; diamond: renal cell, n=3; closed circle: other (adrenocortical, chordoma, lung, medullary thyroid, neuroendocrine, ovarian, prostate, and spindle cell), n=9; open circle: healthy donors, n=15). Differences were defined by an adjusted p<0.05, the median of groups showing a >50% difference, and a frequency above 0.01% for PBMCs. P value was calculated using the Mann-Whitney test and with Holm adjustment made for multiple comparisons using the number of standard immune cell types with a frequency above 0.01% of PBMCs (n=9). For refined subsets, Holm adjustment was made using the number of subsets within each standard subset with a frequency above 0.01% for PBMCs (n=29 for CD4+ T cells, 25 for CD8+ T cells, 5 for Tregs, 14 for NK cells, 3 for NKT cells, 4 for B cells, 2 for cDCs, 3 for pDCs and 15 for MDSCs). Figure adapted from Lepone.18 ALC, absolute lymphocyte count; BATF, basic leucine zipper ATF-like transcription factor; cDC, conventional dendritic cells; CTLA-4, cytotoxic T lymphocyte-associated protein-4; GI, gastrointestinal; gMDSCs, granulocytic myeloid-derived suppressor cells; NK, natural killer; PBMCs, peripheral blood mononuclear cells; pDC, plasmacytoid DC; PD-L1, programmed cell death ligand-1; Tregs, regulatory T cells.
jitc-2020-002087supp002.pdf (12.8KB, pdf)
jitc-2020-002087supp003.pdf (86.4KB, pdf)
Soluble factors
The mainstay of analyses of immune responses of patients with COVID-19 will be detection of SARS-CoV-2 virus or viral RNA, and the detection of antiviral antibody responses. Other soluble factors that should be analyzed in a comprehensive manner are the spectrum of cytokines and chemokines from sera or plasma. There are other soluble factors, however, detected in sera or plasma that may also aid in defining immune responses to SARS-CoV-2; these include the detection of soluble (s) CD27, sCD40L, sPD-L1, sPD-1 and soluble granzyme. These assays are being used to monitor patients’ responses in various cancer immunotherapy trials. sCD27, for example, is preferentially derived from activated CD4+ T cells, and greater levels are seen in apparently healthy individuals versus cancer patients.21 31 Some studies have shown that immunotherapy can increase sCD27 in sera.31 sCD40L is a functional trimer that is shed from activated T lymphocytes and more likely from platelets, and has also been investigated in autoimmune disease.32 Evidence has been provided that higher levels of sCD40L are seen in some cancer patients and that this may have an immunosuppressive effect.33 Studies have also shown that the ratio of sCD27:sCD40L may be indicative of a therapeutic benefit.17
These soluble factors are already proving to be important biomarkers in COVID-19, reflecting the need to develop effective antiviral immunity without tipping the balance towards toxic inflammation. An investigation of soluble checkpoints in patients with confirmed COVID-19 infection found that increased baseline levels of sIDO, s4-1BB, sTIM-3, and sCD27 were associated with higher disease severity rates and longer ICU stays.34 This may reflect complex dysregulation of the T-cell responses needed to overcome SARS-CoV-2 infection through a combination of hyperactivation followed by exhaustion.28 A study of inflammatory cytokines in over 1400 hospitalized patients found initial levels of both IL-6 and TNF-α to be independent predictors of disease severity and mortality.35 While elevated sCD40L may be associated with immunosuppression and possible impairment of antiviral responses, CD40L is also critical in humoral immunity.36 In that context, lower levels may negatively impact antiviral antibody responses. Further investigation of these soluble factors on both humoral and cellular immune responses may provide further predictive biomarkers as well as insights for vaccine and therapeutic development.
