Significance
Distinguishing the effects of light and sugars in photoautotrophic cells is challenging. The circadian system is a regulatory network that integrates light and metabolic signals and controls rhythmic physiology and growth. Our experimental approach has defined a light-independent, sugar-regulated transcriptome in Arabidopsis and revealed reactive oxygen species (ROS) as a prominent feature. ROS are byproducts of photosynthetic metabolism and oscillate with circadian rhythms but have not previously been demonstrated as inputs to the plant circadian oscillator. Our data suggest a role for superoxide as a rhythmic sugar signal which acts in the evening and affects circadian gene expression and growth.
Keywords: circadian, superoxide, sugar, redox, ROS
Abstract
Plants must coordinate photosynthetic metabolism with the daily environment and adapt rhythmic physiology and development to match carbon availability. Circadian clocks drive biological rhythms which adjust to environmental cues. Products of photosynthetic metabolism, including sugars and reactive oxygen species (ROS), are closely associated with the plant circadian clock, and sugars have been shown to provide metabolic feedback to the circadian oscillator. Here, we report a comprehensive sugar-regulated transcriptome of Arabidopsis and identify genes associated with redox and ROS processes as a prominent feature of the transcriptional response. We show that sucrose increases levels of superoxide (O2–), which is required for transcriptional and growth responses to sugar. We identify circadian rhythms of O2–-regulated transcripts which are phased around dusk and find that O2– is required for sucrose to promote expression of TIMING OF CAB1 (TOC1) in the evening. Our data reveal a role for O2– as a metabolic signal affecting transcriptional control of the circadian oscillator in Arabidopsis.
Plant metabolism is inextricably linked to daily photoperiodic cycles because of the requirement of light for photosynthesis. Anticipation and adaptation to changing light availability enables plants to optimize metabolism according to their immediate environment. Plant metabolism responds to environmental cues, such as light, temperature, and biotic and abiotic stress, by diverse mechanisms (1).
Plant cells require signaling mechanisms to sense carbon and energy status and adjust metabolism. Snf1 RELATED KINASE 1 (SnRK1) and TARGET OF RAPAMYCIN 1 (TOR1) are counteracting signaling hubs which are activated under low and replete carbon status, respectively (2, 3). Trehalose-6-phosphate (T6P) is an essential signaling sugar which indicates carbon status and acts through SnRK1 (4, 5).
Circadian clocks are an endogenous timekeeping mechanism, which regulate rhythms of physiology and metabolism and control responses to environmental signals according to the time of day (6). The core circadian oscillator in Arabidopsis is a network of transcription factors composed of Myb-like genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and REVIELLE (RVE) expressed at dawn; PSEUDO RESPONSE REGULATOR (PRR) genes expressed through the day, including TIMING OF CAB 1 (TOC1) at dusk; and the Evening Complex in the night. The phase and amplitude of gene expression and protein levels are responsive to environmental cues, and they, in turn, coordinate the regulation of thousands of genes.
There is extensive transcriptional and post-transcriptional control of photosynthetic metabolism by the circadian clock, and there is metabolic feedback on the circadian oscillator. Elevated SnRK1 activity under carbon limitation lengthens the circadian period, and sucrose shortens the period by T6P-SnRK1 acting on the oscillator gene PRR7 (7–9). The period also responds to glucose by a TOR-dependent mechanism (10). In continuous dark, circadian rhythms rapidly dampen but can be sustained by the addition of sugars. This effect of sugar requires GIGANTEA (GI), a clock protein which is stabilized by sucrose in the evening (11). Sugars can also reinitiate transcriptional rhythms in dark-adapted seedlings, setting the phase according to the time of sugar application (8, 12), but the mechanism in unknown.
Redox state and levels of reactive oxygen species (ROS), which are tightly linked to metabolism, are also associated with circadian rhythms in plants. There are circadian rhythms of hydrogen peroxide (H2O2) and NADP(H)+ in Arabidopsis (13, 14). Circadian rhythms of peroxiredoxin oxidation have been detected across Kingdoms (15). These rhythms of redox state and associated ROS are generally considered as outputs of rhythmic metabolism controlled by the circadian clock (13) or even independent of the circadian oscillator (15). The defense hormone salicylic acid perturbs redox state and affects gating of immune response, dependent on the redox-sensitive transcription factor NON-EXPRESSOR OF PATHOGENESIS 1 (NPR1) (14). However, there is presently no clear evidence of a role for redox signals as a mechanism of metabolic feedback to the circadian oscillator in plants.
Distinguishing sugar and light signals can be challenging in photosynthetic cells since it is likely that sugar signaling will be activated in the light. Recent advances in our understanding of the impact of metabolic signaling to the plant circadian clock have relied on experiments in low light or darkness (7, 8, 10–12, 16). Here, we use an experimental approach based on the previous observation that sugar can activate the expression of circadian clock genes in dark-adapted seedlings to define a light-independent, sugar-regulated transcriptome in Arabidopsis (8, 12). We compare the response of the transcriptome to sucrose in the dark and inhibition of photosynthesis in the light and identify redox and ROS processes as a prominent feature of transcriptional responses to sugars. We demonstrate that superoxide (O2–) can act as a signal to alter gene expression and growth in response to sucrose. This O2– signal acts to promote transcription of circadian oscillator genes in the evening. These reveal that ROS can function as metabolic signals affecting circadian rhythms in Arabidopsis.
