Significance
The same gene is often regulated differently in response to stress in even closely related plant species. Directly measuring stress-responsive gene expression can be financially and logistically challenging in nonmodel species. Here, we show that models trained using data on which genes respond to cold in one species can predict which genes will respond to cold in related species, even when the training and target species vary in their degree of tolerance to cold. The prediction models we used require only genomic sequence and gene models. As a result, data from well-studied model species may be used to predict which genes will respond to stress in less-studied species with sequenced genomes.
Keywords: transcriptional regulation, comparative genomics, machine learning, cold stress
Abstract
Although genome-sequence assemblies are available for a growing number of plant species, gene-expression responses to stimuli have been cataloged for only a subset of these species. Many genes show altered transcription patterns in response to abiotic stresses. However, orthologous genes in related species often exhibit different responses to a given stress. Accordingly, data on the regulation of gene expression in one species are not reliable predictors of orthologous gene responses in a related species. Here, we trained a supervised classification model to identify genes that transcriptionally respond to cold stress. A model trained with only features calculated directly from genome assemblies exhibited only modest decreases in performance relative to models trained by using genomic, chromatin, and evolution/diversity features. Models trained with data from one species successfully predicted which genes would respond to cold stress in other related species. Cross-species predictions remained accurate when training was performed in cold-sensitive species and predictions were performed in cold-tolerant species and vice versa. Models trained with data on gene expression in multiple species provided at least equivalent performance to models trained and tested in a single species and outperformed single-species models in cross-species prediction. These results suggest that classifiers trained on stress data from well-studied species may suffice for predicting gene-expression patterns in related, less-studied species with sequenced genomes.
The genomes of over 300 plant species have been sequenced to date. Ambitious efforts are under way to sequence the genomes of up to 10,000 plant and algae species by 2023 (1). Even members of closely related groups of species can be adapted to different environments and exhibit different degrees of tolerance for different stresses. The panicoid grasses are a clade of approximately 3,000 plant species, including several domesticated crops. While panicoid grasses grow in and are adapted to a wide range of environments, many of the most agriculturally and economically important species, including maize (Zea mays subspecies ssp. mays) and sorghum (Sorghum bicolor), were originally domesticated at tropical latitudes and are not cold-tolerant. For these crops, the low temperatures in the spring and autumn constrain the length of the growing season and pose a major limit to total agricultural production. While the majority of panicoid grasses are native to the tropics or subtropics (2), a number of lineages have evolved to grow in temperate environments where cold and freezing temperatures occur annually. For instance, miscanthus (Miscanthus giganteus), a cold-tolerant relative of maize and sorghum that is native to temperate environments, exhibits substantially higher total photosynthetic productivity per year than these crops due to its longer growing season and reduced susceptibility to photoinhibition at chilling temperatures (3). Thus, the clade contains a complex mixture of cold-tolerant species, such as foxtail millet (Setaria italica) and switchgrass (Panicum virgatum) (4, 5), and cold-sensitive species, including maize, sorghum (S. bicolor), proso millet (Panicum miliaceum), and pearl millet (Pennisetum glaucum).
Plants have evolved a variety of physiological, biochemical, and transcriptional regulatory mechanisms to sense and respond to abiotic stress (6). The repeated acquisition and/or loss of cold tolerance within the panicoid grasses provides an opportunity to better understand the biochemical and evolutionary mechanisms responsible for changes in temperature tolerance. However, the patterns of gene-expression variation in response to cold stress are not conserved across species (7, 8) or even between genotypes within the same species (9). The modulation of transcriptional regulation in response to abiotic stress often requires synchronous actions among cis-regulatory elements (e.g., promoter and enhancer), trans-regulatory elements (e.g., transcription factor and regulating RNA), transposable elements, and epigenetic regulators (e.g., DNA methylation and chromatin structure) (6, 9–11). One explanation for the rapid divergence of cold-responsive transcriptional regulation between orthologous genes is that new insertions of transposable elements appear to have the potential to induce the cold-responsive expression of nearby genes (11–13). It is likely that the rewiring of transcriptional regulation plays a significant role in how different plant lineages adapt independently to low-temperature stress.
Here, we demonstrate that, even though orthology is not an effective predictor of transcriptional responses to cold stress across even closely related species, it is possible to train supervised classification models using data from one species to predict which genes will respond to cold stress in another species. The usefulness of supervised classification algorithms has been demonstrated for a range of biological applications, such as distinguishing gene models with the potential for expression (14), inferring human gene expression based on a mouse model (15), predicting functional annotations of individual gene models from functional genomic data (16), distinguishing genes involved in specialized or primary metabolism (17), and predicting posttranslational modification sites (18). In this study, we generated transcriptional data from four closely related species: foxtail millet, pearl millet, switchgrass, and proso millet (Fig. 1A). Importantly, models used to predict which genes would transcriptionally respond to cold stress provided equivalent prediction accuracy when trained using only features calculated from the genome and gene-model annotations as when trained by using larger feature sets that included evolutionary, chromatin, and population diversity features. Models trained in one species using only features calculated from genome-sequence assemblies and gene-model annotations could be used to make effective predictions in a second species. This cross-species prediction method provides an effective means of predicting which genes will transcriptionally respond to cold stress without the need to generate new expression datasets under equivalent conditions for each species. With the growing number of sequenced plant genomes, the ability to predict transcriptionally responded genes to stresses based on data from genome-sequence assemblies will lower the barriers to investigating the basis of widespread variation in stress tolerance across the plant kingdom.
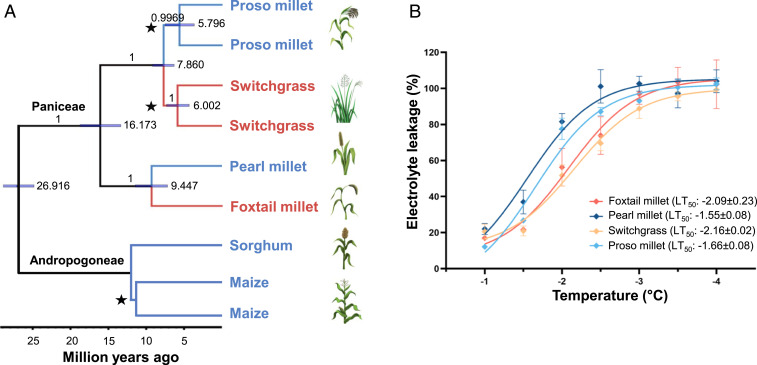
Fig. 1.
Phylogenetic and phenotypic relationships between foxtail millet, pearl millet, switchgrass, and proso millet. (A) Species tree for the six species investigated in this study. Branches shown in red are relatively cold-tolerant compared to branches shown in blue. Branch supports are Bayesian posterior probabilities; node bars show 95% highest posterior density of node age; and the scale bar represents millions of years ago. The whole-genome duplication events are marked by stars and indicate that the species contains two subgenomes. Maize was not included during species-tree analysis, and the divergence time between maize and sorghum was calibrated to 11.9 million years ago (22). (B) Electrolyte leakage from nonacclimated leaves frozen to a range of different temperatures. Curves were fitted by using nonlinear regression with a sigmoidal dose–response model. LT50 values are the concentrations that give half-maximal effects (23). Error bars indicate SEM from at least three replicate measurements.
Results
Cold-Responsive Genes and Gene-Expression Patterns Vary Among Related Species.
