Abstract
Background
Epigenetics may play an important role in the pathogenesis of lung diseases. However, little is known about the epigenetic factors that influence impaired gas exchange at the lung.
Aim
To identify the epigenetic signatures of the diffusing capacity of the lung measured by carbon monoxide uptake (the diffusing capacity of the lung for carbon monoxide (DLCO)).
Methods
An epigenome-wide association study (EWAS) was performed on diffusing capacity, measured by carbon monoxide uptake (DLCO) and per alveolar volume (VA) (as DLCO/VA), using the single-breath technique in 2674 individuals from two population-based cohort studies. These were the Rotterdam Study (RS, the “discovery panel”) and the Framingham Heart Study (FHS, the “replication panel”). We assessed the clinical relevance of our findings by investigating the identified sites in whole blood and by lung tissue specific gene expression.
Results
We identified and replicated two CpG sites (cg05575921 and cg05951221) that were significantly associated with DLCO/VA and one (cg05575921) suggestively associated with DLCO. Furthermore, we found a positive association between aryl hydrocarbon receptor repressor (AHRR) gene (cg05575921) hypomethylation and gene expression of exocyst complex component 3 (EXOC3) in whole blood. We confirmed that the expression of EXOC3 in lung tissue is positively associated with DLCO/VA and DLCO.
Conclusions
We report on epigenome-wide associations with diffusing capacity in the general population. Our results suggest EXOC3 to be an excellent candidate, through which smoking-induced hypomethylation of AHRR might affect pulmonary gas exchange.
Short abstract
Epigenetic changes, including smoking-induced hypomethylation, may affect pulmonary gas exchange https://bit.ly/3k4ZdvH
Introduction
Tests for diffusing capacity of the lung for carbon monoxide (DLCO) and DLCO per alveolar volume (VA) (as DLCO/VA) provide a quantitative measure of gas exchange in the lung. In addition to its utility in the diagnosis and monitoring of lung diseases such as emphysema and pulmonary fibrosis, measure of gas exchange is an independent predictor of mortality in chronic obstructive pulmonary disease (COPD) patients [1]. DLCO and DLCO/VA measurements are influenced by environmental factors [2] and exposure to such factors (e.g. smoking, occupation related compounds and air pollution) decreases the level of gas exchange in the lungs [3].
While DLCO and DLCO/VA are heritable traits, the fraction of variation in these lung function tests unexplained by genetic variation suggests an important role for environmental interactions [4]. Although genetics play an important role, epigenetic mechanisms such as DNA methylation are important for regulation of gene expression but also essential in understanding the interplay between genes, environment and disease [5, 6]. Like DLCO and DLCO/VA, DNA methylation is also influenced by smoking [7, 8]; however, the question remains as to whether DNA methylation is associated with gas exchange independently of smoking.
Little is known about the effects of DNA methylation on lung function [9–11] and epigenome-wide association studies (EWAS) of pulmonary gas exchange are lacking. Therefore, we aimed to perform an EWAS in order to investigate which epigenetic variants are related to the phenotypic variation in DLCO and DLCO/VA. In addition, we examined the impact of the discovered epigenetic variants on gene expression in the lungs.
Methods
Study populations
This EWAS study encompassed a discovery study embedded within the Rotterdam Study (RS) and a replication study embedded within the Framingham Heart Study (FHS). The RS is an ongoing prospective population-based cohort study in Rotterdam, The Netherlands. The design has been previously described [12, 13]. Briefly, the RS includes four sub-cohorts. For this study, data from the third visit of the second sub-cohort (RSII-3) and the second visit of the third sub-cohort (RSIII-2) was used. The discovery panel consisted of 659 participants from a random subset of 747 individuals of European descent with methylation and diffusing capacity data available.
Replication of the identified CpG sites was performed in 2114 individuals from the FHS offspring cohort with methylation and diffusing capacity data available. The design of the FHS has been described extensively before [14]. The replication analyses were focused on offspring cohort participants of European descent who attended the eighth exam (2005–2008).
In both cohorts we only included participants >45-years old with blood cell counts and batch effects data available.