Results
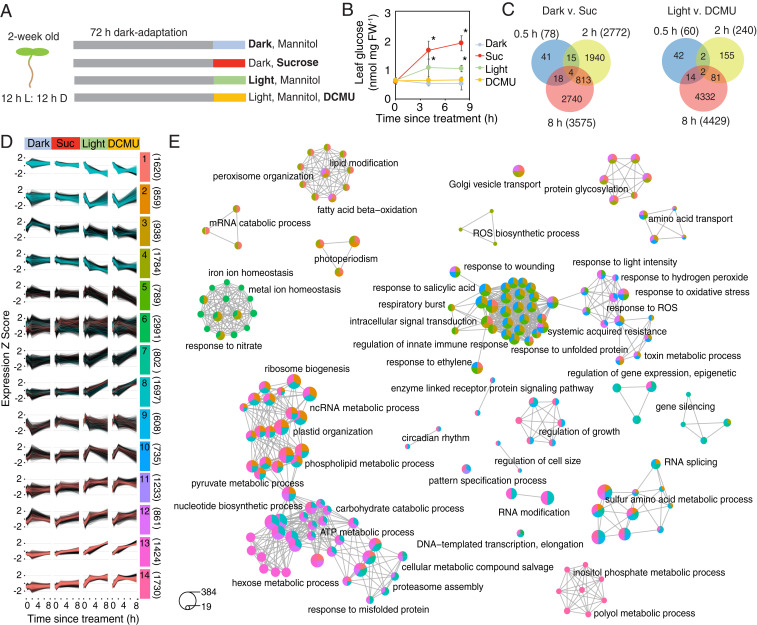
To identify transcripts that are regulated by sugars in the presence and absence of light and photosynthesis, we designed an RNA sequencing (RNA-Seq) experiment based on the previous observation that sugars can reinitiate transcriptional circadian rhythms in dark-adapted Arabidopsis seedings (8, 12). Two-wk-old wild-type (Col-0) seedings were grown in the dark for 72 h to dampen circadian rhythms and establish a stabilized C starvation state. At subjective dawn, dark-adapted seedlings were transferred to media containing 10 mM mannitol (osmotic control) or sucrose and maintained in the dark or transferred to media-containing 10 mM mannitol with or without 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of photosynthesis, and grown in the light. The four treatments provide conditions of no sugar/no light (Dark), sugar/no light (Suc), sugar/light (Light), and light/no sugar (DCMU) (Fig. 1A). We confirmed that seedling glucose content increased in the Suc and Light treatments but not in the Dark or DCMU treatments (Fig. 1B). To capture both early and late transcriptional responses within the timeframe of a typical photoperiod, shoot tissue was harvested at subjective dawn (0 h) and 0.5, 2, and 8 h after the treatments and prepared for RNA-Seq.
Fig. 1.
A light-independent sugar-regulated transcriptome of Arabidopsis. (A) The 2-wk-old seedlings were grown in the dark for 72 h and then transferred to 10 mM mannitol (Dark) or sucrose (Suc) in the dark, into the light with 10 mM mannitol (Light), or 20 µM DCMU and 10 mM mannitol (DCMU). Shoot tissue was collected at 0, 0.5, 2, and 8 h for RNA-Seq. (B) Leaf glucose content in seedlings grown as in A (means ± SD, n = 3; *P < 0.05 from Dark; Bonferroni-corrected t test). (C) Venn diagrams of differentially expressed genes at each time point in samples collected in the dark (Left) or light (Right). (D) Expression trajectories of 14 clusters of coexpressed genes identified by variational Bayesian–Gaussian mixture model. Pink and blue lines indicate genes identified as up- and down- or down- and up-regulated by sucrose/DCMU, respectively. The number of genes within each cluster are in parentheses. (E) GO enrichment maps of the top 15 terms in each cluster in D. Node colors correspond to the cluster(s) represented in D. Node sizes are proportional to the number of genes. Selected nodes are labeled with significantly enriched, representative GO terms for each network. See Dataset S4 for the fully annotated networks.
We detected 5,571 Suc-regulated genes that were differentially expressed between Dark and Suc treatments and 4,628 DCMU-regulated genes differentially expressed between Light and DCMU (Fig. 1C and Dataset S1). The quantification of gene expression by RNA-Seq was corroborated for 31 representative transcripts by qRT-PCR with a strong positive correlation (R2 = 0.91) (SI Appendix, Fig. S1). The overlap of differentially expressed genes between time points was relatively low (Fig. 1C), suggesting the sampling design captures a wide dynamic range of the transcriptional response. A comparison of our list of Suc-regulated genes to published microarray datasets (17, 18) indicated that we have captured a more extensive sugar-regulated transcriptome (SI Appendix, Fig. S2A).
To identify genes that are regulated by sugar, independent of light availability, we generated a list of genes that were up-regulated by Suc in the dark and down-regulated by DCMU in the light (sugar-activated; 927) or down-regulated by Suc in the dark and up-regulated by DCMU in the light (sugar-repressed; 1,117) (Dataset S2 and SI Appendix, Fig. S3). The sugar-activated genes were enriched for Gene Ontology (GO) terms related to protein and cell wall synthesis (SI Appendix, Fig. S3A). Sugar-repressed genes were enriched for GO terms related to light signaling, circadian rhythm, and sugar metabolism (SI Appendix, Fig. S3 B and C). We compared our list of all 2,042 sugar-regulated genes to published lists of genes regulated by SnRK1 and TOR, which are two major energy signaling hubs (2, 3). There was significant overlap with both datasets, but 1,080 sugar-regulated genes were unique to this study (SI Appendix, Fig. S3D), including 929 genes represented on the Arabidopsis Genome (ATH1) microarrays. These unique genes could represent responses either upstream or independent of SnRK1- and TOR-mediated signaling. Among the most significantly enriched GO terms in this list was response to oxygen-containing compound and circadian rhythm (SI Appendix, Fig. S3E).
