Abstract
Background
Although a diagnosis of infectious diseases is essential for timely treatment, the performance of diagnostic tests has been hardly evaluated due to variable results that are influenced by multiple factors in different conditions. In the present study, the performance of the Alinity i system, which is a newly developed immunoassay to diagnose infectious diseases, was evaluated.
Methods
We evaluated the precision, linearity, correlation, and carryover of 16 analytes (HAV Ab IgG, HBsAg, HBeAg, anti‐HBc, anti‐HBe, anti‐HBs, anti‐HCV, HIV Ag/Ab, EBV VCA IgM, EBV VCA IgG, EBV EBNA IgG, CMV IgM, CMV IgG, Toxoplasma IgG, Rubella IgG, and Syphilis TP) of Alinity i by comparison with ARCHITECT i2000SR system following the rationale of the Clinical and Laboratory Standards Institute (CLSI).
Results
For quantitative tests, the coefficients of variation (CV) % of repeatability and intermediate precision were between 0% and 4.18%. The coefficients of the linearity (r 2) over a widely tested analytical range were ≥ 0.990 and the correlation between Alinity i and the ARCHITECT i2000SR system was strong (r ≥ 0.994). For qualitative tests, the agreement between Alinity i and the ARCHITECT i2000SR system was excellent (kappa coefficient 1) with 100% sensitivity and specificity. Carryover rates for all analytes were less than 1.0% (−0.11% ~ 0.21%).
Conclusion
The Alinity i system showed good analytical performance and favorable comparability with the ARCHITECT i2000SR. It could be suitable as a routine immunoassay analyzer for screening and diagnosis of infectious disease.
Keywords: Alinity i system, analytical performance, comparison study, immunoassay, infectious disease
For both qualitative and quantitative measurements, the Alinity i system showed good analytical precision and excellent agreement with ARCHITECT i2000SR system. Alinity i system would be an excellent routine immunoassay analyzer for screening and diagnosing infectious disease.

1. INTRODUCTION
Diagnosis of infectious disease is necessary for the timely treatment of patients, screening of asymptomatic individuals, surveillance, and epidemiological investigation. 1 The diagnostic tests for these infectious diseases detect the presence of the pathogens themselves, antigens, or antibodies against them. The test results should be appropriately evaluated to determine whether these tests are accurate and reliable under certain conditions. 2 In particular, because the results of serologic tests can be influenced by multiple variables in different conditions, 3 the performance evaluation for the test is essential before reporting the results to clinicians.
Immunoassays are bioanalytical methods to measure the concentration of an analyte through the reaction of an antigen and an antibody. Among these methods, the chemiluminescence detection method is a versatile and ultrasensitive tool that can simultaneously detect a broad range of molecules in clinical diagnosis and has been widely used with complete automation and the development of technology and related materials. 4 However, the equipment using the chemiluminescence detection method and related materials differs from laboratory to laboratory, resulting in difficulty of evaluation for analytical precision, reproducibility, and reliability, so validation of the method under certain conditions is necessary. 5
Most diagnostic tests of infectious diseases are performed in a qualitative manner. By applying a cutoff or ordinal scale to the quantitative results, converted qualitative results reveal discontinuous and reduced information and the result near the cutoff shows high uncertainty. 6 , 7 Validation for these qualitative tests is not as easy as that for quantitative tests and only limited analytes not related to infectious disease has been evaluated. In present study, we aimed to validate the performance of Alinity i, which is a newly developed immunoassay platform, under routine clinical laboratory conditions and to compare the results of Alinity i with those of ARCHITECT i2000SR system. The evaluation was conducted in accordance with objective recommendations for analytical performance (Clinical and Laboratory Standards Institute).
2. MATERIALS AND METHODS
2.1. General information
The analytical performances were evaluated for the Alinity i by comparison with ARCHITECT i2000SR system (Abbott Laboratories, IL, USA). A total of 16 analytes were selected: HAV Ab IgG(signal/cutoff (S/CO)), HBsAg (S/CO), HBeAg (S/CO), anti‐HBc (S/CO), anti‐HBe (S/CO), anti‐HBs (mIU/mL), anti‐HCV (S/CO), HIV Ag/Ab (S/CO), EBV VCA IgM (S/CO), EBV VCA IgG (S/CO), EBV EBNA IgG (S/CO), CMV IgM (relative light units, RLU), CMV IgG (AU/mL), Toxoplasma IgG (IU/mL), Rubella IgG (IU/mL), and Syphilis TP (S/CO). Among them, anti‐HBs (mIU/mL), CMV IgG (AU/mL), Toxoplasma IgG (IU/mL), and Rubella IgG (IU/mL) are quantitative tests, and the remaining analytes are qualitative tests. For evaluation of compatibility, a total of 800 samples were derived from healthy adults and patients with positive results for various infectious diseases from December 2018 to December 2019. This study was approved by the Institutional Review Board for human‐based research of Seoul National University (IRB No. 1810‐080‐980).
