Abstract
The aim of the present study was to address method-dependent implications during the quantification of viable Campylobacter coli cells on meat over time. Traditional colony counting on selective and non-selective culture media along with an optimized viability real-time PCR utilizing propidium monoazide-quantitative PCR (PMA-qPCR), spheroplast formation and an internal sample process control (ISPC), were comparatively evaluated for monitoring the survival of C. coli on fresh lamb meat during refrigeration storage under normal atmospheric conditions. On day zero of three independent experiments, lamb meat pieces were artificially inoculated with C. coli and then stored under refrigeration for up to 8 days. Three meat samples were tested on different days and the mean counts were determined per quantification method. An overall reduction of the viable C. coli on lamb meat was observed regardless of the applied quantification scheme, but the rate of reduction followed a method-dependent pattern, the highest being observed for colony counting on modified charcoal cefoperazone deoxycholate agar (mCCDA). Univariate ANOVA indicated that the mean counts of viable C. coli using PMA-qPCR were significantly higher compared to Columbia blood agar (CBA) plating (0.32 log10 cell equivalents, p = 0.015) and significantly lower when mCCDA was compared to CBA plating (0.88 log10 CFU, p < 0.001), indicating that selective culture on mCCDA largely underestimated the number of culturable cells during the course of meat storage. PMA-qPCR outperformed the classical colony counting in terms of quantifying both the culturable and viable but non-culturable (VBNC) C. coli cells, which were generated over time on meat and are potentially infectious and equally important from a public health perspective as their culturable counterparts.
Keywords: Campylobacter, meat, viability PCR, propidium monoazide, viable but non-culturable
Introduction
Campylobacter spp. are ubiquitous in nature and successfully colonize the gastrointestinal tract of warm-blooded animals including food-producing animals (cattle, sheep, pigs and poultry), usually without causing symptoms. Slaughter has been recognized as a crucial stage for fresh meat contamination by thermophilic Campylobacter spp. that persists across the meat production chain and represents a public health risk (Lazou et al., 2014a,b; Shange et al., 2019). Campylobacteriosis remains the most commonly reported zoonotic gastrointestinal illness in the European Union (EU) since 2005, commonly related to domestically acquired infection via the consumption and handling of raw or undercooked foods of animal origin, particularly poultry meat (EFSA and ECDC, 2019). Human cases are attributed mainly to Campylobacter jejuni (C. jejuni) followed by Campylobacter coli, however, subspecies information is provided only for approximately half of the confirmed cases in the EU since these two subspecies are rarely differentiated in routine diagnostics, although risk factors for human infection may differ between them (Gillespie et al., 2002; Kärenlampi et al., 2007; Roux et al., 2013; EFSA and ECDC, 2019). Nowadays, prevalence, virulence potential and health burden of C. coli are regarded substantial and much greater than previously thought (Du et al., 2018; Gomes et al., 2018); it is being increasingly isolated from various samples of animal origin, including meat and offal of small ruminants (Kärenlampi et al., 2007; Løvdal et al., 2011; Voidarou et al., 2011; Nobile et al., 2013; Lazou et al., 2014a,b; Pedonese et al., 2017; Di Giannatale et al., 2019), whereas, a tendency towards increased antimicrobial resistance of its isolates has been recently observed compared to C. jejuni (Voidarou et al., 2011; Du et al., 2018; García-Sánchez et al., 2018; Di Giannatale et al., 2019). Data regarding the survival of C. coli cells on fresh meat, other than poultry, during refrigeration in the domestic environment are scarce, although cross-contamination incidents are likely to occur, since consumer awareness regarding the risk of food contamination by Campylobacter in the household remains deficient (Hansson et al., 2018).
A laborious and time-consuming procedure lasting several days is required for the in vitro detection, enumeration and biochemical confirmation of Campylobacter colony-forming units (CFU) according to the ISO 10272-2 standard colony-count technique, which is the only universally accepted, adequately validated, and standardized method (Elizaquível et al., 2014; Zeng et al., 2016; ISO 10272-2:2017, 2017). Another major drawback for the routine implementation and universal acceptance of classical plate counting is the inability to detect viable but non-culturable (VBNC) cells. The VBNC state is regarded as an adaptive response of bacterial cells to stressful environmental conditions, which render them unable to proliferate on typical culture media, including enrichment media designed to resuscitate injured cells (Baffone et al., 2006; Oliver, 2010; Oh et al., 2015). Campylobacters have been reported to enter the VBNC state when exposed to low temperatures and atmospheric oxygen (e.g., chilling chambers); however, VBNC bacteria remain capable of switching to the infectious state once inside the human host (Oliver, 2010; Chaisowwong et al., 2012; Stingl et al., 2012; Li et al., 2014; Oh et al., 2015). Although the reliable detection of both culturable and VBNC cells is equally important in view of public health inferences, culturable Campylobacter bacteria in any given sample can be detected and quantified by traditional plate counting in contrast to their VBNC counterparts that will remain undetected (Chaisowwong et al., 2012; Elizaquível et al., 2014; Krüger et al., 2014; Zeng et al., 2016). Nevertheless, the rapid and reliable detection and quantification of viable Campylobacter directly in samples complies with the modern requirements of research and food industry, especially regarding products with limited shelf life, such as fresh meat, but the utilization of culture-based methods is hampered by the aforementioned shortcomings (Elizaquível et al., 2014; Krüger et al., 2014; Duarte et al., 2015; Papić et al., 2017).
Real-time PCR (quantitative PCR; qPCR) could serve as an alternative method for the rapid, high throughput, and specific detection of foodborne pathogens in general and Campylobacter in particular in food samples, but its inability to differentiate between the amplified DNA of viable and dead cells results in indefinite information regarding the pertained public health risk (Fittipaldi et al., 2012; Elizaquível et al., 2014; Zeng et al., 2016). In order to overcome this impediment, viability PCR (v-PCR) utilizing qPCR and nucleic acid intercalating dyes has been developed to provide rapid quantification combined with viability information (Nocker et al., 2006). This technique is based on the viability criterion of cell membrane integrity, which operates as a physical barrier against the entrance of DNA-intercalating dyes, principally propidium monoazide (PMA), into live bacterial cells (both culturable and nonculturable). The theoretical mode of action includes the selective entrance of PMA into dead bacterial cells via their compromised-membranes and the DNA cleavage upon photoactivation that prohibits its subsequent amplification during qPCR (Nocker et al., 2006; Wagner et al., 2008; Varma et al., 2009). However, applications of v-qPCR protocols with PMA (PMA-qPCR) on meat samples have encountered various challenges and limitations attributed to a rather conditional and not absolute suppression of qPCR signals originating from dead Campylobacter bacteria (false-positive signals) due to a complex set of parameters including experimental, target and sample features (Nocker et al., 2006; Pacholewicz et al., 2013; Krüger et al., 2014; Duarte et al., 2015; Vondrakova et al., 2018). The development of a PMA-qPCR assay utilizing spheroplast formation as a pretreatment for enhancing the selective entrance of PMA to dead cells and an internal sample process control (ISPC), unique for any given sample, that could address false-positive signals originating from dead cells, has only recently been achieved using C. coli as a bacterial model (Lazou et al., 2019).
The aim of the present study was to address method-dependent implications during the quantification of viable C. coli cells on meat over time. The aforementioned PMA-qPCR utilizing spheroplast formation and ISPC, along with traditional colony counting on selective and non-selective solid media were comparatively evaluated for monitoring the survival of C. coli on fresh lamb meat during refrigeration storage under normal atmospheric conditions.
