Abstract

While the discovery of immune checkpoint inhibitors has led to robust, durable responses in a range of cancers, many patients do not respond to currently available therapeutics. Therefore, an urgent need exists to identify alternative mechanisms to augment the immune-mediated clearance of tumors. Hematopoetic progenitor kinase 1 (HPK1) is a serine-threonine kinase that acts as a negative regulator of T-cell receptor (TCR) signaling, to dampen the immune response. Herein we describe the structure-based discovery of isofuranones as inhibitors of HPK1. Optimization of the chemotype led to improvements in potency, selectivity, plasma protein binding, and metabolic stability, culminating in the identification of compound 24. Oral administration of 24, in combination with an anti-PD1 antibody, demonstrated robust enhancement of anti-PD1 efficacy in a syngeneic tumor model of colorectal cancer.
Keywords: Hematopoetic progenitor kinase 1, HPK1, MAP4K1, cancer immunotherapy
Immunotherapy has revolutionized the treatment of cancer and offered new hope to patients. In particular, the introduction of checkpoint inhibitors that target the PD-1 and CTLA-4 pathways have, in many cases, led to robust, durable responses and increases in overall survival across a range of tumor histologies.1,2 Checkpoint inhibitors block inhibitory signals that would otherwise reduce the ability of T-cells to clear the tumor. Despite these advances, only a fraction of patients respond to currently available therapeutics. Therefore, there is an urgent need to identify medicines that target alternative signaling pathways to augment T-cell response.
Hematopoetic progenitor kinase 1 (HPK1 or MAP4K1) is a serine-threonine kinase that is expressed predominantly in cells of hematopoetic lineage.3 In T-cells, HPK1 acts as a negative regulator of T-cell receptor (TCR) signaling to dampen the immune response.4,5 Activation of the TCR recruits HPK1 to the plasma membrane, where it is activated via phosphorylation of Tyr381, Ser171, and Thr165. Once activated, HPK1 phosphorylates Ser376 of the adaptor protein SLP76, destabilizing the TCR signaling complex and inhibiting T-cell activation and proliferation. In vitro, T-cells derived from HPK1-deficient mice5 and HPK1 kinase-dead mice4,6 showed enhanced proliferation in response to TCR stimulation with anti-CD3 relative to wild type mice. Furthermore, kinase-dead mice were characterized by enhanced antitumor immunity in syngeneic tumor models, both in the absence and presence of anti-PD-L1. Importantly, neither knockout mice nor kinase-dead mice exhibited lethal inflammation in contrast with mice lacking other negative regulators such as CTLA-4. Taken together, this data suggests the potential utility of a small molecule HPK1 inhibitor in the treatment of cancer.
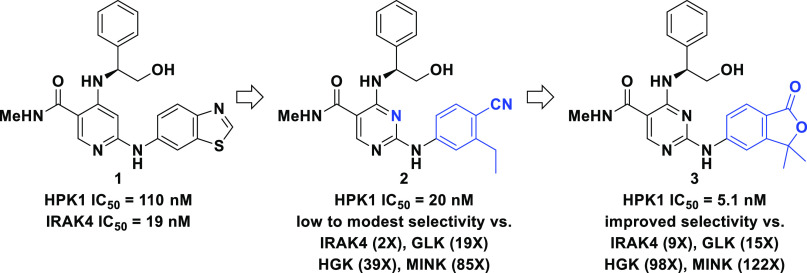
Mining of historical kinome selectivity data at Bristol Myers Squibb led to the identification of 1 (Figure 1), a compound prepared for our previously disclosed IRAK4 inhibitor program.7 This compound demonstrated very promising potency in our caliper-based HPK1 inhibition assay (HPK1 IC50 = 110 nM). When tested in a panel of >250 kinase assays, this compound demonstrated significant off-target kinase activity, inhibiting 25 kinases with an IC50 less than 1 μM. Unsurprisingly, its most potent activity was against IRAK4, where it showed more potent inhibition than to HPK1 (IRAK4 IC50 = 19 nM). Extensive structure–activity relationship studies were undertaken around this chemotype with a focus on improving kinase selectivity. Compound 2 emerged from this work as a lead compound, demonstrating improved potency against HPK1 (HPK1 IC50 = 20 nM) and at least 100-fold selectivity against 87.1% of the kinome (233 kinases tested). Despite these advances, compound 2 required additional optimization prior to advancement. Compound 2 retained potent IRAK4 inhibition (IC50 = 38 nM) and fell short of our goal of 100-fold selectivity against >90% of the kinome. Also, given the diverse roles that MAP4K kinases play in immune cell signaling, immune responses, and inflammation,8 we required high selectivity over other MAP4K family members (MAP4K2–6) such that we would not diminish the beneficial effects of HPK1 inhibition and would increase confidence that any observed in vivo effects were on-target. Whereas selectivity against MAP4K2 (GCK; 240×) and MAP4K5 (KHS; 550×) was high, additional optimization of family selectivity would be required as 2 exhibited low selectivity against MAP4K3 (GLK; 19×) and modest selectivity against MAP4K4 (HGK; 39×) and MAP4K6 (MINK; 85×).