2.2. Method
2.2.1. Precision
The analytical precision of quantitative tests was evaluated according to the Clinical and Laboratory Standards Institute (CLSI) guidelines EP15−A3. 8 Three levels of quality control materials were used for quantitative tests. The verification was conducted by using each of five replicates of the same quality control materials and performed during 5‐day evaluation periods. The values of repeatability and intermediate precision were compared with those claimed by the manufacturer, which were obtained based on the CLSI guidelines EP05‐A3 (two or three levels of quality control materials were evaluated in duplicate on two separate runs for 20 days). 9
2.2.2. Linearity
The linearity for the quantitative tests was represented according to the CLSI guidelines EP06‐A. 10 For each analyte, two patient samples with high (H) and low (L) concentration were mixed at ratios of 4H, 1L + 3H, 2L + 2H, 3L + 1H, and 4L. We measured five levels with four replicates. The linearity was depicted, and the deviation was calculated by polynominal regression analysis. The results were acceptable if the percentage of error was within the total allowable error, defined as 30%, suggested by the manufacturer.
2.2.3. Method comparison
The Alinity i and ARCHITECT i2000SR system were compared for quantitative and qualitative tests. For quantitative test, comparison was performed based on the CLSI guidelines EP09‐A3. 11 Each of fifty serum samples, spanning most clinically relevant linear range, was tested using both analyzer in duplicate. Correlation coefficient (r), the slope, and intercept were calculated by the Deming regression and mean bias was calculated by Bland–Altman plot. For qualitative tests, based on CLSI guidelines EP12‐A2, 12 kappa equation, the positive and negative, and total agreement with 95% CI between the Alinity i and ARCHITECT i2000SR system were calculated.
2.2.4. Carryover
Carryover was evaluated by using patient samples of high and low concentrations with four replicates at each levels according to the CLSI guidelines EP10‐A3. 13 The carryover rate was calculated by the equation: [L1‐(L3 + L4)/2 × 100/[(H2 + H3)/2‐(L3 + L4)/2]. The acceptable carryover rate was less than 1.0%. 14
2.3. Statistics
To access precision, linearity, method comparison, and carryover, all analysis were performed by using EP Evaluator Release 11 (David G. Rhoads Assoc., Kennett Squre, PA, USA) and Medcalc software (Frank Schoonjans, Mariakerke, Belgium). If p‐value was less than 0.05, it was considered statistically significant.
3. RESULTS
3.1. Precision
For low, medium, and high level of 4 quantitative analytes (anti‐HBs, CMV IgG, Toxoplasma IgG, and Rubella IgG), the percent coefficient of variation (%CV) of repeatability and intermediate precision were between 0% and 4.18%. Most of %CV showed lower than the limits claimed by the manufacturer except the intermediate precision of medium level for anti‐HBs (verified estimated and manufacturer's claim: 3.5% vs. 2.9%) and the repeatability and intermediated precision of medium level for Rubella IgG (verified estimate and manufacturer's claim: 4.2% vs. 3.3%; 4.2% vs. 4.0%, respectively) (Table 1).
TABLE 1.
