Abstract
Background
Diffuse large B‐cell lymphoma (DLBCL) is the most common category of non‐Hodgkin lymphoma (NHL). However, the underlying molecular mechanism of DLBCL remains unclear.
Methods
Real‐time PCR and Western blot analysis were performed to assess the expression of ubiquitin‐specific peptidase 21 (USP21) or enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2). CCK8 assay and cell death staining were carried out to examine the role of USP21 in cell proliferation and cell death, respectively.
Results
We found that the deubiquitinase USP21 was highly expressed in the DLBCL lymphoid tissue. The expression of USP21 promoted DLBCL cell proliferation, while it had no obvious effect on cell death. In addition, we found that USP21 regulated cell proliferation via cysteine 221, the catalytic site of USP21. Furthermore, we identified that USP21 could stabilize EZH2, a protein required for germinal center formation and lymphoma formation.
Conclusion
The deubiquitinase USP21 promotes cell proliferation by maintaining the EZH2 protein level in DLBCL.
Keywords: cell proliferation, deubiquitination, diffuse large B‐cell lymphoma, EZH2, USP21
USP21 is enriched in diffuse large B‐cell lymphoma (DLBCL) and is beneficial for tumor cell proliferation. Mechanistically, USP21 promotes cell growth by maintaining EZH2 protein level. Our data suggest that USP21 can act as a promising therapeutic target for DLBCL.
1. INTRODUCTION
Diffuse large B‐cell lymphoma (DLBCL) is the most common category of non‐Hodgkin lymphoma (NHL), accounting for approximately 30% of NHL cases worldwide. 1 , 2 , 3 DLBCL is more prevalent in elderly patients and tumor mass growing one or more lymph nodes and extranodal sites. 2 , 4 , 5 The high incidence rate and heterogeneity of DLBCL have attracted extensive attention. 6 , 7 , 8 Hence, it is important to reveal the underlying molecular mechanism of the origins and development of DLBCL for medical diagnosis and treatment.
USP21, a member of the ubiquitin‐specific protease (USP) family, plays particularly crucial roles in regulating cellular signaling and disease development. 9 , 10 , 11 , 12 USP21 is not only localized at microtubule to regulate microtubule dynamics but also posit on centriole to regulate primary cilium formation. 13 , 14 , 15 , 16 , 17 Moreover, USP21 plays a vital role in stem cell differentiation by regulating the polyubiquitination of Nanog. 18 , 19 USP21 can modulate cell cycle progression by deubiquitinating Forkhead box M1 (FOXM1) in basal‐like breast cancer. 20 However, the role of USP21 in DLBCL development has not been clarified.
In our study, we find that the USP21 mRNA level is enriched in DLBCL patients, which suggests that USP21 was associated with DLBCL growth. USP21 knockdown or overexpression in the DLBCL cell line shows that USP21 promotes cell proliferation. Furthermore, we then identify USP21 modulates the protein level of EZH2, a key regulatory gene of DLBCL growth.
2. MATERIALS AND METHODS
2.1. Cell culture
The DLBCL cell line SU‐DHL‐4 and A20 were acquired from the American Tissue Culture Collection (ATCC). The cells were grown in RPMI 1640 medium (BI) supplemented with 10% fetal bovine serum (BI) and 100 U/ml Penicillin/Streptomycin (Sigma‐Aldrich) at 37℃ in a 5% CO2 humidified atmosphere.
2.2. Plasmid and cell transfection
Full‐length USP21 was amplified by PCR from the cDNA library and cloned into pcDNA3.1 or pEGFP‐N1 vectors. The USP21 siRNAs (#1: 5′‐GCUAGAAGAACCUGAGUUA‐3′; #2: 5′‐GAGCUGUCUUCCAGAAAUA‐3′) were synthesized by RiboBio. Cell transfection was performed as previously described. 21
2.3. CCK‐8 assay
Cell counting kit‐8 (CCK‐8) assay was used to measure cell viability and was performed as previously described. 22 Cells were inoculated into a 96‐well plate, with 1 × 104 cells in each well, after transfection for 48 h. 10 μl CCK‐8 was added into the well, and then, cells were incubated for 2 h at 37℃. Then, OD values at 450 nm and 630 nm of the cells were detected by a microplate reader.