Figure 1.
Evolution of kinome mining hit 1 to isofuranone lead 3.
At the time of this work, no X-ray crystal structures of HPK1 had been published, and we had not yet been able to generate a construct of the protein that yielded crystals, either alone or bound to a ligand.9−11 Instead, we relied on a homology model based on an X-ray crystal structure of the structurally related kinase, MST1 (PDB: 3COM; Figure 2). Docking of compound 2 into that homology model suggested that the pyrimidine, the aniline NH, and the amide NH formed hydrogen bonds to the extended hinge region of HPK1, consistent with the binding of this chemotype to IRAK4.7 Further examination of the homology model suggested the possibility of an intramolecular hydrogen bond between the amide oxygen and the phenylglycinol NH which may serve to orient the phenyl and hydroxymethyl substituents. When so oriented, the hydroxy substituent is well-positioned to form a hydrogen bond with the side chain of Asp101. Further, this model suggested the potential for the nitrile to form hydrogen bonds with Lys46 and/or the backbone NH of Asp155, placing the ethyl substituent into a pocket created by Lys46, Val31, and Tyr28. Examination of this homology model revealed that the ethyl substituent of 2 did not completely fill this pocket, suggesting that a larger substituent might occupy it fully and thereby confer selectivity against all kinases without the capacity to accommodate larger substituents. This hypothesis led to the design of 3 (Figure 1), a compound that fulfilled the requirement of increased steric bulk in the targeted pocket and featured a carbonyl in place of the nitrile. Surprisingly, despite the different geometries associated with the carbonyl (trigonal) and the nitrile (linear), molecular modeling suggested that the nitrogen and oxygen atoms occupied nearly identical positions and that both projected a lone pair of electrons capable of satisfying the putitive hydrogen bonding interactions (Figure 2 overlay). We were delighted to find that 3 was characterized by a 4-fold improvement in potency (HPK1 IC50 = 5.1 nM). While there was a slight erosion in GLK selectivity (15× vs 19×), selectivity against IRAK4, HGK, and MINK was improved.
Figure 2.

Docking of 2 and 3 into HPK1 homology model.
Our work in this new series began by surveying the effect of amide substitution on HPK potency and selectivity (Table 1). Extension of the nitrogen by one or two carbons (compounds 4, 6) gave a slight reduction in HPK1 potency, as did α-branching (5). Formation of the tertiary amide (7) led to ablation of all kinase activity, suggesting the need for a hydrogen bond donor. Deletion of the nitrogen substituent to give the primary amide (8) also reduced potency slightly. This brief survey of amide substituents gave no indication that kinase selectivity could be significantly enhanced using this approach. Therefore, amide bioisosteres were also examined. Replacement of the amide with a carboxylic acid (9), which would be deprotonated at physiologic pH, led to a dramatic reduction in potency and, again, suggested the importance of a hydrogen bond donor at this site. However, replacement of the amide with an ethyl ester (10), an amide isostere which lacks a hydrogen bond donor, was nearly equipotent to the parent N-methylamide. This raises the possibility that the loss of activity of tertiary amide 7 may not be due to the loss of its hydrogen bond donor, but instead may be due to the increased steric demand of that substituent. The ester improved selectivity against IRAK4 (46×), but offered little selectivity against GLK (11×). In contrast, replacement of the amide with an isoxazole (11) retained all of the HPK1 activity of 3 while increasing selectivity against both IRAK4 (182×) and GLK (23×).
Table 1. Structure–Activity Relationship of Amides and Amide Bioisosteres.