Both maize and sorghum are sensitive to cold stress (4, 7, 19, 20). Reports of the differences in the degrees of low-temperature tolerance among Paniceae species are sparse and varied, although switchgrass is extremely tolerant of cold and freezing, at least under some conditions (4, 5, 21). Cold tolerance can vary substantially, depending on treatment, developmental stage, and acclimation (as reviewed in ref. 4). Here, we grew seedlings of four Paniceae species under controlled conditions and assayed freezing tolerance at the three-leaf stage using an in vitro electrolyte leakage assay resulting from cell breakage to quantify the extent of damage. When not previously acclimated to stress conditions, switchgrass and foxtail millet seedlings showed slower rates of electrolyte leakage when challenged with progressively greater freezing stress compared to pearl millet and proso millet seedlings grown and tested under the same conditions (Fig. 1B). Therefore, low-temperature tolerance is not monophyletic within the Paniceae and could reflect the parallel adaptation of different lineages within the grass tribe to temperate climates (Fig. 1).
Changes in gene expression induced by cold stress were assayed by using paired control and stress treatment RNA-sequencing (RNA-seq) datasets collected from foxtail millet, pearl millet, switchgrass, and proso millet at the three-leaf stage 0.5, 1, 3, 6, 16, and 24 h after the onset of cold stress. The number of identified cold-responsive genes in each species increased with increased duration of cold stress in general. Overlap in the identities of cold-responsive genes identified at different time points ranged from 20 to 80% (SI Appendix, Fig. S1). Among genes showing cold-responsive changes in messenger RNA (mRNA) abundance, at least 47% were not syntenically conserved among the four species (Dataset S1, Tab 1 and SI Appendix, Fig. S2A). The number of nonsyntenic genes that responded transcriptionally to cold stress was more variable across species compared to syntenic genes (SI Appendix, Fig. S2A). Syntenic orthologous genes and promoters are derived from a single common ancestral gene and promoter of the most recent common ancestor of the species being studied. However, despite this shared evolutionary history, a gene responding transcriptionally to cold stress in one species was not a good predictor of whether syntenic orthologous genes in related species would also respond to cold stress in the same treatment at the same developmental stage (SI Appendix, Fig. S2B). This low conservation of transcriptional responses across conserved genes in related species is consistent with the results of a previous comparison of the transcriptional responses of maize and sorghum to cold stress (7) and the variation in transcriptional responses to cold stress between different alleles of the same gene in maize (9).
Supervised Classification Models Can Accurately Predict Cold-Responsiveness.
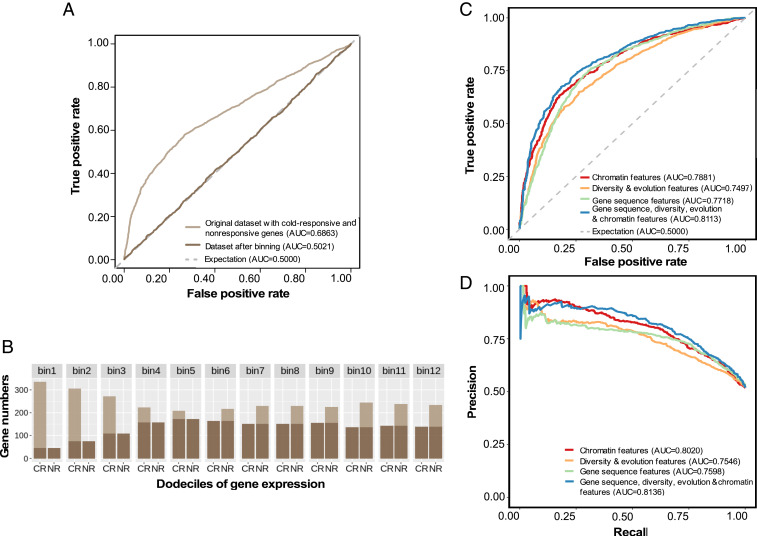
Stress-responsive transcriptional regulation of a given gene cannot be predicted efficiently by using data from orthologous genes in related species. However, perhaps specific features or properties of the gene itself can be used to predict whether its expression will respond to cold stress. We first evaluated this approach in maize, as many different types of feature data are available for all or nearly all gene models in this plant (16). One potential factor that could confound efforts to predict differential gene expression is that the average gene-expression level itself is a reasonably good predictor of whether or not a gene will be identified as showing statistically significant differential expression. In the current study, the areas under the receiver operating characteristic curves (AUC-ROCs) for predicting differential expression solely based on average expression levels varied from 0.48 to 0.70 for the six species tested (Fig. 2A and SI Appendix, Fig. S3A). Average gene-expression levels can be predicted reasonably well based on genomic features (24). The association between average gene expression and the odds of a gene being identified as differentially expressed, combined with the observation that average gene-expression levels themselves can be predicted reasonably well based on genomic features (24), suggests that on uncontrolled data, models trained to predict differential gene expression could achieve significant performance simply from learning to predict average gene-expression level (Fig. 2A). We employed a gene-binning strategy where genes were divided into 12 bins (dodeciles) based on average expression levels and subsampled to ensure equal representation of cold-response and cold-nonresponsive genes within each dodecile (Fig. 2 A and B and SI Appendix, Figs. S3B and S4). This effort to control for gene-expression level was motivated by concerns about bias, which might be introduced by statistical power to detect differential gene expression. After binning and subsampling, the prediction of which genes would be differentially expressed based solely on average expression values produced AUC-ROCs of approximately 0.50, i.e., equal to the null expectation for balanced data (Fig. 2A and SI Appendix, Fig. S3A). In addition, we performed a modified version of the gene-family guided splitting strategy proposed by Washburn et al. (24), to avoid obtaining misleadingly high accuracy values that can result when prediction models learn gene-family-specific features (SI Appendix, Fig. S4). Evaluation of parallel models trained with and without controls for gene-expression level and evolutionary relationships among genes provided greater performance (Dataset S1, Tab 2). Depending on the specific use case, this additional performance could be seen as beneficial, or it could represent an example of undesirable data leakage between training and testing datasets. In this study, the choice was made to employ both controls for all experiments.
Fig. 2.
Predicting cold-responsive genes in maize. (A and B) Baseline expression control. (A) Ability to predict whether a gene will be differentially expressed in response to cold based solely on baseline expression level before and after controlling for variation in gene-expression level. (B) Distribution of average FPKM values of cold-responsive genes (CR) and nonresponsive genes (NR), and training sets resampled from genes in 12 bins with balanced gene-expression levels (darker color). (C) ROC curves showing the performance of different maize models trained to predict cold-responsive gene expression using different types of features to describe genes. (D) PR curves showing the performance of different maize models trained to predict cold-responsive gene expression using different types of features to describe genes.