2.4. Cell death detection
Generally, PI can enter into the dead cells, but cannot enter into the viable cells. 23 After 48 h of cell transfection, the cells were inoculated into 96 well plates. PI (10 μg/ml) was then added into the wells, and cells were incubated for 10 min at 37℃. All images were captured by Leica Inverted Microscope.
2.5. Real‐time PCR
Total RNA was harvested and isolated from DLBCL patient tissue using TRIzol reagent (Invitrogen). Reverse transcription was performed as previously described. 24 All patients were informed that their tissue sample would be used for research before this study was carried out. RNA was reverse transcripted to synthesize cDNA using a reverse transcription kit (Takara). 25 The relative expression of USP21 mRNA was normalized against GAPDH using SYBR Premix Ex Taq enzyme (Thermo Fisher Scientific) and LightCycler® 480 Instrument II (Roche Life Science). The primer sequences were listed as follows: USP21 forward primer: 5′‐GCAGGATGCCCAAGAGTT‐3′; USP21 reverse primer: 5′‐GCAGGGACAGGTCACAAAA‐3′; GAPDH forward primer: 5′‐TGTGTCCGTCGTGGATCTGA‐3′; GAPDH reverse primer: 5′‐CACCACCTTCTTGATGTCATCATAC‐3′.
2.6. Western blot
Western blot was performed as previously described. 26 Total protein was harvested and extracted from cells using cell lysis buffer (1 mM EDTA, 150 mM NaCl, 50 mM Tris‐HCL,1% NP‐40, 3% glycerol). 27 The protein was separated by sodium dodecyl sulfate polyacrylamide‐gel electrophoresis (SDS‐PAGE) and transferred to PVDF membranes (Millipore). After blocking for 1 h with 5% skim milk in TBST buffer, the membranes were incubated with indicated primary antibodies at 4℃ overnight or 1 h at room temperature and then probed with HRP‐conjugated secondary antibodies. Chemiluminescence detection was performed with ECL reagents (Millipore).
2.7. Statistical analysis
All statistical analyses were performed with the GraphPad Prism 8 statistical software, and the statistical significance was analyzed by Student's t test. The data were represented as the mean ± SEM, and the difference was considered significant at *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3. RESULTS
3.1. USP21 is enriched in DLBCL tissues and cell lines
To explore the effect of USP21 on DLBCL, we firstly investigated the Gene Expression Profiling Interactive Analysis (GEPIA) database and found that USP21 is highly expressed in tumor samples compared with paired normal tissues (http://gepia.cancer‐pku.cn/detail.php?gene=USP21) (Figure 1A). Furthermore, the survival curve of the patients with high expression of USP21 or low expression of USP21 was examined, which shows high expression of USP21 correlated well with the low survival probability of the patients (Figure 1B). Then, we examined the mRNA expression level of USP21 in DLBCL tumor tissues, which was identified higher than adjacent normal tissues (Figure 1C). Consistently, the expression level of USP21 protein in DLBCL was higher than other normal cells (Figure 1D). Thus, these results illustrated that USP21 is enriched in DLBCL.
FIGURE 1.
