Significance
Central carbon metabolism has the overlapping functions of converting substrate into biomass and the extraction and storage of chemical energy. These metabolic pathways are rewired by cancer cells to selectively support increased biomass demands. This reprogramming is a potential therapeutic target if a molecular-level understanding of the relevant control processes can be attained. Through genomic editing of an HCT116 colonic tumor cell line, we uncover metabolic supervision by a single pair of cell-signaling enzymes, the PPIP5Ks. Our global measurements of steady-state metabolite levels, plus isotope tracing analysis, show in detail how HCT116 cells recruit PPIP5Ks to select specific metabolic pathways to provide adequate precursor supply for biomass production. Crucially, in the absence of PPIP5K activity, HCT116 tumorigenesis is reduced.
Keywords: inositol pyrophosphates, PPIP5K, nucleotide synthesis, pentose phosphate pathway, serine–glycine–one-carbon metabolism
Abstract
Identification of common patterns of cancer metabolic reprogramming could assist the development of new therapeutic strategies. Recent attention in this field has focused on identifying and targeting signal transduction pathways that interface directly with major metabolic control processes. In the current study we demonstrate the importance of signaling by the diphosphoinositol pentakisphosphate kinases (PPIP5Ks) to the metabolism and proliferation of the HCT116 colonic tumor cell line. We observed reciprocal cross talk between PPIP5K catalytic activity and glucose metabolism, and we show that CRISPR-mediated PPIP5K deletion suppresses HCT116 cell proliferation in glucose-limited culture conditions that mimic the tumor cell microenvironment. We conducted detailed, global metabolomic analyses of wild-type and PPIP5K knockout (KO) cells by measuring both steady-state metabolite levels and by performing isotope tracing experiments. We attribute the growth-impaired phenotype to a specific reduction in the supply of precursor material for de novo nucleotide biosynthesis from the one carbon serine/glycine pathway and the pentose phosphate pathway. We identify two enzymatic control points that are inhibited in the PPIP5K KO cells: serine hydroxymethyltransferase and phosphoribosyl pyrophosphate synthetase, a known downstream target of AMP-regulated protein kinase, which we show is noncanonically activated independently of adenine nucleotide status. Finally, we show the proliferative defect in PPIP5K KO cells can be significantly rescued either by addition of inosine monophosphate or a nucleoside mixture or by stable expression of PPIP5K activity. Overall, our data describe multiple, far-reaching metabolic consequences for metabolic supervision by PPIP5Ks in a tumor cell line.
Metabolic rewiring is a crucial hallmark of cancer. Tumor cells consistently reprogram metabolic pathways to cope with both genetic alterations and environmental challenges that threaten cell survival and proliferation (1, 2). These adaptations also provide tumor cells with an appropriate supply of energy to maintain anabolic pathways to serve an elevated proliferative capacity. A key goal of research in the field is to gain mechanistic understanding of how cancer cell metabolism becomes repro-grammed in order that new therapeutic approaches can be designed (2). Although tumor metabolism exhibits considerable heterogeneity, there is a growing belief that it may be susceptible to targeting members of a finite set of signal transduction pathways that interface directly with metabolic control processes (2). It is this concept that has led us to study regulation of cancer cell metabolism by a unique group of cellular signals known as the inositol pyrophosphates (PP-InsPs), which include 5-InsP7 and InsP8 (3, 4) (Fig. 1A). The “energetic” diphosphate groups in these PP-InsPs are functionally significant features that distinguish them from other members of the inositol phosphate (InsP) signaling family. These diphosphate groups have two separate mechanisms of action: they dictate ligand specificity in allosteric control process, and they can nonenzymatically transphosphorylate proteins (4–7).
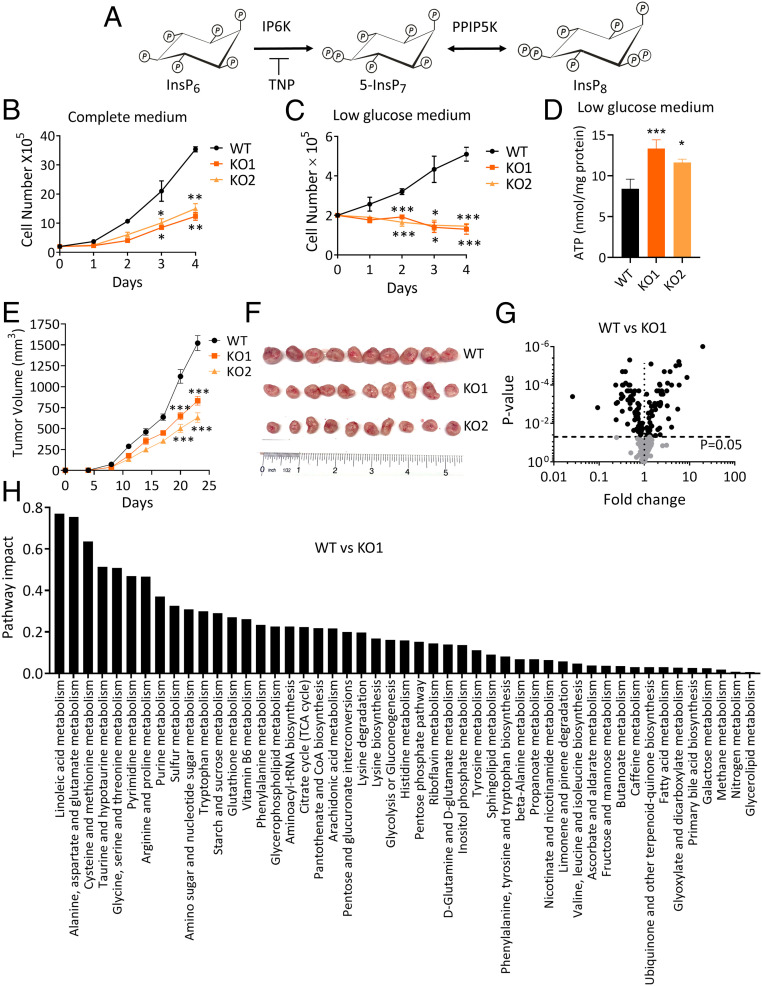
Fig. 1.