DNA methylation
DNA was extracted from whole peripheral blood (stored in ethylenediamine tetraacetic acid (EDTA) tubes) by standardised salting out methods. Genome-wide DNA methylation levels were measured using the Illumina Human Methylation 450 K array (Illumina, San Diego, CA, USA). In summary, samples (500 ng of DNA per sample) were first treated with bisulfite using the Zymo EZ-96 DNA methylation kit (Zymo Research, Irvine, CA, USA). Subsequently, samples were hybridised to the arrays according to the standardised protocols. The methylation percentage of a CpG site was presented as an effect estimate (β) value ranging between zero (no methylation) and one (full methylation). Data processing was performed in the Genetic Laboratory of the Dept of Internal Medicine, Erasmus University Medical Centre, Rotterdam. The Netherlands. Quality control was performed using Genome Studio version 2011.1, methylation module version 1.9.0 (Illumina). For each probe, individuals with methylation levels higher than three-times the interquartile range (IQR) were excluded. In both studies, cross-reactive and polymorphic probes (probes with an underlying single-nucleotide polymorphism (SNP) at the CpG site) were excluded, in addition to probes within 10 bp of the single-base extension (minor allele frequency >1% in European ancestry (EUR) 1000 Genomes Project data) [15]. Additionally, probes with a missing rate >20% at p<0.01 were excluded in the Framingham cohort. In total, 363 387 CpGs were included.
DLCO and DLCO/VA
DLCO (mmol·min−1·kPa−1) and VA (L) were measured by the single-breath technique using a Master Screen PFT Pro (CareFusion, San Diego, CA, USA) in accordance with European Respiratory Society (ERS)/American Thoracic Society (ATS) guidelines [16]. DLCO/VA (mmol·min−1·kPa−1·L−1) was calculated by dividing the DLCO by VA. Analyses were restricted to participants with two interpretable and reproducible measurements of DLCO and DLCO/VA. Two measurements were considered reproducible if the difference between the first and second DLCO measurement ((highest value–lowest value)/highest value) was ≤10% and the difference between the first and second DLCO/VA measurement was ≤15%.
Covariates
Covariates were selected based on known association with DNA methylation and diffusing capacity, and included age, sex, smoking status, weight, height and batch effects, as well as array number and position. Smoking status was obtained using a questionnaire. Cigarette pack years were computed as duration of smoking (years) multiplied by the number of smoked cigarettes, divided by 20.
Functional analysis
We used data from the Biobanking and Biomolecular Research Infrastructure (BBMRI) atlas [17] to identify methylation-gene expression associations (so called “expression quantitative trait methylation (eQTM)”). The BBMRI atlas includes all association studies performed by the BBMRI-NL consortium. We queried the database (http://bbmri.researchlumc.nl/atlas/#query) for association between the significant CpGs and differential gene expression and downloaded the original data tables (http://bbmri.researchlumc.nl/atlas/#data) to learn more about the direction of the estimated effects.
Gene expression in lung tissue
Lung resection specimens were obtained from 92 patients, of which 78 were from surgery for solitary pulmonary tumours (Ghent University Hospital, Ghent, Belgium) and 14 were from explant lungs of end-stage COPD patients undergoing lung transplantation (University Hospital Gasthuisberg, Leuven, Belgium). Lung tissue samples at maximum distance from the pulmonary lesions and without signs of retro-obstructive pneumonia or tumour invasion were collected by a pathologist. None of the patients operated on for malignancy were treated with neo-adjuvant chemotherapy. Written informed consent was obtained from all subjects. This study was approved by the medical ethical committees of Ghent University Hospital (2011/14) and the University Hospital Gasthuisberg (S51577).
RNA was extracted with the miRNeasy Mini kit (Qiagen, Hilden, Germany) from total lung tissue blocks submersed in RNA-later (Qiagen). cDNA was obtained by the miScript II RT kit (Qiagen), following manufacturer's instructions. Expression of target genes ADGRG6 (GPR126) and reference genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase-1 (HPRT-1) and succinate dehydrogenase complex flavoprotein subunit A (SDHA) was analysed using gene expression assay (TaqMan, Applied Biosystems, Forster City, CA, USA). Real-time PCR reactions were set up in duplicate using diluted cDNA under identical amplification conditions for each of the target and reference genes. A standard curve, derived from serial dilutions of a mixture of all samples, was included in each run. The amplification conditions were as follows: 10 min at 95 °C and 60 cycles of 95 °C for 10 s and 60 °C for 15 s. Amplification was performed using a LightCycler 96 detection system (Roche, Basel, Switzerland). Data were processed using the standard curve method. Expression of target genes was corrected by a normalisation factor calculated based on the expression of three reference genes, according to the guidelines and theoretical framework previously described [18].