A set of features was assembled for each maize gene, including gene-sequence features, chromatin features, and diversity/evolutionary features (16) (Dataset S1, Tab 3). Either the complete or a subset of features were used to train random-forest models separately (25). Of results, three metrics were used to evaluate the model performance, including AUC-ROC, area under precision recall curve (AUPRC), and F1 score (the value calculated from precision and recall). The complete set of features performed the best (AUC-ROC = 0.81, AUPRC = 0.81, and F1 = 0.72 for 90% training data; AUC-ROC = 0.79, AUPRC = 0.77, and F1 = 0.70 for 10% holdout test data) to predict which genes would exhibit differential expression in response to cold stress and which would not (Fig. 2 C and D and SI Appendix, Fig. S5). Models trained with subsets of features did not match the accuracy of the combined model. A model trained using only features that can be extracted from genomic sequence data was able to predict which genes would exhibit differential expression in response to cold stress and which would not in modestly lower performance (AUC-ROC = 0.77, AUPRC = 0.76, and F1 = 0.70 for 90% training data; AUC-ROC = 0.72, AUPRC = 0.71, and F1 = 0.62 for 10% holdout test data) (Fig. 2 C and D and SI Appendix, Fig. S5).
Unlike the combined model, which requires data obtained using a range of specialized sequencing techniques, as well as resequencing data from diverse populations, the pure genomic feature model can be applied to any species with a sequenced genome and annotated gene models. We scored the same set of genomic sequence-derived features for each gene model in foxtail millet, pearl millet, switchgrass, and proso millet and trained the species-specific random-forest prediction models for each of the four species. The performance of models trained in foxtail millet (mean AUC-ROC = 0.85, AUPRC = 0.82, and F1 = 0.76), pearl millet (mean AUC-ROC = 0.86, AUPRC = 0.86, and F1 = 0.77), switchgrass (mean AUC-ROC = 0.77, AUPRC = 0.75, and F1 = 0.67), and proso millet (mean AUC-ROC = 0.85, AUPRC = 0.83, and F1 = 0.76) was comparable to the performance in maize (Fig. 3 and SI Appendix, Fig. S6).
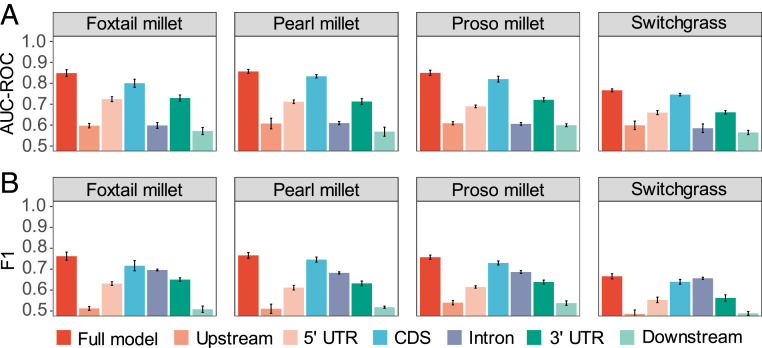
Fig. 3.
Performance in predicting cold-responsive gene expression across four grasses using DNA-sequence features. (A) AUC-ROCs achieved by models trained using DNA sequence features calculated from all gene-related regions within specific species. Error bars indicate SE across four models trained with differentially subset training and testing data. (B) F1 scores achieved by models trained using DNA sequence features calculated from all gene-related regions within specific species. Error bars indicate SE across four models trained with differentially subset training and testing data.
Cold stress can disrupt the circadian clock in plants (26). One potential explanation for the ability of the models trained on genomic sequence features to predict which genes were responding transcriptionally to cold stress would be if the model were learning features associated with diurnal cycling of gene expression. However, no significant differences in the amplitude of diurnal cycling were observed between true-positive (TP) and false-negative (FN) genes in maize (P = 0.86, Mann–Whitney U) and foxtail millet (P = 0.28, Mann–Whitney U), and in sorghum, a significant difference was observed in the opposite direction with higher diurnal amplitudes among FN genes than TP genes (P = 2.4e-2, Mann–Whitney U; median raw amplitude is 6.28 in FN and 4.81 in TP) (SI Appendix, Fig. S7), and this outcome was robust when a single outlier with extremely low amplitude was removed from the analysis. No consistent tendency was observed toward higher prediction accuracy among genes which exhibited significant diurnal cycling than among those which did not exhibit significant diurnal cycling (SI Appendix, Fig. S8).
Models trained by using data from on DNA sequence data in different species to predict transcriptonal responses exhibited similar trends in terms of feature importance. The CG and AA dinucleotide content of coding sequence (CDS) regions ranked as the two most important features distinguishing cold-responsive genes from genes that did not transcriptionally respond to cold stress in most models. A complete list of the 20 features estimated to be most important in models trained independently for each species is provided as Dataset S1, Tab 4. Based on the results from models trained using only the subset of sequence features calculated from specific gene regions: CDS, intron, 5 untranslated region (UTR), 3 UTR, and upstream and downstream regions, it appears that features calculated from the CDS, 5 UTR, and 3 UTR provided more useful information for building predictive models than did features calculated from intron, upstream, or downstream regions (Fig. 3A and SI Appendix, Fig. S6). CDS-only models consistently performed the best of any of the single sequence context models, but they did not exceed the prediction accuracy of the full model in any of the four species tested. Employing an alternate metric, F1 score, which balances sensitivity and precision, models trained by using intron-only features exceeded the performance of 5 UTR-only or 3 UTR-only models (Fig. 3B). However, the combined model employing features calculated from every gene region tended to outperform models trained using only subsets of features with any performance metric (AUC-ROC, AUPRC, or F1).
Models Trained in One Species Can Predict Which Genes Will Be Cold-Responsive in Another.
Because the same sequence features can be calculated for genes in different species, it is possible to evaluate how well cold-responsive gene expression can be predicted in one species based on only information about which genes did and did not respond to cold in another species. Single-species models trained in the six species (foxtail millet, pearl millet, switchgrass, proso millet, sorghum, or maize) to predict which genes would respond transcriptionally to cold were evaluated by using separate holdout test data from each of the six species (Fig. 4). The accuracy with which cold-responsive gene expression was predicted by models trained in one species and evaluated by using data from another was comparable or modestly lower than the accuracy of within-species prediction. Predictions using species that were more closely related were not obviously consistently superior to predictions using species that share common cold-stress phenotypes (sensitivity or tolerance) (Fig. 4).
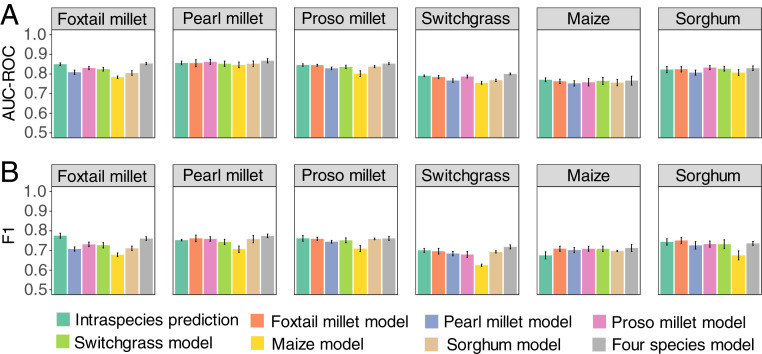
Fig. 4.
Predicting cold-responsive genes in one species using models trained in another species. Performance of species-specific prediction models and a four-species model when applied to predicting which genes will respond to cold stress in another species employing the same set of features calculated from DNA sequence information. Model performance was assessed by using both AUC-ROC values (A) and F1 scores (B). In both panels, error bars indicate SE among five values obtained from five models trained with differentially subset training and testing data. All predictions shown here, including intraspecies predictions, were made by using a common cross-species prediction framework, including the use of holdout test data from the same gene families (Materials and Methods).