To define the temporal characteristics of the complete transcriptome dataset, we performed clustering analysis of expression of 18,071 genes across all 53 samples using variational Bayesian–Gaussian mixture models (Fig. 1D and Dataset S3). We opted for 14 clusters as a tradeoff between maximizing the explained variance and producing meaningful clusters (SI Appendix, Fig. S4 and Fig. 1D). Several clusters were associated with either sugar-repressed (clusters 1 through 4) or sugar-activated (clusters 11 through 14) genes (Fig. 1D). We searched for enriched GO terms within each cluster (Dataset S3) and summarized these using an enrichment map of the top 15 terms within each cluster (Fig. 1E and Dataset S4). Some highly enriched GO term networks were specific to one or two clusters, such as inositol phosphate processes in cluster 13 or circadian rhythm and growth in clusters 8 and 13. Other enrichment GO term networks represent four or five clusters. The largest of these networks included terms associated with metabolism of sugars, nucleotides and phospholipids, chloroplast function, and proteostasis. The second largest enrichment network included terms associated with ROS metabolism and signaling, metabolic stress, and immune responses.
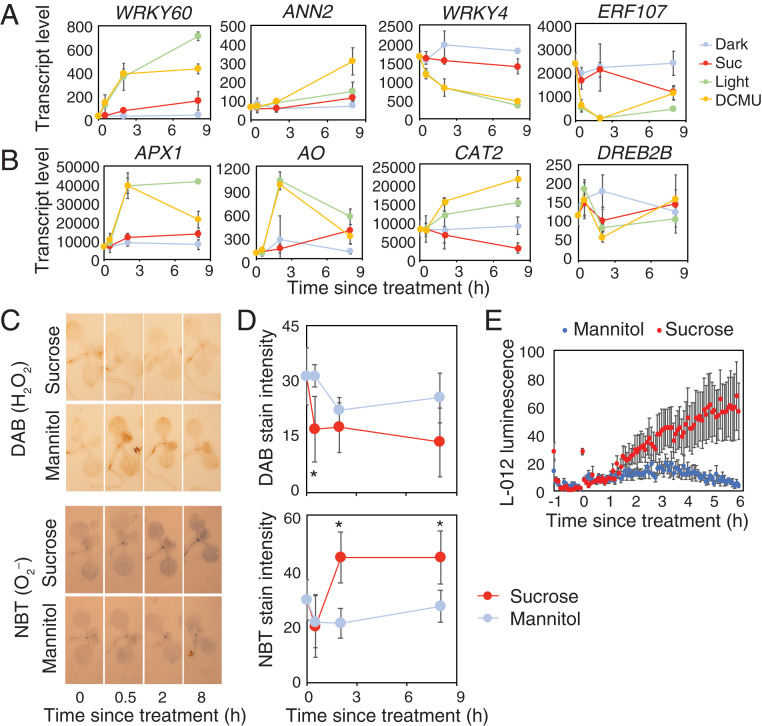
Since GO terms associated with ROS appear to be a strong feature of the complete dataset, we hypothesized that ROS might be contributing to transcriptional responses to sugar. Indeed, response to oxygen-containing compound was the most significantly enriched GO term among all 2,042 sugar-regulated genes and among Suc-regulated genes at 2 h (SI Appendix, Fig. S2B). Within the former, 195 genes are associated with this GO term, including ANNEXIN 2 (ANN2) and six WRKY transcription factor genes (Fig. 2A and Dataset S5). We also identified 95 sugar-regulated genes previously reported as ROS-responsive (19), including ASCORBATE PEROXIDASE 1 (APX1) and CATALASE 2 (CAT2) (Fig. 2B and Dataset S5).
Fig. 2.
Sucrose promotes superoxide production and ROS-regulated transcripts in dark-adapted seedlings. Transcript levels of representative ROS-associated genes identified as sugar-regulated from RNA-Seq that are (A) from the GO class “responsive to oxygen-containing compound” or (B) identified from a previous study (19) (means ± SD, n = 3). (C) Histochemical stains for hydrogen peroxide (DAB) and superoxide (NBT) in 10-d-old, dark-adapted Col-0 seedlings treated with 30 mM mannitol or sucrose. (D) DAB and NBT stain intensity in seedlings grown as in C (means ± SD, n = 6; *P < 0.05 from mannitol; Bonferroni-corrected t test). (E) L-012 luminescence in dark-adapted Col-0 treated with 30 mM mannitol or sucrose (means ± SEM, n = 6).
To test whether treatment of Arabidopsis seedlings with sucrose affects production of ROS in dark-adapted seedlings, we used histochemical stains for H2O2 and O2– (Fig. 2 C and D). Treatment of dark-adapted seedlings with sucrose led to a decrease in staining for H2O2 within 30 min. By contrast, sucrose treatment of dark-adapted seedlings increased stain for O2– within 2 h compared to mannitol controls. The elevated nitroblue tetrazolium (NBT) stain was observed throughout the shoot, including hypocotyl, cotyledons, and leaves. To corroborate this observation, we used an L-012 luminescence assay, which does not discriminate between H2O2 and O2– but provides better temporal resolution of ROS production than histochemical stains. Consistent with the NBT stains for O2–, we detected elevated L-012 luminescence within 2 h in sucrose-treated seedlings compared to mannitol-treated controls (Fig. 2E). Presumably, this assay underestimates the difference in O2– production since the signal in sucrose-treated seedlings will be the sum of the reduced H2O2 and the increased O2– (Fig. 2C). The ROS response detected in both the histochemical and luminescent assays is concomitant with the timing of the transcriptional response associated with ROS-related genes that we detected after 2 h (Fig. 2 A and B, SI Appendix, Fig. S2B, and Dataset S1).