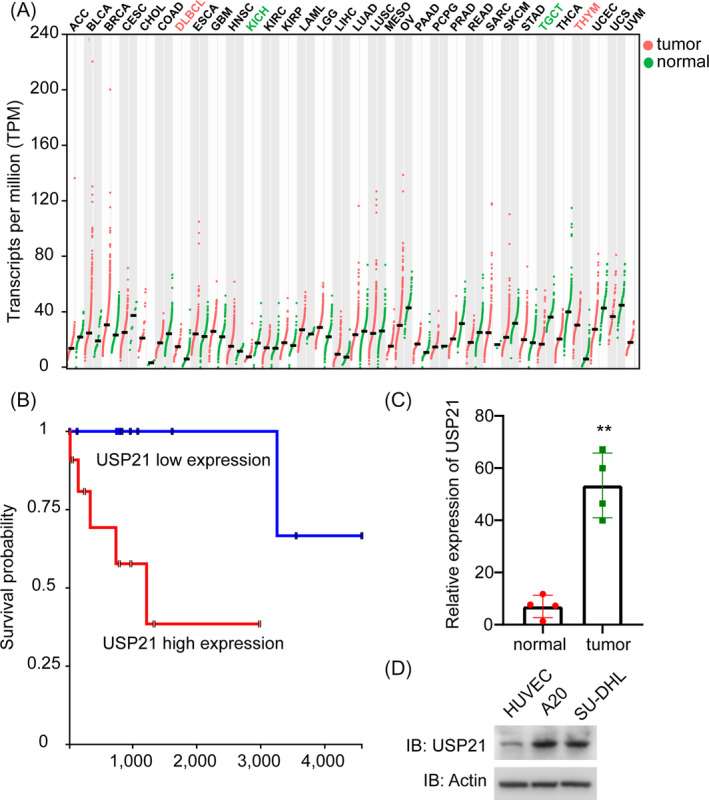
USP21 is highly expressed in DLBCL tissues and cell lines. (A) Database analysis shows that USP21 is enriched in DLBCL cases. (B) Kaplan‐Meier method analysis shows that USP21 expression correlates with the survival curve. (C) The mRNA level of USP21 in adjacent normal tissue and tumor tissue. **p < 0.01. Error bars indicate ±SEM. (D) The protein expression of USP21 in DLBCL cell lines (A20 and SU‐DHL‐4) and normal cell lines (HUVEC)
3.2. USP21 knockdown restrains cell proliferation in DLBCL cells
Then, we induced depletion of USP21 using siRNA to explore the role of USP21 in tumor progression of DLBCL (Figure 2A). USP21 knockdown dramatically decreased cell proliferation rate examined by CCK8 assay (Figure 2B). To further illustrate whether USP21 affects cell death, we examined the percentage of dead cells (PI‐positive) (Figure 2C). USP21 depletion did not interfere with cell death compared to control siRNA (Figure 2D). Therefore, USP21 stimulates cell proliferation but cannot cause cell death.
FIGURE 2.
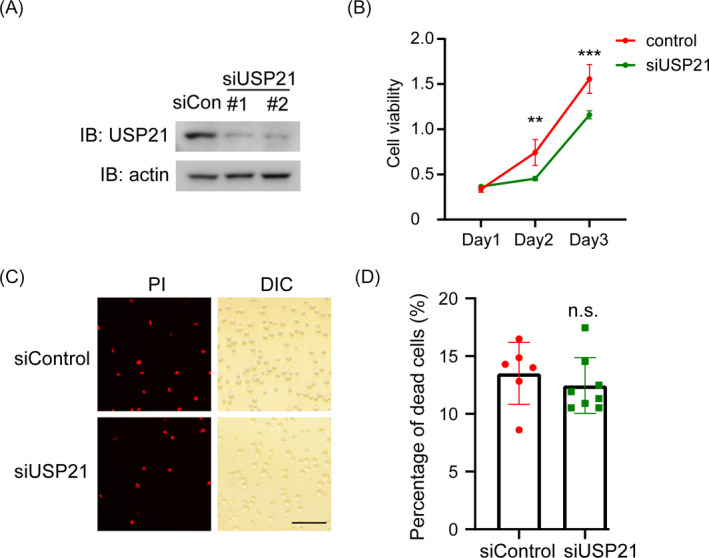
USP21 knockdown restrains cell proliferation. (A) Western blots show USP21 protein expression in SU‐DHL‐4 cells after transfection with control or USP21 siRNAs for 48 h. (B) Cell proliferation of SU‐DHL‐4 cells transfected with USP21 siRNA were measured by CCK8 assay. The siRNA was scaled down to siUSP21#1 only. (C) Representative images of Propidium Iodide (PI)‐positive cells (Red) representing dead cells and DIC representing total cells. Scale bar, 100 μm. The siRNA was scaled down to siUSP21#1 only. (D) Quantification the number of dead cells. n.s., not significant; **p < 0.01; ***p < 0.001. Error bars indicate ±SEM
3.3. The deubiquitinating activity of USP21 is required for DLBCL cell proliferation
USP21 acts as an efficient deubiquitinating enzyme. To examine the deubiquitinating activity of USP21 on DLBCL growth, we overexpressed USP21 wild‐type or catalytically inactive mutant C221A (CA) in SU‐DHL‐4 cells (Figure 3A). We found that overexpression of USP21 dramatically increased cell proliferation, while USP21‐CA could not (Figure 3B). In accordance with the USP21 knockdown results, there is no difference between USP21 expression and USP21‐CA expression in inducing cell death (Figure 3C,D). These data demonstrate that USP21 promotes cell proliferation via its deubiquitinating active site.