KO of PPIP5Ks from HCT116 cells induces broad changes in cell metabolism and tumorigenesis. (A) The pathway of PP-InsP synthesis in mammalian cells. (B and C) Growth curves of WT HCT116 cells (black symbols) and two independent PPIP5K KO clones, KO1 (dark orange symbols) and KO2 (light orange symbols) cultured in media in which the initial glucose concentration is either high (17.5 mM) or low (initially 0.2 to 0.3 mM; see SI Appendix, Fig. S1 B and C). (D) Cellular ATP levels in cells cultured in low-glucose medium for 24 h. (E) Tumor volume of xenografts obtained from WT and PPIP5K KO cells growing in mice. (F) Representative pictures of tumor xenografts. (G) Volcano plots of metabolites of WT cells and PPIP5K KO1 cells cultured in low-glucose media. (H) Network-based pathway analysis of statistically significant changes in metabolite levels in PPIP5K KO1 cells compared to WT cells cultured in low-glucose medium (data for PPIP5K2 KO2 cells are shown in SI Appendix, Fig. S3). Student’s t test was used to determine statistical significance of differences between means (*P < 0.05, **P < 0.01, ***P < 0.001).
InsP8 is the final product of the PP-InsP synthetic pathway, where its levels and hence its signaling strength are controlled by the PPIP5Ks: an evolutionarily conserved family of cell-signaling enzymes with mutually antagonistic 5-InsP7 1-kinase and InsP8 1-phosphatase activities (8, 9). Human cells express type 1 and type 2 isoforms of the PPIP5Ks (4). We previously studied their signaling functions by their CRISPR-mediated knockout (KO) from the HCT116 colonic epithelial tumor cell line (3). In that study, we determined that elimination of InsP8 from cells reduced their rate of growth, even though they are hypermetabolic in the sense that adenosine triphosphate (ATP) levels were elevated, due to more active glycolysis and oxidative phosphorylation. This seemed an anomalous result since this enhanced production of cellular energy would normally be expected to support the biosynthetic requirement for cell proliferation. To pursue this apparent paradox, we sought to obtain a detailed description of the supervisory role of the PPIP5Ks upon tumor cell metabolism, specifically with regard to the metabolic basis for the slower rate of proliferation. For these experiments, we used liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS), and we show that PPIP5Ks regulate key aspects of global anabolic/catabolic balance; PPIP5K KO cells exhibit a dramatic reduction in precursor supply for nucleotide synthesis, which we demonstrate to account for the proliferative defect. We define the metabolic basis by which a cancer cell line recruits PPIP5Ks to maintain nucleotide synthesis, and we propose this information provides a direction for cancer treatment.
Results
KO of PPIP5Ks Has Global Effects upon Cell Metabolism.
Our interest in understanding how PP-InsP metabolism interfaces with metabolic homeostasis (4, 8) previously led us to determine that CRISPR-directed knockout of PPIP5Ks (PPIP5K KO) from the HCT116 tumor cell yields an apparently paradoxical phenotype: a reduction in the rate of proliferation despite enhanced production of cellular energy (ref. 3 and Fig. 1B). The latter experiments were performed using regular culture media with a “high” (17.5 mM) glucose concentration. However, the metabolic demands of human tumors reduces the glucose concentration in their microenvironment to <1 mM (10). We have now mimicked this metabolically critical aspect (i.e., <1 mM; see Materials and Methods) by using culture medium in which the only source of glucose was that provided by the serum (11).
We have obtained data from two independently generated PPIP5K KO clonal lines, which we have named KO1 and KO2. Under these low-glucose conditions, the impact of the PPIP5K KO upon cell proliferation is especially dramatic: The cell numbers diminished throughout the course of these experiments (Fig. 1C). In further experiments we slightly increased the initial glucose concentration by either 1 or 2 mM, and the PPIP5K KO growth-inhibited phenotype was still evident (SI Appendix, Fig. S1A). In these experiments we also monitored the concentration of medium glucose and found no difference between wild-type (WT) and PPIP5K KO cultures (SI Appendix, Fig. S1 B and C); the decrease in PPIP5K KO cell numbers could mask a faster rate of glycolysis. It is also interesting that the elevated level of ATP that is a characteristic of HCT116 PPIP5K KO cells (3) remains evident in low-glucose conditions (Fig. 1D).
Furthermore, decreased tumorigenic capacity of the PPIP5K KO cells is suggested by reduced tumor volumes in mice and their reduced ability to form colonies in soft agar or on a solid surface (Fig. 1 E and F and SI Appendix, Fig. S1 D and E).
It may be interesting when creating additional PPIP5K KO tumor cell lines to investigate how widespread is a growth inhibited phenotype that is exacerbated by low extracellular glucose, although it should not be anticipated that all cancer cell types will respond to a PPIP5K KO in a similar manner. We have noted that PPIP5K KO HEK cells grow more slowly than WT cells in low-glucose media (SI Appendix, Fig. S1F).
PPIP5K KO cells have higher steady-state levels of 5-InsP7 than WT cells (3). Thus, it is interesting that the cell growth phenotype of PPIP5K KO cells was not rescued by treatment with N2-(m-trifluorobenzyl), N6-(p-nitrobenzyl) (TNP) to inhibit IP6K-mediated 5-InsP7 synthesis (12); in fact, TNP reduced the proliferative capacity of both WT and PPIP5K KO cells (SI Appendix, Fig. S1G). We conclude that the impaired growth rate of PPIP5K KO cells is not due to their elevated 5-InsP7 levels.
We also studied the impact of glutamine on cell growth. Elimination of glutamine (normally 2.5 mM) from the cell culture medium has a less profound impact upon proliferation of the PPIP5K KO line than glucose limitation (SI Appendix, Fig. S1H and Fig. 1C), implying normal operation of the tricarboxylic acid cycle (TCA) cycle.
We also uncovered an additional aspect to the cross talk between PPIP5K and glucose metabolism: the culture of WT cells in the low-glucose condition for 4 h elicited a 60% decrease in the levels of InsP8 (SI Appendix, Fig. S2 A and B). This was a specific effect upon PP-InsP turnover; InsP7 levels were not affected (SI Appendix, Fig. S2C). InsP8 specifically responds to the change in glucose status even before ATP levels are impacted (SI Appendix, Fig. S2D).These data are interesting in the context that PP-InsPs are proposed to be metabolic sensors (4).