Effect of steroids on immune subsets
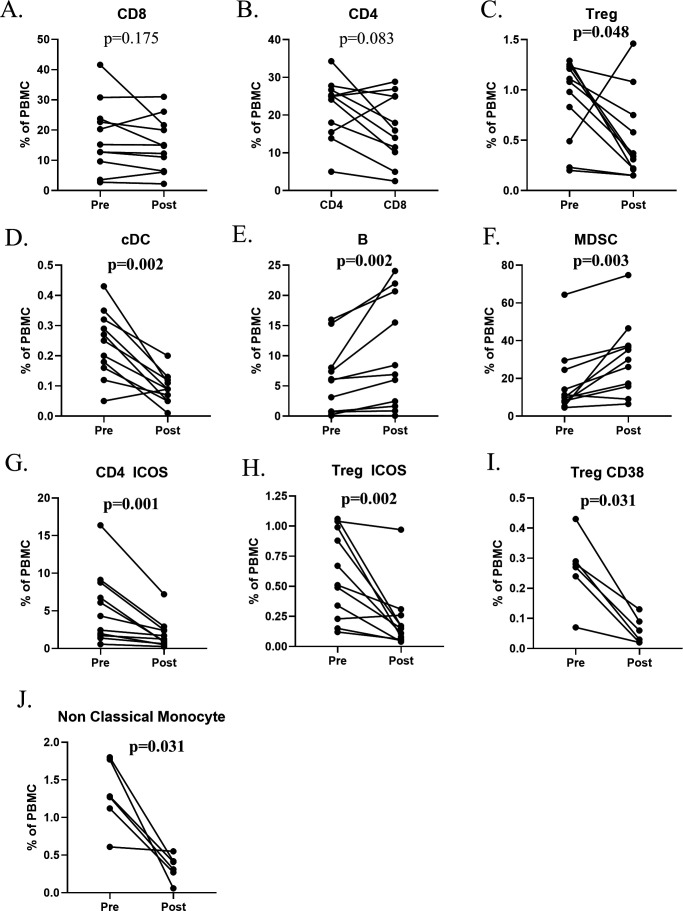
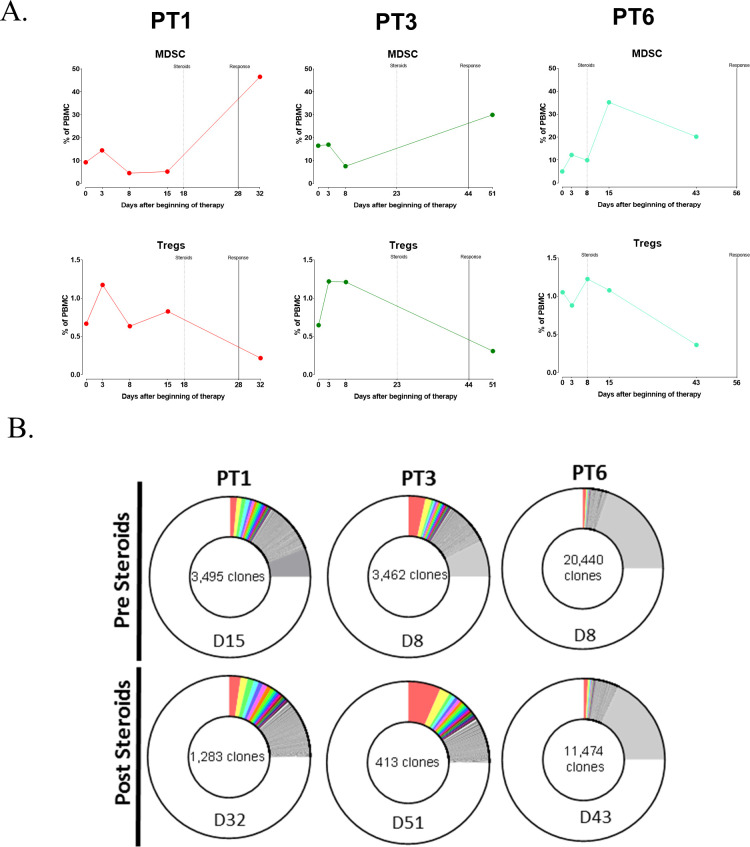
One of the consequences of cancer immunotherapy, especially the use of checkpoint inhibitor MAbs, is the induction of immune-related adverse events in some patients. These patients are often treated with corticosteroids such as dexamethasone or prednisone. Naïve lymphocytes are typically most susceptible to glucocorticoid effects with increases in the neutrophil to lymphocyte ratio (NLR) caused by both a rapid decrease in lymphocytes and a later sustained increase in neutrophils.37 Figure 4 shows the effects on various immune subsets of cancer patients treated with corticosteroids. As seen in figure 4A, there is little effect on CD8+ T cells, but profound effects involving decreases in CD4+ T cells and Tregs, as well as on cDCs in most patients (figure 4B–D); on the other hand, one observed an increase in B cells and MDSCs poststeroid treatment (figure 4E, F). Examples of changes seen in the more refined immune subsets are seen in figure 4G–J. This effect of corticosteroids is further exemplified in three patients receiving the anti-PDL1 MAb avelumab. The increase in MDSCs and simultaneous decrease in Tregs is seen poststeroid treatment (figure 5A). Additional changes in T-cell diversity are also observed poststeroid treatment with increases in T-cell clonality seen in all three patients evaluated (figure 5B).
