Abstract
Background
Mitochondrial-derived peptides (MDPs) such as MOTS-c and humanin have been studied for their cytoprotective functions. In mice, humanin-encoding Mtrnr2 is a mitochondrial pseudogene, and the humanin-like peptide is encoded by the nuclear Gm20594 gene. However, endogenous tissue-specific expression profiles of Gm20594 have not yet been identified.
Methods
Mtrnr1 and Gm20594 expression was profiled via reverse transcription using only oligo(dT) primers from tissues of C57BL6/J mice. To analyze altered expression upon mitochondrial biogenesis, C2C12 myocytes and brown adipocytes were differentiated. Mitochondrial DNA copy numbers were quantified for normalization.
Results
Both Mtrnr1 and Gm20594 were highly expressed in brown adipose tissue. When normalized against mitochondrial content, Mtrnr1 was identified as being highly expressed in the duodenum, followed by the jejunum. In models of mitochondrial biogenesis, both Mtrnr1 and Gm20594 were upregulated during myocyte and brown adipocyte differentiation. Increased Mtrnr1 expression during brown adipocyte differentiation remained significant after normalization against mitochondrial DNA copy number, whereas myocyte differentiation exhibited biphasic upregulation and downregulation in early and late phases, respectively.
Conclusion
Nuclear-encoded Gm20594 showed similar expression patterns of mitochondrial-encoded Mtrnr1. Brown adipose tissue presented the highest basal expression levels of Gm20594 and Mtrnr1. When normalized against mitochondrial DNA copy number, gut tissues exhibited the highest expression of Mtrnr1. Upregulation of Mtrnr1 during mitochondrial biogenesis is independent of mitochondrial content.
Keywords: Mitochondrial-derived peptide, Mtrnr1, Gm20594, Gene expression, Mitochondria
INTRODUCTION
The mitochondrial genome contains genes involved in the electron transport chain and tRNA- and rRNA-encoding loci. Recently, novel open reading frames (ORFs) within rRNA regions have been identified. These are mitochondrial-derived peptides (MDPs) referred to as a mitochondrial open reading frame of the 12S rRNA type-c (MOTS-c) [1] within 12s rRNA (Mtrnr1) and humanin [2] and small humanin-like peptides (SHLP1-6) [3] within 16s rRNA (Mtrnr2). MDPs are preserved among species, but the first amino acid for humanin in mice is not methionine, but rather threonine, showing that murine Mtrnr2 is a pseudogene. Instead, a peptide with a sequence similar to that of human humanin is encoded by Gm20594, which is a nuclear gene [4]. This might be a result of nuclear mitochondrial DNA segments (NUMT), which translocate mitochondrial genes into the nucleus [5].
MDPs have been studied for their cytoprotective roles. For example, MOTS-c exerts beneficial effects to counter metabolic stress [6]. Humanin exerts anti-apoptotic effects in cardio- and neuroprotection [7]. So far, major research surrounding MDPs has focused on their biological effects and mechanisms [6,8,9], or on amino acid-substituted derivatives, including S14G-humanin (HNG) and colivelin, to maximize humanin functions [10,11].
However, endogenous tissue-specific expression, as well as the regulation of MDPs, have not been well studied. Several human studies have reported blood levels of MDPs in diabetes [12], pre-eclampsia [13], obstructive sleep apnea [14], and Alzheimer’s disease [15], which implies that MDPs act as mitokines [16,17]. However, the tissues responsible for synthesizing a major proportion of circulating blood MDPs have not been identified. Moreover, as in the case of Mtrnr2 NUMT in mice, there are many MTRNR2-like genes in the human nucleus, MTRNR2L1 to MTRNR2L13, which encode very similar amino acid sequences that might cross-react with mitochondrial MTRNR2-encoded humanin [4,18]. Another method for detecting MDPs is reverse transcription-polymerase chain reaction (RT-PCR) of MDP genes, but technical concerns remain. Because MDPs are expressed within rRNA regions, conventional methods that use random hexamer primers for reverse transcription cannot distinguish whether the resultant cDNA is mRNA- or rRNA-templated.
Gm20594 is a predicted gene that has not been studied previously. In this study, we compared the mRNA levels of mitochondrial Mtrnr1 and nuclear Gm20594 in mouse tissues for the first time using oligo(dT) for reverse transcription. We also checked the expression levels of MDP transcripts in models of mitochondrial biogenesis.
MATERIALS AND METHODS
1. Animal experiments
The experimental protocols for this study were approved by the Institutional Animal Care and Use Committee (IACUC) at Yonsei University College of Medicine, Wonju, Korea (IACUC approval No. YWC-200907-2). Male C57BL/6J mice were purchased from Orient Bio (Seoul, Korea) and housed in a climate-controlled facility with a 12-h light- dark cycle and free access to food and water. Mice at 20 weeks of age were anesthetized with tribromoethanol (Sigma, St Louis, MO, USA), followed by cervical dislocation. Mouse tissues were harvested and immediately snap-frozen.
