Significance
Coat proteins play a central role in the intracellular transport pathways by coupling two main functions: bending the membrane to generate transport carriers and binding to cargoes for their sorting into these carriers. Studies thus far have mostly solved the structure of coat proteins in solution, but their functional form requires assembly on the membrane into protein complexes. Here, we have pursued cryo-EM to reveal in molecular detail how SNX1 assembles on the membrane to deform the membrane. When compared to a previously solved retromer-SNX complex, our elucidation also suggests how retromer affects SNX in this complex as well as the intermediary stages of this coat assembly.
Keywords: helical reconstruction, coat complex, membrane deformation, SNX1, cryoelectron microscopy
Abstract
The sorting nexin (SNX) family of proteins deform the membrane to generate transport carriers in endosomal pathways. Here, we elucidate how a prototypic member, SNX1, acts in this process. Performing cryoelectron microscopy, we find that SNX1 assembles into a protein lattice that consists of helical rows of SNX1 dimers wrapped around tubular membranes in a crosslinked fashion. We also visualize the details of this structure, which provides a molecular understanding of how various parts of SNX1 contribute to its ability to deform the membrane. Moreover, we have compared the SNX1 structure with a previously elucidated structure of an endosomal coat complex formed by retromer coupled to a SNX, which reveals how the molecular organization of the SNX in this coat complex is affected by retromer. The comparison also suggests insight into intermediary stages of assembly that results in the formation of the retromer-SNX coat complex on the membrane.
Sorting nexins (SNXs) exist as a large family of proteins defined by the presence of a PX (phox homology) domain (1, 2). Members of this family have been found to act as coat proteins in endosomal pathways that include recycling from endosomes to the plasma membrane and retrieval from endosomes to the Golgi complex (3, 4). Defects in these transport processes is associated with various neurologic disorders including Alzheimer’s disease, Parkinson’s disease, and Down’s syndrome (5, 6).
Coat proteins assemble into complexes on the membrane to initiate intracellular transport pathways by coupling two main functions: bending the membrane to generate transport carriers and binding to cargoes for their sorting into these carriers (7). Retromer, a trimeric complex consisting of Vps26, Vps29, and Vps35, has been found to couple with different SNXs to form multiple endosomal coat complexes, in which select members of the SNX family act in membrane deformation while retromer acts in cargo recognition (8–17). Recently, a detailed molecular view of this functional cooperation has been achieved by elucidating the structure of a retromer-SNX complex on the membrane (18).
Notably, it has been further discovered recently that an endosomal coat complex can be formed with only SNX members. SNX1/2 have been found to heterodimerize with SNX5/6 to form the endosomal SNX–BAR sorting complex for promoting exit 1 (ESCPE-1) complex, in which SNX1/2 are proposed to act in membrane deformation while SNX5/6 act in cargo recognition (19). As such, a key question has become whether SNX that acts in membrane deformation in this type of coat complex would be organized similarly on the membrane, as previously elucidated for SNX in the context of a retromer-SNX complex (18).
One of the best characterized mechanisms of membrane deformation involves proteins that possess the BAR (Bin/Amphiphysin/Rvs) domain. This domain has been shown to undergo homodimerization to form a banana-shaped structure, which can impart membrane curvature through a scaffolding mechanism that involves electrostatic interactions between the positive charges lining the concave side of the curved BAR dimer and the negative charges that line the surface of the membrane bilayer. In some cases, the BAR domain can deform the membrane through a second mechanism, which involves the formation of an amphipathic helix that inserts into one leaflet of the membrane bilayer to generate bilayer asymmetry in driving membrane curvature (20, 21).
Besides the PX domain, SNX1 also possesses a BAR domain. However, studies have found that its BAR domain is not sufficient in driving membrane deformation. Instead, the PX domain as well as the linker region between the BAR and PX domains are also needed (22, 23). As such, a key goal has been to achieve a better understanding of how the various parts of SNX1 contribute to its ability to deform the membrane.