FIGURE 3.
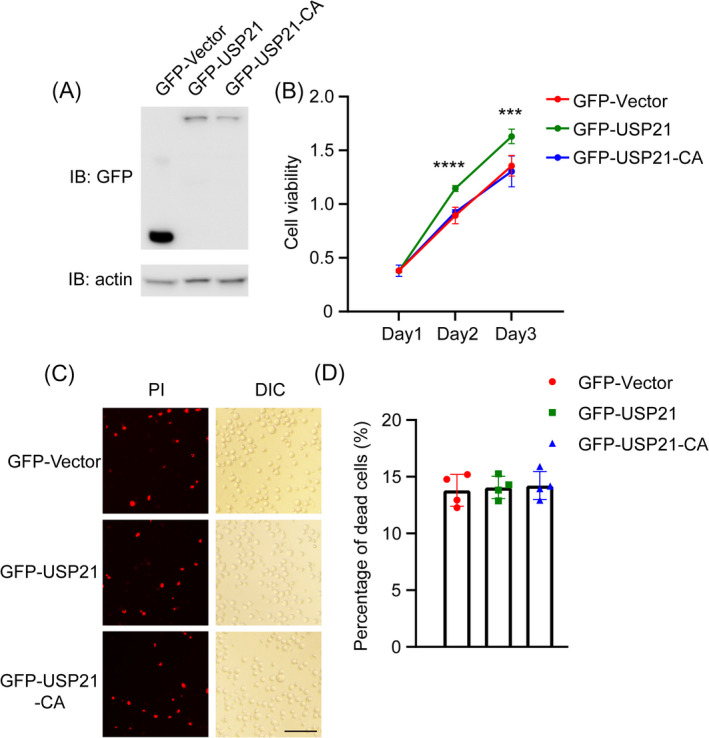
USP21 promotes cell proliferation via Cys‐221. (A) Overexpression of GFP‐Vector, GFP‐USP21, and GFP‐USP21‐C221A in SU‐DHL‐4 cells. (B) Cell proliferation of SU‐DHL‐4 cells transfected with GFP‐Vector, GFP‐USP21, and GFP‐USP21‐C221A were measured by CCK8 assay. (C) Representative images of Propidium Iodide (PI)‐positive cells (Red) representing dead cells and DIC representing total cells. Scale bar, 100 μm. (D) Quantification of dead cells after transfection with GFP‐Vector, GFP‐USP21, or GFP‐USP21‐C221A. ***p < 0.001, ****p < 0.0001, Error bars indicate ±SEM
3.4. USP21 regulates the DLBCL cells proliferation by maintaining EZH2
Previous studies illustrated that EZH2 was necessary for the DLBCL development. 28 , 29 To correlate USP21 with EZH2 in DLBCL, we overexpressed USP21 in SU‐DHL‐4 cells and found that the protein level of EZH2 was increased upon USP21 overexpression (Figure 4A,B). In contrast, the EZH2 protein level was decreased in USP21‐depleted cells (Figure 4C,D). Therefore, these results demonstrated that USP21 can maintain the protein level of EZH2 in DLBCL cells.
FIGURE 4.