Figure 4.
Change in standard parental immune cell types and refined subsets after corticosteroids. Cancer patients (n=11) enrolled in immunotherapy trials received moderate- to high-dose corticosteroids (prednisone, n=6; methylprednisolone and prednisone, (n=4); or dexamethasone, methylprednisolone, and prednisone, n=1) for the development of immune-related adverse events. (A–F) Changes in standard parental immune cell types after corticosteroids. (G–J) Representative graphs are shown for notable refined subsets that changed with corticosteroids. Significant changes were defined by a p value <0.05, a median difference poststeroid versus presteroids >0.05% of PBMCs, and at least half of evaluated patients having a >25% change. The panels used for refined subsets reflecting maturation/functional status of subsets were slightly different for the various immunotherapy trials, so certain subsets were not tested in all patients (n=6 for Treg CD38 and non-classical monocytes). MDSCs, myeloid-derived suppressor cells; PBMCs, peripheral blood mononuclear cells; Tregs, regulatory T cells.
Figure 5.
The effect of steroids on peripheral immune subsets and TCR diversity in patients with thymoma and thymic epithelial carcinoma treated with avelumab and receiving corticosteroids for the treatment of developed immune-related adverse events. (A) An increase in MDSCs and decrease in Tregs was observed in clinical responders who developed autoimmune adverse events and were treated with corticosteroids. Dashed line denotes timing of steroids and solid line indicates time of clinical response. (B) TCR diversity (measured by the metric of repertoire size) was reduced after corticosteroids; values indicate the number of individual clonotypes comprising the top 25th percentile by ranked molecule count after sorting by abundance. The day (D) PBMCs were assessed for TCR diversity (with respect to avelumab treatment) before and after corticosteroids is indicated. The different colors are used to represent individual clonotypes. Figure adapted from Rajan.24 MDSCs, myeloid-derived suppressor cells; PBMCs, peripheral blood mononuclear cells; TCR, T-cell receptor; Tregs, regulatory T cells.
These lymphocyte subset changes induced by corticosteroids may also be relevant to patients with COVID-19. Patients with COVID-19 who have an elevated NLR have a worse prognosis.38 While glucocorticoids can improve survival in patients who require respiratory support due to the severe inflammation caused by the SARS-CoV2 virus, their use earlier on in viral infections may be counterproductive.39–41 In addition, CD4+ T cell help is required for antibody responses to coronavirus infections; thus the decreases in CD4+ T cells caused by glucocorticoid use could be counterproductive early on in the disease. Furthermore, high doses of corticosteroids are associated with prolonged viral shedding and decreased clearance of SARS-CoV2.42
Perspective
The multiple compartments of the adaptive and innate cellular immunome should be interrogated to help define which patients may benefit most from preventive SARS-CoV-2 vaccines and in determining which factors better define the severity of both primary and secondary infections with SARS-CoV-2. Thus, in addition to the development of preventive vaccines primarily designed to induce antibodies to the SARS-CoV-2 spike protein, emphasis should be placed on vaccines designed to also enhance cellular immune responses; these cellular immune responses should both be directed to the spike protein and to internal components of the SARS-CoV-2 virion. The nucleocapsid N protein of SARS-CoV-2 may well be more highly conserved than the spike protein; this may be of importance in limiting the severity of primary and secondary infections if SARS-CoV-2 variants in the spike proteins arise. The cell-based and soluble factor assays described here may also be employed in the analysis of the effects of various antivirals and other potential anti-SARS-CoV-2 therapeutics on the cellular immunome, and in the study of the pathogenesis of SARS-CoV-2 infections.
Acknowledgments
The authors also thank Debra Weingarten for her editorial assistance in the preparation of this manuscript.