2. Cell culture
The AML12 mouse hepatocyte cell line was cultured in DMEM/F-12 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), a mixture of 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 5 ng/mL selenium, and 40 ng/mL dexamethasone. C2C12 mouse myoblasts were cultured in high glucose DMEM (Gibco) supplemented with 10% FBS. When the cells reached 90% confluence, they were induced to differentiate by incubation in DMEM containing 5% horse serum for 8 days. Brown adipocyte cells were cultured in DMEM supplemented with 10% calf serum. After reaching 95-98% confluence, the cells were induced to differentiate using DMEM with 10% FBS, 1 nM triiodothyronine (T3), 5 μg/ml insulin, 0.5 mM isobutylmethylxanthine, 2 μg/mL dexamethasone, and 125 μM indomethacin. After 2 days, the medium was replaced with DMEM containing 10% FBS, 1 nM T3, and 5 μg/ml insulin. After this, medium with insulin and T3 was refreshed every 2 days until day 8.
3.Nucleic acid extraction and reverse transcription
Total RNA and DNA were isolated from mouse tissues and cells using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. SuperScript III First-Strand Synthesis system (Invitrogen, Carlsbad, CA, USA) was used for reverse transcription of 1 μg of total RNA using oligo(dT) primers with or without random hexamers.
4. Quantitative polymerase chain reaction (qPCR)
Gene expression and quantification of mitochondrial copy number were analyzed by real-time qPCR using Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. The primer sequences used in this study are listed in Table 1. Relative gene expression was analyzed by the △△Ct method using Rplp0 as an internal control. Relative mitochondrial gene copy number was calculated using relative quantification of mitochondrial Nd6 levels from DNA, with nuclear Gapdh as an internal control.
Table 1.
Sequences of primers used in this study
| Gene | Sequence (5’ to 3’) |
|---|---|
| Rplp0 | TCGTTGGAGTGACATCGTCT |
| TAGTTGGACTTCCAGGTCGC | |
| Mtrnr1 | GACACCTTGCCTAGCCACAC |
| TGGCTGGCACGAAATTTACC | |
| Gm20594 | CTGCCTGCCCAGTGACTAAA |
| AGCCATTCATGCTAGTCCCTA | |
| Nd6 | GCTACCCCAATCCCTCCTTC |
| TCTTGATGGTTTGGGAGATTGGT | |
| Gapdh | GAACTCCTCATGGGTCTGTAGTG |
| TGTTGTGGTACGTGCATAGCTG | |
| Myod | TGAGCAAAGTGAATGAGGCCTTCG |
| AGAGCCTGCAGACCTTCGATGTA | |
| Myh4 | GTCCTTCCTCAAACCCTTAAAGT |
| GGTGCACCTCTTGTGTTACC | |
| Cd36 | GGCCAAGCTATTGCGACAT |
| CAGATCCGAACACAGCGTAGA | |
| Fabp4 | CACCGCAGACGACAGGAAG |
| GCACCTGCACCAGGGC | |
| Ucp1 | CTTTGCCTCACTCAGGATTGG |
| ACTGCCACACCTCCAGTCATT |
5. Statistics
Data are shown as means ± standard error of the mean. Data were analyzed using SPSS (version 25.0; IBM Corp., Armonk, NY, USA). Student’s t-test and analysis of variance were used to compare gene expression data. Differences were considered statistically significant at p-values < 0.05.
RESULTS
1. Testing RT primers for the amplification of MDP genes
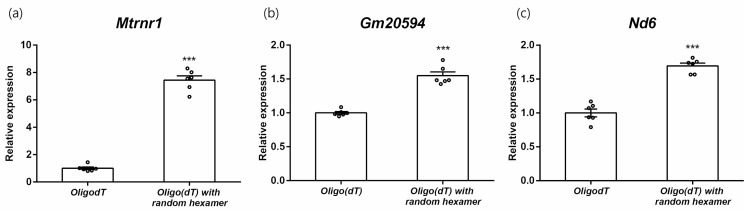
First, we checked cDNA detection levels using different reverse transcription primers. We compared two primers: oligo(dT) alone and oligo(dT) with random hexamers (Fig. 1a). Relative expression levels of Mtrnr1 were increased using oligo(dT) with random hexamers. As controls, nuclear Gm20594 and mitochondrial Nd6, which do not share sequence with other RNAs, were analyzed (Fig. 1b, 1c). Although a significant decrease in the expression of Gm20594 and Nd6 was also detected, the fold changes involved were small, less than that for Mtrnr1. Because primers used in reverse transcription reactions are in excess relative to template RNAs, this result showed that conventional methods using oligo(dT) and random hexamers cannot distinguish cDNA from actual peptide-coding mRNA and rRNA of Mtrnr1. Therefore, we used oligo(dT) alone for reverse transcription in this study.