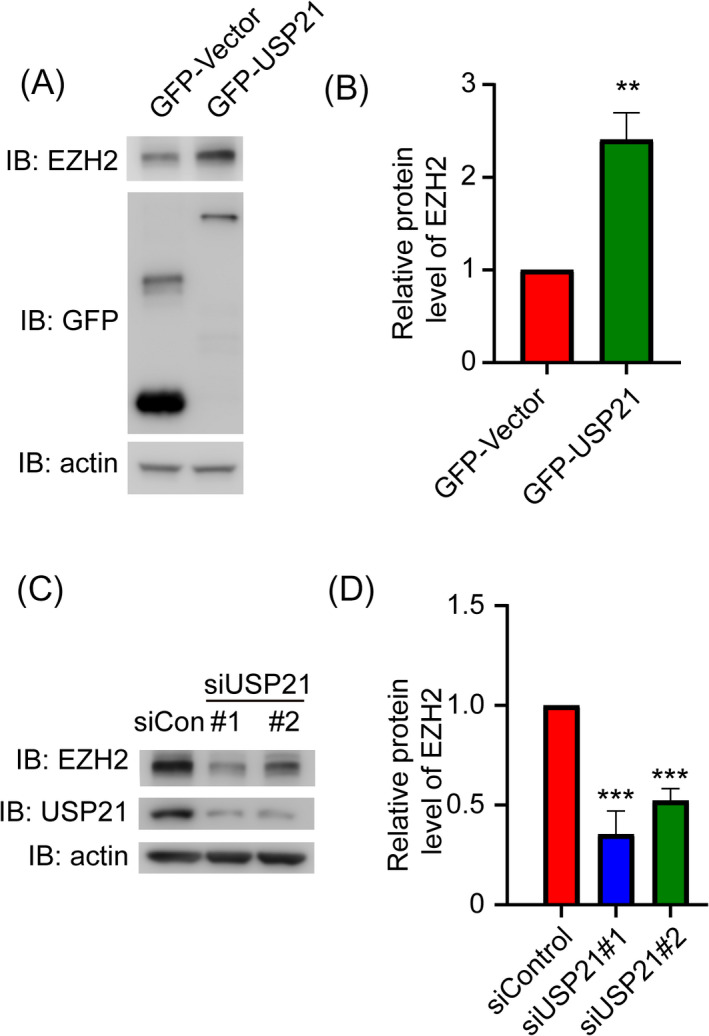
USP21 maintains EZH2 protein levels in SU‐DHL‐4 cells. (A) Western blots show EZH2 expression level in SU‐DHL‐4 cells after transfection with GFP‐Vector or GFP‐USP21 for 48 h. (B) Quantification of the EZH2 protein level upon overexpression of GFP‐Vector or GFP‐USP21. (C) Western blots show EZH2 expression level in SU‐DHL‐4 cells after transfection with control or siUSP21 for 48 h. (D) Quantification of the EZH2 protein level upon transfection with control or siUSP21. **p < 0.01; ***p < 0.001. Error bars indicate ±SEM
4. DISCUSSION
DLBCL is one of the common categories of NHL, which is a hazard for human health. 30 According to relevant literature, there are about 25,000 new cases of DLBCL diagnosed in China each year, which accounts for 11–13% of diagnosed malignancies. 31 , 32 Therefore, novel therapeutic targets are urgently needed for DLBCL patients in China. Here, we find that the USP21 mRNA level is enriched in DLBCL patient lymphoid tissue, in accordance with the data in the GEPIA database. Besides, the USP21 protein expression level in DLBCL cell lines is identified to be higher than that in normal cell lines, which verifies the mRNA result in the patient.
Ubiquitination acts as one of the post translational modifications of protein, which eliminates misfolded proteins and regulates different signaling pathways and cell functions. 33 , 34 , 35 The ubiquitination level of protein relies on the synergistical effects of its ubiquitinases and deubiquitinases. 36 , 37 In recent years, an increasing number of studies have reported that the deubiquitinating enzymes (DUBs) play important roles in cancer development and therapeutic resistance. 38 , 39 , 40 , 41 For example, USP15 upregulates the TGF‐β pathway to promote cell proliferation in glioblastoma pathogenesis. 42 Kruppel‐like factor 5 (KLF5) is stabilized by the DUB BRCA1‐associated Protein 1 (BAP1), promoting breast cancer cell proliferation. 43 The proteasomal cysteine deubiquitinase inhibitor b‐AP15 inhibits proteasome DUB activities and induces cell apoptosis in DLBCL. 30 However, the role of specific DUB in DLBCL development is rarely reported. We identify that knockdown of USP21 inhibits cell proliferation, and overexpression of USP21 promotes cell growth. Moreover, USP21‐induced oncogenesis is dependent on its deubiquitinating activity.
EZH2, the key regulatory protein in DLBCL, modulates cell proliferation and other cell functions. 28 , 44 GSK126, the specific inhibitor of EZH2, can silence transcription through trimethylation of histone H3 lysine 27 (H3K27me3) to depress cancer cells proliferation. 45 USP21 could suppress EZH2 ubiquitination and thus promotes cell proliferation and metastasis in bladder carcinoma. 11 In our study, we identified that USP21 can maintain the EZH2 protein level in DLBCL cells, which may due to deubiquitination and protein stabilization of EZH2 by USP21. Therefore, USP21 may be supposed to modulate DLBCL oncogenesis through this underlying mechanism.