Structural studies, such as those involving crystallography and single-particle electron microscopy (EM), have been advancing a molecular understanding of coat proteins (24), including components of endosomal coats (17, 19, 22, 25–27). Notably, however, these approaches solve protein structures in solution, but the functional form of coat proteins involves their association with the membrane. In this study, we have pursued cryo-EM to reveal how SNX1 is organized on the membrane to explain its ability to deform the membrane. The result advances a molecular understanding of how an endosomal coat that contains only SNXs generates transport carriers. Moreover, by comparing our SNX1 structure to the previously solved retromer-SNX structure (18), we delineate the extent to which the molecular organization of SNX on the membrane is affected by the presence of retromer. This comparison also suggests insight into intermediary stages of coat assembly that form the retromer-SNX complex on the membrane.
Results
Membrane Binding and Tubulation by SNX1.
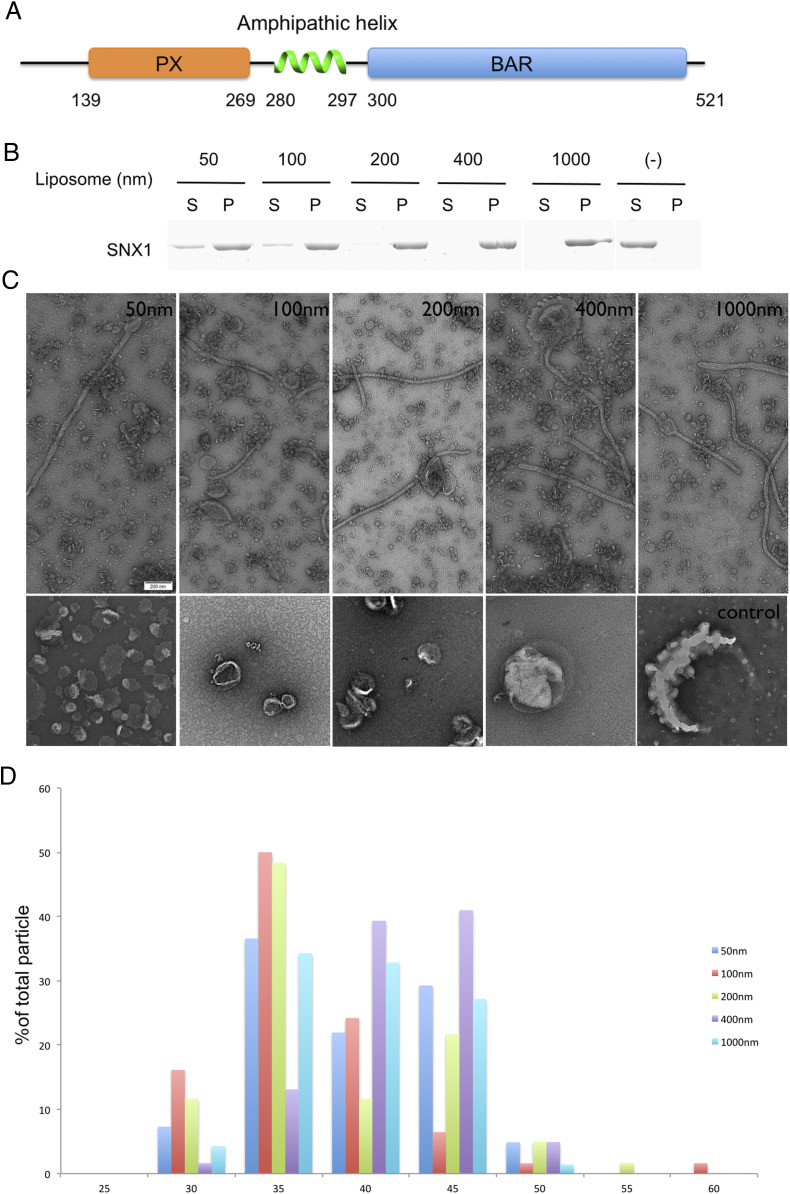
We initially generated liposomes that contain the major phospholipids of organellar membrane. To this mixture, we also added phosphatidylinositol 3-phosphate (PI3P), as it is enriched on endosomal membranes and has been shown previously to promote membrane tubulation by SNX1 (22, 28, 29). The generated liposomes were then incubated with full-length SNX1 (Fig. 1A) and analyzed for membrane recruitment. We found that SNX1 binds to liposomes independent of their size, which ranged from 50 nm to 1 μm in diameter (Fig. 1B). We also confirmed that the full-length SNX1 induces membrane tubulation, as visualized by negative-stain EM (Fig. 1C).