A model trained by using data from the four Paniceae species (foxtail millet, pearl millet, switchgrass, and proso millet) exhibited either equivalent or superior performance to within-species predictions in foxtail millet, proso millet, switchgrass, and pearl millet when assessed by using mean AUC-ROC, AUPRC, or F1 score (Fig. 4; SI Appendix, Fig. S9 and Dataset S1, Tab 5). The performance of this four-species model was also assessed in maize and sorghum, outgroups to the four species used to train the model. The four-species model was able to predict cold-responsive gene expression in maize and sorghum with equivalent performance to models trained with data on cold-responsive gene expression collected directly from those species (Fig. 4; SI Appendix, Fig. S9 and Dataset S1, Tab 5), suggesting that for inferring cold-responsive or nonresponsive genes in a species with only genome assembly and annotation information using models trained in related species may be a practical strategy.
Different models exhibited more similar performance when evaluated in the same species than the same model evaluated using data from different species. Models consistently performed the best in classifying pearl millet, foxtail millet, or proso millet genes as cold-responsive or nonresponsive and generally performed the worst in predictions on data from maize and switchgrass. This pattern would be consistent with the notion that a certain proportion of classification errors resulted from varying amounts of noise in the ground-truth classifications of gene-expression patterns in individual species and/or variation in the accuracy of gene structural annotations used to calculate the sequence features used for prediction.
The analyses presented above all utilized cold-stress gene-expression data generated by a single research group following a common experimental protocol. Four additional cold-stress datasets in maize from two additional studies conducted by independent research groups following different protocols were identified in the literature (referred to as the Minnesota or Gansu datasets; see Materials and Methods for details) (27, 28). Models trained and tested in these outside datasets performed modestly worse than models trained by using the multi-time-point maize dataset in this study (SI Appendix, Fig. S10 and Dataset S1, Tab 6). This decline in performance may be related to the difference between single-time-point and multiple-time-point data, as models trained and tested using data from only individual time points within the time-series gene-expression data generated in this study also performed modestly worse than models trained and tested using the union of differentially expressed genes across time points (SI Appendix, Fig. S11). However, models trained by using either maize-expression data from this study or the combination of foxtail millet, pearl millet, proso millet, and switchgrass data (four-species model) exhibited better or equal performance in predicting which genes would transcriptionally respond to cold stress, relative to the performance of models trained and tested using the Minnesota or Gansu data. The relatively high translatability across datasets generated by different research groups using different protocols suggests that the patterns learned by the models described above are not artifacts of the specific protocol or experimental design employed for data generation in this study, but can indeed translate to independent data. Translatability was lower for the Gansu 16 °C dataset, potentially because different sets of genes respond to different severities of cold stress (28). Successful cross-species predictions for cold-responsive genes between cold-tolerant and cold-sensitive species or between genetically relatively distant species indicate that cold-responsive genes in Panicoideae share a high level of similarity in terms of gene-sequence features. The determinants of gene expression under cold stress are consistent across species at the gene-sequence level, even though only a small proportion of cold-responsive genes were conserved in different species (SI Appendix, Fig. S2). We further conducted k-mean clustering analysis of cold-responsive genes from the four Paniceae species based on gene expression in response to different durations of cold treatment (SI Appendix, Fig. S12 A–D). Genes belonging to each of the four species were distributed across all of the clusters (SI Appendix, Fig. S12E).
Discussion
Several factors have lowered the barriers to generating reference genome sequences for new species, including declining sequencing costs, advances in long-read sequencing technologies, and improvements to genome assembly and annotation algorithms. To date, over 300 plant genomes have been sequenced; in addition, a recent study collected transcriptome data from 1,124 plant species (29). Unfortunately, progress in generating layers of functional genomic data, including RNA-seq data for many of these newly sequenced genomes, has been much slower due to issues ranging from seed dormancy and limited access to wild plant species to difficulties in staging plants or delivering controlled stresses, tissues, and cell types, which require complicated, labor-intensive techniques to sample. As mentioned above, methods used to predict stress-responsive gene expression based on data on orthologous genes in related species have low accuracy and would, in any case, likely miss the changes in gene regulation associated with differences in stress tolerance between related species. Instead, we have demonstrated that supervised classification models trained on gene features, including sets of features that can be calculated solely from genomic sequence data and gene structural annotation, can provide significant accuracy to predict which genes will transcriptionally respond to a specific abiotic stress (cold, in this case). Many stresses are difficult to replicate effectively across different species or laboratories (4); here, we were able to use plants grown in the same laboratory with consistent growth conditions and treatments, and at the same developmental stage. This consistency represented an advantage that undoubtedly contributed to the success of prediction. However, when the models training in this study were evaluated on independent cold-stress gene-expression datasets generated by other research groups using different protocols, they still provided significant predictive performance (SI Appendix, Fig. S10). The success we achieved in prediction based on gene-sequence features greatly expands the potential application of this technique to nonmodel species—including those adapted to extreme environments—for which a reference genome sequence has been generated, but substantial functional genomic datasets are lacking.
Across species, CG dinucleotide content in CDS regions ranked as either the first or second most important feature in most trained prediction models (Dataset S1, Tab 4). Cytosines in CG sites are active targets of methylation and can be involved in regulating gene expression (30), but there is also evidence that CG sites can contribute to the regulation of transcriptional activity independently of DNA methylation (31). C/G content and CG dinucleotide content have been shown to be associated with regions of open chromatin in yeast (32, 33) and Drosophila (34), and CG dinucleotide content also plays a role in models which can predict regulation of gene expression in humans (35). In this study, DNA sequence features calculated from CDS, 5 UTR, and 3 UTR regions tended to provide more useful information content to predictive models than features calculated from intron, upstream, or downstream regions (Fig. 3), while models trained by using only DNA sequence features calculated from introns produced higher F1 scores than did models trained by using features from only 5 UTR or 3 UTR regions (Fig. 3B). The importance of exonic features relative to upstream and downstream regions observed across grasses for cold in this study is the opposite of the pattern reported in a recent study of heat- and drought-responsive genes in Arabidopsis using k-mer features (36). The involvement of UTRs in transcriptional regulation was also observed in a study predicting mRNA expression levels from DNA sequence features in maize and sorghum (24). These different patterns of feature importance may be explained by differences between the approaches employed in different studies to extracting features from raw DNA sequence information, differences in patterns of transcriptional regulation between cold and heat/drought stress, or differences in mechanisms of transcriptional regulation between grasses and crucifers, such as Arabidopsis.
A strikingly low level of conservation of cold responsiveness was observed among syntenic orthologous genes across the species examined in this study (SI Appendix, Fig. S2). The divergence of transcriptional patterns between orthologous genes can result from either trans-regulatory changes or cis-regulatory changes. In a comparison of natural maize haplotypes, cis-regulatory divergence was observed much more frequently than trans-regulatory divergence (9). A model where the high degree of divergence in cold-responsive expression between orthologous genes in related species is indeed primarily due to cis-regulatory changes is consistent with the observation that feature importance was conserved between models trained in different species. Specifically, a median of 70% of the 20 features with the highest importance scores overlapped between models trained in different individual species or the four-species model. In addition, the importance of cis-regulatory changes could explain why the models trained in one species that were successful at predicting cold-responsive gene expression tended to be successful in a second species. However, it is too early to conclude with certainty that features consistently ranked as highly important in multiple models play a causal role in determining whether a gene will transcriptionally respond to cold stress or whether they are simply correlated with this response.