Fig. 1.
Comparisons of cDNA detection levels according to primers of reverse transcription. Expressions of Mtrnr1 (a), Gm20594 (b), and Nd6 (c) of AML12 cells. ***p < 0.001 vs. oligo(dT).
2. Tissue-specific expression of Mtrnr1 and Gm20594
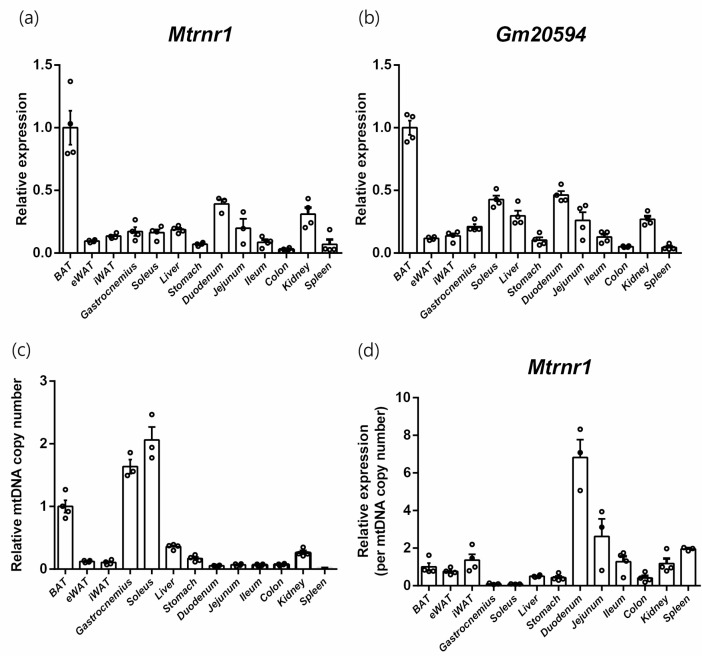
Next, we intended to identify tissue-specific expression profiles of mouse MDPs at the mRNA level. As mitokines, it was predicted that mitochondria-rich organs might have higher MDP expression levels. We found that Mtrnr1 was highly expressed in brown adipose tissue, followed by the duodenum and kidney (Fig. 2a). However, expression levels in skeletal muscles were comparable to those of other tissues. Gm20594 also exhibited similar Mtrnr1 expression patterns (Fig. 2b). Because there are various levels of mitochondrial content according to tissues that might affect the expression of MDPs, we re-analyzed Mtrnr1 expression using relative mitochondrial content levels. Mitochondrial DNA copy number quantified using the ratio of a mitochondrial gene (Nd6) and a nuclear gene (Gapdh) were highest in skeletal muscles, including the soleus and gastrocnemius, followed by brown adipose tissue, liver, and kidney (Fig. 2c). We normalized the expression of Mtrnr1 to mitochondrial DNA copy number (Fig. 2d). When normalized, the duodenum showed the highest expression of Mtrnr1, followed by the jejunum. Other tissues showed similar levels of Mtrnr1 expression, except for low abundance in skeletal muscles, which have relatively higher mitochondrial contents.
Fig. 2.
Tissue-expressional profiles of mouse MDPs. Relative expression of Mtrnr1 (a) and Gm20594 (b) in tissues of C57BL/6 mice with oligo(dT)-only reverse transcription. (c) Relative mitochondrial DNA copy number. (d) Relative expression of Mtrnr1 normalized against relative mitochondrial DNA copy number.