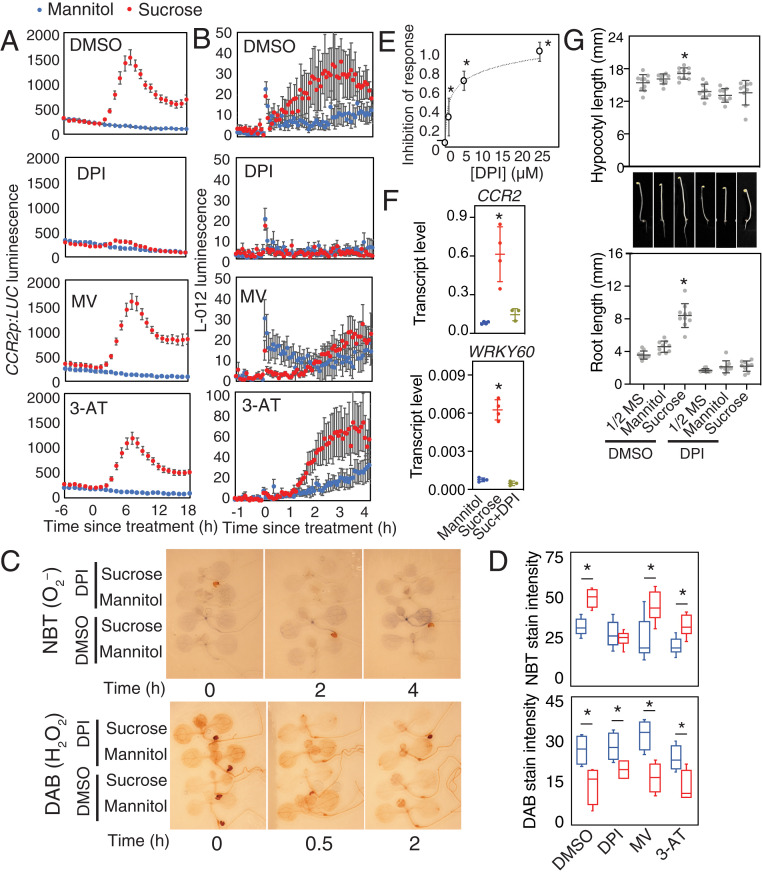
The accumulation of O2– in sucrose-treated seedlings might be a byproduct of increased energy metabolism or could be contributing as a signal to affect transcriptional changes. We looked for chemicals that could inhibit the sucrose-induced production of O2–. Diphenyleneiodonium (DPI) is an inhibitor of NADPH oxidases, which generate O2– at the plasma membrane. Methyl viologen (MV) interferes with electron transport from photosystem I (PS I) and elevates O2–. 3-amino-1,2,4-triazole (3-AT) is a catalase inhibitor which promotes H2O2 accumulation. We tested the effect of these chemicals on induction of a circadian-regulated luciferase reporter for COLD, CIRCADIAN RHYTHM REGULATED 2 (CCR2). DPI strongly inhibited the increase of luciferase luminescence in sucrose-treated, dark-adapted CCR2p:LUC seedlings, whereas MV and 3-AT did not (Fig. 3A). Similarly, DPI, but not MV or 3-AT, also inhibited sucrose-induced L-012 luminescence (Fig. 3B) and histochemical staining for O2– but did not affect sucrose-induced changes in staining for H2O2 (Fig. 3 C and D).
Fig. 3.
Modifiers of superoxide inhibit responses to sucrose. (A) Luciferase luminescence in dark-adapted CCR2p:LUC seedlings treated with 30 mM mannitol or sucrose in the presence of DMSO, 10 µM DPI, 2 µM MV, or 200 µM 3-AT (means ± SEM, n = 6). (B) L-012 luminescence in dark-adapted Col-0 treated as in A (means ± SEM, n = 6). (C) Histochemical NBT stain for O2– and DAB stains for H2O2 in dark-adapted Col-0 seedlings treated with 30 mM mannitol or sucrose in the presence of 0.1% DMSO or 10 µM DPI. (D) Stain intensity in Col-0 seedlings 4 h (NBT) or 0.5 h (DAB) after treatment as in A (n = 6; *P < 0.05; t test). (E) Inhibition of response of luciferase luminescence to 30 mM sucrose in dark-adapted CCR2p:LUC seedlings in the presence of 0 (0.1% DMSO), 1, 5, or 25 µM DPI. (means ± SEM, n = 3; *P < 0.05 from DMSO; Bonferroni-corrected t test). (F) Transcript level of CCR2 and WRKY60 relative to UBQ10 in dark-adapted Col-0 seedlings 8 h after treatment with 30 mM mannitol, sucrose, or sucrose with 10 µM DPI (means ± SD, n = 4; *P < 0.05 from mannitol; Bonferroni-corrected t test. (G) Hypocotyl length and root length of 5-d-old dark-grown Col-0 seedlings grown on 1/2 MS with or without 30 mM mannitol or sucrose, 0.1% DMSO, or 1 µM DPI (means ± SD, n = 10; *P < 0.05 from 1/2 MS; Bonferroni-corrected t test).