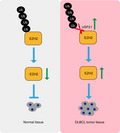
In conclusion, USP21 is enriched in DLBCL and is beneficial for tumor cell proliferation. Mechanistically, USP21 promotes cell growth by maintaining EZH2 protein level. Thus, we propose the model that USP21 and its downstream target EZH2 should be novel chemotherapeutic targets for DLBCL treatment (Figure 5).
FIGURE 5.
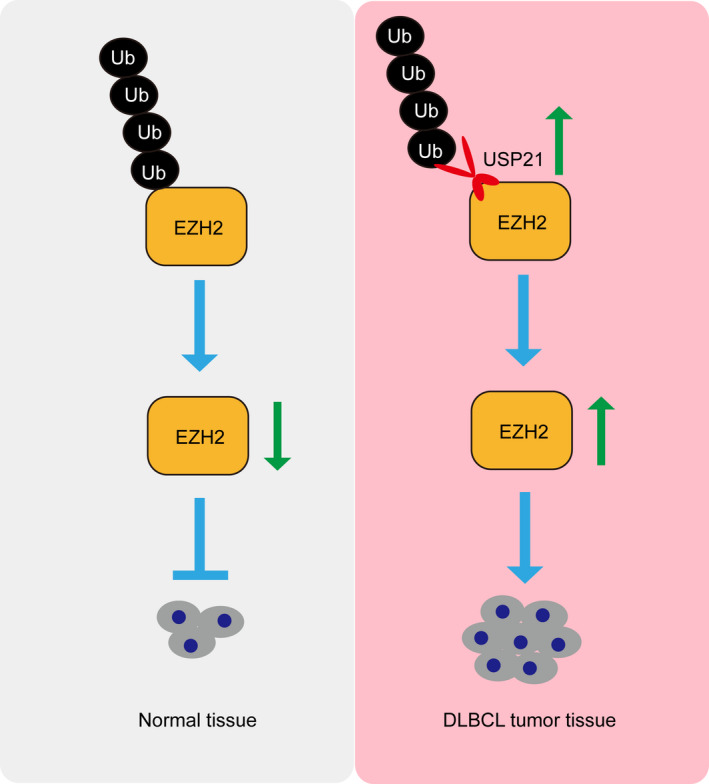
Proposed model for the function of USP21 in oncogenesis of diffuse large B‐cell lymphoma (DLBCL). USP21 is beneficial for DLBCL cell proliferation. Mechanistically, USP21 promotes cell growth by maintaining the EZH2 protein level. Thereby, USP21 can act as a promising therapeutic target for DLBCL
CONFLICT OF INTEREST
The authors declare no competing financial interests.
AUTHOR CONTRIBUTION
M.H., L.X., Z.P., and H.N. performed the experiments; M.H., L.X., Z.P., H.N., L.M., and X.W. analyzed the data; M.H., J.Z., L.M., and X.W. conceived and designed the experiments; and M.H. and X.W. wrote the article.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China [grant number 31701209].