While 11 represented a milestone in the optimization of the series, a number of issues remained to be addressed (Table 2). Antagonism of GLK is expected to be associated with immunosuppression (a phenotype opposite that of HPK1 inhibition), and hence, minimal inhibition of this particular kinase was desired (selectivity > 30×). Also, despite its potent kinase inhibition in the caliper assay, 11 was 100 times less potent in a T-cell SLP76 phosphorylation inhibition assay (EC50 pSLP76 = 400 nM). We reasoned that this modest level of functional activity, coupled to its low free fraction in human serum (0.9% free), would ultimately limit in vivo efficacy. Finally, 11 was subject to moderate-to-high metabolic clearance in NADPH-supplemented mouse liver microsomes (63% remaining after a 10 min incubation) and was therefore unlikely to give adequate exposure during prospective efficacy studies in mice. It was against this backdrop that we sought to balance potency, selectivity, and in vitro ADME properties to deliver a compound suitable for advancement into efficacy studies.
Table 2. Structure–Activity Relationship of 1,2,4-Oxadiazole Substitution.
Percentage remaining after 10 min incubation with mouse liver microsomes.
We began our optimization work with a brief survey of substitutions at C3 of the oxadiazole. It was found that lengthening/substitution of the methyl group (12–14) led to small increases in biochemical potency in the biochemical assay but adversely impacted stability in mouse liver microsomes. Replacement of the methyl with a cyclopropyl gave 15, which was essentially equipotent to 11 but offered improved microsomal stability. Substitution with a 4-fluorophenyl (16) eroded biochemical potency by 7-fold, whereas the 4-pyridyl oxadiazole 17 exhibited improved biochemical inhibition of HPK1. Unfortunately, despite the incorporation of the more polar pyridine, neither protein binding nor pSLP76 potency was improved. Our binding model indicated that this substituent would be projected toward solvent and therefore could not be used to rationalize the origin of this potency increase. We hypothesized that the basic nitrogen was making a favorable interaction with the solvent-exposed surface of the kinase, a hypothesis that we sought to explore further with additional basic functional groups in this region. This approach led to the identification of N-methylpiperidine 18, which demonstrated subnanomolar potency and an improved free fraction. Importantly, these attributes translated into improved cellular potency (pSLP76 IC50 = 31 nM) but further eroded mouse microsomal stability and selectivity against GLK. Suspecting that 18 might be subject to N-demethylation in liver microsomes, we designed quinuclidine 19 to mitigate this issue. Gratifyingly, this compound retained the exquisite potency of 18 and offered improved metabolic stability, but unfortunately, it was nearly equipotent against GLK. In addition to losses in GLK selectivity, both 18 and 19 demonstrated substantial inhibition in a hERG binding assay (IC50 = 2.5 and 0.57 μM, respectively). Due to the loss of GLK selectivity and the introduction of hERG inhibition associated with incorporation of basic amines at C3, we sought alternative strategies to drive increases in potency and free fraction.
Matched pair analysis has shown that 1,3,4-oxadiazoles show decreased lipophilicity relative to otherwise identical 1,2,4-oxadiazoles due to their increased dipole moments.12 Therefore, we opted to employ them in this series and were pleased to find that the methyl-substituted 1,3,4-oxadiazole 20 retained the potency of the identically substituted 1,2,4-oxadiazole 11 (Table 3). As hoped, the reduced lipophilicity led to an increase in free fraction from 0.9% to 2.0% free in human. While there was some loss in IRAK4 selectivity, GLK selectivity was nearly double that observed for the corresponding 1,2,4-oxadiazole. We incorporated many of the preferred fragments from our work in the 1,2,4-oxadiazole series and were surprised that earlier structure–activity relationship (SAR) trends were not recapitulated in this closely related set of analogues. Whereas larger substituents had led to slight potency increases in the earlier series, the opposite trend was apparent among the 1,3,4-oxadiazoles. For instance, cyclopropane 21 was about half as potent as methyl 20. It was also surprising that incorporation of a basic piperidine (22) or quinuclidine (23) reduced potency, again opposite to what had been observed in the 1,2,4-oxadiazole series. For instance, incorporation of the quinuclidine gave a 7-fold reduction in potency (vs a 20-fold increase for the analogous change in the 1,2,4-oxadiazole). Given the apparent inverse correlation between size and biochemical potency, we prepared unsubstituted oxadiazole 24. While this compound did not further increase biochemical potency, it did retain the potency of 20 while offering a modest increase in selectivity against GLK (54×). This compound demonstrated appreciable human free fraction (2.6% free), moderate metabolic clearance in human and mouse liver microsomes, and the most potent inhibition of T-cell SLP76 phosphorylation of any compound in this series. Selectivity was improved against all MAP4K family members, including GCK (390×), GLK (54×), HGK (466×), KHS (395×), and MINK (577×). Furthermore, when tested in a panel of >300 kinase inhibition assays, 24 was >100-fold selective against 92% of kinases evaluated.