Materials and Methods
General Experimental Scheme
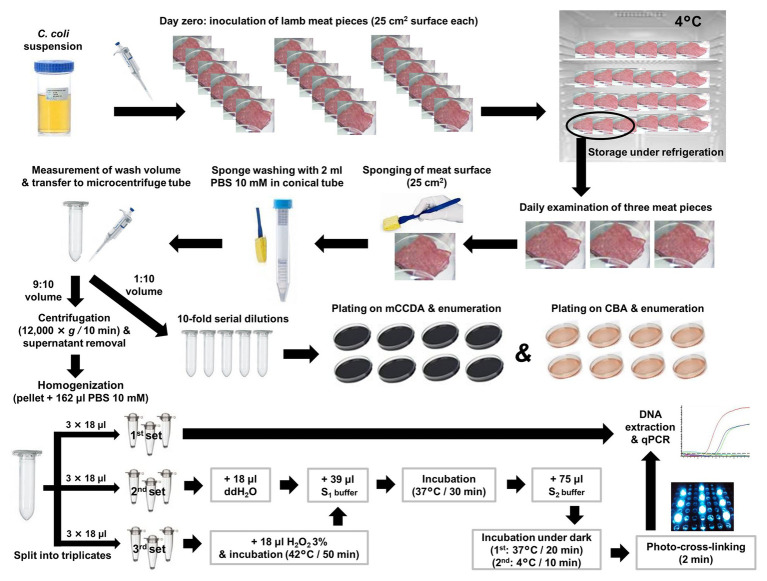
Three independent experiments were performed at different time frames, following the general experimental scheme as illustrated in Figure 1. On day 0 of each experiment, individual lamb meat pieces were inoculated with a defined C. coli population and then placed in a refrigerator set at 4°C. Temperature was recorded daily and three individual meat pieces were examined at each time-point, in order to quantify the average viable C. coli count by plating on two culture media and by using PMA-qPCR. In the first experiment, the average viable C. coli count of three samples was quantified daily for a week (in total, 21 meat pieces), at the end of which meat spoilage was visually evident. Based on the results of the first experiment, viable C. coli counts were quantified every other day from day 0 to 8 during the second and third experiment (in total, 15 meat pieces per experiment).
Figure 1.
General experimental scheme for the quantification of culturable, total, and viable C. coli cells on artificially inoculated lamb meat pieces using plate counting, qPCR (control) and PMA-qPCR, respectively. S1 buffer: 10 mM Phosphate Buffer Saline (PBS; pH 7.0); 0.5 M Sucrose; 1.0 mg/ml Lysozyme; 50 mM EDTA. S2 buffer: 10 mM PBS (pH 7.0); 50 mM EDTA; 100 μM PMA. ddH2O: double-distilled water.
Preparation of Campylobacter coli Suspension
On each experimental occasion, C. coli ATCC® 43478™ (LGC Standards GmbH, Wesel, Germany), previously stored at −80°C in nutrient broth No. 2 (Oxoid, Basingstoke, England) supplemented with 5% lysed horse blood (Oxoid) and 20% glycerol (BDH VWR, Lutterworth, England), was recovered on Columbia blood agar (CBA; bioMérieux, Marcy-l’Étoile, France), then isolated on modified charcoal cefoperazone deoxycholate agar (CCDA; Oxoid) and subcultured once more on CBA. Plate incubation was always performed at 41.5 ± 1°C for 24 h under microaerobic conditions (GENbox microaer and GENbox jar, bioMérieux). For the generation of pure C. coli suspensions, one pure colony from CBA was inoculated in 100 ml of cation-adjusted Mueller-Hinton broth (CAMHB; BBL™ BD, Franklin Lakes, NJ) following incubation at 37 ± 1°C for 20 ± 2 h under microaerobic conditions. The growth state of the bacterial culture was approximately estimated at the end of the incubation by measuring the optical density (OD) at 600 nm using a spectrophotometer (BioPhotometer®; Eppendorf, Hamburg, Germany). The estimated count was confirmed by subjecting three aliquots of 100 ml to qPCR, as described below, and by performing 10-fold serial dilutions in buffered peptone water (BPW, LabM, United Kingdom) followed by spread plating on double CBA plates.
Artificial Inoculation of Lamb Meat and Sampling
For each experiment, a deboned lamb leg was purchased from the retail market and tested for the presence of C. coli as follows: a representative sample of 25 g of meat was homogenized in 225 ml of BPW; subsequently, five aliquots of 1 ml each were subjected to DNA extraction and qPCR. Once the C. coli-free status of the lamb meat was confirmed, meat pieces (25 cm2 total surface) were generated (n = 21 in the first experiment; n = 15 in the second and third experiments) by using sterile lancet and forceps and placed individually in sterile petri dishes, without applying any method of decontamination of the autochthonous microbiota. The samples for each experiment were inoculated on day 0 by uniformly applying 100 μl of a pure C. coli suspension in CAMHB (as described previously) on their surface, and then stored at 4°C. The first three meat samples were tested following 4 ± 1 h of storage at 4°C (allowed time for adherence of the bacterial cells to the meat surface). A sterile cellulose sponge (Whirl-Pak® Speci-Sponge®, Nasco, NY, United States), cut aseptically (0.5 cm × 1.0 cm) and pre-moistened with phosphate-buffered saline (PBS) 10 mM (Sigma-Aldrich, Steinheim, Germany), was used to sample each meat surface (Figure 1) by 10 horizontal, 10 vertical, and 10 diagonal swabbings. The sponge was then transferred in a conical centrifuge tube (15 ml, Falcon®, NY, United States) containing 2 ml of PBS 10 mM and homogenized for 60 s, using a vortex shaker (IKA®-Werke GmbH & Co. KG, Staufen, Germany). Subsequently, the sponge was drained by squeezing it against the tube wall and discarded. The volume of the remaining wash suspension was determined by a calibrated adjustable micropipette (Eppendorf Research® plus G, variable 100–1000 μl, NY, United States) while being transferred to a microcentrifuge tube (2.0 ml, Kisker Biotech GmbH & Co, Steinfurt, Germany). One tenth of the wash suspension was used for 10-fold serial dilutions and culture-based enumeration, whereas the remaining quantity (nine parts out of 10) was subjected to qPCR-based quantification (Figure 1).