Furthermore, the restoration of glucose levels to 10 mM quickly rescued PPIP5K-mediated InsP8 synthesis (SI Appendix, Fig. S2B). Significantly, the addition of metabolically inert l-glucose did not restore InsP8 levels, indicating that levels of this PP-InsP can sense glucose metabolism. Indeed, knockdown of individual enzymes in glucose metabolism pathways (hexokinase, glyceraldehyde 3-phosphate dehydrogenase [GAPDH], glucose-6-phosphate dehydrogenase [G6PD], and phosphoglycerate dehydrogenase [PHGDH]) also significantly decreased InsP8 levels in WT cells (SI Appendix, Fig. S2 E–I). These data underscore our conclusion that there is dynamic cross talk between PPIP5K activity and glucose metabolism.
We hypothesized that a global metabolomic analysis might reveal information relevant to the basis for the growth-inhibited phenotype of the PPIP5K KO cells. Therefore, we determined steady-state metabolite levels in low-glucose cultured cells by LC-HRMS (13, 14). Volcano plots (Fig. 1G and SI Appendix, Fig. S3A) and pathway analyses (Fig. 1H and SI Appendix, Fig. S3B) dramatically illustrate the extent of the global metabolic changes to multiple pathways that occur in PPIP5K KO tumor cells (Dataset S1). Network-based pathway analysis did not uncover a significant change to the activity of the TCA cycle upon KO of PPIP5K in cells cultured in low-glucose conditions (Fig. 1H and SI Appendix, Figs. S3B and S4A). Nevertheless, we noted that among the metabolic pathways that are perturbed by the PPIP5K KO (Fig. 1H), there are several known to directly affect tumor cell proliferation, including nucleotide metabolism, the pentose–phosphate pathway, serine–glycine–one-carbon (SGOC) metabolism, and central carbon metabolism through glycolysis (15–19). We therefore undertook a deeper analysis of these metabolomic data.
De Novo Nucleotide Biosynthesis Is Inhibited in PPIP5K KO Cells.
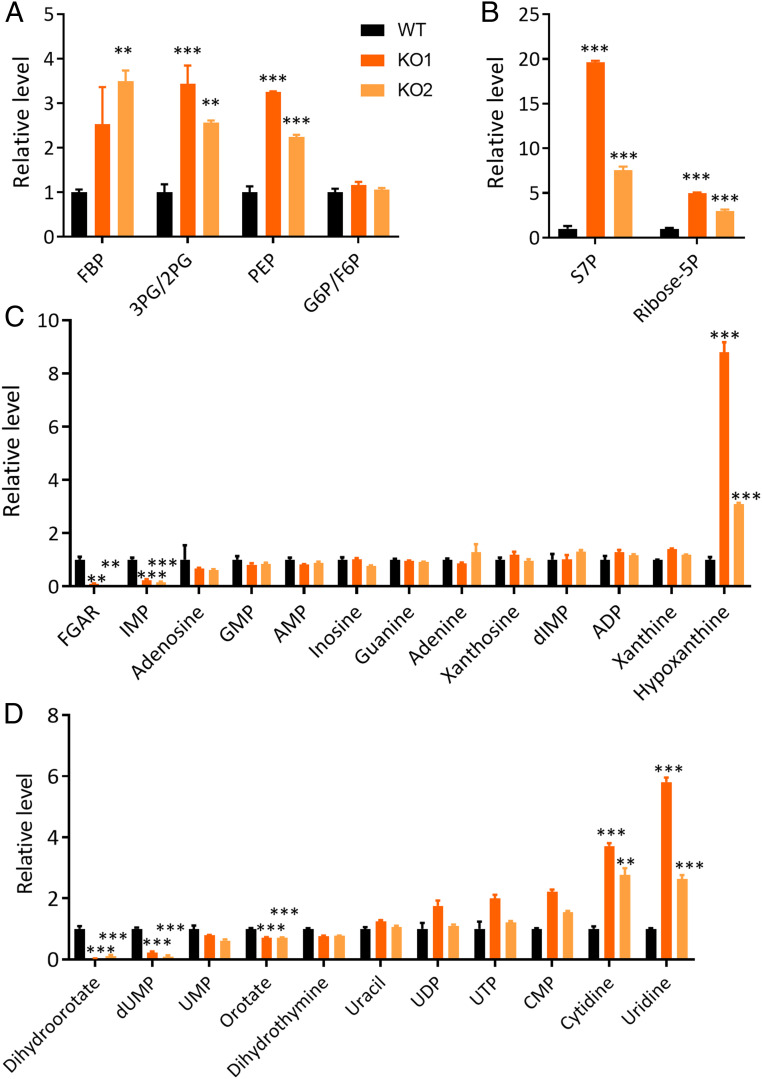
Our data show that the PPIP5K KO led to elevated steady-state levels of several glycolytic metabolites: fructose bisphosphate, 3-phosphoglycerate/2-phosphoglycerate, and phosphoenolpyruvate (Fig. 2A and Dataset S1). Moreover, we found that the PPIP5K KO substantially increased levels of two intermediates in the pentose phosphate pathway, namely, ribose-5-phosphate and sedoheptulose-7-phosphate (Fig. 2B). Furthermore, compared to WT cells, the PPIP5K KO cells contained lower levels of several key metabolites related to the SGOC cycle, including glutathione, homocysteine, and S-adenosyl homocysteine (SI Appendix, Fig. S4B), which could affect redox status and methylation of DNA and histones.
Fig. 2.