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Sant M, Allemani C, Tereanu C, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood. 2010;116(19):3724‐3734. [DOI] [PubMed] [Google Scholar]
- 2. Yang J, Li D, Zhou J. Histone deacetylase 6 as a therapeutic target in B cell‐associated hematological malignancies. Front Pharmacol. 2020;11:971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yan B, Liu Y, Bai H, et al. HDAC6 regulates IL‐17 expression in T lymphocytes: implications for HDAC6‐targeted therapies. Theranostics. 2017;7(4):1002‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martelli M, Ferreri AJ, Agostinelli C, Di Rocco A, Pfreundschuh M, Pileri SA. Diffuse large B‐cell lymphoma. Crit Rev Oncol Hematol. 2013;87(2):146‐171. [DOI] [PubMed] [Google Scholar]
- 5. Wang Y, Link BK, Witzig TE, et al. Impact of concurrent indolent lymphoma on the clinical outcome of newly diagnosed diffuse large B‐cell lymphoma. Blood. 2019;134(16):1289‐1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;66(6):443‐459. [DOI] [PubMed] [Google Scholar]
- 7. Armand P, Oki Y, Neuberg DS, et al. Detection of circulating tumour DNA in patients with aggressive B‐cell non‐Hodgkin lymphoma. Br J Haematol. 2013;163(1):123‐126. [DOI] [PubMed] [Google Scholar]
- 8. Ageberg M, Rydström K, Lindén O, Linderoth J, Jerkeman M, Drott K. Inhibition of geranylgeranylation mediates sensitivity to CHOP‐induced cell death of DLBCL cell lines. Exp Cell Res. 2011;317(8):1179‐1191. [DOI] [PubMed] [Google Scholar]
- 9. Nijman SM, Luna‐Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773‐786. [DOI] [PubMed] [Google Scholar]
- 10. Peng L, Hu Y, Chen D, Jiao S, Sun S. Ubiquitin specific peptidase 21 regulates interleukin‐8 expression, stem‐cell like property of human renal cell carcinoma. Oncotarget. 2016;7(27):42007‐42016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y, Zhou B, Chen D. USP21 promotes cell proliferation and metastasis through suppressing EZH2 ubiquitination in bladder carcinoma. Onco Targets Ther. 2017;10:681‐689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou P, Song T, Sun C, et al. USP21 upregulation in cholangiocarcinoma promotes cell proliferation and migration in a deubiquitinase‐dependent manner. Asia Pac J Clin Oncol. 2020. 10.1111/ajco.13480 [DOI] [PubMed] [Google Scholar]
- 13. Urbé S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome‐ and microtubule‐associated functions. Mol Biol Cell. 2012;23(6):1095‐1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu F, Li T, Sui Y, et al. O‐GlcNAc transferase regulates centriole behavior and intraflagellar transport to promote ciliogenesis. Protein Cell. 2020;11(11):852‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen M, Wang J, Yang Y, et al. Redox‐dependent regulation of end‐binding protein 1 activity by glutathionylation. Sci China Life Sci. 2020. 10.1007/s11427-020-1765-6 [DOI] [PubMed] [Google Scholar]
- 16. Yang Y, Chen M, Li J, et al. A cilium‐independent role for intraflagellar transport 88 in regulating angiogenesis. Sci Bull. 2020. 10.1016/j.scib.2020.10.014 [DOI] [PubMed] [Google Scholar]
- 17. Yu F, Guo S, Li T, et al. Ciliary defects caused by dysregulation of O‐GlcNAc modification are associated with diabetic complications. Cell Res. 2019;29(2):171‐173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin J, Liu J, Chen C, et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat Commun. 2016;7:13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X, Yao Y, Ding H, et al. USP21 deubiquitylates Nanog to regulate protein stability and stem cell pluripotency. Signal Transduct Target Ther. 2016;1:16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arceci A, Bonacci T, Wang X, et al. FOXM1 deubiquitination by USP21 regulates cell cycle progression and paclitaxel sensitivity in basal‐like breast cancer. Cell Rep. 2019;26(11):3076.e3076‐3086.e3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie W, Li D, Dong D, et al. HIV‐1 exposure triggers autophagic degradation of stathmin and hyperstabilization of microtubules to disrupt epithelial cell junctions. Signal Transduct Target Ther. 2020;5(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen M, Li Y, Liu Z, et al. Exopolysaccharides from a Codonopsis pilosula endophyte activate macrophages and inhibit cancer cell proliferation and migration. Thorac Cancer. 2018;9(5):630‐639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duan Y, Zhang X, Yang L, et al. Disruptor of telomeric silencing 1‐like (DOT1L) is involved in breast cancer metastasis via transcriptional regulation of MALAT1 and ZEB2. J Genet Genomics. 2019;46(12):591‐594. [DOI] [PubMed] [Google Scholar]