The Precision for Quantitative Tests obtained by the Alinity i System
| Analyte | Level | N | Mean | Verified estimate | Manufacturer's claim | ||
|---|---|---|---|---|---|---|---|
|
Repeatability CV (%) |
Intermediate precision CV (%) |
Repeatability CV (%) |
Intermediate precision CV (%) |
||||
| Anti‐HBs (mIU/mL) | Low | 25 | 0.0 | NA | NA | NA | NA |
| Medium | 25 | 15.1 | 2.0 | 3.5 | 2.0 | 2.9 | |
| High | 25 | 78.3 | 1.4 | 1.4 | 2.0 | 3.0 | |
| CMV IgG (AU/mL) | Low | 25 | 0.1 | NA | NA | NA | NA |
| Medium | 25 | 31.9 | 2.2 | 3.4 | 3.6 | 5.9 | |
| High | 25 | 156.1 | 2.1 | 2.1 | 2.5 | 5.2 | |
| Toxoplasma IgG (IU/mL) | Low | 25 | 0.0 | NA | NA | NA | NA |
| Medium | 25 | 6.4 | 1.3 | 1.9 | 2.5 | 4.2 | |
| High | 25 | 111.7 | 2.7 | 3.8 | 3.4 | 6.8 | |
| Rubella IgG (IU/mL) | Low | 25 | 0.0 | NA | NA | NA | NA |
| Medium | 25 | 25.5 | 4.2 | 4.2 | 3.3 | 4.0 | |
| High | 25 | 296.9 | 3.4 | 3.4 | 4.3 | 5.6 | |
Abbreviations: anti‐HBs, hepatitis B surface antibody; CMV, cytomegalovirus; CV, coefficient of variation; N, number; NA, not applicable.
3.2. Linearity
For four quantitative analytes (anti‐HBs, CMV IgG, Toxoplasma IgG, and Rubella IgG), the linearity was shown in Table 2. All correlation coefficients (r 2) for four analytes were ≥ 0.990, representing satisfied linearity ranges.
TABLE 2.
Linearity for Quantitative Tests obtained by the Alinity i System
| Analyte | Test range | Observed linear range | Slope | Intercept | r 2 | Recovery (%) |
|---|---|---|---|---|---|---|
| Anti‐HBs (mIU/mL) | 2.0‐1000.0 | 2.26‐870.74 | 1.012 | −5.636 | 0.998 | 95.6‐103.3 |
| CMV IgG (AU/mL) | 1.1‐250 | 1.4‐236.9 | 1.010 | 0.312 | 0.999 | 97.8‐103.5 |
| Toxoplasma IgG (IU/mL) | 0.2‐200 | 0‐171.8 | 0.983 | 0.781 | 0.995 | 93.8‐105.9 |
| Rubellar IgG (IU/mL) | 0.5‐500 | 0.4‐500 | 1.027 | 5.402 | 0.992 | 100‐109.9 |
3.3. Method comparison
In the method comparison between the Alinity i and ARCHITECT i2000SR system, all quantitative analytes showed a very strong correlation (r ≥ 0.994) based on Deming regression. The slope representing constant bias ranged from 0.956 to 1.006. The intercepts representing proportional bias showed significant difference from 0 for anti‐HBs (6.579) and CMV IgG (5.400) (Table 3). Mean bias based on Bland–Altman plots was between −0.6 and 4.5. The samples near the cutoff values showed lesser bias than the samples far from the cutoff values (Figure 1). For qualitative tests (HAV Ab IgG, HBsAg, HBeAg, anti‐HBc, anti‐HBe, anti‐HCV, HIV Ag/Ab, EBV VCA IgM, EBV VCA IgG, EBV EBNA IgG, CMV IgM, and Syphilis TP), the Alinity i system presented excellent agreement with the ARCHITECT i2000SR system (positive, negative and total agreement = 100%; kappa coefficient = 1), showing 100% sensitivity and specificity (Table 4).
TABLE 3.
Comparison between Alinity i and ARCHITECT i2000SR System in Quantitative Results
| Analyte | Test ranges |
Correlation coefficient (r) |
Slope | Intercept* |
|---|---|---|---|---|
| Anti‐HBs (mIU/mL) | 0.0‐963.1 | 0.996 | 0.982 (0.956‐1.007) | 6.579 (−1.268 to 14.427) |
| CMV IgG (AU/mL) | 0.2‐628.8 | 0.994 | 0.977 (0.946‐1.008) | 5.400 (0.180 to 10.630) |
| Toxoplasma IgG (IU/mL) | 0.0‐1772.6 | 1.000 | 1.006 (1.005‐1.007) | −0.280 (−0.440 to − 0.120) |
| Rubella IgG (IU/mL) | 0.1‐70.5 | 0.997 | 0.956 (0.936‐0.976) | 0.140 (−0.310 to 0.700) |
Abbreviations: anti‐HBs, hepatitis B surface antibody; CMV, cytomegalovirus.