Selected changes in levels of steady-state metabolites following KO of PPIP5Ks from HCT116 cells. (A) Relative integrated peak intensities for glycolysis pathway metabolites in WT HCT116 cells (black bars), PPIP5K KO1 cells (dark orange bars), and PPIP5K KO2 cells (light orange bars) following culture in low-glucose media for 24 h: FBP (fructose-1,6-bisphosphate), 3PG/2PG (3-phosphoglycerate/2-phosphoglycerate), PEP (phosphoenolpyruvate), and G6P/F6P (glucose-6-phosphate/fructose-6-phosphate). (B) Relative integrated peak intensities for pentose phosphate pathway metabolites: S7P (sedoheptulose-7-phosphate) and ribose-5P (ribose-5-phosphate). (C) Relative integrated peak intensities for purine biosynthetic precursors: FGAR (phosphoribosyl-N-formylglycineamide), IMP, GMP, AMP, dIMP (deoxyinosine monophosphate), ADP (adenosine diphosphate), and ATP. (D) Relative integrated peak intensities for pyrimidine biosynthetic precursors: dUMP (deoxyuridine monophosphate), UMP, UDP (uridine diphosphate), UTP (uridine triphosphate), and CMP (cytidine monophosphate). The complete analysis is provided in Dataset S1. Student’s t test was used to determine statistical significance of differences between means (**P < 0.01, ***P < 0.001).
Our analysis of metabolites in the purine and pyrimidine nucleotide synthesis pathways (Fig. 2 C and D) showed that, compared to WT cells, the PPIP5K KO cells contained lower levels of several precursors for de novo nucleotide synthesis: phosphoribosyl-N-formylglycineamide, dihydroorotate, and orotate. Levels of the purine inosine monophosphate (IMP) were strongly reduced by the PPIP5K KO, despite the downstream products adenosine monophosphate (AMP) and guanine monophosphate (GMP) being unaffected (Fig. 2 C and D). The PPIP5K KO cells also accumulated metabolites in nucleotide salvage pathways: hypoxanthine, uridine, and cytidine (Fig. 2 C and D). These data indicate a disrupted balance of nucleotide synthesis. To further investigate this effect, we used an alternate metabolomic approach.
13C-Tracing Analysis of Central Carbon Metabolism.
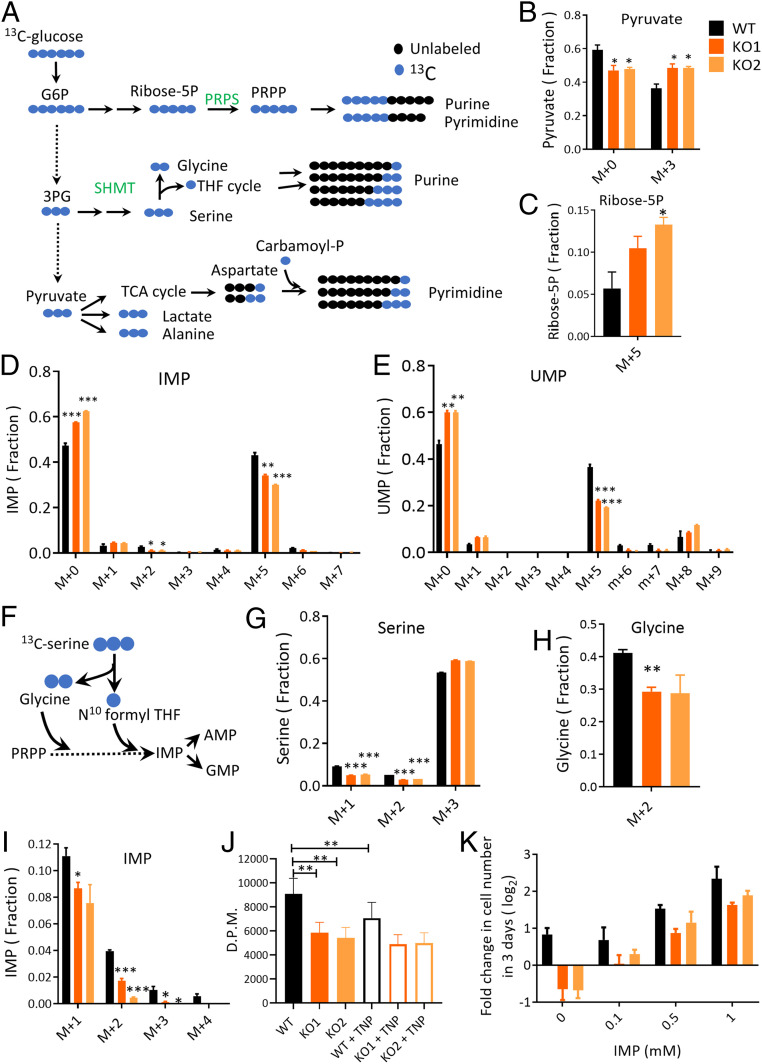
We performed direct metabolite labeling analysis to further define the mechanism by which PPIP5K KO compromises nucleotide synthesis, through isotope tracing of cells cultured in medium supplemented with uniformly labeled glucose ([U-13C] glucose; Fig. 3A and Dataset S2). We found that the fraction of isotope-labeled glucose-6-phosphate/fructose-6-phosphate was decreased, while the [U-13C]-labeled fractions of pyruvate, lactate, and alanine were all significantly elevated in PPIP5K KO cells (Fig. 3 A and B and SI Appendix, Fig. S5 A–C), suggesting an increased glycolytic rate in PPIP5K KO cells. Our labeling protocol also allows us to measure relative rates of glucose incorporation into pathways that are involved in purine and pyrimidine synthesis (Fig. 3A). For example, glucose provides the carbon structure for the purine precursor IMP and pyrimidine uridine monophosphate (UMP) via the ribose-5-phosphate that is generated by the pentose phosphate pathway. We obtained evidence of a stimulated incorporation of 13C label into glucose-derived ribose-5-phosphate, albeit only in PPIP5K KO2 cells (Fig. 3C). On the other hand, there was a consistent reduction in the 13C-labeling of IMP and UMP in both PPIP5K KO clonal lines, particularly the M+5-labeled isotopologues (Fig. 3 D and E). AMP, the downstream product of IMP, also showed reduced 13C-labeling (SI Appendix, Fig. S5D), indicating its inhibited de novo synthesis even though the total level remained intact (Fig. 2C). The reduced level of M+5-labeled isotopologue incorporated into nucleotides led us to further investigate if the PPIP5K KO affected provision of ribose-5-phosphate for nucleotide synthesis by the phosphoribosyl pyrophosphate synthetase (PRPS) (20). We found elevated phosphorylation of the PRPS1 oligomers in PPIP5K KO cells compared to WT cells (SI Appendix, Fig. S6A), which is reported to be associated with inhibition of this enzyme (20). Assays of PRPS activity in cell-free lysates are also consistent with its activity being reduced in PPIP5K KO cells versus WT cells (SI Appendix, Fig. S6B).