Footnotes
Twitter: @gulleyj1
RND and JLM contributed equally.
JLG and JS contributed equally.
Contributors: JS and JLG contributed to the conception of the manuscript. RND, JLM, MG, NJT, Y-TT, JS and JLG all contributed to the content and writing of the manuscript. All authors read and approved the final manuscript.
Funding: This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 2020;41:355–9. 10.1016/j.it.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walls AC, Park Y-J, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181:281–92. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020;181:1489–501. e15. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun J, Loyal L, Frentsch M, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 2020;587:270–4. 10.1038/s41586-020-2598-9 [DOI] [PubMed] [Google Scholar]
- 5.Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 2020;5. 10.1126/sciimmunol.abd2071. [Epub ahead of print: 26 06 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020;584:457–62. 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 7.Ferretti AP, Kula T, Wang Y, et al. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity 2020;53:1095–107. 10.1016/j.immuni.2020.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kared H, Redd AD, Bloch EM, et al. CD8+ T cell responses in convalescent COVID-19 individuals target epitopes from the entire SARS-CoV-2 proteome and show kinetics of early differentiation. bioRxiv 2020. 10.1101/2020.10.08.330688. [Epub ahead of print: 08 Oct 2020]. [DOI] [Google Scholar]
- 9.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020;183:158–68. e14. 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson EJ, Rouphael NG, Widge AT, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med 2020;383:2427–38. 10.1056/NEJMoa2028436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folegatti PM, Ewer KJ, Aley PK, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020;396:467–78. 10.1016/S0140-6736(20)31604-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin U, Muik A, Derhovanessian E, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020;586:594–9. 10.1038/s41586-020-2814-7 [DOI] [PubMed] [Google Scholar]
- 13.Rice A, Verma M, Shin A. A next generation bivalent human Ad5 COVID-19 vaccine delivering both spike and nucleocapsid antigens elicits Th1 dominant CD4+, CD8+ T-cell and neutralizing antibody responses. bioRxiv 2020:2020.07.29.227595. [Google Scholar]
- 14.Hoek KL, Samir P, Howard LM, et al. A cell-based systems biology assessment of human blood to monitor immune responses after influenza vaccination. PLoS One 2015;10:e0118528. 10.1371/journal.pone.0118528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivison S, Malek M, Garcia RV, et al. A standardized immune phenotyping and automated data analysis platform for multicenter biomarker studies. JCI Insight 2018;3:e121867. 10.1172/jci.insight.121867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leipold MD, Newell EW, Maecker HT. Multiparameter phenotyping of human PBMCs using mass cytometry. Methods Mol Biol 2015;1343:81–95. 10.1007/978-1-4939-2963-4_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donahue RN, Lepone LM, Grenga I, et al. Analyses of the peripheral immunome following multiple administrations of avelumab, a human IgG1 anti-PD-L1 monoclonal antibody. J Immunother Cancer 2017;5:20. 10.1186/s40425-017-0220-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lepone LM, Donahue RN, Grenga I, et al. Analyses of 123 peripheral human immune cell subsets: defining differences with age and between healthy donors and cancer patients not detected in analysis of standard immune cell types. J Circ Biomark 2016;5:5. 10.5772/62322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilusic M, Heery CR, Collins JM, et al. Phase I trial of HuMax-IL8 (BMS-986253), an anti-IL-8 monoclonal antibody, in patients with metastatic or unresectable solid tumors. J Immunother Cancer 2019;7:240. 10.1186/s40425-019-0706-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farsaci B, Donahue RN, Grenga I, et al. Analyses of pretherapy peripheral immunoscore and response to vaccine therapy. Cancer Immunol Res 2016;4:755–65. 10.1158/2326-6066.CIR-16-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heery CR, Ibrahim NK, Arlen PM, et al. Docetaxel alone or in combination with a therapeutic cancer vaccine (PANVAC) in patients with metastatic breast cancer: a randomized clinical trial. JAMA Oncol 2015;1:1087–95. 