Statistical analysis
Methylation probes were tested for association with DLCO or DLCO/VA using a linear regression model in the RS and a linear mixed model in the FHS (with a random effect to account for familial relationships in the FHS cohort). Surrogate variable analysis was used to adjust for inflation in the effect estimates by batch effect in the FHS. The first model (model M1) included age, sex, current smoking, former smoking, white blood cells, array number and position. In the second model (model M2), we additionally adjusted for weight and height. Model M2 was used for all sensitivity analyses. A false discovery rate (FDR) of 5% was used to correct for multiple testing (a FDR<0.05 was considered statistically significant). We tested the association between mRNA expression of aryl hydrocarbon receptor repressor (AHRR) and exocyst complex component 3 (EXOC3) genes and gas exchange in a linear regression adjusting for all covariates used in model M2. For these analyses, outliers (mean±three standard deviations (sd)) were excluded and mRNA levels were log transformed to make sure the normality assumption was fulfilled. A p-value of less than 0.05 was considered statistically significant for gene expression analysis.
Results
General characteristics
The general characteristics of the study populations are shown in table 1. Mean age was 67.4±5.9 years in the RS and 65.6±8.4 years in the FHS. Mean levels of DLCO and DLCO/VA were similar in both study populations.
TABLE 1.
General characteristics in the Rotterdam Study (RS) population (the “discovery panel”) and the Framingham Heart Study (FHS) population (the “replication” panel)
| Characteristic | Study | |
| RS (n=659) | FHS (n=2114) | |
| Age years | 67.4±5.90 | 65.6±8.38 |
| Female sex | 369 (56) | 1159 (55) |
| Current smokers | 70 (11) | 151 (7) |
| Ex-smokers | 379 (58) | 1186 (56) |
| Weight kg | 79.9±13.94 | 79.5±17.83 |
| Height cm | 169.8±9.16 | 167.3±9.54 |
| FEV1 L | 2.72 (0.71) | 2.68 (0.75) |
| % predicted FEV1 | 102.85 (22.82) | 97.94 (16.94) |
| FVC L | 3.56 (0.90) | 3.68 (0.98) |
| % predicted FVC | 108.93 (16.92) | 101.39 (15.09) |
| FEV1/FVC % | 76.65 (7.33) | 72.89 (6.86) |
| DLCO# mmol·min−1·kPa−1 | 7.81±1.64 | 7.47±2.13 |
| DLCO/VA# mmol·min−1·kPa−1·VA−1 | 1.52±0.23 | 1.42±0.23 |
Data are presented as mean±sd (for continuous variables) or n (%) (for dichotomous variables). FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; DLCO: diffusing capacity of the lung for carbon monoxide; VA: alveolar volume; sd: standard deviation. #: DLCO was corrected for haemoglobin levels in the RS. Interpretable spirometry was available in the RS (n=625) and FHS (n=2114).
Discovery phase
Data from the RS was used in the discovery phase of both EWAS for DLCO and DLCO/VA.
EWAS of DLCO
No statistically significant associations were found between DNA methylation and DLCO in analyses adjusted for age, sex, smoking status, white blood cell counts and batch effects (model M1), nor in analyses additionally adjusted for weight and height (model M2). However, cg05575921 (gene: AHRR, chromosome: five) was suggestively associated with DLCO in both models (FDRM1=0.07; FDRM2=0.09).
EWAS of DLCO/VA
In model M1, we identified one CpG site associated with DLCO/VA: cg05575921 (gene: AHRR, chromosome: five; FDR=0.017). In model M2, we identified two CpG sites associated with DLCO/VA: cg05575921 (gene: AHRR, chromosome: five; FDR=0.047) and cg05951221 (closest gene: ALPG, chromosome: two; FDR=0.047). Sensitivity analyses of the top associations, performed by additionally adjusting for smoking-related CpGs, forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC) or in never smokers, are presented in the supplementary material.
Replication phase
We replicated our findings using data from the FHS. The statistically significant associations between DNA methylation and DLCO/VA are summarised in table 2.
TABLE 2.