We used the transcriptional response of CCR2p:LUC to generate a dose–response curve of inhibition by DPI. This response was inhibited by 30% at 1 µM DPI and by >70% at concentrations above 5 µM (Fig. 3E). Similar dose-dependent effects were also observed for two other NADPH oxidase inhibitors, VAS2870 (20) and apocynin (21), but not for the xanthine dehydrogenase inhibitor allopurinol (22) (SI Appendix, Fig. S5). We confirmed that DPI also inhibited sucrose induction of CCR2 and WRKY60 transcripts by qRT-PCR (Fig. 3F) as well as WRKY11p:β-GLUCURONIDASE (GUS) and WRKY30p:GUS reporters (SI Appendix, Fig. S6). Thus, DPI effectively inhibits transcriptional regulation of multiple sugar-regulated genes.
DPI could be inhibiting transcriptional responses to sugar in our assay by affecting uptake of sucrose, altered sugar metabolism, or inhibition of sugar sensing or signaling. We measured soluble sugars glucose, fructose, and sucrose in sucrose-treated dark-adapted seedlings in the presence of dimethyl sulfoxide (DMSO; control) or DPI. We did not detect a difference from controls for any sugar within 8 h of sucrose treatment (SI Appendix, Fig. S7), suggesting that inhibition of sugar uptake or sucrose catabolism cannot account for the dramatic inhibition of the transcriptional response by DPI.
Since DPI can inhibit transcriptional responses to sugar, we sought to establish whether DPI also affects other sugar-regulated processes in Arabidopsis. Seed germination in both dormant and nondormant seeds is inhibited by exogenous sugar, acting through abscisic acid-dependent pathways (23). Similar to sucrose, DPI also inhibits germination (24) (SI Appendix, Fig. S8). If DPI inhibits germination by the same pathway as sucrose, we expected that their effects would be nonadditive. However, the effect of DPI on inhibition of germination was detected both with and without sucrose in dormant and nondormant seeds (SI Appendix, Fig. S8). This suggests that DPI does not affect the regulatory pathways through which sucrose inhibits seed germination.
Sugars promote growth. To test the effect of DPI on growth promotion by sucrose, we measured effects on hypocotyl elongation and root growth in dark-grown seedlings. This growth assay enables quantification of effects of sugar on cell elongation in the hypocotyl and cell division in the root in the absence of light signals. Seedlings growing on media containing DPI had slightly reduced hypocotyl length and root length in control media, and DPI strongly attenuated the positive effects of sucrose on both hypocotyl and root length (Fig. 3G). These data suggest that DPI inhibits the signaling or metabolism of sucrose to promote cell elongation and cell division.
NADPH oxidases are encoded by a family of 10 RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) genes in Arabidopsis. We tested whether rboh mutants had altered ROS production in dark-adapted seedlings using L-012 luminescence assays. Both the rbohb and rbohc mutants had a similar response to sucrose as wild type, but rboha mutants and rbohd rbohf double mutants had reduced L-012 luminescence (SI Appendix, Fig. S9A), similar to wild type treated with DPI, VAS2890, or apocynin (SI Appendix, Fig. S5B). We also tested whether rboh mutants had altered growth responses to sucrose (SI Appendix, Fig. S9B). The rbohd rbohf double mutant had reduced root and hypocotyl length on control media compared to wild type, but growth was still responsive to sucrose in the mutant. Stimulation of hypocotyl growth by sucrose was reduced in the rboha mutant compared to wild type, but stimulation of root growth was unaffected. Thus, although we detected small growth effects in the mutants, none of those tested were able to phenocopy the effect of DPI. Similarly, the transcriptional response of CCR2 or WRKY60 to sucrose in dark-adapted seedlings was not reduced in rboh mutants (SI Appendix, Fig. S9C). These suggest that there is residual O2– accumulation in these mutants sufficient to elicit a response and that there is genetic redundancy in the molecular targets of DPI contributing to these sugar responses.
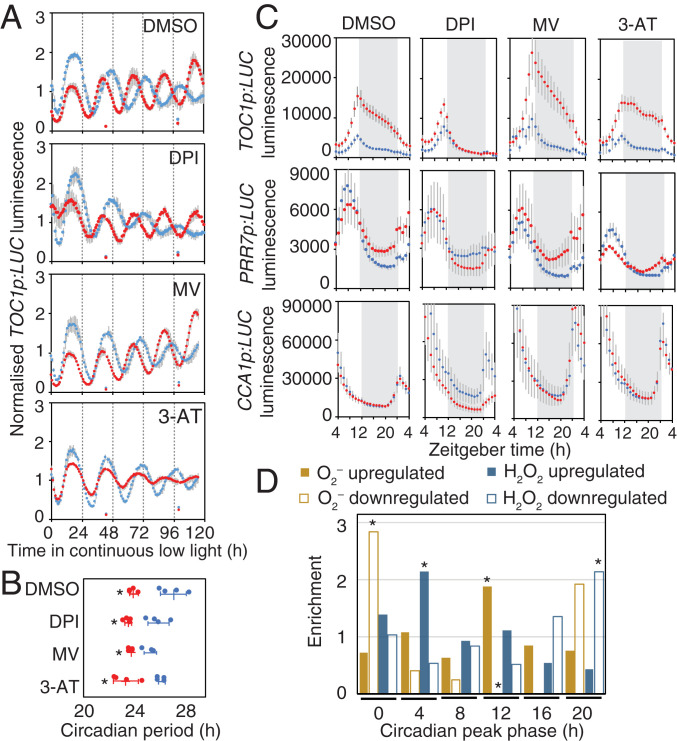
Sugars affect period of circadian rhythms (8), and the circadian clock contributes to rhythms of ROS homeostasis (13). We tested the effect of DPI, MV, and 3-AT on circadian rhythms in media with or without sucrose. We measured circadian rhythms of TOC1p:LUC in continuous low light (10 µmol · m−2 · s−1) because the effect of exogenous sucrose on circadian rhythms is more pronounced in these conditions (8). The circadian period was significantly shorter in seedlings grown on sucrose compared to mannitol for all ROS modifiers, similar to the DMSO control (Fig. 4 A and B). This suggests that these chemicals did not affect the adjustment of period by exogenous sucrose.