Fig. 1.
SNX1 is sufficient for membrane binding and tabulation. (A) Domain organization of SNX1 (NP_062701.2). PX, Phox homology domain; BAR domain. (B) Binding of SNX1 to liposomes of varying sizes (as indicated) is assessed by coprecipitation assay. Supernatant, S; Pellet, P. (C) Negative-stain EM visualizing liposomes incubated either with (first row) or without (second row) SNX1 (scale bar, 200 nm). The diameters of the liposomes are also indicated. (D) Statistical histogram of the diameters of tubules generated by SNX1 is plotted as a percentage of all tubules; n = 41 for 50 nm liposomes, n = 62 for 100 nm liposomes, n = 60 for 200 nm liposomes, n = 61 for 400 nm liposomes, and n = 70 for 1,000 nm.
The diameters of membrane tubules were similar regardless of the initial size of the liposome (Fig. 1D). Specifically, tubules with a diameter of 39.6 ± 5.0 nm were generated from liposomes of 50 nm diameter (n = 41), tubules with a diameter of 36.6 ± 5.4 nm from liposomes of 100 nm diameter (n = 62), tubules with a diameter of 38.5 ± 6.5 nm from liposomes of 200 nm diameter (n = 60), tubules with a diameter of 41.7 ± 3.9 nm from liposomes of 400 nm diameter (n = 61), and tubules with a diameter of 39.0 ± 4.4 nm from liposomes of 1,000 nm diameter (n = 70). These measurements corresponded well to those previously observed for liposome tubulation by SNX1 (22). Thus, different from the properties of an F-BAR protein, whose ability to induce membrane tubulation is dependent on liposome size (30), SNX1 can act on a wide range of liposome sizes to induce membrane tubules that have relatively constant diameters.
Cryo-EM and Three-Dimensional Structure of Coated Tubules Containing SNX1.
To gain further insight into how SNX1 induces membrane tubulation, we next pursued cryo-EM. After generating mixed populations of tubules, the samples were flash frozen and then EM micrographs were obtained. Images revealed that the outer surfaces of tubulated liposomes were coated with SNX1 (Fig. 2A). Consistent with a previous report (22) and also our measurements of these tubules by negative-stain EM (Fig. 1D), the diameters of these tubules were in the range of 35 to 48 nm (SI Appendix, Fig. S1A).