Epigenome-wide associations between genome-wide DNA methylation and diffusing capacity of the lung for carbon monoxide (DLCO)/alveolar volume (VA)
| Model | CpG | Chromosome | Position | Gene | RS | FHS | ||||
| Effect estimate | Standard error | FDR | Effect estimate | Standard error | FDR | |||||
| M1 | cg05575921 | 5 | 373 378 | AHRR | 0.065 | 0.012 | 0.017 | 0.037 | 0.0055 | 8.72×10−6 |
| M2 | cg05575921 | 5 | 373 378 | AHRR | 0.064 | 0.012 | 0.047 | 0.040 | 0.0057 | 1.56×10−6 |
| M2 | cg05951221 | 2 | 233 284 402 | ALPG# | 0.049 | 0.0093 | 0.047 | 0.030 | 0.0051 | 3.60×10−4 |
RS: Rotterdam Study (the “discovery panel”); FHS: Framingham Heart Study (the “replication panel”); FDR: false discovery rate; M1: model adjusted for age, sex, current smoking, former smoking, white blood cell count and batch effect; M2: model adjusted for age, sex, current smoking, former smoking, weight, height, white blood cell count and batch effect; #: closest gene (distance >8 Kbp).
Functional analysis
We used the BBMRI atlas to identify eQTMs using the identified DLCO/VA-associated CpG sites (cg05575921 and cg05951221). Cg05575921 (AHRR gene region) was significantly associated with the expression of the EXOC3 gene (effect estimate=0.15, standard error=0.039, FDR=3.9E-04) and no eQTM was retrieved between AHRR and AHRR expression. No eQTM was retrieved for cg05951221.
Sensitivity analyses
Although we adjusted for smoking in our analyses, residual confounding might still play a role. To further disentangle the potential role of smoking we performed two sensitivity analyses. First, we adjusted the analyses of the lead associations for well-known smoking CpGs with the highest explained variance according to the publication by Zeilingen et al. [8]. This resulted in our adjusting the analyses of the lead associations for each other and for another CpG (cg06126421, chromosome six). Secondly, we performed the analysis in a subset of 207 non-smokers. The results of the sensitivity analyses are presented in supplementary tables S1–S4. The results suggest that smoking partially explains the observed associations, supporting the idea that the effect of smoking on diffusing capacity is mediated by DNA methylation and that there is the potential for additional smoking-independent effects.
Gene expression in lung tissue
Based on the results of the previous section, we were interested in the association between EXOC3 and DLCO or DLCO/VA independent of smoking. As AHRR is strongly associated with smoking, we also investigated the association between AHRR and DLCO or DLCO/VA after adjustment for smoking. Therefore, mRNA was extracted from lung resection specimens of 92 patients who underwent surgery for lung transplantation or solitary pulmonary tumours (including 48 patients with COPD and 44 patients without COPD) and mRNA expression of AHRR and EXOC3 in lung tissue was examined using quantitative real-time PCR. General characteristics of this study population are given in table 3. Two outliers were excluded for AHRR and four for EXOC3.
TABLE 3.
Characteristics of study individuals (n=92) for lung mRNA analysis by real-time PCR
| Characteristics | Never smokers (n=18) | Smokers without COPD (n=26) | COPD GOLD II (n=34) | COPD GOLD III–IV (n=14) |
| Sex male/female | 6/12# | 19/7# | 31/3# | 8/6# |
| Age years | 65 (56–70) | 63 (55–70) | 66 (58–69)§ | 56 (54–60)¶,+,§ |
| Smoking status | ||||
| Current smoker | na | 16 | 22 | 0 |
| Ex-smoker | na | 10 | 12 | 14 |
| Smoking pack-years | na | 28 (15–45)¶ | 45 (40–60)¶,§ | 30 (25–30)¶,+,§ |
| DLCO mmol·min−1·kPa−1 | 21.58 | 23.30 | 17.22¶ | 2.9¶,+,§ |
| % predicted DLCO | 90.00 | 80.00 | 67.00¶ | 35.00¶,+,§ |
| DLCO/VA mmol·min−1·kPa−1·VA−1 | 4.64 | 3.89¶ | 3.51¶ | 0.87¶,+,§ |
| % predicted DLCO/VA | 102.85 | 91.00¶ | 87.00¶ | 58.50¶,+,§ |
| % predicted FEV1 | 102.00 | 95.00 | 68.00¶,§ | 25.50¶,+,§ |
| FEV1/FVC % | 78.00 | 75.00 | 56.00¶,§ | 32.00¶,+,§ |
Data are presented as n, n/n or median (IQR). COPD: chronic obstructive pulmonary disease; GOLD: Global Initiative for Chronic Obstructive Lung Disease; na: not applicable; DLCO: diffusing capacity of the lung for carbon monoxide; VA: alveolar volume; FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; IQR: interquartile range. #: p<0.001 (by Fisher's exact test); ¶: p<0.05 versus never smokers (by Mann–Whitney U-test); +: p<0.05 versus COPD GOLD II (by Mann–Whitney U-test); §: p<0.05 versus smokers without COPD (by Mann–Whitney U-test).