Fig. 3.
Isotope tracing of metabolites in WT and PPIP5K KO HCT116 cells. (A) Schematic of [U-13C]-glucose labeling of purines and pyrimidines in WT and PPIP5K KO cells. (B–E) HCT116 cells (black bars), PPIP5K KO1 cells (dark orange bars), and PPIP5K KO2 cells (light orange bars) were cultured in low-glucose media for 24 h with 13C-glucose. The “fraction” depicted in these panels is that proportion of a specific metabolite that is either nonlabeled (i.e., M+0) or a 13C-labeled isotopologue (i.e., M+n). B shows the fractions of M+0 and M+3 pyruvate isotopologues, and C shows the fractions of the M+5 ribose-5P isotopologue. D and E show fractions of all isotopologues of IMP and UMP, respectively. (F) Schematic of [U-13C] serine labeling of purines. (G–I) Cells were cultured in low-glucose media for 24 h with [U-13C]-serine, and G, H, and I show fractions of isotopologues of serine, glycine, and IMP, respectively. (J) WT and PPIP5K KO cells were cultured in the presence or absence of 10 µM TNP for 18 h and then incubated in low-glucose media for 3 h; cell lysates were then prepared, and SHMT enzymatic activity was assayed. (K) Cell growth with supplementation of IMP at the indicated concentrations. Student’s t test was used to determine statistical significance of differences between means (*P < 0.05, **P < 0.01, ***P < 0.001).
Phosphorylation-mediated inhibition of PRPS1 is reported to be promoted by activation of AMPK (20). Indeed, we found that the degree of AMPK phosphorylation was enhanced in PPIP5K KO cells compared to WT cells (SI Appendix, Fig. S7A). AMPK activation in PPIP5K KO cells occurs independently of any change to the AMP/ATP ratio (SI Appendix, Fig. S7 B and C), which was determined by LC-HRMS, in which we used 15N-labeled internal standards. Thus, PPIP5Ks and/or InsP8 appear not to be acting through the canonical energy-sensing pathway to activate AMPK, and their effects may instead be mediated by the glucose-sensing pathway or another of the noncanonical AMPK regulators (21, 22). This activation of AMPK is not due to increased levels of 5-InsP7 in PPIP5K KO cells as neither knockdown of IP6K2 [the major IP6K isoform in HCT116 cells (23)] nor TNP treatment altered AMPK phosphorylation (SI Appendix, Fig. S7 D–G). Using S6K phosphorylation as a readout, we did not find any impact upon the mTORC1 pathway of elevated AMPK activity in PPIP5K KO cells (SI Appendix, Fig. S7H). Nevertheless, compound C, the AMPK kinase activity inhibitor, partially rescued cell growth of PPIP5K KO cells (SI Appendix, Fig. S7I), suggesting that an AMPK-mediated inhibition of PRPS contributes to inhibited cell proliferation of PPIP5K KO cells in low-glucose conditions.
Glucose carbons also incorporate into IMP through glycine and 10-formyl-tetrahydrafolate from the serine synthesis pathway, while UMP can also be derived through aspartate derived from the TCA cycle (Fig. 3A). [U-13C]-glucose incorporation into aspartate and TCA cycle intermediates was not impacted by the PPIP5K KO (SI Appendix, Fig. S8 A and B). In contrast, enrichment of [13C] from glucose into both serine and glycine was dramatically reduced in KO cells, indicating decreased SGOC activity (SI Appendix, Fig. S8 C and D). This could also contribute to reduced nucleotide synthesis (Fig. 3F).
To further investigate whether a perturbed SGOC pathway contributes to the reduced de novo purine synthesis in PPIP5K KO cells, we performed [U-13C] serine tracing to track serine incorporation into purines (Fig. 3F and Dataset S2). The similarity of 13C labeling in the intracellular M+3 serine fraction in WT and PPIP5K KO cells (Fig. 3G) indicates that serine uptake was not impacted by the PPIP5K KO. However, in the PPIP5K KO1 cells there was a statistically significant reduction in serine incorporation into glycine (Fig. 3H). On the other hand, in both PPIP5K KO lines there was a consistent decrease in M+1 and M+2 13C labeling of the serine fractions, which are reversibly synthesized from labeled or unlabeled glycine and one-carbon units (Fig. 3 G and H). These data confirm the impaired activity of the SGOC in PPIP5K KO cells. Furthermore, the M+1- to M+4-labeled IMP fractions which derive from the SGOC were also decreased in PPIP5K KO cells (Fig. 3I). This is consistent with less glucose derived M+2 isotopologue of IMP from the SGOC (Fig. 3D). Fractions of all labeled isotopologues of AMP were reduced in PPIP5K KO cells, confirming inhibited de novo purine synthesis from the SGOC (SI Appendix, Fig. S8E).
We next investigated the activity of serine hydroxymethyltransferases (SHMTs) which catalyze the serine/glycine interconversion, upon which cancer cell proliferation depends (17, 24–27). There was not an effect of the PPIP5K KO upon expression levels of either cytosolic SHMT1 or mitochondrial SHMT2 (SI Appendix, Fig. S9A). However, using previously published methods (28–30), we assayed the total SHMT enzymatic activity and found it to be significantly reduced in PPIP5K KO cells (Fig. 3J and SI Appendix, Fig. S9B). Furthermore, TNP treatment and IP6K2 knockdown both reduced SHMT activity in WT cells but not PPIP5K KO cells, suggesting that loss of InsP8 is responsible for the inhibited SHMT activities (Fig. 3J and SI Appendix, Fig. S9B). In addition, we observed reduced expression levels of methylenetetrahydrofolate dehydrogenase 1 (MTHFD1), methylenetetrahydrofolate dehydrogenase 1-like (MTHFD1L) and PHGDH in PPIP5K KO cells, which could contribute to the impaired SGOC activity (SI Appendix, Fig. S9A). The latter effects are also not due to increased 5-InsP7 in PPIP5K KO cells, as the expression levels of MTHFD1 and PHGDH are not affected by TNP treatment or by IP6K2 knockdown (SI Appendix, Fig. S9 C and D). Further evidence in support of a deficiency in the SGOC pathway in PPIP5K KO cells comes from the failure of exogenous serine to rescue these cells’ defective growth, even though serine supports the proliferation of WT cells (SI Appendix, Fig. S10A), as shown previously for cancer cell lines (17). Overall, our isotope enrichment analysis indicates that the PPIP5K KO results in reduced de novo nucleotide synthesis through the pentose phosphate pathway and the SGOC pathway.