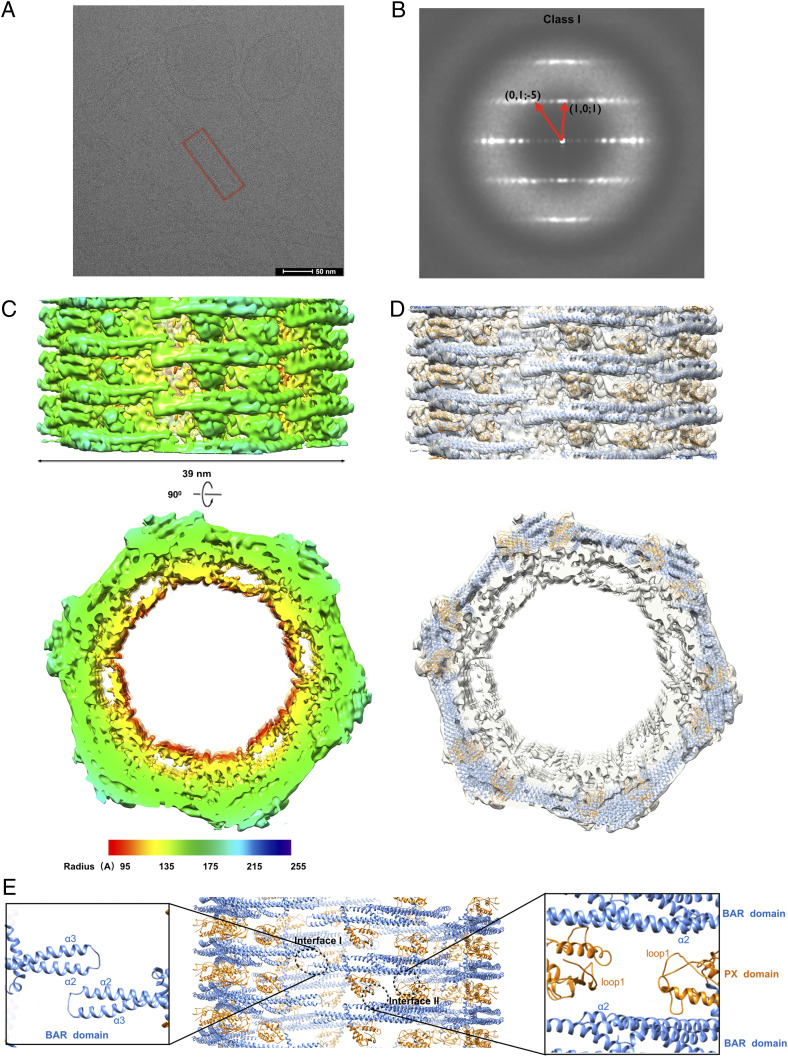
Fig. 2.
Cryo-EM and three-dimensional reconstructions of SNX1 coated tubules. (A) Raw cryo-EM micrographs of tubules coated with SNX1. A regular tube is boxed in red (scale bar, 50 nm). (B) Helical diffraction patterns of tubules exemplifying class I. (C) Cryo-EM map of a class I tubule, with the side view on the top and cross-section view on the bottom, with a lower threshold in cross-section view to show the inner membrane more clearly. The map is colored according to the cylinder radius from red to blue. (D) The structural models of SNX1 in cartoon representation are fitted into the map. The PX domain and BAR domain are colored in gold and blue, respectively. (E) Structural model of the SNX1 helical assembly for the class I tubules. Interface I and II are indicated by black circles, with zoom-in views also shown.
We next segmented these tubules for more detailed analysis. The spiral feature of some class averages suggested helical symmetry for the packing of SNX1 on tubules. This was further confirmed based on the layer-line diffraction patterns of select well-ordered tubules. Two types of helical diffraction patterns were observed and indexed, designated as class I and class II (Fig. 2B and SI Appendix, Fig. S1B). Using the iterative helical real-space reconstruction approach (31, 32), we determined the cryo-EM map of SNX1-coated tubules with a resolution of 9.0 Å for class I and 10.0 Å for class II, as assessed by Fourier Shell Correlation (FSC 0.143) between two independent maps reconstructed from the even- and odd-half dataset (SI Appendix, Table S1 and Fig. S1C). The layer-line diffraction patterns of cryo-EM maps also matched well with the ones from the original micrographs (SI Appendix, Fig. S1D). In an effort to improve the map, we also pursued the package RELION3.0 (33). However, no further significant improvement of resolution was observed (SI Appendix, Fig. S2).