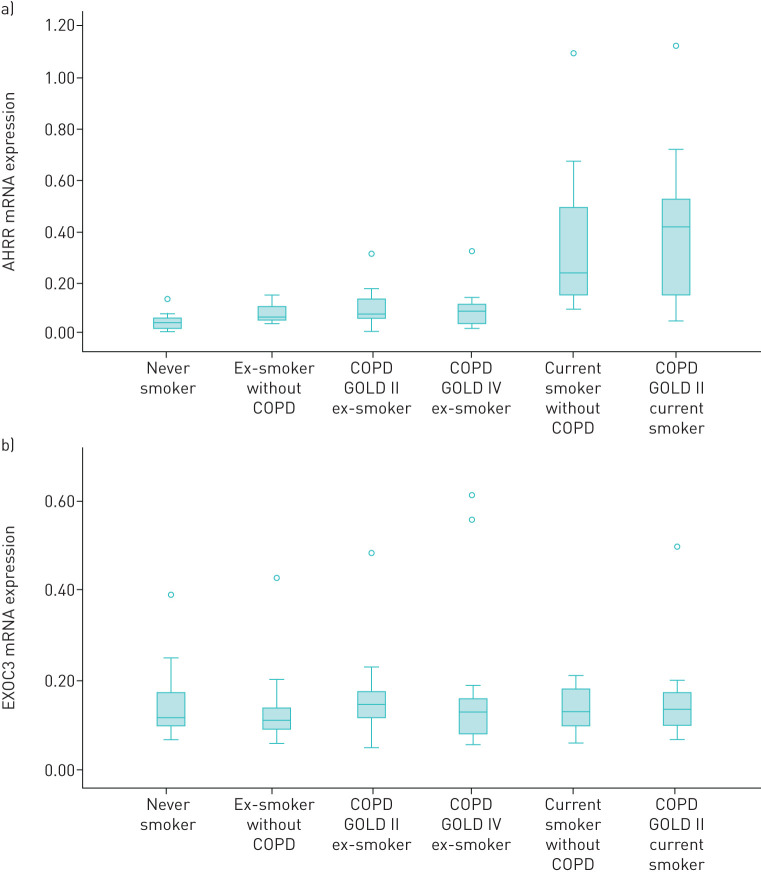
The expression levels of the AHRR and EXOC3 genes stratified by smoking status and COPD status are shown in figure 1. mRNA levels of AHRR were significantly higher in current smokers compared to never smokers or former smokers, while mRNA levels of EXOC3 were not significantly different between groups.
FIGURE 1.
Human lung tissue mRNA expression of (a) the aryl hydrocarbon receptor repressor (AHRR) gene and (b) the exocyst complex component 3 (EXOC3) gene stratified by smoking and chronic obstructive pulmonary disease (COPD) status. GOLD: Global Initiative for Chronic Obstructive Lung Disease.
Regression analysis revealed no significant association between mRNA expression of AHRR and DLCO or DLCO/VA after adjustment for age, sex and smoking status in model M1, or after additional adjustment for weight and height in model M2.
On the other hand, mRNA expression of EXOC3 showed no statistically significant association with DLCO or DLCO/VA in model M1; however, the association became significant after additional adjustment for weight and height in model M2 (supplementary table S1).
Discussion
In this EWAS, we investigated the epigenetic signature of DLCO and DLCO/VA as a measure of gas exchange in the lung. We observed two CpG sites with DLCO/VA epigenome-wide associations in the RS and those were replicated in the FHS. Additional analysis revealed a potential pathway through which methylation of AHRR might influence gas exchange.
This is the first EWAS on DLCO. Few studies have investigated the effect of DNA methylation on lung function and these were restricted to spirometry measures (FEV1 and FVC). Bell et al. [9] investigated age-related phenotypes, including FEV1 and FVC, in 172 twin females and reported one association at chromosome 11 in the WT1 gene region. Another study by Marioni et al. [10] performed an EWAS on FEV1 in 920 individuals and did not find epigenome-wide associations. Qui et al. [11] performed an EWAS on FEV1, FVC and COPD and found many associations, but the analyses were not adjusted for differences in white blood cell count.