3. MDP expression in myocyte differentiation
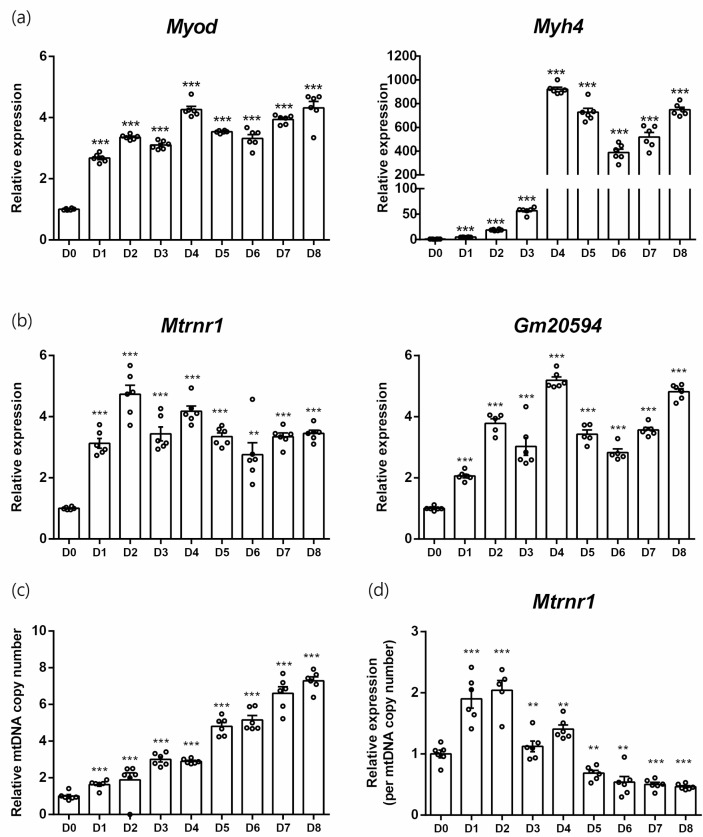
Because the expression of Mtrnr1 varied according to mitochondrial content, we investigated the expression profiles of MDPs in models of mitochondrial biogenesis. First, C2C12 myoblast differentiation was used [19]. During the 8 days of differentiation, markers of myocytes such as Myod and Myh4 showed significantly increased levels (Fig. 3a). After 1 day of differentiation, the expression of Mtrnr1 and Gm20594 was increased, which was sustained during the remainder of the induction period (Fig. 3b). We observed a gradual increase in mitochondrial content during the differentiation of myoblasts into myocytes (Fig. 3c). Because the expression levels of Mtrnr1 in the later period of differentiation did not increase further, the mitochondrial content-normalized expression of Mtrnr1 exhibited a biphasic pattern, increasing in the early period, followed by a decrease at the later period (Fig. 3d).
Fig. 3.
MDP expressions in myocyte differentiation. (a) Relative expressions of myocyte differentiation markers from differentiation day 0 to 8. (b) Relative expression of Mtrnr1 and Gm20594. (c) Relative mitochondrial DNA copy numbers. (d) Relative expression of Mtrnr1 normalized against relative mitochondrial DNA copy numbers. **p < 0.01; ***p < 0.001 vs. D0.
4. MDP expression in brown adipocyte differentiation
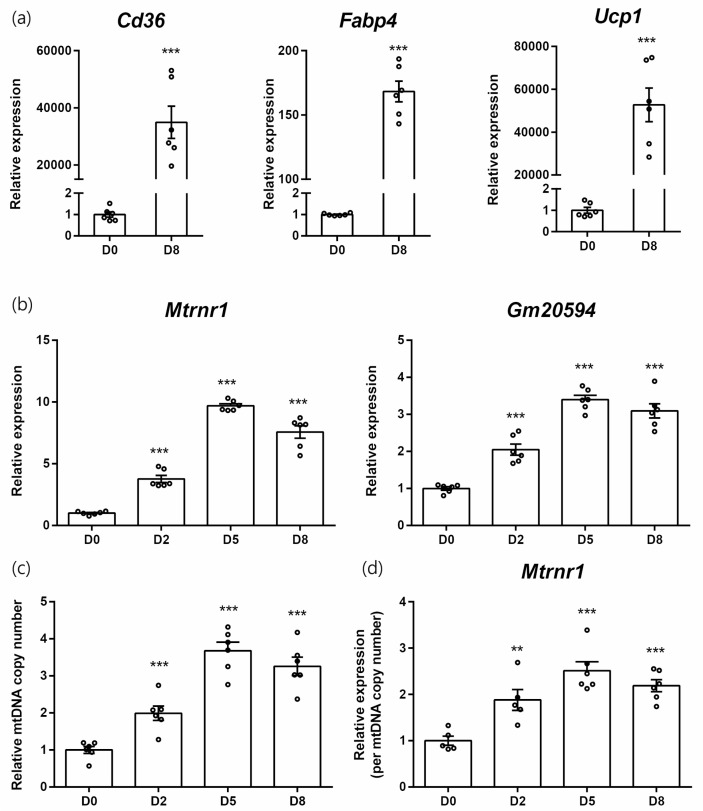
We used another model of mitochondrial biogenesis involving the differentiation of brown adipocytes [20]. After differentiation, brown adipocytes showed increased expression of adipocyte markers such as Cd36 and Fabp4, as well as Ucp1, a functional brown adipocyte marker (Fig. 4a). As in the case of C2C12, differentiation of brown adipocytes was accompanied by upregulation of Mtrnr1 and Gm20594 (Fig. 4b). Mitochondrial DNA copy number presented the same pattern, with lower fold changes (Fig. 4c). Therefore, Mtrnr1 expression, normalized against mitochondrial copy number, was upregulated during the differentiation of brown adipocytes (Fig. 4d).
Fig. 4.
MDP expressions in brown adipocyte differentiation. (a) Relative expressions of brown adipocyte differentiation markers from differentiation day 0 and 8. (b) Relative expression of Mtrnr1 and Gm20594. (c) Relative mitochondrial DNA copy numbers. (d) Relative expression of Mtrnr1 normalized against relative mitochondrial DNA copy numbers. **p < 0.01; ***p < 0.001 vs. D0.