10.1001/jamaoncol.2015.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heery CR, Madan RA, Stein MN, et al. Samarium-153-Edtmp (Quadramet®) with or without vaccine in metastatic castration-resistant prostate cancer: a randomized phase 2 trial. Oncotarget 2016;7:69014–23. 10.18632/oncotarget.10883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heery CR, O'Sullivan-Coyne G, Madan RA, et al. Avelumab for metastatic or locally advanced previously treated solid tumours (javelin solid tumor): a phase 1A, multicohort, dose-escalation trial. Lancet Oncol 2017;18:587–98. 10.1016/S1470-2045(17)30239-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajan A, Heery CR, Thomas A, et al. Efficacy and tolerability of anti-programmed death-ligand 1 (PD-L1) antibody (Avelumab) treatment in advanced thymoma. J Immunother Cancer 2019;7:269. 10.1186/s40425-019-0723-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss J, Heery CR, Kim JW, et al. First-In-Human phase I trial of a tumor-targeted cytokine (NHS-IL12) in subjects with metastatic solid tumors. Clin Cancer Res 2019;25:99–109. 10.1158/1078-0432.CCR-18-1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss J, Heery CR, Schlom J, et al. Phase I trial of M7824 (MSB0011359C), a bifunctional fusion protein targeting PD-L1 and TGFβ, in advanced solid tumors. Clin Cancer Res 2018;24:1287–95. 10.1158/1078-0432.CCR-17-2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 2020;11. 10.3389/fimmu.2020.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol 2020;20:529–36. 10.1038/s41577-020-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng H-Y, Zhang M, Yang C-X, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020;17:541–3. 10.1038/s41423-020-0401-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masselli E, Vaccarezza M, Carubbi C, et al. Nk cells: a double edge sword against SARS-CoV-2. Adv Biol Regul 2020;77:100737. 10.1016/j.jbior.2020.100737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Jochems C, Anderson AM, et al. Soluble CD27-pool in humans may contribute to T cell activation and tumor immunity. J Immunol 2013;190:6250–8. 10.4049/jimmunol.1300022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlom J, Jochems C, Gulley JL, et al. The role of soluble CD40L in immunosuppression. Oncoimmunology 2013;2:e22546. 10.4161/onci.22546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Jochems C, Talaie T, et al. Elevated serum soluble CD40 ligand in cancer patients may play an immunosuppressive role. Blood 2012;120:3030–8. 10.1182/blood-2012-05-427799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong Y, Wang Y, Wu X, et al. Storm of soluble immune checkpoints associated with disease severity of COVID-19. Signal Transduct Target Ther 2020;5:192. 10.1038/s41392-020-00308-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Valle DM, Kim-Schulze S, Huang H-H, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020;26:1636–43. 10.1038/s41591-020-1051-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol 2000;67:2–17. 10.1002/jlb.67.1.2 [DOI] [PubMed] [Google Scholar]
- 37.Marté JL, Toney NJ, Cordes L, et al. Early changes in immune cell subsets with corticosteroids in patients with solid tumors: implications for COVID-19 management. J Immunother Cancer 2020;8:e001019. 10.1136/jitc-2020-001019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020;323:1061–9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brun-Buisson C, Richard J-CM, Mercat A, et al. Early corticosteroids in severe influenza A/H1N1 pneumonia and acute respiratory distress syndrome. Am J Respir Crit Care Med 2011;183:1200–6. 10.1164/rccm.201101-0135OC [DOI] [PubMed] [Google Scholar]
- 40.Horby P, Mafham M, Linsell L. Effect of hydroxychloroquine in hospitalized patients with COVID-19: preliminary results from a multi-centre, randomized, controlled trial. medRxiv 2020:2020.07.15.20151852. [Google Scholar]
- 41.Yang J-W, Fan L-C, Miao X-Y, et al. Corticosteroids for the treatment of human infection with influenza virus: a systematic review and meta-analysis. Clin Microbiol Infect 2015;21:956–63. 10.1016/j.cmi.2015.06.022 [DOI] [PubMed] [Google Scholar]
- 42.Li S, Hu Z, Song X. High-dose but not low-dose corticosteroids potentially delay viral shedding of patients with COVID-19. Clin Infect Dis 2020:ciaa829. 10.1093/cid/ciaa829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002087supp001.pdf (118.8KB, pdf)
jitc-2020-002087supp002.pdf (12.8KB, pdf)
jitc-2020-002087supp003.pdf (86.4KB, pdf)