Table 3. Structure–Activity Relationship of 1,3,4-Oxadiazole Substitution.
Percentage remaining after 10 min incubation with human or mouse liver microsomes.
Binding of 24 to HPK1 was confirmed by cocrystallization.13 The X-ray crystal structure was remarkably similar to that predicted by the homology model, showing the predicted hinge contacts between N3 of the pyrimidine and the aniline NH (Figure 3). Homology modeling of secondary amide 3 had predicted a third hydrogen bond with Gly95 on the extended hinge. It is interesting to note that the oxadiazole of 24, which lacks a hydrogen bond donor, was found to reside in very close proximity to Gly95, suggesting that it retains significant van der Waals contacts with that residue. Additionally, it was found that the carbonyl of the isofuranone formed a hydrogen bond with Asp155, but not with Lys46.
Figure 3.

X-ray crystal structure of 24 bound to HPK1 (PDB: 7KAC).
Following intravenous administration (1 mg/kg) to mice, 24 was characterized by moderate plasma clearance (43 mL/min/kg) and a large volume of distribution (4.4 L/kg). After oral administration (20 mg/kg), the Cmax was 5.3 μM and the AUC0–24h was 19 μM·h. The calculated oral bioavailability based on these pharmacokinetics studies was approximately 100%. Supported by these favorable properties, 24 was advanced into a colorectal syngeneic tumor model (MC38, murine colon adenocarcinoma). An oral dosing regimen (100 mg/kg twice daily) was selected to approach coverage of the mouse whole blood pSLP76 IC50 (6 μM) at 24 h. Previous studies using HPK1 kinase-dead mice had demonstrated only a modest effect on tumor growth inhibition (relative to wild-type controls) in the absence of anti-PD1 but showed enhanced efficacy in combination with PD-1 blockade.4 Therefore, this study compared three groups: 24 in combination with anti-PD1, anti-PD1 alone, and an isotype control antibody (Figure 4). In this study, 24 enhanced the efficacy of anti-PD1 treatment, garnering a 100% cure rate vs a 20% cure rate with anti-PD1 alone.14 Importantly, 24 was well-tolerated and did not produce any significant changes in body weight.
Figure 4.

Efficacy in the MC38 syngeneic tumor model. Compound 24 was administered orally (100 mg/kg twice daily) through day 28. Tumor volumes were monitored for 58 days.
All HPK1 inhibitors disclosed herein can be prepared as outlined in Scheme 1.15 The convergent synthesis begins with a Suzuki–Miyaura coupling of bromide 25 with the pinacol ester of 2-isopropenyl boronic acid to afford styrene 26. Mukaiyama hydration of 26 proceeded smoothly in the presence of oxygen and tris(dipivaloylmethanato)manganese(III) to give the semistable acyclic tertiary alcohol, which upon treatment with sodium bicarbonate underwent smooth cyclization onto the pendant nitrile to give isofuranone 27. Installation of the amino alcohol was accomplished through a highly regioselective SNAr reaction of (S)-phenylglycinol with commercially available dichloropyrimidine 28 under basic conditions. Replacement of the remaining chloride with amino-isofuranone 27 proceeded smoothly using acid catalysis to afford intermediate 10 in 91% yield.
Scheme 1.

Reagents and conditions: (a) 2-isopropenyl boronic acid pinacol ester, PdCl2(dppf)(CH2Cl2), K2CO3, 80 °C, 93%; (b) O2, Mn(dpm)3, PhSiH3, 0 °C then NaHCO3 in DMF/H2O, 85%; (c) DIEA, CH3CN, 84%; (d) 4 M HCl, NMP, 80 °C, 91%; (e) (i) NaOH, H2O/EtOH, 100%; (ii) R′RNH, BOP reagent, DIEA, DMF; (f) R(CNH)NHOH, NaOEt, 3 Å mol sieves, EtOH, 70 °C; (g) (i) hydrazine, EtOH, 70 °C, 99%; (ii) RCO2H, EDC, HOBt, DIEA, DMF, 40 °C then TsCl, TEA, 55 °C.