DISCUSSION
As MDPs exert beneficial cytoprotective effects against aging and metabolic stress [21-23], most studies have targeted exogenous treatments with MDPs or MDP-derivatives. However, in terms of therapeutic approaches, exogenous applications raise a concern regarding compliance because these peptides cannot be administered orally. Therefore, enhancing the expression or function of endogenous MDPs may be a more suitable strategy, as well as a physiologic strategy. Profiling endogenous MDP expression should be the first step in this approach. However, most studies reporting MDP expression have focused on levels in blood that cannot reveal tissue-specific profiles. Here, we present endogenous MDP expression levels in mouse tissues, especially humanin-like nuclear-encoded Gm20594, for the first time.
After MDPs were recognized as beneficial mitokines, many approaches regarding the use of MDPs as biomarkers for several diseases have been developed. For example, decreased blood MOTS-c levels have been shown in obese children [24] and patients with coronary endothelial dysfunction [25]. Moreover, MDPs are not only considered as candidate biomarkers, but also for their predictive value, such as lower serum SHLP2 levels linked to increased prostate cancer risk [26]. Because blood levels of MDPs are determined by a balance between release (and synthesis) and degradation (uptake and excretion), simple determination of blood levels only present current overall status, which is neither dynamic nor tissue-specific. For example, it has been reported that plasma MOTS-c levels decrease with aging, but are increased in skeletal muscle [27]. Conversely, plasma humanin levels are higher in patients with chronic kidney disease, whereas humanin levels in skeletal muscle were diminished [28]. Considering that skeletal muscle is expected to have abundant MDPs due to high mitochondrial content, these results indicate the limitation of current MDP expressional profile studies. Therefore, tissue-specific profiling of MDP synthesis and degradation is needed to resolve these discrepancies.
Tissue-specific detection of MDPs has been attempted at both the protein and mRNA levels. Humanin can be detected using immunofluorescence staining or immunohistochemistry in human muscle fibers and blood vessels [29,30]. However, together with other antibody-based techniques such as western blotting [31] or enzyme-linked immunosorbent assays [12,32], there should be serious precautions regarding the use of anti-MDP antibodies, especially against humanin. There are many nuclear-encoded MTRNR2-like genes that produce humanin-like peptides, which have only one (MTRNR2L8), two (MTRNR2L1), or four (MTRNR2L6) different amino acids compared to mitochondrial-encoded humanin [18]. Because cross-reactivity involving humanin and all humanin-like peptides with commercial humanin antibodies has not been completely confirmed to date, we still cannot definitively distinguish the sources (nuclear or mitochondrial) of peptides that are recognized as humanins. In mice, although there is only one gene (and one corresponding peptide) for humanin, some studies used antibodies against rattin (rat homolog to humanin) [33] or HNG [34] for detection, which have many mismatches to Gm20594- expressed humanin. Collectively, antibody-based detection of MDPs should provide precise data concerning cross- reactivity.
Some studies have used RT-qPCR for the detection and quantification of MDP expression [35]. The expression profile at the mRNA level can present the current status of synthesis and rule out cross-reactivity. However, detection of MDPs at the mRNA level can be another target of concern because of their unusual genomic structures. Because the ORFs of MDPs are located within rRNA genes, conventional total RNA extraction and reverse transcription using random hexamers cannot distinguish mRNA-templated cDNA from rRNA-templated cDNA. We compared the detection of MDP cDNA using different primers for reverse transcription. Oligo(dT) primer resulted in decreased detection of Mtrnr1 compared to oligo(dT) and random hexamers. However, this was not the case for Nd6, which does not share sequence with other RNAs or nuclear-encoded Gm20594. Thus, studies presenting MDP mRNA levels should indicate the specific primer(s) used. Alternatively, RNA extraction using size exclusion techniques can minimize cross-detection [27,36].
We profiled tissue-specific expression of Mtrnr1 and Gm20594 mRNA levels in mice. Initially, mitochondria-rich organs were expected to show increased levels of Mtrnr1 expression due to high mitochondrial content. Although we found that brown adipose tissue exhibited high expression of MDPs, skeletal muscle showed relatively low expression. Because tissues were harvested without any interventions, such as exercise training or diet modification, basal endogenous MDP expression in muscle seems to be low. These results agree with the findings of a previous study revealing low levels of humanin in isolated mouse extensor digitorum longus muscle, which were elevated after ex vivo muscular contractions [36]. On the contrary, brown adipose tissue might represent a major target for enhancing endogenous MDP expression for therapeutic strategies. Here, after normalization against mitochondrial DNA copy number, the duodenum showed the highest expression of Mtrnr1, followed by the jejunum. The duodenum was the second-highest Gm20594-expressing organ; therefore, further studies concerning the functions of MDPs in gut tissues are needed to expand the understanding of the physiological roles of MDPs.