Materials and Methods
Plant Material, Growth, and Stress Conditions.
For three of the six species tested, we employed the same genotype that had been sequenced to assemble the reference genome for that species: maize (Z. mays ssp. mays genotype B73), sorghum (S. bicolor genotype BTx623), and foxtail millet (S. italica genotype Yugu1). For the three other species, we were unable to employ the reference genotype and used another variety instead: switchgrass (P. virgatum genotype kanlow), proso millet (P. miliaceum genotype earlybird US Department of Agriculture [USDA] PI 578073), and pearl millet (P. glaucum synonym Cenchrus americanus genotype USDA PI 583800). For maize and sorghum, gene-expression data and details about growth conditions and stress treatments were described in Zhang et al. (2017) (7). Seeds were planted in standard potting mix (40% Canadian peat, 40% coarse vermiculite, 15% masonry sand, and 5% screened topsoil) in a Percival growth chamber (Percival model E-41L2) under 111 molm-2s-1 light intensity, 60% relative humidity, and a 12-h/12-h day/night cycle at 29 °C during the day and 23 °C at night. To target the approximately three-leaf stage in the different species, planting dates were staggered to allow cold-stress treatments to be performed simultaneously for batches of seedlings from multiple species: Foxtail millet, pearl millet, switchgrass, and proso millet seedlings were subjected to cold-stress treatment at 12, 10, 17, and 14 d after planting, respectively. Seedlings at the desired growth stage were divided, with one-half of each variety transferred to a growth chamber maintained at 6 °C and the other half used as the control. The seedlings were always transferred to cold-stress treatment at the end of the 12-h day cycle. Paired samples were collected from control and cold-stress treatments at 0.5, 1, 3, 6, 16, and 24 h after the onset of cold stress. Each sample was a pool of all above-ground tissue from at least three individual seedlings. Samples were collected from three independent biological replicates grown and harvested on separate dates.
Electrolyte-Leakage Analysis.
Plants used for electrolyte-leakage analysis were harvested from nonacclimated plants at the same growth stage and conditions described above. Leaf tissue was harvested from pearl millet and proso millet by using a 5-mm punch, and three punches were tested per sample. For switchgrass and foxtail millet, the narrow leaf blades prevented the even application of the 5-mm punch; instead, six 5-mm leaf sections were cut with a razor blade and pooled for each sample. Efforts were made to ensure that equivalent portions of the leaf were included in each replicate, and only the midsection of each leaf was used, avoiding the stalk or tip. All leaf samples were immersed in sterile water with a resistivity of 18.2 M at 25 °C. All conductivity measurements were performed with an Accumet 200 conductivity meter (Fisher Scientific, probe: catalog no. 13-620-101). Initial readings were collected from samples incubated at 0 °C for 30 min in a precooled chiller (initial measurement). After prechilling, a small ice crystal was added to each sample to initiate ice nucleation. After nucleation, individual samples were incubated in the chiller at a rate of –0.5 °C per 0.5 h, and samples were removed when the temperature reached –1 °C, –1.5 °C, –2 °C , –2.5 °C, –3 °C, –3.5 °C, and –4 °C. The samples were thawed at 4 °C in a cooling water bath for 2 to 4 h, incubated at room temperature for 30 min, and mixed on an orbital shaker at 250 to 300 rpm for an additional 20 min at room temperature. At this point, the conductivity of the water was measured (treatment measurement). Finally, each sample was incubated at 65 °C for 30 min and shaken for 20 min before a final conductivity reading was taken for each sample (final measurement). Percent electrolyte leakage for each sample was calculated by using the formula (treatment measurement – initial measurement)/(final measurement – initial measurement). The temperature of 50% electrolyte leakage (LT50) for each set of samples was defined to be the value of the log of 50% of maximum electrolyte conductivity for a sigmoidal curve fit to the percent leakage values calculated at different temperatures, based on the initial and final measurements. Curves were fit to percent electrolyte-leakage value points by using the sigmoidal dose–response model provided by the software package GraphPad Prism (version [v]8.1.2) following the protocol outlined by Thalhammer et al. (23).
Generating RNA-Seq Data and Identifying Cold-Responsive Genes.
RNA isolation and library construction were performed as described by Zhang et al. (2017) (7). Sequencing was conducted at the Illumina Sequencing Genomics Resources Core Facility at Weill Cornell Medical College with 1 × 50 bp (SE) run on the HiSeq2500 platform. Raw sequencing data from maize and sorghum with the same experimental design and cold treatment were previously deposited at the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/bioproject) under accession no. PRJNA344653 (7). The raw reads were quality-filtered, and adaptors were removed from the data with the sequence-preprocessing tool Trimmomatic (v0.38) (37) (MINLEN = 36, LEADING = 3, TRAILING = 3, SLIDINGWINDOW = 4,15). The trimmed reads were mapped to the corresponding reference genome for each species by using GSNAP (38) (v2018-03-25) (-B 4 -N 1 -n 2 -Q -nofails format = sam). Genome assemblies of S. italica (v2.2) (39), P. virgatum (v4.1) (Department of Energy Joint Genome Institute [DOE-JGI], phytozome.jgi.doe.gov/), Z. mays (APGv4) (40), and S. bicolor (v3.1.1) (41) were downloaded from Phytozome v12.1. Genome assemblies for P. miliaceum and P. glaucum were downloaded from NCBI (42) and the Gigascience Database (dx.doi.org/10.5524/100192) (43), respectively. Samtools (v1.9) (44) was used to convert the raw Sequence Alignment Map (SAM) output from GSNAP to sorted Binary Alignment Map (BAM) files. Fragments per kilobase of transcript per million mapped reads (FPKM) values were calculated by using sorted BAM files with cufflinks (v2.2) (45). Genes were classified as expressed if their averaged FPKM values at all time points under both treatment and control conditions were 1 (14). HTSeq (v 0.6.1) was used to extract the number of reads in each RNA-seq library that were mapped to annotated exons of each gene in each species using union mode (46). Read counts were used to identify cold-responsive genes by comparing the expression of genes in treatment vs. control samples, with differentially expressed genes defined as having adjusted P value 0.05 and absolute log2 of fold change at any of the six time points using DESeq2 (47). Nonresponsive genes were defined as those meeting the definition of expressed genes with absolute log2 of fold change of between treatment and control value at all time points. For data collected in a single time point per species, cold-responsive genes were defined as adjusted P value 0.05 and absolute log2 of fold change , and nonresponsive genes were defined as absolute log2 of fold change of between treatment and control value . Raw sequencing data generated in this study were deposited into NCBI (BioProject accession no. PRJNA650146).
Quantifying Gene Features.
Genomic features of foxtail millet, switchgrass, maize, and sorghum were scored by using the corresponding gff annotation file and the mRNA transcript that was scored as primary for each individual gene model. For pearl millet and proso millet, instead, all annotated genes were scored due to the lack of primary transcript information. Annotation of UTR sequences was inconsistent across species. In pearl millet and proso millet, UTR annotations were absent, while in maize, sorghum, switchgrass, and foxtail millet, only a partial set of genes included UTR annotations. When UTRs were present, their median lengths were approximately 200 bp (5 UTR) and 350 bp (3 UTR). These lengths were standardized for all species. The frequencies of all individual nucleotides (4 features) and dinucleotides (16 features) were calculated for each of six regions: the CDS, intron, estimated 5 UTR, estimated 3 UTR, 1 Kb upstream of the 5 UTR starting site, and 1 Kb downstream of the 3 UTR ending site (SI Appendix, Fig. S4). Overall, 120 features were scored for each gene. The code used to calculate these features has been deposited in the Bitbucket repository (https://bitbucket.org/shanwai1234/coldgenepredict/src/master/).