Selection of appropriate conditions allowed diversion of ethyl ester 10 into a variety of amides and amide bioisosteres. Basic saponification afforded the carboxylic acid in quantitative yield. BOP-mediated coupling of the acid to a variety of amines gave the corresponding amides (30) in high yields. Alternately, treatment of ethyl ester 10 with an appropriately substituted N-hydroxyimidamide in the presence of sodium ethoxide led directly to the corresponding 1,2,4-oxadiazole (31) in a single chemical step. To access 1,3,4-oxadiazoles, ester 10 could be directly converted to the hydrazide by treatment with hydrazine in ethanol. EDC-mediated condensation of the hydrazide with a variety of carboxylic acids, followed by dehydration using tosyl chloride, afforded the corresponding 1,3,4-oxadiazoles (32).
In conclusion, the development of an HPK1 homology model enabled the structure-based discovery of a series of potent and selective HPK1 inhibitors. This homology model suggested that an isofuranone could serve as a bioisostere for a cyanophenyl ring and led to the identification of 3. Diversification of a late-stage intermediate permitted the exploration of SAR to assist in optimizing the series for potency, selectivity, plasma protein binding, and metabolic stability. These studies culminated in the identification of 24, a potent HPK1 inhibitor with excellent family and kinome selectivity. The high oral bioavailability of this compound facilitated its advancement into an MC38 syngeneic tumor model where it demonstrated robust efficacy in combination with anti-PD1.
Acknowledgments
We thank the Department of Chemical Synthesis for the scale-up of intermediates. We also thank Spectrix and Discovery Analytical Sciences for final compound purification and characterization.
Glossary
Abbreviations
- GCK
germinal center kinase
- GLK
germinal center kinase-related protein kinase
- HGK
hepatocyte progenitor kinase-like/germinal center kinase-like kinase
- HPK1
hematopoetic progenitor kinase 1
- IRAK4
interleukin-1 receptor-associated kinase 4
- KHS
kinase homologous to SPS1/STE20
- MAPK
mitogen-activated protein kinase
- MC38
murine colon adenocarcinoma 38
- MINK1
misshapen-like kinase 1
- MST1
macrophage-stimulating protein 1
- PDB
Protein Data Bank
- pSLP76
phospho-SH2 domain-containing leukocyte protein of 76 kDa
- SLP76
SH2 domain-containing leukocyte protein of 76 kDa
- TCR
T-cell receptor
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00660.
Details for the synthesis and characterization of 3–24; kinase selectivity data for 24; experimental details for in vitro assays and murine tumor model (PDF)
The authors declare no competing financial interest.
Notes
The coordinates of HPK1:24 were deposited under PDB ID 7KAC.
Supplementary Material
References
- Ribas A.; Wolchok J. D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350. 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkona S.; Diamandis E. P.; Blasutig I. M. Cancer immunotherapy: the beginning of the end of cancer?. BMC Med. 2016, 14, 73. 10.1186/s12916-016-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M.; Fagerberg L.; Hallström B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson Å.; Kampf C.; Sjöstedt E.; Asplund A.; Olsson I.; Edlund K.; Lundberg E.; Navani S.; Szigyarto C. A.; Odeberg J.; Djureinovic D.; Takanen J. O.; Hober S.; Alm T.; Edqvist P. H.; Berling H.; Tegel H.; Mulder J.; Rockberg J.; Nilsson P.; Schwenk J. M.; Hamsten M.; von Feilitzen K.; Forsberg M.; Persson L.; Johansson F.; Zwahlen M.; von Heijne G.; Nielsen J.; Pontén F. Tissue-based map of the human proteome. Science 2015, 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Hernandez S.; Qing J.; Thibodeau R. H.; Du X.; Park S.; Lee H.