We found upregulation of MDPs and mitochondrial DNA copy number during the differentiation of C2C12 myocytes, and mitochondrial DNA copy number-normalized Mtrnr1 expression showed an initial increase followed by a decrease in the later stages of differentiation. This biphasic pattern appears to be due to altered mitochondrial content. Humanin expression in muscles was increased during exercise training [35,36]. Although exercise induces mitochondrial biogenesis, different expression patterns in myocyte differentiation and exercise might be the result of other myokines or alterations in metabolites that affect MDP expression. In the process of brown adipocyte differentiation, MDP expression was upregulated, even after normalization against increased DNA copy number. Considering the case of C2C12 differentiation, in which mitochondria-normalized MDP expression was biphasic, this upregulation of Mtrnr1 again implies that brown adipocytes are the highest MDP- expressing sites. In addition, MDP expression results from differentiation, and MOTS-c from Mtrnr1 itself can induce brown adipogenesis because exogenous MOTS-c treatment promotes white fat browning and brown fat activation during cold exposure [37]. Therefore, functional associations between MDPs and brown adipogenesis may represent targets for further studies regarding adipocyte biology.
Another notable finding in this study was the very similar pattern of Mtrnr1 and Gm20594 expression. Mitochondrial transcription is initiated by heavy- and light-strand promoters [38]. Although mouse Mtrnr2 is a pseudogene, nuclear-encoded Gm20594 is a NUMT derivative of Mtrnr2. Because very little is known regarding Gm20594, the Gm20594 promoter sequence or transcription factors involved in its expression remain unidentified. Additional studies are needed to determine whether regulatory mechanisms are also preserved in the nucleus as they are in the genomic sequence. Some studies have revealed situations that modulate MDP expression. For example, growth hormone and the insulin-like growth factor-1 axis reduce humanin expression [34] and hyperbaric oxygen therapy increases serum humanin levels [39].
CONCLUSION
This study presented the tissue-specific endogenous expression of mouse Mtrnr1 and Gm20594 using oligo(dT) RT primers. Brown adipose tissue showed the highest expression, while MDPs in gut tissues were the most abundant when normalized against mitochondrial DNA copy number. In models of mitochondrial biogenesis in myocytes and brown adipocytes, mitochondrial Mtrnr1 and nuclear Gm20594 showed increased expression along with mitochondrial DNA copy number. However, the upregulation of Mtrnr1 was independent of mitochondrial content. This study represents preliminary, but fundamental, findings involving Gm20594 expression and tissue-specific research surrounding MDPs.
ACKNOWLEDGEMENTS
This work was supported by the Yonsei University Wonju Campus Future-Leading Research Initiative of 2019-52- 0052.
Footnotes
CONFLICTS OF INTERESTS
None to declare.
REFERENCES
- 1.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–54. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Proc Natl Acad Sci (USA) 2001;98:6336–41. doi: 10.1073/pnas.101133498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N, Cohen P. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging (Albany NY) 2016;8:796–809. doi: 10.18632/aging.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodzioch M, Lapicka-Bodzioch K, Zapala B, Kamysz W, Kiec-Wilk B, Dembinska-Kiec A. Evidence for potential functionality of nuclearly-encoded humanin isoforms. Genomics. 2009;94:247–56. doi: 10.1016/j.ygeno.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Puertas MJ, Gonzalez-Sanchez M. Insertions of mitochondrial DNA into the nucleus-effects and role in cell evolution. Genome. 2020;63:365–74. doi: 10.1139/gen-2019-0151. [DOI] [PubMed] [Google Scholar]
- 6.Merry TL, Chan A, Woodhead JST, Reynolds JC, Kumagai H, Kim SJ, Lee C. Mitochondrial-derived peptides in energy metabolism. Am J Physiol Endocrinol Metab. 2020;319:E659–66. doi: 10.1152/ajpendo.00249.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazafa A, Batool A, Ahmad S, Amjad M, Chaudhry SN, Asad J, Ghuman HF, Khan HM, Naeem M, Ghani U. Humanin: A mitochondrial-derived peptide in the treatment of apoptosis-related diseases. Life Sci. 2021;264:118679. doi: 10.1016/j.lfs.2020.118679. [DOI] [PubMed] [Google Scholar]