We next considered that SNX1/2 form heterodimers with SNX5/6, with SNX1/2 being responsible for membrane bending and SNX5/6 acting in cargo binding (19). Thus, we also examined a heterodimer composed of SNX1 and SNX6. This heterodimer also binds liposomes (SI Appendix, Fig. S3A) and induces membrane tubulation (SI Appendix, Fig. S3B). The diameters of the membrane tubules were mainly in the range from 30 to 50 nm (SI Appendix, Fig. S3C), which are similar to those seen for tubules induced by the SNX1 homodimer. Notably, however, tubules induced by the SNX1/SNX6 heterodimer exhibit greater irregularity, having variable diameters along the length of the tubule, suggesting that the incorporation of a nonmembrane bending SNX member (SNX6) contributes to the irregular tubules observed. At the practical level, this variability also rendered the iterative helical real-space reconstruction approach unfeasible. Thus, when also considering that our goal is to understand at the molecular level how a prototypic SNX achieves membrane bending, we pursued further studies focusing on the SNX1 homodimer.
Helical Lattices of SNX1 on Tubular Membrane.
The class I tubules formed by the SNX1 homodimer have an outer diameter of 39 nm with a protein-coat thickness of 7 nm (Fig. 2C and Movie S1). The membrane beneath the protein coating could not be clearly discerned, which could be attributed to the close interaction between the membrane layer and the protein coating (which will be further discussed below). However, by lowering the threshold for the membrane layer, we could observe a more distinct membrane boundary in discerning the inner diameter of the coated tubules (SI Appendix, Fig. S4). At the higher threshold set for the protein coating, the secondary structural elements of the SNX1 assembly could be clearly resolved (Fig. 2C and Movie S1). When compared to class I tubules, class II tubules exhibited different helical symmetries and a larger diameter of 43 nm (SI Appendix, Figs. S1B and S5A). Nevertheless, the overall helical packing of SNX1 on the membrane in class II tubules was similar to that seen for class I tubules (SI Appendix, Fig. S5 A and B).
The BAR domain of SNX1 has been crystalized previously (Protein Data Bank [PDB] 4FSZ) (22), which shows a dimer that adopts a banana-shaped structure typical of a conventional BAR domain. However, this structure, which was solved in solution, was too highly curved to be docked onto our cryo-EM density map, suggesting that the BAR domain of SNX1 adopts a lower curvature upon binding to the membrane. Thus, we sought to fit the structural model of SNX1 generated by i-TASSER (34–36) and molecular dynamics flexible fitting (MDFF) (37) into the cryo-EM map.
In the reconstructed class I tubules, there is one SNX1 dimer forming an asymmetric unit of the helical packing. The dimer is oriented with its concave surface facing the membrane and the BAR domains mediating the dimerization, while the PX domains are positioned at the opposite ends of the dimer (Fig. 2D and Movie S1). Inspection of the SNX1 helical packing suggested two main interfaces of protein–protein interaction. Along the same helical row, two adjacent SNX1 dimers interact with each other through short lateral contacts formed by the α2 and α3 helices and the connecting loop between these two helices in the BAR domain (Fig. 2E). This set of interactions is referred to hereafter as “Interface I,” which is more similar to that previously seen for the structure of N-BAR proteins, such as endophilin A1 and B1 (38, 39), rather than that of a F-BAR protein, such as CIP4 (40) (SI Appendix, Fig. S6). The other major protein–protein interaction, referred to hereafter as “Interface II,” involves contacts between adjacent helical rows, which results in the crosslinking of these parallel rows. This interaction is mediated through the β-sheets and loops of the PX domain from one dimer residing in one helical row interacting with the α2 helix of the BAR domain from another dimer residing in an adjacent helical row (Fig. 2E).
In class II tubules, there is also one SNX1 dimer forming an asymmetric unit of the helical packing (SI Appendix, Fig. S5B). Moreover, the main interfaces are similar to those seen for class I tubules (SI Appendix, Fig. S5C). However, in contrast to class I tubules, the number of dimers in one complete helical turn in class II tubules is seven as compared to six dimers seen for one complete helical turn in class I tubules (Fig. 2D and SI Appendix, Fig. S5B). This difference could be attributed to flexibility in the short lateral interactions in Interface I, which allows the SNX1 lattice structure to slide or tilt on the membrane surface in accommodating different curvature states. Overall, however, the SNX1 lattice structure seen for the two classes of tubules is quite similar, as reflected by the structural rmsd between class I and class II being 1.9 Å for 364 C-α when superimposing these two classes (SI Appendix, Fig. S5D).