In this EWAS, we discovered and confirmed two epigenome-wide associations, one CpG site in the AHRR gene, which encodes a repressor protein of the aryl hydrocarbon receptor (AHR) and a second CpG site on chromosome 2 (2q37.1). These CpG sites are well described as being strongly associated with smoking behaviour [7, 8, 19]. Cg05575921 is located in the AHRR gene region and is involved in the metabolism of smoking-released chemicals, where the AHRR gene suppresses the function of the AHR gene, which is responsible for the regulation of smoking-related substances through a negative feedback loop [20]. Although the interaction between the genetics and DNA methylation is complex, it is believed that AHRR hypomethylation inhibits the translation of the gene by preventing transcription factors from binding to the promotor regions [21, 22]. We hypothesise that AHRR hypomethylation by smoking might interfere with the elimination of the smoking-related substances from the body.
In addition to the association with smoking, AHRR has also recently been associated with impaired lung function. Bojesen et al. [19] have shown that hypomethylation of cg05575921 is associated with smoking-related phenotypes such as COPD, COPD exacerbations and lung cancer. Similarly, Kodal et al. [23] have reported that hypomethylation of cg05575921 is associated with low lung function, steeper lung function decline and respiratory symptoms. Finally, Imboden et al. [24] and Morrow et al. [25] have confirmed the CpG association with smoking and have suggested its potential use as a biomarker for predicting smoking-associated morbidity and mortality.
A recent study by Li et al. [20] has proved the causal effect of smoking on DNA methylation. This information eliminates the possibility for smoking to be an intermediate in the association between DNA methylation of smoking-related probes and lung function. Alternatively, smoking can be a confounding factor for which we can adjust in our models. However, residual confounding is still possible as assessment of smoking status is self-reported [26]. Therefore, the question remains: does residual confounding by smoking explain the whole effect in the association between smoking-related probes and lung function? Or is there any significant effect of DNA methylation on those smoking-related probes independent of smoking? Both scenarios lead to very interesting hypotheses and might have important clinical implications. In the case where smoking explains the entire association, the hypothesis might be (as proposed by Li et al. [20] and Kodal et al. [23]) that the association between hypomethylation of smoking-related probes and decreased lung function might reflect the smoking-induced damage to the lungs through methylation of the smoking-related probes. In other words, DNA methylation might act as an intermediate in the effect of smoking on smoking-related diseases. The second scenario would be that smoking affects DNA methylation, but that the effect of the hypomethylation of the smoking-related probes on lung function is (partly) independent of smoking. In this case, hypomethylation of the smoking-related probes might provide clinically relevant information beyond the effect of smoking on lung function. Unfortunately, it is difficult to investigate these hypotheses in cross-sectional observational studies. Nevertheless, our sensitivity analyses (which either additionally adjusted for smoking probes in the analysis or performed the main analysis in never smokers) revealed interesting results, whereby smoking explained only part of the association. We hypothesise that smoking as well as independent effects might both play a role at the same time in the epigenetic signature of diffusing capacity of the lungs. However, finding any association in never smokers is still no proof that smoking-related probes are independently associated with lung function, as passive smoking or the unmeasured effects of air pollution might still bias these associations.
Regardless of the role of smoking in the association between hypomethylation of AHRR and decreased pulmonary gas exchange, in this study we propose a pathway through which AHRR might affect lung function. We have observed that the hypomethylation of the cg05575921 site in AHRR is associated with decreased expression of the EXOC3 gene in whole blood. Subsequently, we have been able to link decreased expression of EXOC3 in human lung tissue to decreased gas exchange in the lungs.
EXOC3, previously known as SEC6, is located on chromosome 5 upstream of the AHRR gene and downstream of the SLC9A3 gene. EXOC3 is part of the exocyst protein complex and the protein encoded by it is involved in post-Golgi trafficking and is essential for biogenesis of epithelial cell surface polarity [27–29]. Variation in EXOC3 is linked to variability in the clinical phenotype of subjects with cystic fibrosis [30, 31]. This makes EXOC3 an interesting candidate for elucidation regarding its role in membrane pathology and gas exchange.