For maize, additional nongenomic sequence features were scored as detailed by Dai et al. (16). Briefly, the epigenetic features included DNA methylation (quantified separately in the CG, CHG, and CHH contexts), three histone modifications (H3K4me3, H3K27me3, and H3K27ac), and open chromatin (quantified by Assay for Transposase-Accessible Chromatin using sequencing [ATAC-seq]) (48). Diversity and evolutionary features included genomic evolutionary rate profiling scores (49), presence–absence variations frequency, orthologous gene in close relatives, synonymous mutation rate (Ks), nonsynonymous mutation rate (Ka), Ka/Ks value, minor allele frequency (MAF) distributions, and single-nucloetide polymorphism (SNP) density features. MAF distributions and SNP density were calculated from the maize 282 association panel with data downloaded from Panzea (https://www.panzea.org/) (50). Ka and Ks values for maize genes were calculated based on orthologous genes in maize, sorghum, and foxtail millet, and the resulting values were obtained from previous work (51). A syntenic gene list for Z. mays, S. bicolor, S. italica, S. viridis, O. sativa, B. distachyon, and O. thomaeum was downloaded from Figshare (dx.doi.org/10.6084/m9.figshare.3113488.v1) (7). Any missing values in the nonsequence-based feature set for maize were imputed by using the median value for that feature across all genes. Feature datasets were preprocessed by using the scale and center transformation methods of the “preProcess()” function in the R package “caret” (v 6.0-80) (52). Summaries of feature values for foxtail millet, pearl millet, proso millet, switchgrass, sorghum, and maize are deposited at the Bitbucket repository (https://bitbucket.org/shanwai1234/coldgenepredict/src/master/).
Binning of Cold-Responsive and Nonresponsive Genes.
A binning method was used to reduce the bias of baseline gene expression and to balance the number of genes in the cold-responsive and nonresponsive datasets for supervised machine-learning classification. The joint set of all cold-responsive and nonresponsive genes was sorted and segmented into 12 bins (dodeciles) based on average expression value. Within each dodecile, all genes of the less abundant class (either cold-responsive or nonresponsive) were included as potential data points for training and testing, while the more abundant class was randomly subsampled to provide equal numbers of cold-responsive and nonresponsive genes within that particular dodecile.
Gene-Family Clustering.
Protein sequences of the six species (maize, sorghum, foxtail millet, pearl millet, proso millet, and switchgrass) were clustered into families by using the Markov Cluster (MCL) Algorithm as described (24, 53). Pairwise similarity of the protein sequence encoded by the primary annotated transcript of each gene in the six species was quantified by using the e-value reported by BLASTP (54). Gene families were defined by using OrthoMCL clustering with an inflation index of 1.5 (55). If a gene was not assigned to any gene family, it was treated as a single-member gene family.
Random-Forest Training, Classification, and Evaluation.
Two approaches were taken to generating holdout test data, one for within-species prediction and another for cross-species prediction. These approaches differed only in the order different controls were applied. For within-species and cross-species prediction, cold-responsive and nonresponsive genes were first balanced by using the expression-binning method (SI Appendix, Fig. S4). Afterward, for within-species prediction, the data were further subsampled to include only one gene per gene family, considering only those genes remaining after balancing by expression bin. These subsampled data were then split into training/validation data (90% of remaining genes) and holdout test data (10% of genes), ensuring that different gene families were never represented in training and testing data simultaneously. For cross-species predictions, after genes were balanced in each species based on expression bins, whole gene families were partitioned into training/validation (90%) and testing data (10%). After this partitioning, training data were generated by subsampling genes from the relevant training species, and testing data were generated by subsampling genes from the relevant testing species. This modified approach ensured that pairs of orthologs or recently diverged homologs were never represented in training and testing data simultaneously and that models trained or evaluated in different species were still being trained with equivalent sets of genes and evaluated on equivalent test sets to aid comparability. For purposes of comparison, the cross-species approach was also employed to generate “intraspecies” results in Fig. 4.
For both approaches, models were trained by using the random-forest algorithm as implemented by the “rf” method in the R package “caret” (v 6.0-80). Models were trained by using 10-fold cross-validation of 81% of the total data employed for training and 9% for validation. The performance of the final model was assessed by using the 10% holdout test data unless otherwise stated. Two parameters—ntree, the number of trees to grow, and mtry, the number of variables randomly sampled as candidates at each split—were optimized for each model by using a grid-search strategy. The minimum size of terminal nodes was set as one (nodesize = 1), and the growth of trees was not constrained (maxnodes = NULL). For each model, receiver operating characteristic (ROC) and precision-recall (PR) curves and the area under each type of curve were calculated by using the R package PRROC (56). Confusion matrices was produced for each model to calculate TPs (also referred to as recall/sensitivity), true negatives (TNs), false positives (FPs), and FNs. Precision (TP/(TP + FP)) and specificity (TN/(TN + FP)) were calculated by using the R package “caret” (v 6.0-80). F1 score was calculated as the harmonic mean of precision and recall. Codes used to train each model, the associated datasets, and trained models were deposited in a Bitbucket repository (Data Availability).
Evaluation of Model Performance on Independent Datasets.
Two additional published maize cold-stress datasets were used to assess how well the trained models performed on data generated by other research groups in other parts of the world. Data on gene expression in the third leaf of 13-d-old maize seedlings grown in Minnesota under control and 6 °C conditions were obtained from NCBI with accession ID of PRJNA657262 (27). Data on gene expression in the second leaf of maize seedlings at the three-leaf stage grown in Gansu, China, under control conditions and three levels of cold stress (4 °C, 10 °C, and 16 °C) were obtained from NCBI with accession ID of PRJNA645274 (28). Raw sequence reads were aligned, read counts per gene were obtained, and differential gene-expression analysis was conducted by using the same protocols described above. The four sets of cold-responsive and nonresponsive genes were treated as additional “species” and assessed by using the cross-species approach to partitioning training/validation and testing data described in the previous section.
Impact of Diurnal Gene Expression on Prediction Accuracy.
Data on diurnal patterns of gene expression were available for three of the six species analyzed in this study: maize, sorghum, and foxtail millet (57). The dataset employed here consists of samples collected every 3 h over a course of 72 h and classified into cycling and noncycling genes by using JTK_Cycle analysis. JTK_Cycle was also employed to estimate the amplitude of cycling for each gene. Results from ref. 57 were converted from v5b of the maize reference genome to vAPGv4 by using a published conversion list provided by the Maize Genetics and Genomics Database.