- 8.Popov LD. Mitochondrial peptides-appropriate options for therapeutic exploitation. Cell Tissue Res. 2019;377:161–5. doi: 10.1007/s00441-019-03049-z. [DOI] [PubMed] [Google Scholar]
- 9.Rochette L, Meloux A, Zeller M, Cottin Y, Vergely C. Role of humanin, a mitochondrial-derived peptide, in cardiovascular disorders. Arch Cardiovasc Dis. 2020;113:564–71. doi: 10.1016/j.acvd.2020.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Chiba T, Yamada M, Hashimoto Y, Sato M, Sasabe J, Kita Y, Terashita K, Aiso S, Nishimoto I, Matsuoka M. Development of a femtomolar-acting humanin derivative named colivelin by attaching activity-dependent neurotrophic factor to its N terminus: characterization of colivelin-mediated neuroprotection against Alzheimer's disease-relevant insults in vitro and in vivo. J Neurosci. 2005;25:10252–61. doi: 10.1523/JNEUROSCI.3348-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y, Feng Y, Zou Y, Wang F, Liu H, Liu C, Zhang Y. [Gly14]-humanin restores cathepsin D function via FPRL1 and promotes autophagic degradation of Ox-LDL in HUVECs. Nutr Metab Cardiovasc Dis. 2020;30:2406–16. doi: 10.1016/j.numecd.2020.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Ramanjaneya M, Bettahi I, Jerobin J, Chandra P, Abi Khalil C, Skarulis M, Atkin SL, Abou-Samra AB. Mitochondrial-Derived Peptides Are Down Regulated in Diabetes Subjects. Front Endocrinol (Lausanne) 2019;10:331. doi: 10.3389/fendo.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolakopoulos P, Tzimagiorgis G, Goulis DG, Chatzopoulou F, Zepiridis L, Vavilis D. Serum humanin concentrations in women with pre-eclampsia compared to women with uncomplicated pregnancies. J Matern Fetal Neonatal Med. 2018;31:305–11. doi: 10.1080/14767058.2017.1285885. [DOI] [PubMed] [Google Scholar]
- 14.Baylan FA, Yarar E. Relationship between the mitochondria-derived peptide MOTS-c and insulin resistance in obstructive sleep apnea. Sleep Breath. 2021 doi: 10.1007/s11325-020-02273-0. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Conte M, Sabbatinelli J, Chiariello A, Martucci M, Santoro A, Monti D, Arcaro M, Galimberti D, Scarpini E, Bonfigli AR, Giuliani A, Olivieri F, Franceschi C, Salvioli S. Disease-specific plasma levels of mitokines FGF21, GDF15, and Humanin in type II diabetes and Alzheimer's disease in comparison with healthy aging. Geroscience. 2020 doi: 10.1007/s11357-020-00287-w. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong CQY, Tang BL. A Mitochondrial Encoded Messenger at the Nucleus. Cells. 2018;7(8):105. doi: 10.3390/cells7080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conte M, Martucci M, Chiariello A, Franceschi C, Salvioli S. Mitochondria, immunosenescence and inflammaging: a role for mitokines? Semin Immunopathol. 2020;42:607–17. doi: 10.1007/s00281-020-00813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yen K, Lee C, Mehta H, Cohen P. The emerging role of the mitochondrial-derived peptide humanin in stress resistance. J Mol Endocrinol. 2013;50:R11–9. doi: 10.1530/JME-12-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remels AH, Langen RC, Schrauwen P, Schaart G, Schols AM, Gosker HR. Regulation of mitochondrial biogenesis during myogenesis. Mol Cell Endocrinol. 2010;315:113–20. doi: 10.1016/j.mce.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Butow RA, Bahassi EM. Adaptive thermogenesis: orchestrating mitochondrial biogenesis. Curr Biol. 1999;9:R767–9. doi: 10.1016/S0960-9822(00)80008-1. [DOI] [PubMed] [Google Scholar]
- 21.Kim SJ, Miller B, Kumagai H, Silverstein AR, Flores M, Yen K. Mitochondrial-derived peptides in aging and age-related diseases. Geroscience. 2020 doi: 10.1007/s11357-020-00262-5. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon C, Sun JL, Jeong JH, Jung TW. Humanin attenuates palmitate-induced hepatic lipid accumulation and insulin resistance via AMPK-mediated suppression of the mTOR pathway. Biochem Biophys Res Commun. 2020;526:539–45. doi: 10.1016/j.bbrc.2020.03.128. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Gao H, Zhou H, Liu Q, Qi Z, Zhang Y, Zhang J. The role of mitochondria-derived peptides in cardiovascular disease: Recent updates. Biomed Pharmacother. 2019;117:109075. doi: 10.1016/j.biopha.2019.109075. [DOI] [PubMed] [Google Scholar]