DNA Extraction and qPCR Quantification
After centrifugation of the 9:10 volume of the wash suspension (12,000 × g for 10 min), the supernatant containing any extracellular DNA was discarded, and the pelleted bacterial cells were obtained in 162 μl PBS 10 mM. This suspension was then homogenized and nine aliquots (18 μl each) were generated and transferred to equal number of transparent PCR tubes (0.2 ml; Kisker Biotech GmbH & Co). The treatments of an optimized PMA-qPCR utilizing spheroplast formation (induced by lysozyme and EDTA) and ISPC, as described by Lazou et al. (2019), were applied. In brief, the first set of thee aliquots was directly subjected to DNA extraction and qPCR in order to quantify the total C. coli cell equivalents (quantification controls). The second set was subjected to spheroplast formation and PMA treatment, and the third set underwent inactivation by 1.5% hydrogen peroxide (H2O2) at 42°C for 50 min, followed by spheroplast formation and PMA treatment. The final concentration of EDTA in each aliquot subjected to spheroplasting was 25 mM. All aliquots were finally subjected to DNA extraction and qPCR (Figure 1). A protocol previously described and evaluated for Campylobacter (Lazou et al., 2014b), was used for the extraction of genomic DNA. In brief, each aliquot was mixed with 100 μl of “Lysis buffer I” (50 mM Tris-HCl, 50 mM EDTA, 4 M Guanidinium hydrochloride-GuHCl, 10 mM CaCl2, 1% Triton X-100, and 2% N-lauroyl-sarcosine, pH 7.5) and 25 μl of proteinase K (0.56 mg; New England Biolabs, Ipswich, MA, United States), and mixtures were incubated at 56°C for 1 h. Following this step, 250 μl of “Lysis buffer II” (50 mM Tris-HCl, 25 mM EDTA, 8 M GuHCl, 3% Triton X-100, and 3% N-lauroyl-sarcosine, pH 6.3) were added and mixtures were incubated at 70°C for 10 min. Absolute ethanol (250 μl) was added to the lysates, and each mixture was passed through a silica column (FT-2.0; Kisker Biotech GmbH & Co) by centrifugation (8,000 × g). Columns were washed three times, once with “Wash buffer I” (25 mM Tris-HCl, 4 M GuHCl, and 50% ethanol, pH 6.6), and twice with “Wash buffer II” (10 mM Tris-HCl, 0.1 M NaCl, and 80% ethanol, pH 6.6), followed by elution in 80 μl TE buffer.
The qPCR assay, as described by Lazou et al. (2019), targeting the serine hydroxymethyltransferase (glyA) single-copy gene (one genome copy represents one cell equivalent) was applied. Briefly, the primers used were Cc-F (5'-TGTAAAACCAAAGCTTATCGTGTGC-3') and Cc-R (5'-AGTCCAGCAATGTGTGCAATG-3'), along with the Cc-FAM TaqMan probe (5'-6-FAM-AGCTCCAACTTCATCCGCAATCTCTCT-BHQ1-3'). The 50 μl qPCR reaction was comprised by 1× ThermoPol® DF reaction buffer (New England Biolabs), 0.2 μM of each primer, 0.4 μM of the probe, 0.2 mM of each dNTP, 2.5 mM MgSO4, 4 U HotStarTaq DNA polymerase (Qiagen, Hilden, Germany), and 15 μl of DNA extract. Cycling conditions were as follows: 95°C for 15 min, followed by 45 cycles in two steps: (i) 95°C for 30 s and (ii) 60°C for 50 s. Fluorescence was measured at the end of each cycle. The qPCR assay exhibited a linear range of 100–107 copies per reaction (R2 > 0.99), with a PCR efficiency of 93.4% and an LOD of 7.046 copies per reaction (95% CI). The standard curve was described by the equation y = 10[(41.763 ‐ Ct)/3.491)], where y stands for the genome copies per qPCR reaction (Lazou et al., 2019).
Each obtained Ct value during qPCR was transformed to cell equivalents based on the standard curve and the corresponding dilution factors with regard to the original sample. In particular, the C. coli count (y) per meat surface (25 cm2) was calculated by the following formula, taking into account the previously described standard curve equation (Lazou et al., 2019):
y = (10 [(41.763-Ct)/3.491] × a × b × c) /d
where,
Ct = the Cycle threshold.
a = the ratio DNAtotal/DNAassay, with DNAtotal accounting for the total extracted pure DNA volume (80 μl) and DNAassay for the volume of the DNA template (15 μl) used per qPCR assay (80 μl/15 μl = 5.33).
b = the total number of aliquots of the bacterial pellet homogenate that were subjected to qPCR (b = 9).
c = the reduction coefficient of the C. coli cells that were present in the 9:10 volume of the wash suspension with respect to the original wash suspension (c = 1.11).
d = the recovery (%) of C. coli from the meat surface during swabbing (mean value of the total genome copies of the quantification controls divided by the mean value of the total genome copies of the original C. coli suspension inoculated on meat, as obtained by qPCR).
The three individual cell equivalent counts corresponding to the three meat pieces examined per day were used for the calculation of the average cell equivalent count on lamb meat for that particular day in each experiment. The mean count of the second set of triplicate aliquots (PMA-treated spheroplasts) minus the mean count of the third set of triplicate aliquots (ISPC) was considered as the viable cell equivalent count obtained by PMA-qPCR for each meat sample. The artificial inoculation of lamb meat pieces with a qPCR-defined C. coli population (quantified as cell equivalents) and the inclusion of quantification controls enabled a direct calculation of the recovery (%) of campylobacters by the applied non-destructive method of swabbing.
Culture-Based Quantification
For the culture-based quantification, one tenth of the washed suspension was subjected to 10-fold serial dilutions in BPW (Merck) and 100 μl of each dilution were inoculated on double plates both of mCCDA, which is the solid selective medium of first choice according to the standard method for the enumeration of Campylobacter spp. (ISO 10272-2:2017, 2017), and of the non-selective CBA (no CBA plates were used in the first experiment) followed by incubation at 41.5 ± 1°C for 24 to 48 h under microaerobic conditions (Figure 1). The plates were initially checked after the first 24 h of incubation to assess the extent of colony growth on the agar surface and to enumerate the presumptive Campylobacter colonies at this stage, followed by incubation under microaerobic conditions for another day and final enumeration at 48 h. The aforementioned qPCR assay was applied for the confirmation of 10 presumptive Campylobacter colonies on each selected plate and the weighted mean was used for the calculation of CFU (Jarvis, 2016). For the quantification of the viable C. coli on each meat surface (CFU/25 cm2), the reduction coefficient (c = 10) and the recovery (d) were considered, as described previously. The three obtained CFU counts corresponding to the three meat pieces examined per day were used for the calculation of the average CFU count on lamb meat for each particular day in each experiment.
Statistical Analysis
All counts of viable C. coli on lamb meat, as calculated per day by each applied method, were log10-transformed to be further analyzed. Microsoft Office Excel 2010 (Microsoft Corporation, Redmond, Washington, United States) was used for the generation of linear regression equations, which were subsequently used for the calculation of the average log-rank reduction of viable campylobacters per quantification method in each experiment.
Univariate ANOVA (SPSS v21, IBM) were used to assess the effect of quantification method (3 levels) on the log-transformed counts of viable C. coli in lamb meat, adjusting for the fixed effects of experimentation number (3 levels) and testing day (4 levels) as presented below:
Cbabc = mabc + Ma + Eb + Dc + eabc
where,
Cbabc = Log-transformed value of viable C. coli population per 25 cm2 of lamb meat surface.
mabc = overall mean.
Ma = Quantification method of viable C. coli (a = 3 levels: 1 = PMA-qPCR, 2 = mCCDA, and 3 = CBA).
Eb = Experiment number (b = 3 levels: 1st, 2nd and 3rd experiment).
Dc = Testing day (c = 4 levels: day 0, 2, 4, and 6).
eabc = SE.
Statistical significance was set at 0.05 level.