- 24. Yan B, Xie S, Liu Y, et al. Histone deacetylase 6 modulates macrophage infiltration during inflammation. Theranostics. 2018;8(11):2927‐2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie S, Zhang L, Dong D, et al. HDAC6 regulates antibody‐dependent intracellular neutralization of viruses via deacetylation of TRIM21. J Biol Chem. 2020;295(42):14343‐14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie S, Wu Y, Hao H, et al. CYLD deficiency promotes pancreatic cancer development by causing mitotic defects. J Cell Physiol. 2019;234(6):9723‐9732. [DOI] [PubMed] [Google Scholar]
- 27. Yang Y, Hao H, Wu X, et al. Mixed‐lineage leukemia protein 2 suppresses ciliary assembly by the modulation of actin dynamics and vesicle transport. Cell Discov. 2019;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neves Filho EH, Hirth CG, Frederico IA, Burbano RM, Carneiro T, Rabenhorst SH. EZH2 expression is dependent on MYC and TP53 regulation in diffuse large B‐cell lymphoma. Apmis. 2020;128(4):308‐315. [DOI] [PubMed] [Google Scholar]
- 29. Brach D, Johnston‐Blackwell D, Drew A, et al. EZH2 inhibition by Tazemetostat results in altered dependency on B‐cell activation signaling in DLBCL. Mol Cancer Ther. 2017;16(11):2586‐2597. [DOI] [PubMed] [Google Scholar]
- 30. Ollila TA, Olszewski AJ. Extranodal diffuse large B cell lymphoma: molecular features, prognosis, and risk of central nervous system recurrence. Curr Treat Options Oncol. 2018;19(8):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miao X, Wu Y, Wang Y, et al. Y‐box‐binding protein‐1 (YB‐1) promotes cell proliferation, adhesion and drug resistance in diffuse large B‐cell lymphoma. Exp Cell Res. 2016;346(2):157‐166. [DOI] [PubMed] [Google Scholar]
- 32. Thompson CA, Ghesquieres H, Maurer MJ, et al. Utility of routine post‐therapy surveillance imaging in diffuse large B‐cell lymphoma. J Clin Oncol. 2014;32(31):3506‐3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lai KP, Chen J, Tse WKF. Role of deubiquitinases in human cancers: potential targeted therapy. Int J Mol Sci. 2020;21(7):2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tian M, Zhu R, Ding F, Liu Z. Ubiquitin‐specific peptidase 46 promotes tumor metastasis through stabilizing ENO1 in human esophageal squamous cell carcinoma. Exp Cell Res. 2020;395(1):112188. 10.1016/j.yexcr.2020.112188:112188 [DOI] [PubMed] [Google Scholar]
- 35. Ran J, Liu M, Feng J, et al. ASK1‐mediated phosphorylation blocks HDAC6 ubiquitination and degradation to drive the disassembly of photoreceptor connecting cilia. Dev Cell. 2020;53(3):287‐299. [DOI] [PubMed] [Google Scholar]
- 36. Mofers A, Pellegrini P, Linder S, D'Arcy P. Proteasome‐associated deubiquitinases and cancer. Cancer Metastasis Rev. 2017;36(4):635‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Critchley WR, Pellet‐Many C, Ringham‐Terry B, Harrison MA, Zachary IC, Ponnambalam S. Receptor tyrosine kinase ubiquitination and de‐ubiquitination in signal transduction and receptor trafficking. Cells. 2018;7(3):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao Z, Zhang P, Ma L. The role of deubiquitinases in breast cancer. Cancer Metastasis Rev. 2016;35(4):589‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qian X, Li S, Yang Z, Zhang J. The long non‐coding RNA HLNC1 potentiates hepatocellular carcinoma progression via interaction with USP49. J Clin Lab Anal. 2020;34:e23462–e23470. 10.1002/jcla.23462:e23462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farshi P, Deshmukh RR, Nwankwo JO, et al. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin Ther Pat. 2015;25(10):1191‐1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song T, Zhou J. Primary cilia in corneal development and disease. Zool Res. 2020;41(5):495‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eichhorn PJ, Rodón L, Gonzàlez‐Juncà A, et al. USP15 stabilizes TGF‐β receptor I and promotes oncogenesis through the activation of TGF‐β signaling in glioblastoma. Nat Med. 2012;18(3):429‐435. [DOI] [PubMed] [Google Scholar]
- 43. Qin J, Zhou Z, Chen W, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bisserier M, Wajapeyee N. Mechanisms of resistance to EZH2 inhibitors in diffuse large B‐cell lymphomas. Blood. 2018;131(19):2125‐2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lue JK, Amengual JE. Emerging EZH2 inhibitors and their application in lymphoma. Curr Hematol Malig Rep. 2018;13(5):369‐382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