Fig. 4.
Modifiers of superoxide affect modulation of circadian rhythms by sucrose. (A) Normalized luciferase luminescence in TOC1p:LUC seedlings in continuous low light with 30 mM mannitol (blue) or sucrose (red) in the presence of 0.1% DMSO or 10 µM DPI, 2 µM MV, or 200 µM 3-AT (means ± SD, n = 4). (B) Circadian period estimates of luciferase luminescence in TOC1p:LUC seedlings in A (means ± SD, n = 4; *P < 0.05 from mannitol; Bonferroni-corrected t test). (C) Luciferase luminescence in TOC1p:LUC, PRR7p:LUC, and CCA1p:LUC seedlings for 24 h in light/dark treated as in A (means ± SD, n = 4). (D) Phase of rhythmic O2–- and H2O2-responsive transcripts in continuous light. Values are enrichment (observed/expected) of up- and down-regulated genes in each 4-h phase window (*P < 0.01; χ2).
Sugars also affect the amplitude of circadian rhythms (11). Luciferase signal is dramatically elevated in TOC1p:LUC seedlings transferred to media containing sucrose compared to mannitol (Fig. 4 A and C). This transcriptional response does not require GI (SI Appendix, Fig. S10), a clock protein which is post-transcriptionally regulated by sucrose (11). The effect of sucrose in TOC1p:LUC seedlings was strongly attenuated in the presence of DPI, elevated in the presence of MV, and unaffected by 3-AT (Fig. 4C), which is consistent with the effects of these compounds on O2– levels. The effects of DPI and MV were particularly pronounced during the night and were not observed in CCA1p:LUC or PRR7p:LUC seedlings (Fig. 4C), suggesting O2– acts on specific components of the oscillator.
Since the effects of DPI and MV differed between the morning-phased CCA1p:LUC and PRR7p:LUC and evening-phased TOC1p:LUC, we wondered whether this might reflect a global pattern of O2– on transcriptional rhythms. We used a set of previously reported O2–- and H2O2-responsive transcripts (19) to determine their phases in continuous light from a published RNA-Seq dataset (25). The distribution of phases of transcripts up- and down-regulated by O2– or H2O2 deviated significantly from expectations (Fig. 4D and Dataset S5). The phases of transcripts up-regulated by H2O2 were enriched several hours after subjective dawn, and down-regulated transcripts were enriched before subjective dawn. This is consistent with the reported role of CCA1 in driving rhythms of H2O2, which peak in the early morning (13). By contrast, the phase of transcripts up-regulated by O2–, which included TOC1, GI, PRR5, and LUX, were enriched around subjective dusk. About 20% of these genes are direct TOC1 targets (26) (Dataset S5). Transcripts down-regulated by O2–, including LHY and RVE8, were enriched around subjective dawn. This suggests that H2O2 and O2– production or signaling are antiphased and is consistent with a role of O2– contributing to promoting oscillations of circadian transcripts in the evening.
Discussion
We have identified ROS-regulated genes as a prominent feature in the response of the Arabidopsis transcriptome to sugars in both dark and light (Fig. 1). The transcriptional response to sucrose in dark-adapted seedlings coincides with an increase in ROS levels, including O2– (Fig. 2). Both the accumulation of O2– and transcriptional response to sucrose were strongly attenuated in seedlings treated with DPI, a chemical inhibitor of flavoenzymes including NADPH oxidases (Fig. 3). DPI also inhibited the promotion of hypocotyl elongation and root growth by sucrose, demonstrating a broader impact of the ROS signal in sugar responses. Finally, we found that DPI inhibited the effect of sucrose on the evening expressed TOC1 and identified a highly significant antiphasing of rhythmic transcripts that are up- and down-regulated by O2– to dusk and dawn, respectively (Fig. 4). This is different from the redox effects of salicylic acid on both morning and evening genes (14). Thus, we propose that O2– functions as a metabolic signal associated with sugar levels, which acts positively on the circadian oscillator in the evening. An association between cellular sugar status and redox state has been long recognized in the context of metabolism and oxidative stress (27), but our data provide evidence of a role for O2– as a dynamic sugar signal affecting daily rhythms of gene expression. This effect of sugar on the oscillator appears to be distinct from the T6P/SnRK1-mediated effect on the period via transcriptional regulation of PRR7 (7) (Fig. 4) and the post-transcriptional control of GI (11) (SI Appendix, Fig. S9), revealing an additional layer of metabolic control of circadian rhythms in plants.