Results
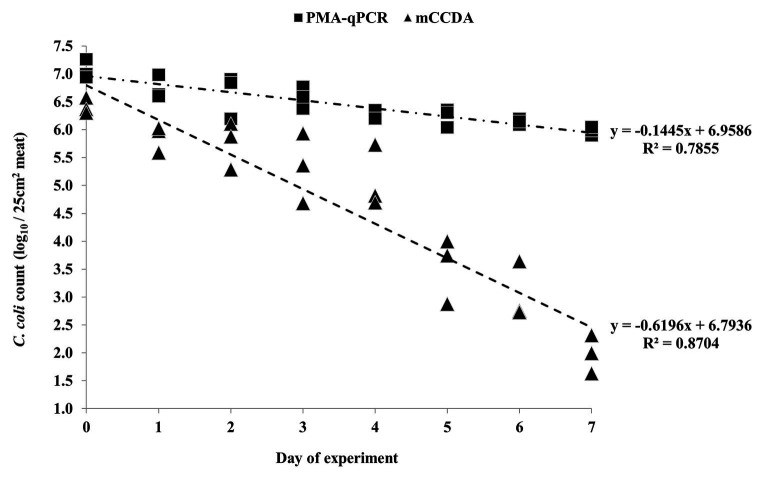
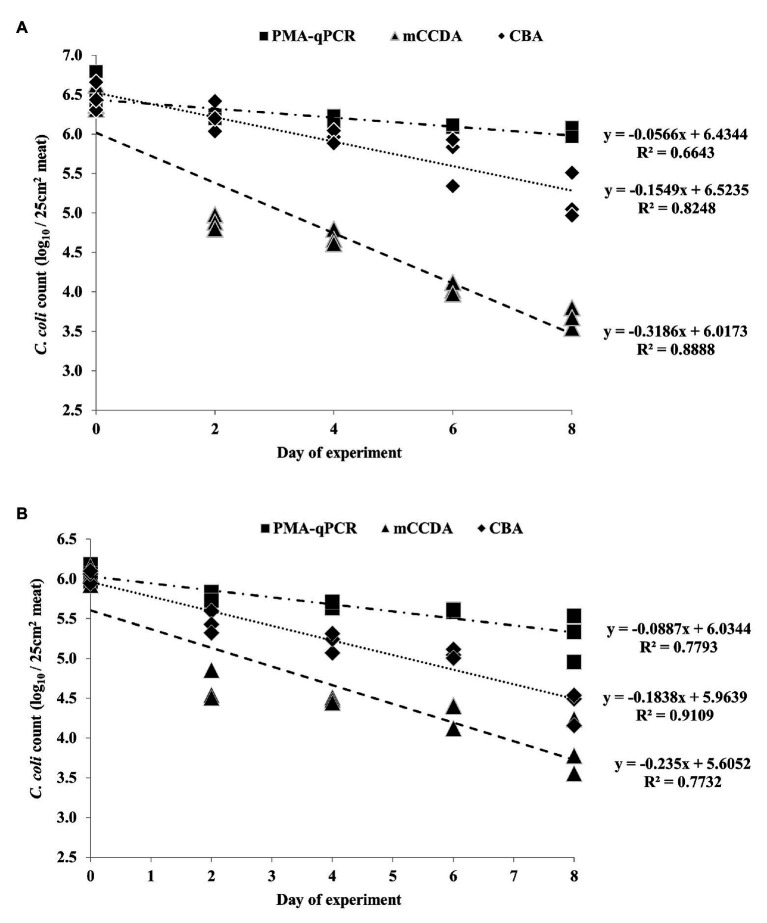
All the applied quantification methods indicated an overall reduction of viable C. coli bacteria under refrigeration storage of lamb meat (Figures 2, 3). Although the initially inoculated C. coli population on the lamb meat pieces was random in each experiment, the difference of the average viable counts among the quantification methods on day 0 was not statistically significant (p > 0.05) in all experiments (Table 1). However, the observed degree of reduction of the viable campylobacters over time was method-dependent; the most dramatic being observed for colony counting on mCCDA and the least for PMA-qPCR. In particular, the average count (three lamb meat samples) of viable C. coli obtained by PMA-qPCR were 1.42–3.99 log10 higher than those obtained by mCCDA plating on the last day of monitoring as regards the three independent experiments. Interestingly, the difference in the culturable C. coli cells between the two solid media peaked over time to result on the last day in CBA yielding higher average counts by 0.54–1.51 log10 compared to mCCDA among the three experiments (Table 1).
Figure 2.
Quantification of C. coli on three lamb meat pieces per day during the first independent experiment, as obtained by PMA-qPCR (log10 cell equivalents/25 cm2 meat surface) and plating on mCCDA (log10 CFU/25 cm2 meat surface). Each dot represents one lamb meat sample. The linear regression equations and R2 values are shown in the diagram.
Figure 3.
Quantification of C. coli on three lamb meat pieces per day during the second (A) and third (B) independent experiments, as obtained by PMA-qPCR (log10 cell equivalents/25 cm2 meat surface), and plating on mCCDA (log10 CFU/25 cm2 meat surface) and CBA (log10 CFU/25 cm2 meat surface). Each dot represents one lamb meat sample. The corresponding linear regression equations and R2 values are shown in the diagram.
Table 1.
Average daily count (±SD) of viable Campylobacter coli cells per day of experiment on lamb meat (surface 25 cm2) as obtained by propidium monoazide-quantitative PCR (PMA-qPCR; log10 cell equivalents), Columbia blood agar (CBA) plating (log10 CFU) and modified charcoal cefoperazone deoxycholate agar (mCCDA) plating (log10 CFU).
| 1st Experiment | 2nd Experiment | 3rd Experiment | ||||||
|---|---|---|---|---|---|---|---|---|
| DAY | Quantification method | Quantification method | Quantification method | |||||
| PMA-qPCR | mCCDA | PMA-qPCR | mCCDA | CBA | PMA-qPCR | mCCDA | CBA | |
| 0 | 7.06 ± 0.17 | 6.42 ± 0.14 | 6.52 ± 0.24 | 6.43 ± 0.16 | 6.47 ± 0.18 | 6.07 ± 0.11 | 6.05 ± 0.12 | 6.04 ± 0.08 |
| 1 | 6.73 ± 0.21 | 5.86 ± 0.24 | ||||||
| 2 | 6.64 ± 0.39 | 5.76 ± 0.42 | 6.22 ± 0.03 | 4.88 ± 0.09 | 6.22 ± 0.19 | 5.79 ± 0.06 | 4.63 ± 0.19 | 5.45 ± 0.14 |
| 3 | 6.57 ± 0.19 | 5.32 ± 0.63 | ||||||
| 4 | 6.27 ± 0.07 | 5.08 ± 0.56 | 6.19 ± 0.06 | 4.69 ± 0.09 | 5.96 ± 0.08 | 5.66 ± 0.04 | 4.48 ± 0.03 | 5.21 ± 0.12 |
| 5 | 6.23 ± 0.17 | 3.54 ± 0.59 | ||||||
| 6 | 6.14 ± 0.05 | 3.04 ± 0.52 | 6.10 ± 0.02 | 4.04 ± 0.07 | 5.70 ± 0.32 | 5.61 ± 0.02 | 4.31 ± 0.16 | 5.05 ± 0.06 |
| 7 | 5.97 ± 0.08 | 1.98 ± 0.34 | ||||||
| 8 | 6.01 ± 0.06 | 3.67 ± 0.13 | 5.17 ± 0.29 | 5.28 ± 0.29 | 3.86 ± 0.35 | 4.40 ± 0.21 | ||
Based on the generated linear regression equations (Figures 2, 3), the average daily log-rank reduction rate (mean ± SD) of viable C. coli on lamb meat in all three experiments was 0.10 ± 0.044, 0.17 ± 0.020, and 0.39 ± 0.202 log10 cell equivalents or CFU regarding PMA-qPCR, plating on CBA and mCCDA, respectively. Likewise, the calculated overall log-rank reduction (mean ± SD) of viable C. coli on lamb meat in all three experiments was 0.77 ± 0.36, 1.36 ± 0.16, and 3.21 ± 1.58 log10 cell equivalents or CFU for PMA-qPCR, and plating on CBA and mCCDA, respectively.