An Amphipathic Helix Is Required for Membrane Remodeling.
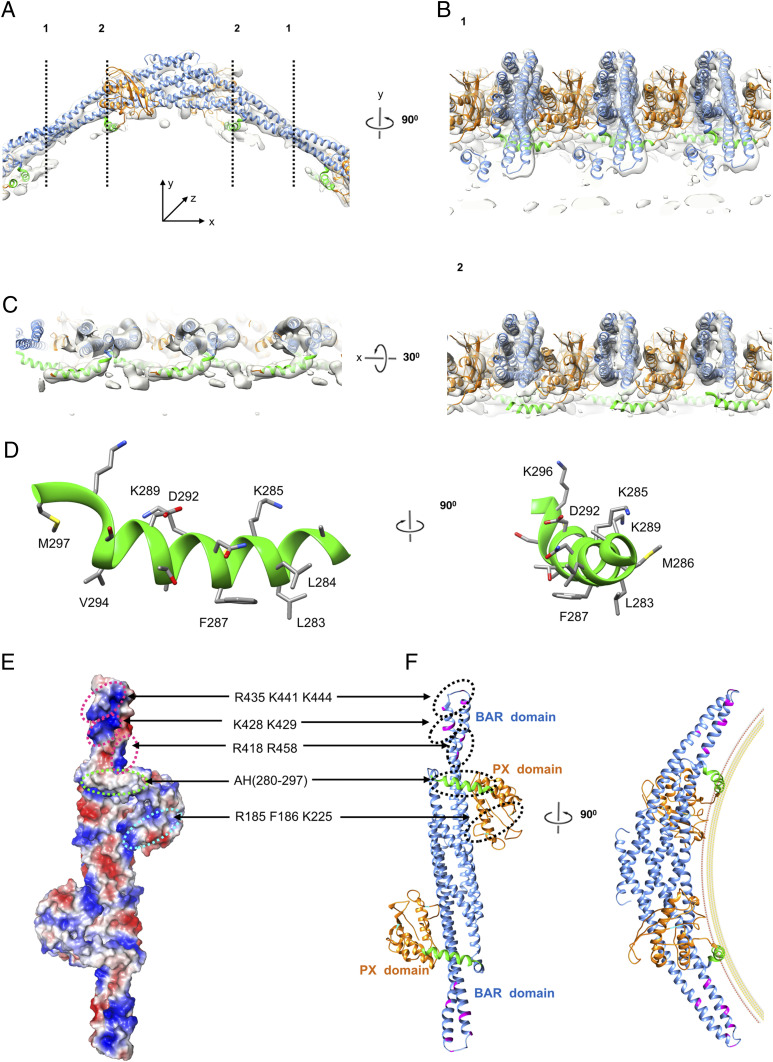
A remarkable finding is that the membrane under the SNX1 lattice could not be distinctly discerned. There is an obvious density on the concave surface of the SNX1 dimer that abuts closely to the membrane, which is to some extent integrated into the membrane (Fig. 3A). Based on the ability to clearly identify other nearby protein densities, the membrane-abutting region of SNX1 likely involves the linker region that connects the BAR and PX domains. Sequence analysis predicts that a portion of this linker region could form an amphipathic helix (SI Appendix, Fig. S7), which has been suggested previously to act in membrane remodeling by SNX proteins (22, 41). As such, we next sought to fit the predicted amphipathic helix onto the cryo-EM map. This resulted in an excellent match (Fig. 3 B and C). Multiple views of the reconstruction further confirmed the likely presence of the amphipathic helix. In cross-section 1, the amphipathic helix appears buried in the membrane, which is even more clearly viewed in cross-section 2 (Fig. 3B), and also from the view of the membrane (Fig. 3C). Further consistent with this region forming an amphipathic helix, one side of the helix is predicted to have mainly polar/charged residues, while the other side is predicted to have mainly hydrophobic residues (Fig. 3D).