DPI is a potent inhibitor of NADPH oxidases, which generate extracellular O2– at the plasma membrane activated by intracellular signals (28). We observed reduced sucrose-activated ROS production and modest growth phenotypes in rboha and rbohd rbohf mutants, but the transcriptional response to sucrose was similar to wild type (SI Appendix, Fig. S8). Notwithstanding that the five rboh mutants examined here represent over 90% of total RBOH gene expression (Dataset S1), the subtle phenotypes in the rboh mutants compared to DPI-treated seedlings probably reflects functional redundancy within this gene family. This will be challenging to verify, since higher order mutants would be expected to be lethal. It is possible that effects of DPI on O2–-mediated responses to sugar can be attributed to inhibition of other flavoenzymes. For example, in photosynthetic organisms, DPI inhibits O2– production from xanthine dehydrogenases, glutathione reductases, and mitochondrial NAD(P)H dehydrogenases (29–31). However, the similar effects of VAS2890 and apocynin, but not allopurinol, on sugar responses support the role of NADPH oxidases (SI Appendix, Fig. S5).
MV interferes with electron transport from PS I, as well as in mitochondria (32), and leads to the accumulation of O2–, so the opposite effects on transcriptional responses might be expected compared to DPI. MV was unable to induce a transcriptional response in CCR2p:LUC seedlings without sucrose (Fig. 3A), which suggests that O2– alone does not activate circadian gene expression or that the site of O2– accumulation in MV-treated seedlings is not sufficient to act as the signal. However, MV elevated the response to sucrose in TOC1p:LUC seedlings (Fig. 4C), suggesting that O2– and sucrose might act synergistically.
O2– is generated in mitochondria, chloroplasts, peroxisomes, and the apoplast (28). O2– is typically scavenged quickly by superoxide dismutases. Elevation of O2– could be due to increased production or reduced scavenging. The increase in O2– triggered by sucrose in dark-adapted seedlings by histochemical stain and L-012 assay was relatively low and slow compared to elicitor-induced respiratory burst (33) but faster than a ROS effect reported for cell wall damage (34). It might be that sucrose generates O2– in specific cell types or subcellular locations, the signal might be diluted in bulk tissues, or our detection methods might have insufficient sensitivity. This might explain why we couldn’t detect L-012 luminescence in rbohd rbohf double mutants (SI Appendix, Fig. S8A). Thus, it will be useful to map the cellular and subcellular location of the O2– signal using the expanding toolset of available redox probes (35–37). This will also provide clearer identity of candidate proteins producing the signal.
Reversible oxidation of redox-sensitive proteins by ROS can alter their activity. In Arabidopsis, redox-sensitive proteins that are oxidized by H2O2 have been identified in most cellular compartments (38). These include plasma membrane receptors (39), glycolytic enzymes (38, 40), which can localize in the nucleus and associate with DNA (41, 42), and transcription factors (43). Thus, localized changes in redox state could affect signaling pathways and gene expression by various mechanisms. Changes in localized O2– concentration could modify protein function indirectly after dismutation to H2O2 or directly by affecting Fe-S proteins (28).
It is experimentally difficult to separate the effects of H2O2, O2–, or other ROS on protein oxidation. Differences in target specificity for ROS might depend on their redox dynamics or subcellular location. H2O2 is regarded as the most likely ROS signal because it is relatively stable compared to the more reactive O2– (28). However, our phase analyses of H2O2 and O2– regulated transcripts indicate clear temporal separation of their effects (Fig. 4). This might reflect differences in spatial organization of oxidative metabolism at different times of day. The mechanism by which sugar-activated O2– production affects gene regulation will depend on its cellular location.
By examining the effects of sugar on the Arabidopsis transcriptome independently of light, we have uncovered a role for redox status, exemplified by accumulation of O2–, that promotes responses to sugar, including growth and circadian rhythms. In contrast to the previously reported association of circadian rhythms of H2O2, which are phased in the morning (13), the O2–-activated transcriptome peaks in the evening and includes core genes within the circadian oscillator. Sugar promotes O2–, which alters gene expression by either an extracellular or intracellular redox signal, which could transmit to the nucleus via signaling or protein localization. We propose that this metabolic signal functions to coordinate rhythmic physiology and growth in response to environmental conditions that affect photosynthetic metabolism.
Materials and Methods
Details of plant materials and growth conditions, RNA-Seq and clustering (44, 45), qRT-PCR, histochemical stains, luminescence assays, and sugar quantification are described in SI Appendix. Primers are listed in Dataset S6.
Supplementary Material
Acknowledgments
We thank Ms. Heather Eastmond (University of York) for technical support and Professor Alex Webb (University of Cambridge) for useful comments on the manuscript. This research was funded by Biotechnology and Biological Sciences Research Council Grant (BB/L021188/1) to M.J.H. and I.A.G., Royal Society Research Grant (RG150144) to M.J.H., The University of Melbourne through the Research Grants Support Scheme to M.J.H., and a Melbourne Research Scholarship to X.L. This research was not funded by the Australian Research Council.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020646118/-/DCSupplemental.
Data Availability
RNA-Seq data have been deposited in European Nucleotide Archive (PRJEB40453). Additional files associated with RNA-Seq analyses are available from Dryad Digital Repository (https://doi.org/10.5061/dryad.v41ns1rv9).