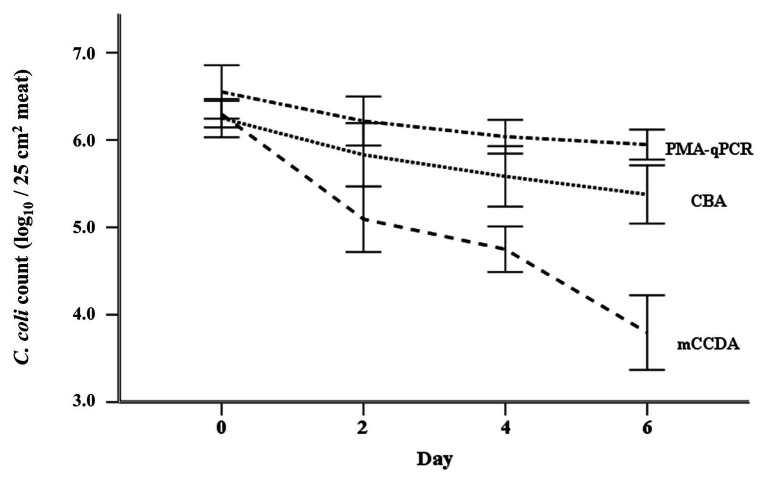
The applied ANOVA accounted for 72.3% of the total variance of the log-rank counts of C. coli, and was found to predict the dependent variable significantly well (p < 0.001). The overall effects of quantification method, experiment number, and test day were significant at p < 0.001 level. In general, in all three experiments, the obtained counts of viable C. coli cells using PMA-qPCR were significantly higher compared to CBA plating (ca. 0.32 log10 cell equivalents, p = 0.015). Similarly, CBA plating counts were significantly higher compared to mCCDA plating (ca. 0.88 log10 CFU, p < 0.001). The statistical model predicted decreasing counts of viable C. coli cells from day 0 to day 6 (Figure 4). Specifically, viable C. coli counts were significantly higher on days 0 (ca. 1.38 log10 cell equivalents, p < 0.001), 2 (ca. 0.70 log10 cell equivalents, p < 0.001), and 4 (ca. 0.44 log10 cell equivalents, p = 0.001) compared to day 6.
Figure 4.
Regression model-predicted curves of the viable C. coli counts (mean ± SD) per applied quantification method (PMA-qPCR, CBA, and mCCDA plating) as regards the first 6 days after the artificial inoculation (day 0) of lamb meat samples.
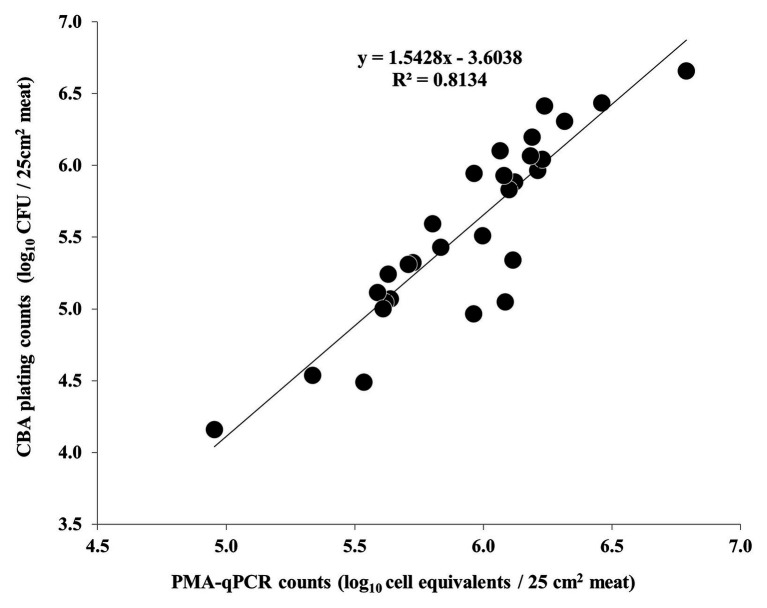
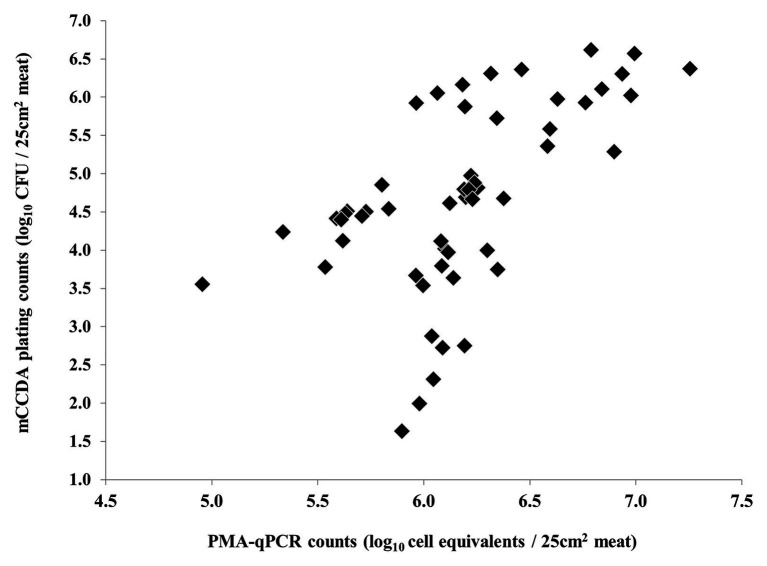
Nonetheless, a strong correlation between the results of PMA-qPCR and plating on CBA was observed regarding the second and third experiments (Figure 5). On the contrary, the counts obtained by PMA-qPCR and plating on mCCDA were not strongly correlated at any time-point during the study (Figure 6).
Figure 5.
Correlation of C. coli log10 cell equivalent counts obtained by PMA-qPCR and log10 CFU counts obtained by plating on CBA during the second and third independent experiments (each dot represents an individual meat sample). The linear regression equation and R2 value are shown in the diagram.
Figure 6.
Non-linear correlation of C. coli log10 cell equivalent counts obtained by PMA-qPCR and log10 CFU counts obtained by plating on mCCDA during all three independent experiments (each dot represents an individual meat sample).
Discussion
The majority of available studies on the detection and/or quantification of Campylobacter on meat are relevant to poultry meat and products thereof. To the best of our knowledge, this is the first study regarding the survival of C. coli on chilled lamb meat in general and the parallel quantification of its viable cells by CFU determination and viability PMA-qPCR quantification in particular.