Restoration of the Proliferation of PPIP5K KO Cells by Exogenous Nucleosides.
We were unable to rescue growth of PPIP5K KO cells by supplementation with either S-adenosyl methionine, N-acetyl-l-cysteine, or glutathione (SI Appendix, Fig. S10 B–E), suggesting that neither oxidative stress nor a decreased level of methylation donors is responsible for the growth defect of PPIP5K KO cells. In contrast, the addition of either nucleosides or IMP significantly rescued the survival and proliferation of PPIP5K KO cells (Fig. 3K and SI Appendix, Fig. S10F). An additional round of metabolomic analyses revealed the major impact of IMP supplementation upon PPIP5K KO cells was to elevate multiple cellular metabolites (a more than twofold increase in 16% of those that were recorded; Dataset S3 and SI Appendix, Fig. S11 A–C). In contrast, only 3% decreased in levels greater than twofold. The cellular IMP level itself increased more than 100-fold after supplementation (SI Appendix, Fig. S12A). In PPIP5K KO cells the levels of the two precursors of de novo pyrimidine synthesis, dihydroorotate and carbamoyl-aspartate, increased more than 20- and 1000-fold, respectively, upon IMP supplementation (SI Appendix, Fig. S12A), indicating a dramatically shifted balance between de novo synthesis and salvage pathways for nucleotide synthesis. The pentose phosphate pathway metabolites ribose-5-phosphate and sedoheptulose-7-phosphate were also elevated significantly by IMP (SI Appendix, Fig. S12B), which could be due to increased flux of the five-carbon sugar back to the pentose phosphate pathway.
Most of the metabolites that showed the largest IMP-mediated increase in levels are components of the nucleotide metabolism: multiple purine and pyrimidine nucleotides and nucleosides (particularly hypoxanthine and inosine; these constituents of salvage pathways increased more than 200-fold) and deoxynucleotides that are required for DNA synthesis (including deoxythymidine triphosphate [dTTP] and deoxyadenosine triphosphate [dATP], which increased 10- to 1000-fold), which could substantially contribute to the rescued cell proliferation of PPIP5K KO cells (SI Appendix, Fig. S12 C and D).
The Kinase Activity of PPIP5K Is Required for Nucleotide Homeostasis and Tumor Cell Proliferation.
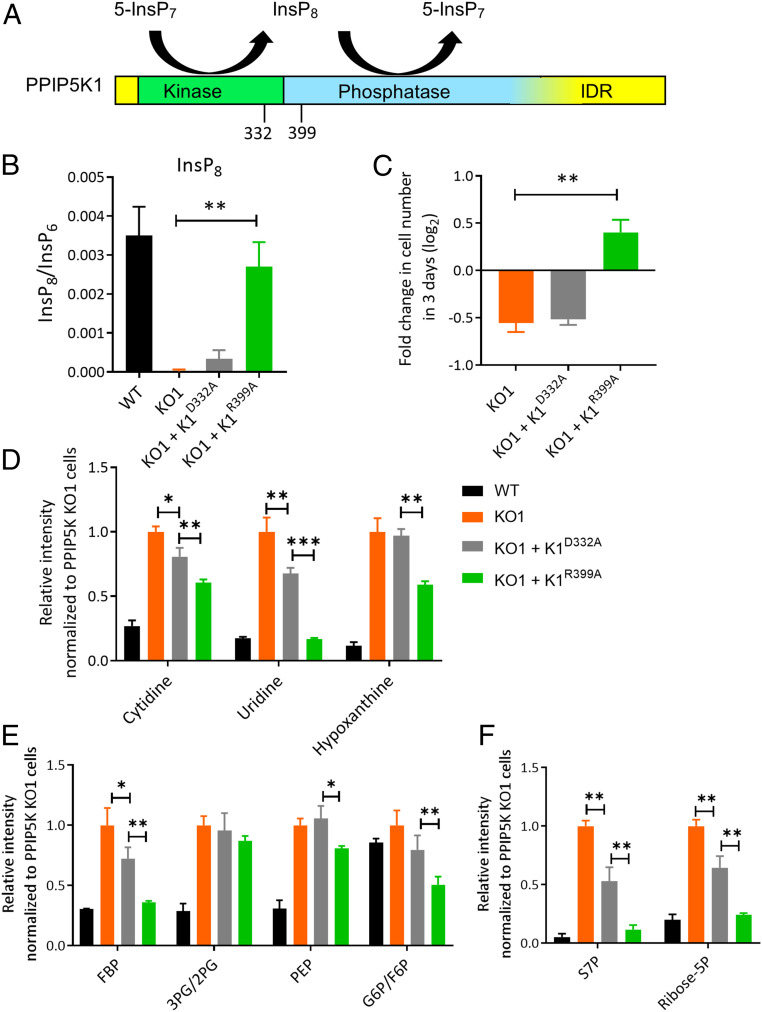
Finally, we investigated whether any of the metabolic rewiring that results from the PPIP5K KO can be corrected by restoration of PPIP5K activity. There are two PPIP5K isoforms, both of which are bifunctional enzymes containing separate kinase and phosphatase domains (Fig. 4A). We were able to stably introduce exogenous PPIP5K1 into the KO cells, but for unknown reasons, we could not stably express PPIP5K2. It should be noted that the catalytic efficiencies of the two isoenzymes are different, such that PPIP5K2 has higher net kinase activity (8). The two PPIP5Ks are also differentially regulated (8), and they exhibit alternative patterns of subcellular localization, so each isoform might individually regulate InsP8 levels in specific subcellular compartments (4). Thus, we did not anticipate that WT PPIP5K1 would fully rescue all aspects of the complex metabolomic phenotypes that result from loss of kinase activities of both PPIP5K1 and PPIP5K2. Therefore, we attempted to compensate through exogenous expression of the phosphatase-dead PPIP5K1R399A construct (Fig. 4A), which has enhanced net kinase activity. Significantly, this genetic maneuver was sufficient to rescue InsP8 levels in the PPIP5K KO cells (Fig. 4B) and substantially improved the rate of proliferation of PPIP5K KO cells in low-glucose medium (Fig. 4C). Expression of the kinase-dead mutant PPIP5K1D332A served as a useful control that failed to rescue either InsP8 levels or cell proliferation (Fig. 4 B and C).