- 24.Du C, Zhang C, Wu W, Liang Y, Wang A, Wu S, Zhao Y, Hou L, Ning Q, Luo X. Circulating MOTS-c levels are decreased in obese male children and adolescents and associated with insulin resistance. Pediatr Diabetes . 2018 doi: 10.1111/pedi.12685. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 25.Qin Q, Delrio S, Wan J, Jay Widmer R, Cohen P, Lerman LO, Lerman A. Downregulation of circulating MOTS-c levels in patients with coronary endothelial dysfunction. Int J Cardiol. 2018;254:23–7. doi: 10.1016/j.ijcard.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Xiao J, Howard L, Wan J, Wiggins E, Vidal A, Cohen P, Freedland SJ. Low circulating levels of the mitochondrial-peptide hormone SHLP2: novel biomarker for prostate cancer risk. Oncotarget. 2017;8:94900–9. doi: 10.18632/oncotarget.20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza RF, Woodhead JST, Hedges CP, Zeng N, Wan J, Kumagai H, Lee C, Cohen P, Cameron-Smith D, Mitchell CJ, Merry TL. Increased expression of the mitochondrial derived peptide, MOTS-c, in skeletal muscle of healthy aging men is associated with myofiber composition. Aging (Albany NY) 2020;12:5244–58. doi: 10.18632/aging.102944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Gidlund EK, Witasp A, Qureshi AR, Soderberg M, Thorell A, Nader GA, Barany P, Stenvinkel P, von Walden F. Reduced skeletal muscle expression of mitochondrial-derived peptides humanin and MOTS-C and Nrf2 in chronic kidney disease. Am J Physiol Renal Physiol. 2019;317:F1122–31. doi: 10.1152/ajprenal.00202.2019. [DOI] [PubMed] [Google Scholar]
- 29.Kariya S, Hirano M, Furiya Y, Sugie K, Ueno S. Humanin detected in skeletal muscles of MELAS patients: a possible new therapeutic agent. Acta Neuropathol. 2005;109:367–72. doi: 10.1007/s00401-004-0965-5. [DOI] [PubMed] [Google Scholar]
- 30.Bachar AR, Scheffer L, Schroeder AS, Nakamura HK, Cobb LJ, Oh YK, Lerman LO, Pagano RE, Cohen P, Lerman A. Humanin is expressed in human vascular walls and has a cytoprotective effect against oxidized LDL-induced oxidative stress. Cardiovasc Res. 2010;88:360–6. doi: 10.1093/cvr/cvq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zacharias DG, Kim SG, Massat AE, Bachar AR, Oh YK, Herrmann J, Rodriguez-Porcel M, Cohen P, Lerman LO, Lerman A. Humanin, a cytoprotective peptide, is expressed in carotid atherosclerotic [corrected] plaques in humans. PLoS One. 2012;7:e31065. doi: 10.1371/journal.pone.0031065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voigt A, Jelinek HF. Humanin: a mitochondrial signaling peptide as a biomarker for impaired fasting glucose-related oxidative stress. Physiol Rep. 2016;4(9):e12796. doi: 10.14814/phy2.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muzumdar RH, Huffman DM, Calvert JW, Jha S, Weinberg Y, Cui L, Nemkal A, Atzmon G, Klein L, Gundewar S, Ji SY, Lavu M, Predmore BL, Lefer DJ. Acute humanin therapy attenuates myocardial ischemia and reperfusion injury in mice. Arterioscler Thromb Vasc Biol. 2010;30:1940–8. doi: 10.1161/ATVBAHA.110.205997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C, Wan J, Miyazaki B, Fang Y, Guevara-Aguirre J, Yen K, Longo V, Bartke A, Cohen P. IGF-I regulates the age-dependent signaling peptide humanin. Aging Cell. 2014;13:958–61. doi: 10.1111/acel.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gidlund EK, von Walden F, Venojarvi M, Riserus U, Heinonen OJ, Norrbom J, Sundberg CJ. Humanin skeletal muscle protein levels increase after resistance training in men with impaired glucose metabolism. Physiol Rep. 2016;4(23):e13063. doi: 10.14814/phy2.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodhead JST, D'Souza RF, Hedges CP, Wan J, Berridge MV, Cameron-Smith D, Cohen P, Hickey AJR, Mitchell CJ, Merry TL. High-intensity interval exercise increases humanin, a mitochondrial encoded peptide, in the plasma and muscle of men. J Appl Physiol (1985) 2020;128:1346–54. doi: 10.1152/japplphysiol.00032.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu H, Tang S, Xue C, Liu Y, Wang J, Zhang W, Luo W, Chen J. Mitochondrial-Derived Peptide MOTS-c Increases Adipose Thermogenic Activation to Promote Cold Adaptation. Int J Mol Sci. 2019;20(10):2456. doi: 10.3390/ijms20102456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shokolenko IN, Alexeyev MF. Mitochondrial transcription in mammalian cells. Front Biosci (Landmark Ed) 2017;22:835–53. doi: 10.2741/4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Wang Q, Qu Z, Yang J, Zhang X, Zhao Y. Protective Effect of Hyperbaric Oxygen Therapy on Cognitive Function in Patients with Vascular Dementia. Cell Transplant. 2019;28:1071–5. doi: 10.1177/0963689719853540. [DOI] [PMC free article] [PubMed] [Google Scholar]