The intercept values in bold are significantly different from 0 by Deming regression.
FIGURE 1.
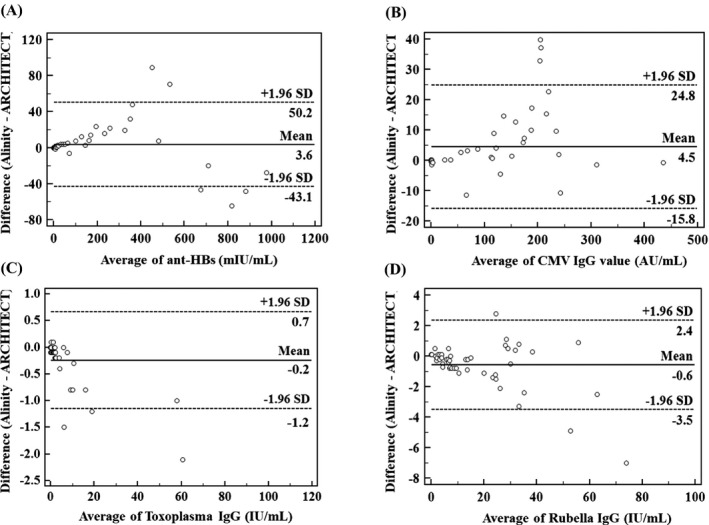
Comparison between Alinity i and ARCHITECT system for (A) anti‐HBs, (B) CMV IgG, (C) Toxoplasma IgG, and (D) Rubella IgG. The solid line represents mean difference and dashed lines indicate the upper and lower 95% confidence limits of difference for both systems
TABLE 4.
Comparison between Alinity i with ARCHITECT i2000SR System in Qualitative Results
| Analyte | Alinity i | ARCHITECT i2000SR | Positive agreement % (95% CI) | Negative agreement % (95% CI) | Total agreement % (95% CI) | Kappa | |
|---|---|---|---|---|---|---|---|
| Reactive | Nonreactive | ||||||
| HAV Ab IgG (S/CO) | Reactive | 25 | 0 | 100 (86.28 ‐ 100.00) | 100 (86.28 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 25 | |||||
| HBsAg (S/CO) | Reactive | 27 | 0 | 100 (87.23 ‐ 100.00) | 100 (85.18 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 23 | |||||
| HBeAg (S/CO) | Reactive | 20 | 0 | 100 (83.16 ‐ 100.00) | 100 (88.43 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 30 | |||||
| Anti‐HBc (S/CO) | Reactive | 28 | 0 | 100 (87.67 ‐ 100.00) | 100 (84.56 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 22 | |||||
| Anti‐HBe (S/CO) | Reactive | 19 | 0 | 100 (82.35 ‐ 100.00) | 100 (88.78 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 31 | |||||
| Anti‐HCV (S/CO) | Reactive | 25 | 0 | 100 (86.28 ‐ 100.00) | 100 (86.28 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 25 | |||||
| HIV Ag/Ab (S/CO) | Reactive | 25 | 0 | 100 (86.28 ‐ 100.00) | 100 (87.23 ‐ 100.00) | 100 (93.15 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 27 | |||||
| EBV VCA IgM (S/CO) | Reactive | 18 | 0 | 100 (81.47 ‐ 100.00) | 100 (89.11 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 32 | |||||
| EBV VCA IgG (S/CO) | Reactive | 32 | 0 | 100 (89.11 ‐ 100.00) | 100 (81.47 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 18 | |||||
| EBV EBNA IgG (S/CO) | Reactive | 25 | 0 | 100 (86.28 ‐ 100.00) | 100 (86.28 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 25 | |||||
| CMV IgM (RLUs) | Reactive | 22 | 0 | 100 (84.56 ‐ 100.00) | 100 (87.67 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 28 | |||||
| Syphilis TP (S/CO) | Reactive | 25 | 0 | 100 (86.28 ‐ 100.00) | 100 (86.28 ‐ 100.00) | 100 (92.89 ‐ 100.00) | 1.00 (1.00 ‐ 1.00) |
| Nonreactive | 0 | 25 | |||||
Abbreviations: Ab, antibody; Ag/Ab, antigen/antibody; anti‐HBc, hepatitis B core antibody; anti‐HBe, hepatitis B envelop antibody; anti‐HCV, hepatitis C virus antibody; CI, confidence interval; CMV, cytomegalovirus; EBNA, Epstein–Barr virus nuclear antigen; EBV, Epstein–Barr virus; HAV, hepatitis A virus; HBeAg, hepatitis B virus envelop antigen; HBsAg, hepatitis B virus surface antigen; RLU, relative light units; S/CO, signal‐to‐cutoff; TP, Treponema pallidum; VCA, viral capsid antigen.