We employed two approaches to assess if circadian or diurnal cycling contributed to the accuracy with which cold-responsive genes could be predicted from DNA sequence features using the framework in the intraspecies model: 1) Based on prediction results, genes in holdout test data were divided into TPs and FNs. Amplitude difference between genes in the TP and FN sets were compared by using a Mann–Whitney U test. 2) Genes in each holdout test data were divided into circadian and noncircadian genes, according to the circadian genes identified in each species (57). The corresponding trained model made predictions on circadian and noncircadian genes separately in each holdout test data. Twenty sets of predictions were generated with different random seeds. For approach 1, TPs and FNs identified across each approach were merged into single nonredundant sets of genes to compare amplitude differences. For approach 2, individual AUC-ROCs, AUPRCs, and F1 scores were recorded for cycling and noncycling genes and compared by using paired t tests.
Identifying Syntenic Orthologs.
CDS data for primary transcripts of S. italica (v2.2) (39) and P. virgatum (v4.1) (DOE-JGI, phytozome.jgi.doe.gov/) were retrieved from Phytozome v 12.1. CDS data for P. miliaceum and P. glaucum were obtained from NCBI (BioProject number PRJNA431363) (42) and the GigaScience Database (dx.doi.org/10.5524/100192) (43), respectively. The software and corresponding settings used to identify syntenic orthologs were as described (7) with minor modifications. The parameter settings for LASTZ (58) were as described (7), except that a 75% sequence identity threshold was used for alignment. The QuotaAlign algorithm was used for further processing with -quota set to 1:1 for comparisons between S. italica and P. glaucum and 1:2 for comparisons between S. italica and P. miliaceum or P. virgatum due to whole-genome duplication in P. miliaceum and P. virgatum. Other parameters used for QuotaAlign and the subsequent polishing procedure were as described (7). The syntenic orthologous pairs between S. italica and S. bicolor were downloaded from Figshare (dx.doi.org/10.6084/m9.figshare.3113488.v1) (7). The Syntenic gene list generated among S. bicolor, S. italica, P. glaucum, P. miliaceum, and P. virgatum is shown in Dataset S1, Tab 7.
Phylogenetic Analysis of Species.
A set of 7,064 gene groups was identified with syntenic ortholog representatives in sorghum, foxtail millet, pearl millet, both subgenomes of proso millet, and both subgenomes of switchgrass (seven total gene copies). Multiple sequence alignments for the annotated CDSs for all seven genes within a group were generated by using MAFFT (v7.149) with the parameter setting L-INS-i (59). Poorly aligned regions after multiple sequence alignment were eliminated by using Gblocks (v0.91b) with the following settings: minimum number of sequences for a conserved position: 9; minimum number of sequences for a flank position: 14; maximum number of contiguous nonconserved positions: 8; minimum length of a block: 10 (60). StarBEAST2 (v0.15.5) (61) implemented in BEAST 2.5.1 (62) employing a Bayesian Markov chain Monte Carlo (MCMC) framework was used to estimate both species trees and divergence dates. Due to the computational intensity of the analyses, an ensemble of 12 separate StarBEAST2 runs was employed, using different sets of 50 loci that were selected randomly (without replacement) from the alignments. Each StarBEAST2 run used analytical population size integration, the uncorrelated lognormal clock model, an HKY nucleotide substitution model with empirical frequencies, gamma category count of 4, and proportion invariant of 0.2. A calibrated yule model was used as a prior for tree topology using the previously estimated divergence time between foxtail millet and sorghum of 26 million y ago as a reference (39), which was derived from the divergence time between rice and Panicodeae at approximately 50 million y ago (63). Two independent runs of 40 million generations (sampled every 5,000) were conducted in each analysis and combined with LogCombiner (v2.5.1) with 20% burn-in for the species tree. Effective sampling sizes and MCMC convergence were examined by using Tracer (v1.7.1) (64). A maximum clade credibility tree was compiled with TreeAnnotator (v2.4.7) after discarding the initial 10% burn-in, and the tree was visualized by using FigTree (v1.4.4) (65).
Clustering and Gene Ontology Enrichment Analyses.
Clustering analysis was performed by using cold-responsive genes from foxtail millet, pearl millet, switchgrass, and proso millet as a whole. The log2 fold values were normalized by row and analyzed by using the R k-means function with 20 groups. Groups with similar expression patterns were further merged into 13 clusters, including 4 early transcriptional response clusters, 4 late transcriptional response clusters, 2 continually changing clusters, and 3 unclassified clusters. The cluster patterns are shown in heat maps and in graphical format. Gene Ontology (GO) annotations were downloaded from phytozome (v12.1) for foxtail millet and switchgrass and from published papers for proso millet (42) and pearl millet (43). GO enrichment analyses of gene sets in each cluster were performed by using GOATOOLS (66) with all annotated genes in the genome as background. GO terms were considered significantly enriched if P 0.05 after controlling for false discovery rate using the Benjamini–Hochberg procedure.
Supplementary Material
Acknowledgments
We thank Nathan M. Springer for a critical reading and helpful suggestions on a draft of this manuscript. This work was supported by USDA National Institute of Food and Agriculture Award 2016-67013-24613 (to J.C.S. and R.L.R.); and by the US Department of Energy, Office of Science, Office of Biological and Environmental Research Program Award DE-SC0020355 (to J.C.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026330118/-/DCSupplemental.
Data Availability
Raw sequencing data for foxtail millet, proso millet, pearl millet, and switchgrass generated in this study are available at NCBI (BioProject accession no. PRJNA650146) (67). Raw sequencing data of maize and sorghum used for model training were previously deposited at the NCBI under accession no. PRJNA344653 (68). Gene expression data of maize seedlings grown in Minnesota under control and cold stress conditions were obtained from NCBI with accession no. PRJNA657262 (69). Data on gene expression in the leaf of maize seedlings grown in Gansu, China, under control conditions and cold stress were obtained from NCBI with accession no. PRJNA645274 (70). Codes used to calculate genomic features per each gene, gene labels and associated features, machine-learning scripts, and trained models are available at the Bitbucket repository (https://bitbucket.org/shanwai1234/coldgenepredict/src/master/).