Fig. 4.
The effect of expression of catalytic mutants of PPIP5K1 upon InsP8 levels, cell growth, and selected metabolites in PPIP5K KO cells. (A) Domain structure showing the catalytic bifunctionality of PPIP5K1; IDR = intrinsically disordered region. The blurring of the border between the phosphatase domain and the IDR depicts uncertainty over its location. (B) Cellular InsP8 levels of WT HCT116 cells (black bars), the corresponding PPIP5K KO1 (orange bars), and PPIP5K KO stably expressing either PPIP5K1D332A (gray bars) or PPIP5K1R399A mutants (green bars). (C) Cell growth of PPIP5K KO1 cells and PPIP5K KO1 cells stably expressing either PPIP5K1D332A or PPIP5K1R399A mutants; all cells were cultured in low-glucose medium for 3 d. (D) Relative integrated peak intensities for metabolites in the nucleotide salvage pathway: cytidine, uridine, and hypoxanthine. Here and in subsequent panels, all data are normalized to those obtained from PPIP5K KO1 cells (hence, the latter all have values of 1). (E) Relative integrated peak intensities for glycolysis-related metabolites: FBP, 3PG/2PG, PEP, and G6P/F6P. (F) Relative integrated peak intensities for the phosphate pathway: S7P and ribose-5P. Data for WT cells are calculated from the results in Dataset S1. Student’s t test was used to determine statistical significance of differences between means (*P < 0.05, **P < 0.01, ***P < 0.001).
We next performed steady-state metabolomic analyses to investigate to what extent exogenous expression of PPIP5K1R399A complements metabolites disturbed by the PPIP5K KO. Intermediates in the TCA cycle were not affected by expression of either PPIP5K1D332A or PPIP5K1R399A (SI Appendix, Fig. S13A and Dataset S4); this is a useful negative control since the TCA cycle was not targeted by the PPIP5K KO (Fig. 1H and SI Appendix, Figs. S3B and S8B). On the other hand, we found that exogenous PPIP5K1 activity reduced levels of three key metabolites in the nucleotide salvage pathway in PPIP5K KO cells: cytidine, uridine, and hypoxanthine; in particular, levels of uridine were returned to those observed in WT cells (Fig. 4D).
It is significant that expression of PPIP5K1R399A reduced the levels of two glycolytic intermediates that show increased levels in PPIP5K KO cells (i.e., fructose bisphosphate and phosphoenolpyruvate; Figs. 2A and 4E). However, expression of PPIP5K1R399A in KO cells did not change the levels of 3-phosphoglycerate/2-phosphoglycerate (Fig. 4E). Additionally, PPIP5K1R399A expression reduced steady-state levels of glucose 6-phosphate/fructose 6-phosphate, even though steady-state levels of these metabolites were not elevated by the PPIP5K KO (Figs. 2A and 4E). Furthermore, PPIP5K1R399A expression did not rescue the levels of the metabolites related to the SGOC that are reduced in PPIP5K KO cells (SI Appendix, Fig. S13B). But as mentioned above, there are several reasons why PPIP5K1 may not rescue all complex metabolomic phenotypes arising from the KO of both PPIP5K1 and PPIP5K2.
In any case, arguably the more remarkable finding concerning the expression of PPIP5K1R399A in PPIP5KO cells is the restoration of the levels of ribose-5-phosphate and sedoheptulose-7-phosphate to those of WT cells (Fig. 4F). These data speak to the success of exogenous PPIP5K activity in restoring the pentose phosphate pathway as a source of material for de novo nucleotide synthesis as part of a partial restoration of anabolic/catabolic balance in HCT116 cells. It is of further significance that kinase-dead PPIP5K1D332A was always less effective than PPIP5K1R399A at rescuing metabolite levels (Fig. 4 D–F). These data underscore the importance of kinase activity of PPIP5Ks to metabolic regulation while not excluding the possibility of additional noncatalytic, scaffolding contributions.
Concluding Comments.
Our study provides a detailed description of several key steps in nucleotide metabolic pathways that are supervised by the PPIP5Ks in the HCT116 tumor cell line, thereby extending insight into the complex interface between PP-InsP signaling and metabolic homeostasis. Consequently, our results provide a dramatic description of the metabolic changes in PPIP5K KO cells that underly their growth-inhibited phenotype. Thus, PPIP5Ks could be considered as therapeutic targets in tumor therapy by exploiting the particular significance of these enzymes to the process of cell proliferation in the glucose-limiting conditions of the tumor microenvironment.
Our study reveals that the PPIP5K KO fashions two separate choke points to the diversion of glycolytic carbon units into nucleotide synthesis. First, we identify that inhibition of SHMT in the KO cells reduces the provision of precursor material to nucleotide synthesis from the SGOC pathway (Fig. 3 G, H, and J). This pathway is a known target for cancer therapy (25, 27, 31); our demonstration that PPIP5Ks license the SGOC pathway opens up additional therapeutic options. Second, we demonstrate that the inhibition of PRPS in the KO cells restrains provision of precursor material from ribose-5-phosphate in the pentose phosphate pathway (Figs. 2B and 3C and SI Appendix, Fig. S6 A and B). Inhibition of PRPS is associated with its phosphorylation-dependent oligomerization, and we identify AMPK as the likely upstream protein kinase (SI Appendix, Fig. S7 A and D–G). Regulation of AMPK by PPIP5Ks appears to involve a noncanonical pathway that acts independently of adenine nucleotide status (SI Appendix, Fig. S7 B and C). Moreover, we found that heterologous expression of the kinase activity of PPIP5K1 relieved this restriction on ribose-5-phosphate metabolism in KO cells, thereby identifying the synthesis of InsP8 as regulating the contributions of the pentose phosphate pathway to nucleotide synthesis.