The strengths of this study include its population-based setting and standardised data collection. In addition, since we are not aware of any other population-based studies with information on gas exchange, it provides unique data. Furthermore, it provides additional results from gene expression analysis on lung tissue. The study also has some limitations. First, our discovery panel is relatively small and bigger sample sizes or joint analyses may help in the identification of more CpG sites that might give additional insight into the pathology of gas exchange. To ensure that our study is sufficiently powered to replicate our findings, we opted to use the smaller cohort in the discovery phase, as advocated by Held et al. [32]. Secondly, although we were able to replicate the findings of the lead association, we cannot exclude the possibility of unmeasured confounding by smoking and air pollution. Thirdly, due to the difficulty of obtaining lung samples from healthy subjects, our gene expression analysis included samples from lung cancer and end-stage COPD patients, potentially affecting the gene expression profile. Fourthly, the current study analyses the data in a cross-sectional manner. As the epigenome changes over time, longitudinal data-analyses might be more informative about the epigenetic modifications over time, as this may play a crucial role in the aetiology of decreased gas exchange. Finally, our methylation analyses used blood samples and may therefore not be fully representative of differential methylation in lungs as the target tissue. However, previous EWAS using lung samples have demonstrated that smoking-induced differential methylation in the AHRR locus (cg05575921) is implicated in lung carcinogenesis and have suggested that blood samples can be used as surrogates for lung samples in some methylation analyses [33].
In conclusion, impaired gas exchange is associated with smoking-related epigenetic changes. We propose a pathway through which AHRR hypomethylation might affect gas exchange.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary tables 00567-2020.supplement (174.9KB, pdf)
Acknowledgments
The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. The generation and management of the Illumina 450 K methylation array data (the EWAS data) for the Rotterdam Study was by the Human Genotyping Facility of the Genetic Laboratory of the Dept of Internal Medicine, Erasmus Medical Center, The Netherlands. We thank Michael Verbiest, Mila Jhamai, Sarah Higgins, Marijn Verkerk and Lisette Stolk for their help in creating the methylation database.
Footnotes
This article has supplementary material available from openres.ersjournals.com.
Support statement: This work was supported by a grant from the Fund for Scientific Research Flanders (FWO) (project G035014N). F.M. Verhame is a post-doctoral fellow of the FWO. The Rotterdam Study is funded by the Erasmus Medical Center and Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH) in collaboration with Boston University (contract numbers N01-HC-25195, HHSN268201500001I and 75N92019D00031). The Illumina 450 K methylation array data (the EWAS data) was funded by the Genetic Laboratory of the Dept of Internal Medicine, Erasmus Medical Center and by the Netherlands Organization for Scientific Research (project number 184021007) and made available as a Rainbow Project (RP3; BIOS) of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL). The laboratory work for this investigation was funded by the Division of Intramural Research, NHLBI, NIH (Bethesda, MD, USA) and the NIH Director's Challenge Award (PI: D. Levy). Funding information for this article has been deposited with the Crossref Funder Registry.
Ethical statement: The Rotterdam Study has been approved by the Medical Ethics Committee of the Erasmus Medical Center (registration number MEC 02.1015) and by the Dutch Ministry of Health, Welfare and Sport (Population Screening Act WBO, license number 1071272-159521-PG). The Rotterdam Study has been entered into the Netherlands National Trial Register (NTR; www.trialregister.nl) and into the WHO International Clinical Trials Registry Platform (ICTRP; www.who.int/ictrp/network/primary/en/) under shared catalogue number NTR6831. All participants provided written informed consent to participate in the study and to have their information obtained from treating physicians.
Conflict of interest: N. Terzikhan reports expert consultation for Boehringer Ingelheim GmbH and Novartis.
Conflict of interest: H. Xu has nothing to disclose.
Conflict of interest: A. Edris has nothing to disclose.
Conflict of interest: K.R. Bracke has nothing to disclose.
Conflict of interest: F.M. Verhamme has nothing to disclose.
Conflict of interest: B.H.C. Stricker has nothing to disclose.
Conflict of interest: J. Dupuis reports grants from the National Institutes of Health (NIH) during the conduct of the study.
Conflict of interest: L. Lahousse reports personal fees from AstraZeneca and Chiesi, outside the submitted work and expert consultation for Boehringer Ingelheim GmbH and Novartis.
Conflict of interest: G.T. O'Connor reports grants from the NIH during the conduct of the study.
Conflict of interest: G.G. Brusselle reports personal fees from AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Novartis and Teva, outside the submitted work and is a member of advisory boards for AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Sanofi/Regeneron and Teva.