An overall decline of viable C. coli bacteria has been observed in the current study irrespectively of the applied quantification method. The reduction of viable Campylobacter counts during refrigeration of fresh meat is rather expected, since campylobacters are regarded unable to multiply in temperatures lower than 30°C despite their prolonged survival at 4°C (Jacobs-Reitsma et al., 2008). Data on the survival of C. coli on poultry meat in the concurrent presence of native microbiota have been reported by Oyarzabal et al. (2010) who applied direct plating on mCCDA. In particular, the log-rank decline of C. coli (0.83 ± 0.41 log10 CFU/g) was found to be lower than that of C. jejuni (2.01 ± 0.48 log10 CFU/g) on artificially inoculated chicken meat maintained at 4°C for 14 days. Differences in the survival potential of Campylobacter on poultry meat and corresponding preparations have been observed between different subspecies, as well as between different strains within the same species. The population of two distinct C. coli strains on the skin of fresh poultry meat has been observed to drop by 2.6 ± 0.43 and 3.4 ± 0.07 log10 CFU/cm2 on mCCDA plates over a period of 9 days at 4°C (El-Shibiny et al., 2009). The importance of the food matrix for the survival of campylobacters has been substantiated in previous studies. In particular, differences have been observed in the decline rate of viable campylobacters between different samples originating from the same carcass, for example fresh poultry meat and corresponding skin (Oyarzabal et al., 2010). Meat surface microbiota is thought to reduce the relative pressure of oxygen due to respiration, thereby reducing the oxidative stress level for campylobacters and promoting their survival (El-Shibiny et al., 2009). Moreover, Campylobacter strains appear to exhibit different survival characteristics when studied individually though C. coli has emerged as more aerotolerant than C. jejuni (Karki et al., 2018). Important differences between strains as regards residual viable cells have also been reported in biofilms of Listeria monocytogenes cultivated on stainless steel as detected by PMA-qPCR and epifluorescence microscopy indicating a diverse potential of adaptation and survival within the same species (Brauge et al., 2019). The fact that only one strain of C. coli was used in the current study is a recognized limitation regarding the overall survival potential of this species on lamb meat and, therefore, more studies utilizing various strains of this pathogen are needed to provide better insight in this context.
Apart from the overall reduction of the viable C. coli on lamb meat under refrigeration regardless of the applied quantification scheme, the relative degree of reduction in the current study followed a method-dependent pattern. Although on day zero all methods yielded approximately similar counts of viable C. coli (p > 0.05), an ascending order in the reduction rate was observed in the following days regarding PMA-qPCR, CBA plating and mCCDA plating, respectively. Accordingly, at the end of the monitoring period in all experiments, significantly higher counts (p < 0.05) of viable C. coli cells were obtained by PMA-qPCR compared to CBA plating and by CBA compared to mCCDA plating. These findings can be further interpreted in view of the quantification capacities of each method. Selective factors present in mCCDA, such as antibiotics, have been reported to impose an additional hurdle to already stressed bacteria, thus, reducing the observed CFU counts when compared to non-selective media, such as CBA (Bell et al., 2005). Lower CFU counts of Campylobacter on poultry meat have also been observed on mCCDA plates in comparison to other selective media (Habib et al., 2011). Likewise, in the current study, the observed CFU count of C. coli on lamb meat was higher on CBA plates compared to mCCDA plates on every occasion. However, the C. coli cells that were conceivably transformed to VBNC due to the exposure to the microenvironment of meat, particularly the stressful atmospheric oxygen and refrigeration temperature, were, by definition, unable to form visible colonies and be enumerated on mCCDA and CBA plates. VBNC cell population can be estimated as the difference between culturable cells on solid media and viable cells as determined by PMA-qPCR (Lazou et al., 2019; Lv et al., 2020). Therefore, VBNC bacteria on lamb meat were quantified as viable along with the culturable ones by the PMA-qPCR assay, which resulted in higher counts compared to plating on CBA for each day of quantification during the present study. Higher counts of viable Campylobacter obtained by PMA-qPCR compared to culture on mCCDA plates have also been reported in other studies regarding meat samples (Pacholewicz et al., 2013; Duarte et al., 2015). As regards bias, any potential overestimation of viable C. coli cells due to an insufficient reduction of the signal from dead cells (false positive signals) has been successfully minimized in the present study by the use of the ISPC comprised of inactivated cells subjected to the same treatments and with an equal population as the ones used for enumeration. The importance of developing and utilizing ISPC during the application of PMA-qPCR assays in order to improve cultivation-independent quantification of thermotolerant Campylobacter has been emphasized in previous studies (Lazou et al., 2019; Pacholewicz et al., 2019). A former application of the same PMA-qPCR protocol used in the present study to meat samples artificially inoculated with C. coli revealed no statistical difference between the viable-cell counts obtained by PMA-qPCR and plating on mCCDA but only regarding bacteria in the exponential phase of growth (Lazou et al., 2019).
The unsuitability of CFU count as a reliable parameter for reproducible quantification and assessment of the risk for infection by Campylobacter detected on food products has been previously reported by Krüger et al. (2014). More specifically, a large variability (>2.0 log10) of the CFU of Campylobacter that managed to grow on CBA plates has been observed between the exponential and stationary phase of growth of a single strain. In addition, the comparative enumeration of CFU, the microscopically determined cell counts, and DNA content of C. jejuni and C. coli indicated that colonies were formed by 25.0, 10.0, and 0.3% of bacterial cells at the early stage of the exponential phase, the late stage of the exponential phase, and the stationary phase, respectively. Consequently, classical culture cannot provide quantification data on the total Campylobacter population that originally contaminated a food item during its production and underestimates the potentially infectious population of the pathogen, which includes any VBNC subpopulation present. This fact is further reinforced by the findings of the present study; the CFU counts obtained on mCCDA, which is the solid selective medium for the standard colony-count technique for Campylobacter (ISO 10272-2:2017, 2017), indicated a more dramatic reduction and significantly (p < 0.001) underestimated the viable C. coli compared to the PMA-qPCR method during the storage of lamb meat at 4°C. The application of PMA-qPCR has also highlighted that Campylobacter survival in raw milk can be largely underestimated when relying merely on CFU data (Wulsten et al., 2020). Accordingly, PMA-qPCR has been reported to detect and quantify VBNC bacteria in 90.48% of culture-negative fresh and processed meat samples regarding Staphylococcus aureus, Bacillus cereus, Clostridium perfringens, and Enterobacteriaceae (Abd El-Aziz et al., 2018).
For meat and products thereof, the original contamination with Campylobacter occurs during slaughter whereas the consumer’s exposure to the pathogen takes place at the time of meat preparation and consumption several days later. Important knowledge gaps regarding prevention, control and diagnosis of campylobacteriosis have been recently highlighted by Hansson et al. (2018). Consumer education on domestic hygiene in order to prevent transfer of Campylobacter from raw to ready-to-eat foods has been identified as an emerging gap for the public health domain. Indeed, relevant epidemiological data reveal that a significant portion of the reported cases of foodborne illness is attributed to poor practices of the final consumer regarding handling of contaminated foods in the household (Redmond and Griffith, 2003). The results of the present study indicate the survival and subsequent diminutive reduction of viable C. coli cells on lamb meat under refrigeration storage, which is in accordance with previously reported findings for other types of meat (Jacobs-Reitsma et al., 2008; El-Shibiny et al., 2009; Oyarzabal et al., 2010). The rather slight reduction of the viable C. coli population on lamb meat during storage for several days and until its spoilage, highlights that cross-contamination by infectious campylobacters is likely to occur within the household environment. Therefore, the impact of the applied quantification method on the obtained viable Campylobacter counts entails considerable public health implications especially in view of the multifactorial induction of the VBNC state and the source attribution of campylobacteriosis. The longer the period between the point of food contamination and the time of sampling, the larger the VBNC bacterial population present and the corresponding deviation of the CFU estimate from the potentially infectious Campylobacter cells, particularly if the initial contamination is rather low.