-M.; Xu M.; Oh S.; Navarro A.; Roose-Girma M.; Newman R. J.; Warming S.; Nannini M.; Sampath D.; Kim J. M.; Grogan J. L.; Mellman I. The Kinase Activity of Hematopoietic Progenitor Kinase 1 Is Essential for the Regulation of T Cell Function. Cell Rep. 2018, 25, 80. 10.1016/j.celrep.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Shui J.-W.; Boomer J. S.; Han J.; Xu J.; Dement G. A.; Zhou G.; Tan T.-H. Hematopoetic progenitor kinase 1 negatively regulates T cell receptor signaling and T cell-mediated immune responses. Nat. Immunol. 2007, 8, 84. 10.1038/ni1416. [DOI] [PubMed] [Google Scholar]
- Liu J.; Curtin J.; You D.; Hillerman S.; Li-Wang B.; Eraslan R.; Xie J.; Swanson J.; Ho C.-P.; Oppenheimer S.; Warrack B. M.; McNaney C. A.; Nelson D. M.; Blum J.; Kim T.; Fereshteh; Reily M.; Shipkova P.; Murtaza A.; Sanjuan M.; Hunt J. T.; Salter-Cid L. Critical role of kinase activity of hematopoetic progenitor kinase 1 in anti-tumor immune surveillance. PLoS One 2019, 14, e0212670. 10.1371/journal.pone.0212670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhide R. S.; Keon A.; Weigelt C.; Sack J. S.; Schmidt R. J.; Lin S.; Xiao H.-Y.; Spergel S. H.; Kempson J.; Pitts W. J.; Carman J.; Poss M. A. Discovery and structure-based design of 4,6-diaminonicotinamides as potent and selective IRAK4 inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 4908. 10.1016/j.bmcl.2017.09.029. [DOI] [PubMed] [Google Scholar]
- Chuang H.-C.; Wang X.; Tan T.-H. MAP4K Family Kinases in Immunity and Inflammation. Adv. Immunol. 2016, 129, 277. 10.1016/bs.ai.2015.09.006. [DOI] [PubMed] [Google Scholar]
- For recently published structures of HPK1, see refs (10) and (11).
- Johnson E.; McTigue M.; Gallego R. A.; Johnson T. W.; Timofeevski S.; Maestre M.; Fisher T. S.; Kania R.; Sawasdikosol S.; Burakoff S.; Cronin C. N. Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation. J. Biol. Chem. 2019, 294, 9029. 10.1074/jbc.AC119.007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.; Sneeringer C. J.; Pitts K. E.; Day E. S.; Chan B. K.; Wei B.; Lehoux I.; Mortara K.; Li H.; Wu J.; Franke Y.; Moffat J. G.; Grogan J. L.; Heffron T. P.; Wang W. Hematopoetic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer. Structure 2019, 27, 125. 10.1016/j.str.2018.10.025. [DOI] [PubMed] [Google Scholar]
- Boström J.; Hogner A.; Llinàs A.; Wellner E.; Plowright A. T. Oxadiazoles in Medicinal Chemistry. J. Med. Chem. 2012, 55, 1817. 10.1021/jm2013248. [DOI] [PubMed] [Google Scholar]
- Lau W. L.; Pearce B.; Malakian H.; Rodrigo I.; Xie D.; Gao M.; Marsilio F.; Chang C.; Ruzanov M.; Muckelbauer J. K.; Newitt J. A.; Lipovsek D.; Sheriff S. Using Yeast Surface Display to Engineer a Soluble and Crystallizable Construct of HPK1. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2021, 77, 22. 10.1107/S2053230X20016015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D.; Hillerman S.; Locke G.; Chaudhry C.; Stromko C.; Murtaza A.; Fan Y.; Koenitzer J.; Chen Y.; Briceno S.; Bhadra R.; Duperret E.; Gullo-Brown J.; Gao C.; Zhao D.; Feder J.; Curtin J.; Degnan A. P.; Kumi G.; Wittman M.; Johnson B. M.; Parrish K. E.; Gokulrangan G.; Morrison J.; Quigley M.; Hunt J. T.; Salter-Cid L.; Lees E.; Sanjuan M. A.; Liu J. Enhanced anti-tumor immunity by a novel small molecule HPK1 inhibitor. J. Immunother. Cancer 2021, 9, e001402. 10.1136/jitc-2020-001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo E. M. V.; Chen Y.; Dasgupta B.; Degnan A. P.; Hill M. D.; Kumi G. K.; Mastalerz H. A.; Wittman M. D.; Pearce B. C.; Zhang G.. Furanone compounds useful as HPK1 inhibitors in the treatment of cancer and viral infections and their preparation. WO2019090198, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.