3.4. Carryover
The percent carryover for all 16 analytes were as follows: HAV Ab IgG, −0.11%; HBsAg, 0.00%; HBeAg, 0.00%; anti‐HBc, 0.00%; anti‐HBe, 0.00%; anti‐HBs, 0.00%; anti‐HCV, 0.09%; HIV Ag/Ab, 0.00%; EBV VCA IgM, 0.02%; EBV VCA IgG, 0.00%; EBV EBNA IgG, 0.00%; CMV IgM, 0.21%; CMV IgG, 0.00%; Toxoplasma IgG, 0.00%; Rubella IgG, 0.14%; and Syphilis TP, 0.00%). All carryover rate for quantitative and qualitative analytes were less than 1.0% (−0.11% ~ 0.21%).
4. DISCUSSION
Infectious diseases can exponentially spread from person to person. Rapid diagnostic tests with high sensitivity and specificity not only enable proper treatment but can also prevent the transmission of infectious diseases. 15 However, the performance of high‐volume analyzers for these infectious diseases has hardly been evaluated due to variable results that are influenced by multiple factors in different condition. In present study, we evaluated the performance of the Alinity i system, which is a newly introduced immunoassay system for detecting antigens or antibodies against pathogens.
Regarding quantitative tests, in this study, the repeatability and intermediate precision was less than 5% CV, which is generally considered to be acceptable for clinical application. 16 However, intermediate precision of medium level for anti‐HBs and repeatability and intermediate precision of medium level for Rubella IgG did not meet to the manufacturer's claims. In the previous study, quantitative measurement to assess the response to vaccination, such as anti‐HBs, also showed high discrepancy and CV% among different systems, even standardized against the same international standard. 17
For four quantitative analytes, Alinity i and ARCHITECT i2000SR system showed excellent correlation in Deming regression and Bland–Altman plots. The correlation coefficients for both systems were 0.994 ~ 1.000, with slopes near 1. However, the intercepts of anti‐HBs and CMV IgG were 6.579 and 5.400, respectively, which were significantly different from 0. The mean differences of anti‐HBs and CMV IgG levels of Alinity i compared to those of ARCHITECT i2000SR system were 3.6 mIU/mL and 4.5 AU/mL, respectively. Nevertheless, for samples with near the cutoff values, the differences were close to 0 by Bland–Altman plot, which suggests no clinically significant relevance.
For twelve qualitative analytes, the Alinity i and ARCHITECT i2000SR system showed 100% positive, negative, and total agreement (kappa equation = 1). The cutoff level suggested by the manufacturer seems to give the best discrimination between positive and negative results.
The limitation of this study is that several analytes, including HAV IgM, anti‐HBc IgM, Toxoplasma IgM, Rubella IgM, and HTLV, could not be analyzed due to the difficulty of specimen collection. In addition, we were not able to compare with the method which is considered the gold standard. However, some studies demonstrated that chemiluminescent detection method, instead of gold standard methods, showed excellent sensitivity and specificity for detection of certain analytes. 18 , 19 , 20 , 21 , 22
The performance of high‐volume analyzers for these infectious diseases has not been previously evaluated according to CLSI guidelines. This is the first study to evaluate simultaneously multiple analytes for the screening of infectious diseases in Alinity i system. We evaluated quantitative and qualitative analytes in Alinity i system according to proper CLSI guidelines. Our finding suggested that Alinity i system showed good analytical performance with low imprecision, low carryover, good linearity, and good correlation and equivalent diagnostic performance with the ARCHITECT i2000SR. In conclusion, Alinity i system characterized to have an excellent performance by ensuring reliable measurements for clinical laboratories and would be suitable as a routine immunoassay analyzer for screening infectious diseases.
CONFLICTS OF INTEREST
This study was supported by Abbott Diagnostics. Abbott Diagnostics did not have any role in the study design or data analysis.