References
- 1.Cheng S., et al. , 10KP: A phylodiverse genome sequencing plan. Gigascience 7, giy013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzvelev N., The system of grasses (Poaceae) and their evolution. Bot. Rev. 55, 141–203 (1989). [Google Scholar]
- 3.Dohleman F. G., Long S. P., More productive than maize in the midwest: How does Miscanthus do it?. Plant. Physiol. 150, 2104–2115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raju S. K. K., Barnes A. C., Schnable J. C., Roston R. L., Low-temperature tolerance in land plants: Are transcript and membrane responses conserved? Plant Sci. 276, 73–86 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hope H., McElroy A., Low-temperature tolerance of switchgrass (Panicum virgatum l.). Can. J. Plant Sci. 70, 1091–1096 (1990). [Google Scholar]
- 6.Mickelbart M. V., Hasegawa P. M., Bailey-Serres J., Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., et al. , Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell 29, 1938–1951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Healy T. M., Schulte P. M., Patterns of alternative splicing in response to cold acclimation in fish. J. Exp. Biol. 222, jeb193516 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Waters A. J., et al. , Natural variation for gene expression responses to abiotic stress in maize. Plant J. 89, 706–717 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Cooper G. M., The Cell: A Molecular Approach (Sinauer Associates, 2000). [Google Scholar]
- 11.Makarevitch I., et al. , Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 11, e1004915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisch D., How important are transposons for plant evolution? Nat. Rev. Genet. 14, 49 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Naito K., et al. , Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Sartor R. C., Noshay J., Springer N. M., Briggs S. P., Identification of the expressome by machine learning on omics data. Proc. Natl. Acad. Sci. U.S.A. 116, 18119–18125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brubaker D. K., Proctor E. A., Haigis K. M., Lauffenburger D. A., Computational translation of genomic responses from experimental model systems to humans. PLoS Comput. Biol. 15, e1006286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai X., et al. , Non-homology-based prediction of gene functions in maize (Zea mays ssp. mays). Plant Genome, e20015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore B. M., et al. , Robust predictions of specialized metabolism genes through machine learning. Proc. Natl. Acad. Sci. U.S.A. 116, 2344–2353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He W., Wei L., Zou Q., Research progress in protein posttranslational modification site prediction. Brief. Funct. Genom. 18, 220–229 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Hetherington S. E., He J., Smillie R. M., Photoinhibition at low temperature in chilling-sensitive and-resistant plants. Plant Physiol. 90, 1609–1615 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinnusamy V., Zhu J., Zhu J. K., Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Poudel H. P., Sanciangco M., Kaeppler S., Buell C. R., Casler M., Quantitative trait loci for freezing tolerance in a lowland upland switchgrass population. Front. Plant Sci. 10, 372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swigoňová Z., et al. , Close split of sorghum and maize genome progenitors. Genome Res. 14, 1916–1923 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thalhammer A., Hincha D. K., Zuther E., “Measuring freezing tolerance: Electrolyte leakage and chlorophyll fluorescence assays” in Plant Cold Acclimation, D. Hincha, E. Zuther, Eds. (Methods in Molecular Biology, Humana Press, New York, NY, 2014), vol. 1166, pp. 15–24. [DOI] [PubMed] [Google Scholar]
- 24.Washburn J. D., et al. , Evolutionarily informed deep learning methods for predicting relative transcript abundance from DNA sequence. Proc. Natl. Acad. Sci. U.S.A. 116, 5542–5549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 26.Bieniawska Z., et al. , Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant. Physiol. 147, 263–279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Z., et al. , Genetic and epigenetic variation in transposable element expression responses to abiotic stress in maize, Plant Physiol., 10.1093/plphys/kiab073 (2021). [DOI] [PMC free article] [PubMed]
- 28.Li Y., et al. , Transcriptomic analysis revealed the common and divergent responses of maize seedling leaves to cold and heat stresses. Genes 11, 881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.OTPT Initiative et al. , One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz R. J., Lewis Z. A., Goll M. G., DNA methylation: Shared and divergent features across eukaryotes. Trends Genet. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartl D., et al. , CG dinucleotides enhance promoter activity independent of DNA methylation. Genome Res. 29, 554–563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tillo D., Hughes T. R., G+C content dominates intrinsic nucleosome occupancy. BMC Bioinf. 10, 442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai X., Nagarajan S., Kim K., Lee K., Choi J. K., Regulation of the boundaries of accessible chromatin. PLoS Genet. 9, e1003778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yáñez-Cuna J. O., et al. , Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res. 24, 1147–1156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natarajan A., Yardımcı G. G., Sheffield N. C., Crawford G. E., Ohler U., Predicting cell-type–specific gene expression from regions of open chromatin. Genome Res. 22, 1711–1722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azodi C. B., Lloyd J. P., Shiu S. H., The cis-regulatory codes of response to combined heat and drought stress in Arabidopsis thaliana. NAR Genom. Bioinform. 2, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu T. D., Reeder J., Lawrence M., Becker G., Brauer M. J., “GMAP and GSNAP for genomic sequence alignment: Enhancements to speed, accuracy, and functionality” in Statistical Genomics, E. Mathé, S. Davis, Eds. (Methods in Molecular Biology, Humana Press, New York, NY, 2016), vol. 1418, pp. 283–334. [DOI] [PubMed] [Google Scholar]
- 39.Bennetzen J. L., et al. , Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Schnable P. S., et al. , The B73 maize genome: Complexity, diversity, and dynamics. Science 326, 1112–1115 (2009). [DOI] [PubMed] [Google Scholar]
- 41.McCormick R. F., et al. , The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Zou C., et al. , The genome of broomcorn millet. Nat. Commun. 10, 436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varshney R. K., et al. , Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 35, 969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C., et al. , Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong P., et al. , 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 10, 1497–1509 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Yang J., et al. , Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. 13, e1007019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bukowski R., et al. , Construction of the third-generation Zea mays haplotype map. Gigascience 7, gix134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Z., Qiu Y., Schnable J. C., Genome-phenome wide association in maize and Arabidopsis identifies a common molecular and evolutionary signature. Mol. Plant 13, 907–922 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Kuhn M., Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).27774042 [Google Scholar]
- 53.Van Dongen S. M., “Graph clustering by flow simulation,” Ph.D. thesis, Utrecht University, Utrecht, Netherlands (2000). [Google Scholar]
- 54.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Stoeckert C. J., Roos D. S., OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grau J., Grosse I., Keilwagen J., PRROC: Computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics 31, 2595–2597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai X., et al. , Interspecific analysis of diurnal gene regulation in panicoid grasses identifies known and novel regulatory motifs. BMC Genom. 21, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris R. S., “Improved pairwise alignment of genomic DNA,” Ph.D. Thesis, Pennsylvania State University, State College, PA: (2007). [Google Scholar]
- 59.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Talavera G., Castresana J., Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Ogilvie H. A., Bouckaert R. R., Drummond A. J., StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 34, 2101–2114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouckaert R., et al. , BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolfe K. H., Gouy M., Yang Y. W., Sharp P. M., Li W. H., Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. U.S.A. 86, 6201–6205 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambaut A., Figtree V1. 4. Molecular Evolution, Phylogenetics and Epidemiology (Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK, 2012). [Google Scholar]
- 66.Klopfenstein D., et al. , GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 8, 10872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng Xiaoxi, et al. , Transcriptional responses of Foxtail millet, Proso millet, Pearl millet and Switchgrass to cold stress. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA650146. Deposited 1 August 2020. [Google Scholar]
- 68.Zhang Y.et al. , mays Zea, italica Setaria, Sorghum bicolor. NCBI. https://www.ncbi.nlm.nih.gov//bioproject/PRJNA344653. Deposited 27 September 2016. [Google Scholar]
- 69.Liang Z., et al. , Maize genotypes under cold/heat stress. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA657262. Deposited 14 August 2020. [Google Scholar]
- 70.Li Y., et al. , Transcriptome analysis of cold and heat stresses in maize seedlings. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA645274. Deposited 10 July 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for foxtail millet, proso millet, pearl millet, and switchgrass generated in this study are available at NCBI (BioProject accession no. PRJNA650146) (67). Raw sequencing data of maize and sorghum used for model training were previously deposited at the NCBI under accession no. PRJNA344653 (68). Gene expression data of maize seedlings grown in Minnesota under control and cold stress conditions were obtained from NCBI with accession no. PRJNA657262 (69). Data on gene expression in the leaf of maize seedlings grown in Gansu, China, under control conditions and cold stress were obtained from NCBI with accession no. PRJNA645274 (70). Codes used to calculate genomic features per each gene, gene labels and associated features, machine-learning scripts, and trained models are available at the Bitbucket repository (https://bitbucket.org/shanwai1234/coldgenepredict/src/master/).