Conclusion
In conclusion, the optimized PMA-qPCR outperformed traditional colony counting as regards the monitoring of viable C. coli on lamb meat during refrigeration storage under normal atmospheric conditions. Utilization only of classical plating for the detection and enumeration of viable campylobacters on meat overlooks by definition the presence of any VBNC counterparts, which are potentially infectious and equally important from a public health perspective. Moreover, standard colony counting on selective solid media, such as mCCDA, underestimates even the culturable Campylobacter cells in comparison to non-selective ones, such as CBA. In contrast, viability PMA-qPCR can be a useful tool for the concurrent quantification of both culturable and VBNC campylobacters on meat. Therefore, optimization of molecular techniques, ‘such as PMA-qPCR,’ in order to achieve reliable Campylobacter quantification would considerably reduce the requirements in terms of time and cost. Such efforts would further facilitate a broader Campylobacter risk assessment and successful surveillance throughout all stages in the food supply chain.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
TL, EI, and CD: design of the study. TL and SC: preparation of laboratory media. TL: experiment performance. TL, AG, and CD: data analysis. TL, AG, SC, and CD: manuscript preparation. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research has been co‐financed by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (project code: T1EDK-04393 / MIS:5063368 ‘InnoLambMeat’). Agrifood Research and Innovation Center IKE is acknowledged for the collaboration.
References
- Abd El-Aziz N. K., Tartor Y. H., Gharib A. A. E., Ammar A. M. (2018). Propidium monoazide quantitative real-time polymerase chain reaction for enumeration of some viable but nonculturable foodborne bacteria in meat and meat products. Foodborne Pathog. Dis. 15, 226–234. 10.1089/fpd.2017.2356, PMID: [DOI] [PubMed] [Google Scholar]
- Baffone W., Casaroli A., Citterio B., Pierfelici L., Campana R., Vittoria E. (2006). Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int. J. Food Microbiol. 107, 83–91. 10.1016/j.ijfoodmicro.2005.08.015, PMID: [DOI] [PubMed] [Google Scholar]
- Bell C., Neaves P., Williams A. P. (eds.) (2005). “Conventional microbiological methods I: equipment, basic techniques and obtaining samples” in Food microbiology and laboratory practice. (Oxford: Blackwell Publishing; ), 173–211. [Google Scholar]
- Brauge T., Midelet-Bourdin G., Soumet C. (2019). Viability detection of foodborne bacterial pathogens in food environment by PMA-qPCR and by microscopic observation. Methods Mol. Biol. 1918, 117–128. 10.1007/978-1-4939-9000-9_9 [DOI] [PubMed] [Google Scholar]
- Chaisowwong W., Kusumoto A., Hashimoto M., Harada T., Maklon K., Kawamoto K. (2012). Physiological characterization of Campylobacter jejuni under cold stresses conditions: its potential for public threat. J. Vet. Med. Sci. 74, 43–50. 10.1292/jvms.11-0305, PMID: [DOI] [PubMed] [Google Scholar]
- Di Giannatale E., Calistri P., Di Donato G., Decastelli L., Goffredo E., Adriano D., et al. (2019). Thermotolerant Campylobacter spp. in chicken and bovine meat in Italy: prevalence, level of contamination and molecular characterization of isolates. PLoS One 14:e0225957. 10.1371/journal.pone.0225957, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Wang C., Ye Y., Liu Y., Wang A., Li Y., et al. (2018). Molecular identification of multidrug-resistant Campylobacter species from diarrheal patients and poultry meat in Shanghai, China. Front. Microbiol. 9:1642. 10.3389/fmicb.2018.01642, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A., Botteldoorn N., Coucke W., Denayer S., Dierick K., Uyttendaele M. (2015). Effect of exposure to stress conditions on propidium monoazide (PMA)-qPCR based Campylobacter enumeration in broiler carcass rinses. Food Microbiol. 48, 182–190. 10.1016/j.fm.2014.12.011, PMID: [DOI] [PubMed] [Google Scholar]
- EFSA and ECDC (2019). The European Union one health 2018 Zoonoses report. EFSA J. 17:e05926. 10.2903/j.efsa.2019.5926, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizaquível P., Aznar R., Sánchez G. (2014). Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 116, 1–13. 10.1111/jam.12365, PMID: [DOI] [PubMed] [Google Scholar]
- El-Shibiny A., Connerton P., Connerton I. (2009). Survival at refrigeration and freezing temperatures of Campylobacter coli and Campylobacter jejuni on chicken skin applied as axenic and mixed inoculums. Int. J. Food Microbiol. 131, 197–202. 10.1016/j.ijfoodmicro.2009.02.024, PMID: [DOI] [PubMed] [Google Scholar]
- Fittipaldi M., Nocker A., Codony F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91, 276–289. 10.1016/j.mimet.2012.08.007, PMID: [DOI] [PubMed] [Google Scholar]
- García-Sánchez L., Melero B., Diez A. M., Jaime I., Rovira J. (2018). Characterization of Campylobacter species in Spanish retail from different fresh chicken products and their antimicrobial resistance. Food Microbiol. 76, 457–465. 10.1016/j.fm.2018.07.004, PMID: [DOI] [PubMed] [Google Scholar]
- Gillespie I. A., O’Brien S. J., Frost J. A., Adak G. K., Horby P., Swan A. V., et al. (2002). A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8, 937–942. 10.3201/eid0809.010817, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C. N., Passaglia J., Vilela F. P., Pereira da Silva F., Duque S. S., Falcão J. P. (2018). High survival rates of Campylobacter coli under different stress conditions suggest that more rigorous food control measures might be needed in Brazil. Food Microbiol. 73, 327–333. 10.1016/j.fm.2018.02.014, PMID: [DOI] [PubMed] [Google Scholar]
- Habib I., Uyttendaele M., De Zutter L. (2011). Evaluation of ISO 10272:2006 standard versus alternative enrichment and plating combinations for enumeration and detection of Campylobacter in chicken meat. Food Microbiol. 28, 1117–1123. 10.1016/j.fm.2011.03.001, PMID: [DOI] [PubMed] [Google Scholar]
- Hansson I., Sandberg M., Habib I., Lowman R., Engvall E. O. (2018). Knowledge gaps in control of Campylobacter for prevention of campylobacteriosis. Transbound. Emerg. Dis. 65 Suppl. 1, 30–48. 10.1111/tbed.12870, PMID: [DOI] [PubMed] [Google Scholar]
- ISO 10272-2:2017 (2017). Microbiology of the food chain—horizontal method for detection and enumeration of Campylobacter spp. — Part 2: colony-count technique. [DOI] [PubMed]
- Jacobs-Reitsma W., Lyhs U., Wagenaar J. (2008). “Campylobacter in the food supply” in Campylobacter. eds. Nachamkin Ι., Szymanski C., Blaser J. (Washington, DC: ASM Press; ), 627–644. [Google Scholar]
- Jarvis B. (2016). “Errors associated with colony count procedures” in Statistical aspects of the microbiological examination of foods. eds. Tenney S., Versteeg-Buschuman L. (London, UK.: Academic Press; ), 122–140. [Google Scholar]