Finally, there is significance to PP-InsPs being members of the wider InsP signaling family (32); initially, it was assumed that each of these InsPs might exert their own independent but limited functions. For example, Ins(1,4,5)P3 specifically gates an intracellular calcium channel (33), and Ins(3,4,5,6)P4 specifically inhibits calcium-activated ion channels (34). However, recent attention has focused on a systems-level concept whereby complex sets of cellular responses are dynamically regulated by the collective actions of various InsPs acting as a combinatorial signaling “code” (32). For example, the differential activation of alternative pools of InsPs has been observed under conditions of constitutive G protein-coupled activation (35). In separate studies, multiple InsPs were shown to cooperate in the activation of pronecroptotic mechanisms (36, 37). Against this backdrop, our data now offer an alternate hypothesis that a single species of InsP—the InsP8 produced by PPIP5Ks—can by itself have extensive influence over multiple metabolic processes that influence multifaceted aspects of cellular biology.
Materials and Methods
Experimental details are provided in the SI Appendix, Materials and Methods. Cells were cultured in high- or low-glucose medium as indicated. Metabolites were extracted with methanol and measured by LC-HRMS. Intracellular inositol phosphates were determined by high-performance liquid chromatography (HPLC) analysis of extracts derived from cells that were radiolabeled with [3H]inositol. Intracellular proteins were measured by Western blot. SHMT activity was assayed by measuring transfer of C-1 unit from [3H(G)]-serine into N5N10-methylene tetrahydrofolate. PRPS activity was assayed using a commercial kit from NovoCIB.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, and NCI Intramural Grant ZIA BC 011438 (to J. Luo).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020187118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Cairns R. A., Harris I. S., Mak T. W., Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 (2011). [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis R. J., Chandel N. S., Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu C., et al., KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc. Natl. Acad. Sci. U.S.A. 114, 11968–11973 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shears S. B., Intimate connections: Inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell. Physiol. 233, 1897–1912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty A., The inositol pyrophosphate pathway in health and diseases. Biol. Rev. Camb. Philos. Soc. 93, 1203–1227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wild R., et al., Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Bhandari R., et al., Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U.S.A. 104, 15305–15310 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu C., et al., The significance of the bifunctional kinase/phosphatase activities of diphosphoinositol pentakisphosphate kinases (PPIP5Ks) for coupling inositol pyrophosphate cell signaling to cellular phosphate homeostasis. J. Biol. Chem. 292, 4544–4555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dollins D. E., et al., Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 9356–9364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Romero I. L., Litchfield L. M., Lengyel E., Locasale J. W., Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab. 24, 728–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindl T., Zell- und Gewebekultur (Spektrum Akademischer Verlag, Heidelberg, Germany, ed. 5, 2002). [Google Scholar]
- 12.Padmanabhan U., Dollins D. E., Fridy P. C., York J. D., Downes C. P., Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 284, 10571–10582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Ser Z., Locasale J. W., Development and quantitative evaluation of a high-resolution metabolomics technology. Anal. Chem. 86, 2175–2184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., et al., High-resolution metabolomics with Acyl-CoA profiling reveals widespread remodeling in response to diet. Mol. Cell. Proteomics 14, 1489–1500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Heiden M. G., DeBerardinis R. J., Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locasale J. W., Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labuschagne C. F., van den Broek N. J., Mackay G. M., Vousden K. H., Maddocks O. D., Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Yang M., Vousden K. H., Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Lunt S. Y., et al., Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 57, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian X., et al., Conversion of PRPS hexamer to monomer by AMPK-mediated phosphorylation inhibits nucleotide synthesis in response to energy stress. Cancer Discov. 8, 94–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S. C., Hardie D. G., AMPK: Sensing glucose as well as cellular energy status. Cell Metab. 27, 299–313 (2018). [DOI] [PubMed] [Google Scholar]
- 22.González A., Hall M. N., Lin S. C., Hardie D. G., AMPK and TOR: The yin and yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Koldobskiy M. A., et al., p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc. Natl. Acad. Sci. U.S.A. 107, 20947–20951 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain M., et al., Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman A. C., Maddocks O. D. K., One-carbon metabolism in cancer. Br. J. Cancer 116, 1499–1504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ducker G. S., et al., Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 114, 11404–11409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Cañaveras J. C., et al., SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia 35, 377–388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braman J. C., Black M. J., Mangum J. H., Serine transhydroxymethylase: A simplified radioactive assay; purification and stabilization of enzyme activity employing Affi-Gel blue. Prep. Biochem. 11, 23–32 (1981). [DOI] [PubMed] [Google Scholar]
- 29.Cao J., et al., HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc. Natl. Acad. Sci. U.S.A. 116, 5487–5492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert T., Taylor H. W., Radioactive assay for serine transhydroxymethylase. Anal. Biochem. 13, 80–84 (1965). [Google Scholar]
- 31.Nonaka H., et al., Design strategy for serine hydroxymethyltransferase probes based on retro-aldol-type reaction. Nat. Commun. 10, 876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatch A. J., York J. D., SnapShot: Inositol phosphates. Cell 143, 1030–1030.e1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berridge M. J., Irvine R. F., Inositol phosphates and cell signalling. Nature 341, 197–205 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Shears S. B., The versatility of inositol phosphates as cellular signals. Biochim. Biophys. Acta 1436, 49–67 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Otto J. C., Kelly P., Chiou S. T., York J. D., Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 15653–15658 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dovey C. M., et al., MLKL requires the inositol phosphate code to execute necroptosis. Mol. Cell 70, 936–948.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNamara D. E., et al., Direct activation of human MLKL by a select repertoire of inositol phosphate metabolites. Cell Chem. Biol. 26, 863–877.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.