Nam M, Song DY, Song SH, et al. Performance evaluation of immunoassay for infectious diseases on the Alinity i system. J Clin Lab Anal.2021;35:e23671. 10.1002/jcla.23671
DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCE
- 1. TDR Diagnostics Evaluation Expert Panel , Banoo SBD, Bossuyt P, et al. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol. 2010;8(12 Suppl):S17‐29. [PubMed] [Google Scholar]
- 2. Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis. 2013;57(Suppl 3):S139‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reverberi R, Reverberi L. Factors affecting the antigen‐antibody reaction. Blood Transfus. 2007;5(4):227‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C, Wu J, Zong C, Xu J, Ju H. Chemiluminescent immunoassay and its applications. Chin J Anal Chem. 2012;40(1):3‐10. [Google Scholar]
- 5. Vassault A, Hulin A, Chapuzet E, Arnaud J, Giroud C. Verification/validation of the performances of analytical method. Ann Biol Clin (Paris). 2010;68:247‐294. [DOI] [PubMed] [Google Scholar]
- 6. Ríos A, Barceló D, Buydens L, et al. Quality assurance of qualitative analysis in the framework of the European project 'MEQUALAN'. Accred Qual Assur. 2003;8:68‐77. [Google Scholar]
- 7. White GH, Farrance I. AACB Uncertainty of Measurement Working Group. Uncertainty of measurement in quantitative medical testing: a laboratory implementation guide. Clin Biochem Rev. 2004;25(4):S1‐24. [PMC free article] [PubMed] [Google Scholar]
- 8. CLSI . User verification of precision and estimation of bias; approved guideline‐third edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. CLSI document EP15‐A3. [Google Scholar]
- 9. CLSI . Evaluation of precision of quantitative measurement procedures; approved guideline‐third edition. Wayne, PA. Clinical and Laboratory Standards Institute. 2018. CLSI document EP05‐A3E. [Google Scholar]
- 10. CLSI . Evaluation of the linearity of quantitative measurement procedures: astatistical approach; approved guideline‐second edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2003. CLSI document EP06‐A3. [Google Scholar]
- 11. CLSI . Measurement procedure comparison and bias estimation using patient samples; approved guideline‐third edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2013. CLSI document EP09‐A3. [Google Scholar]
- 12. CLSI . User protocol for evaluation of qualitative test performance; approved guideline‐second edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. CLSI document EP12‐A2. [Google Scholar]
- 13. CLSI . Preliminary evaluation of quantitative clinical laboratory methods; approved guideline‐third edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2019. CLSI document EP10‐A3. [Google Scholar]
- 14. Broughon PM. Carry‐over in automatic analysers. J Automat Chem. 1984;6:94‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang H, Hwang BY, Bueno J. Biomarkers in infectious diseases. Dis Markers. 2018;2018:8509127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sikaris K. Analytical dquality – What should we be aiming for? Clin Biochem Rev. 2008;29(Suppl 1):S5‐10. [PMC free article] [PubMed] [Google Scholar]
- 17. Huzly D, Schenk T, Jilg W, Neumann‐Haefelin D. Comparison of nine commercially available assays for quantification of antibody response to hepatitis B virus surface antigen. J Clin Microbiol. 2008;46(4):1298‐1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khadem‐Ansari MH, Omrani MD, Rasmi Y, Ghavam A. Diagnostic validity of the chemiluminescent method compared to polymerase chain reaction for hepatitis B virus detection in the routine clinical diagnostic laboratory. Adv Biomed Res. 2014;3:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S, Kim JH, Yoon S, Park YH, Kim HS. Clinical performance evaluation of four automated chemiluminescence immunoassays for hepatitis C virus antibody detection. J Clin Microbiol. 2008;46(12):3919‐3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alonso R, López Roa P, Suárez M, Bouza E. New automated chemiluminescence immunoassay for simultaneous but separate detection of human immunodeficiency virus antigens and antibodies. J Clin Microbiol. 2014;52(5):1467‐1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park Y, Park BG, Ha J, Kim HS. Diagnostic performance and comparative evaluation of the Architect, Liaison, and Platelia Epstein‐Barr Virus antibody assays. Ann Lab Med. 2018;38(5):458‐465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mo X, Jin Y, Yang Y, Hu W, Gu W. Evaluation of a new chemiluminescence immunoassay for diagnosis of syphilis. Eur J Med Res. 2010;15(2):66‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