- Kärenlampi R., Rautelin H., Schönberg-Norio D., Paulin L., Hänninen M. L. (2007). Longitudinal study of finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73, 148–155. 10.1128/AEM.01488-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki A. B., Marasini D., Oakey C. K., Mar K., Fakhr M. K. (2018). Campylobacter coli from retail liver and meat products is more aerotolerant than Campylobacter jejuni. Front. Microbiol. 9:2951. 10.3389/fmicb.2018.02951, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger N. J., Buhler C., Iwobi A. N., Huber I., Ellerbroek L., Appel B., et al. (2014). “Limits of control”—crucial parameters for a reliable quantification of viable Campylobacter by real-time PCR. PLoS One 9:e88108. 10.1371/journal.pone.0088108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazou T., Dovas C., Houf K., Soultos N., Iossifidou E. (2014b). Diversity of Campylobacter in retail meat and liver of lambs and goat kids. Foodborne Pathog. Dis. 11, 320–328. 10.1089/fpd.2013.1678, PMID: [DOI] [PubMed] [Google Scholar]
- Lazou T., Houf K., Soultos N., Dovas C., Iossifidou E. (2014a). Campylobacter in small ruminants at slaughter: prevalence, pulsotypes and antibiotic resistance. Int. J. Food Microbiol. 173, 54–61. 10.1016/j.ijfoodmicro.2013.12.011, PMID: [DOI] [PubMed] [Google Scholar]
- Lazou T. P., Iossifidou E. G., Gelasakis A. I., Chaintoutis S. C., Dovas C. I. (2019). Viability quantitative PCR utilizing propidium monoazide, spheroplast formation, and Campylobacter coli as a bacterial model. Appl. Environ. Microbiol. 85, e01499–e01419. 10.1128/AEM.01499-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mendis N., Trigui H., Oliver J. D., Faucher S. P. (2014). The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5:258. 10.3389/fmicb.2014.00258, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løvdal T., Hovda M. B., Björkblom B., Møller S. G. (2011). Propidium monoazide combined with real-time quantitative PCR underestimates heat-killed Listeria innocua. J. Microbiol. Methods 85, 164–169. 10.1016/j.mimet.2011.01.027, PMID: [DOI] [PubMed] [Google Scholar]
- Lv R., Wang K., Feng J., Heeney D. D., Liu D., Lu X. (2020). Detection and quantification of viable but non-culturable Campylobacter jejuni. Front. Microbiol. 10:2920. 10.3389/fmicb.2019.02920, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. G. A., Costantino R., Bianco A., Pileggi C., Pavia M. (2013). Prevalence and pattern of antibiotic resistance of Campylobacter spp. in poultry meat in Southern Italy. Food Control 32, 715–718. 10.1016/j.foodcont.2013.02.011 [DOI] [Google Scholar]
- Nocker A., Cheung C. Y., Camper A. K. (2006). Comparison of propidium monoazide and ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. 10.1016/j.mimet.2006.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Oh E., McMullen L., Jeon B. (2015). Impact of oxidative stress defense on bacterial survival and morphological change in Campylobacter jejuni under aerobic conditions. Front. Microbiol. 6:295. 10.3389/fmicb.2015.00295, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34, 415–425. 10.1111/j.1574-6976.2009.00200.x, PMID: [DOI] [PubMed] [Google Scholar]
- Oyarzabal O. A., Oscar T. P., Speegle L., Nyati H. (2010). Survival of Campylobacter jejuni and Campylobacter coli on retail broiler meat stored at −20, 4, or 12°C and development of Weibull models for survival. J. Food Prot. 73, 1438–1446. 10.4315/0362-028x-73.8.1438, PMID: [DOI] [PubMed] [Google Scholar]
- Pacholewicz E., Buhler C., Wulsten I. F., Kraushaar B., Luu H. Q., Iwobi A. N., et al. (2019). Internal sample process control improves cultivation-independent quantification of thermotolerant Campylobacter. Food Microbiol. 78, 53–61. 10.1016/j.fm.2018.09.017, PMID: [DOI] [PubMed] [Google Scholar]
- Pacholewicz E., Swart A., Lipman L. J., Wagenaar J. A., Havelaar A. H., Duim B. (2013). Propidium monoazide does not fully inhibit the detection of dead Campylobacter on broiler chicken carcasses by qPCR. J. Microbiol. Methods 95, 32–38. 10.1016/j.mimet.2013.06.003, PMID: [DOI] [PubMed] [Google Scholar]
- Papić B., Pate M., Henigman U., Zajc U., Gruntar I., Biasizzo M., et al. (2017). New approaches on quantification of Campylobacter jejuni in poultry samples: the use of digital PCR and real-time PCR against the ISO standard plate count method. Front. Microbiol. 8:331. 10.3389/fmicb.2017.00331, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedonese F., Nuvoloni R., Turchi B., Torracca B., Di Giannatale E., Marotta F., et al. (2017). Prevalence, phenotypic and genetic diversity of Campylobacter in poultry fresh meat and poultry products on retail sale in Tuscany (Italy). Vet. Ital. 53, 29–37. 10.12834/VetIt.836.4107.2, PMID: [DOI] [PubMed] [Google Scholar]
- Redmond E. C., Griffith C. J. (2003). Consumer food handling in the home: a review of food safety studies. J. Food Prot. 66, 130–161. 10.4315/0362-028x-66.1.130, PMID: [DOI] [PubMed] [Google Scholar]
- Roux F., Sproston E., Rotariu O., Macrae M., Sheppard S. K., Bessell P., et al. (2013). Elucidating the aetiology of human Campylobacter coli infections. PLoS One 8:e64504. 10.1371/journal.pone.0064504, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shange N., Gouws P., Hoffman L. C. (2019). Campylobacter and Arcobacter species in food-producing animals: prevalence at primary production and during slaughter. World J. Microbiol. Biotechnol. 35:146. 10.1007/s11274-019-2722-x, PMID: [DOI] [PubMed] [Google Scholar]
- Stingl K., Knüver M. T., Vogt P., Buhler C., Krüger N. J., Alt K., et al. (2012). Quo vadis? – monitoring Campylobacter in Germany. Eur. J. Microbiol. Immunol. 2, 88–96. 10.1556/EuJMI.2.2012.1.12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma M., Field R., Stinson M., Rukovets B., Wymer L., Haugland R. (2009). Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res. 43, 4790–4801. 10.1016/j.watres.2009.05.031, PMID: [DOI] [PubMed] [Google Scholar]
- Voidarou C., Alexopoulos A., Plessas S., Stavropoulou E., Fotou K., Tzora A., et al. (2011). Hygienic quality and antibiotic resistance profile of sliced butchery. Anaerobe 17, 344–350. 10.1016/j.anaerobe.2011.06.001, PMID: [DOI] [PubMed] [Google Scholar]
- Vondrakova L., Turonova H., Scholtz V., Pazlarova J., Demnerova K. (2018). Impact of various killing methods on EMA/PMA-qPCR efficacy. Food Control 85, 23–28. 10.1016/j.foodcont.2017.09.013 [DOI] [Google Scholar]
- Wagner A. O., Malin C., Knapp B. A., Illmer P. (2008). Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Appl. Environ. Microbiol. 74, 2537–2539. 10.1128/AEM.02288-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulsten I. F., Galeev A., Stingl K. (2020). Underestimated survival of Campylobacter in raw milk highlighted by viability real-time PCR and growth recovery. Front. Microbiol. 11:1107. 10.3389/fmicb.2020.01107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Chen Z., Jiang Y., Xue F., Li B. (2016). Advances and challenges in viability detection of foodborne pathogens. Front. Microbiol. 7:1833. 10.3389/fmicb.2016.01833, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.