References
- 1.Herrmann H. A., Schwartz J. M., Johnson G. N., Metabolic acclimation-a key to enhancing photosynthesis in changing environments? J. Exp. Bot. 70, 3043–3056 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Baena-González E., Rolland F., Thevelein J. M., Sheen J., A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Xiong Y., et al., Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M., Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 6849–6854 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes C., et al., The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 162, 1720–1732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydon M. J., Li X., Ting M. K. Y., Temporal control of plant-environment interactions by the circadian clock. Annu. Plant Rev. 2, 505–536 (2019). [Google Scholar]
- 7.Frank A., et al., Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr. Biol. 28, 2597–2606.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haydon M. J., Mielczarek O., Robertson F. C., Hubbard K. E., Webb A. A., Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J., et al., The metabolic sensor AKIN10 modulates the Arabidopsis circadian clock in a light-dependent manner. Plant Cell Environ. 40, 997–1008 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zhang N., et al., Metabolite-mediated TOR signaling regulates the circadian clock in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 25395–25397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haydon M. J., Mielczarek O., Frank A., Román Á., Webb A. A. R., Sucrose and ethylene signaling interact to modulate the circadian clock. Plant Physiol. 175, 947–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalchau N., et al., The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. U.S.A. 108, 5104–5109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai A. G., et al., CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 17129–17134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M., et al., Redox rhythm reinforces the circadian clock to gate immune response. Nature 523, 472–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar R. S., et al., Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shor E., Paik I., Kangisser S., Green R., Huq E., PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol. 215, 217–228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thum K. E., Shin M. J., Palenchar P. M., Kouranov A., Coruzzi G. M., Genome-wide investigation of light and carbon signaling interactions in Arabidopsis. Genome Biol. 5, R10 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osuna D., et al., Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49, 463–491 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Gadjev I., et al., Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangano S., et al., Molecular link between auxin and ROS-mediated polar growth. Proc. Natl. Acad. Sci. U.S.A. 114, 5289–5294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stolk J., Hiltermann T. J., Dijkman J. H., Verhoeven A. J., Characteristics of the inhibition of NADPH oxidase activation in neutrophils by apocynin, a methoxy-substituted catechol. Am. J. Respir. Cell Mol. Biol. 11, 95–102 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Hesberg C., Hänsch R., Mendel R. R., Bittner F., Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: Differential gene expression and enzyme activities. J. Biol. Chem. 279, 13547–13554 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Price J., Li T. C., Kang S. G., Na J. K., Jang J. C., Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 132, 1424–1438 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller K., Carstens A. C., Linkies A., Torres M. A., Leubner-Metzger G., The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 184, 885–897 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Romanowski A., Schlaen R. G., Perez-Santangelo S., Mancini E., Yanovsky M. J., Global transcriptome analysis reveals circadian control of splicing events in Arabidopsis thaliana. Plant J. 103, 889–902 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Huang W., et al., Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Couée I., Sulmon C., Gouesbet G., El Amrani A., Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57, 449–459 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Smirnoff N., Arnaud D., Hydrogen peroxide metabolism and functions in plants. New Phytol. 221, 1197–1214 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Zarepour M., et al., Xanthine dehydrogenase AtXDH1 from Arabidopsis thaliana is a potent producer of superoxide anions via its NADH oxidase activity. Plant Mol. Biol. 72, 301–310 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Diaz J. M., et al., NADPH-dependent extracellular superoxide production is vital to photophysiology in the marine diatom Thalassiosira oceanica. Proc. Natl. Acad. Sci. U.S.A. 116, 16448–16453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts T. H., Fredlund K. M., Møller I. M., Direct evidence for the presence of two external NAD(P)H dehydrogenases coupled to the electron transport chain in plant mitochondria. FEBS Lett. 373, 307–309 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Cui F., et al., Interaction of methyl viologen-induced chloroplast and mitochondrial signalling in Arabidopsis. Free Radic. Biol. Med. 134, 555–566 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Smith J. M., Heese A., Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Methods 10, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denness L., et al., Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer A. J., et al., Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52, 973–986 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Nietzel T., et al., The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2 O2 and thiol redox integration and elucidates intracellular H2 O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. New Phytol. 221, 1649–1664 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Steinbeck J., et al., In vivo NADH/NAD+ biosensing reveals the dynamics of cytosolic redox metabolism in plants. Plant Cell 32, 3324–3345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P., Zhang H., Wang H., Xia Y., Identification of redox-sensitive cysteines in the Arabidopsis proteome using OxiTRAQ, a quantitative redox proteomics method. Proteomics 14, 750–762 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Wu F., et al., Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578, 577–581 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Marchand C. H., et al., Thioredoxin targets in Arabidopsis roots. Proteomics 10, 2418–2428 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Cho Y. H., Yoo S. D., Sheen J., Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Kim S. C., Guo L., Wang X., Nuclear moonlighting of cytosolic glyceraldehyde-3-phosphate dehydrogenase regulates Arabidopsis response to heat stress. Nat. Commun. 11, 3439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Liu W., Zhong H., Zhang H. L., Xia Y., Redox-sensitive bZIP68 plays a role in balancing stress tolerance with growth in Arabidopsis. Plant J. 100, 768–783 (2019). [DOI] [PubMed] [Google Scholar]
- 44.“RNA-seq analysis of Arabidopsis thaliana seedlings treated with sucrose in the dark or inhibited photosynthesis in the light.” European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB40453. Deposited 22 September 2020.
- 45.Haydon M. J., Davey J. W., Román Á., Superoxide is promoted by sucrose and affects amplitude of circadian rhythms in the evening. Dryad. 10.5061/dryad.v41ns1rv9. Deposited 6 February 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been deposited in European Nucleotide Archive (PRJEB40453). Additional files associated with RNA-Seq analyses are available from Dryad Digital Repository (https://doi.org/10.5061/dryad.v41ns1rv9).