References
- 1.Boutou AK, Shrikrishna D, Tanner RJ, et al. Lung function indices for predicting mortality in COPD. Eur Respir J 2013; 42: 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hegewald MJ. Diffusing capacity. Clin Rev Allergy Immunol 2009; 37: 159–166. [DOI] [PubMed] [Google Scholar]
- 3.Fricker M, Goggins BJ, Mateer S, et al. Chronic cigarette smoke exposure induces systemic hypoxia that drives intestinal dysfunction. JCI Insight 2018; 3: e94040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terzikhan N, Sun F, Verhamme FM, et al. Heritability and genome-wide association study of diffusing capacity of the lung. Eur Respir J 2018; 52: 1800647. [DOI] [PubMed] [Google Scholar]
- 5.Shah S, McRae AF, Marioni RE, et al. Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Res 2014; 24: 1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vercelli D. Genetics, epigenetics, and the environment: switching, buffering, releasing. J Allergy Clin Immunol 2004; 113: 381–386. Quiz 7. [DOI] [PubMed] [Google Scholar]
- 7.Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet 2013; 22: 843–851. [DOI] [PubMed] [Google Scholar]
- 8.Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 2013; 8: e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell JT, Tsai PC, Yang TP, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 2012; 8: e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marioni RE, Shah S, McRae AF, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 2015; 44: 1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu W, Baccarelli A, Carey VJ, et al. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am J Respir Crit Care Med 2012; 185: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikram MA, Brusselle GGO, Murad SD, et al. The Rotterdam Study: 2018 update on objectives, design and main results. Eur J Epidemiol 2017; 32: 807–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofman A, Brusselle GG, Darwish Murad S, et al. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol 2015; 30: 661–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kannel WB, Feinleib M, McNamara PM, et al. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol 1979; 110: 281–290. [DOI] [PubMed] [Google Scholar]
- 15.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013; 8: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macintyre N, Crapo RO, Viegi G, et al. Standardisation of the single-breath determination of carbon monoxide uptake in the lung. Eur Respir J 2005; 26: 720–735. [DOI] [PubMed] [Google Scholar]
- 17.Bonder MJ, Luijk R, Zhernakova DV, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet 2017; 49: 131–138. [DOI] [PubMed] [Google Scholar]
- 18.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bojesen SE, Timpson N, Relton C, et al. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax 2017; 72: 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Wong EM, Bui M, et al. Causal effect of smoking on DNA methylation in peripheral blood: a twin and family study. Clin Epigenetics 2018; 10: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986; 321: 209–213. [DOI] [PubMed] [Google Scholar]
- 22.Machin M, Amaral AF, Wielscher M, et al. Systematic review of lung function and COPD with peripheral blood DNA methylation in population based studies. BMC Pulm Med 2017; 17: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodal JB, Kobylecki CJ, Vedel-Krogh S, et al. AHRR hypomethylation, lung function, lung function decline and respiratory symptoms. Eur Respir J 2018; 51: 1701512. [DOI] [PubMed] [Google Scholar]
- 24.Imboden M, Wielscher M, Rezwan FI, et al. Epigenome-wide association study of lung function level and its change. Eur Respir J 2019; 54: 1900457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrow JD, Make B, Regan E, et al. DNA methylation is predictive of mortality in current and former smokers. Am J Respir Crit Care Med 2020; 201: 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shipton D, Tappin DM, Vadiveloo T, et al. Reliability of self reported smoking status by pregnant women for estimating smoking prevalence: a retrospective, cross sectional study. BMJ 2009; 339: b4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grindstaff KK, Yeaman C, Anandasabapathy N, et al. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell 1998; 93: 731–740. [DOI] [PubMed] [Google Scholar]
- 28.Oztan A, Silvis M, Weisz OA, et al. Exocyst requirement for endocytic traffic directed toward the apical and basolateral poles of polarized MDCK cells. Mol Biol Cell 2007; 18: 3978–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol 2014; 15: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorfman R. Modifier gene studies to identify new therapeutic targets in cystic fibrosis. Curr Pharm Des 2012; 18: 674–682. [DOI] [PubMed] [Google Scholar]
- 31.Corvol H, Blackman SM, Boelle PY, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun 2015; 6: 8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Held L. A new standard for the analysis and design of replication studies. J R Stat Soc Ser A Stat Soc 2020; 183: 431–448. [Google Scholar]
- 33.Stueve TR, Li WQ, Shi J, et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum Mol Genet 2017; 26: 3014–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary tables 00567-2020.supplement (174.9KB, pdf)