Fig. 3.
An amphipathic helix on the membrane interaction surface of SNX1. (A) Ribbon model of the SNX1 dimer interacting with the membrane. The colors of the PX and BAR domains are the same as shown in Fig. 2E. The amphipathic helix is highlighted in green. Dashed black lines denote cross sections that are rotated by 90° and shown in B. (B) Corresponding cross sections of the tube along the dashed black lines 1 and 2 in A; shown is the fitness between the structural model and the map with high threshold. (C) The cross section rotated by 30° from that shown in B, which provides the view from the membrane. (D) Ribbon model of the amphipathic helix, with charged surface on one side and hydrophobic surface on the other side and key residues labeled. (E) Electrostatic surface representation on the concave surface of SNX1, with positive charges colored blue and negative charges colored red. Positively charged patches and other areas implicated in membrane binding and tubulation are labeled. (F) A Ribbon model of SNX1 with the location of charged patches in the BAR domain shown in magenta, the amphipathic helix shown in green, and the predicted PIP3 binding site of the PX domain shown in cyan. A hypothetical membrane is depicted based on the diameter (∼40 nm−1) of tubules observed in EM maps.
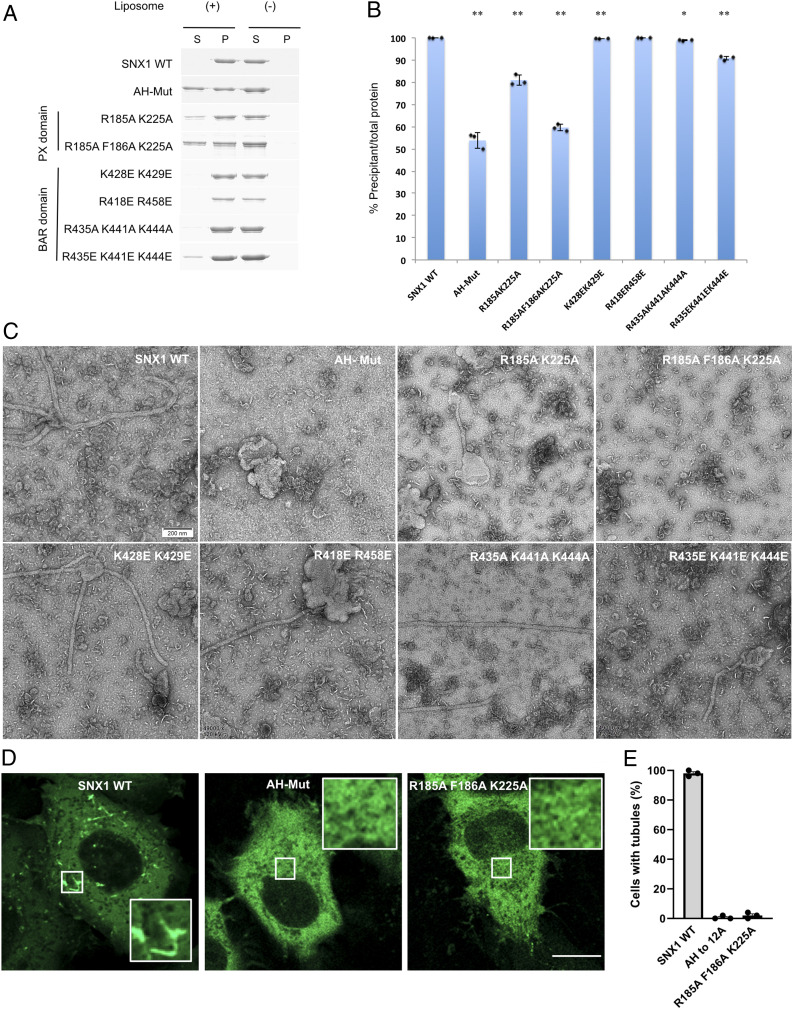
We next pursued functional mutagenesis studies to confirm our structural reconstructions. Initially, we performed membrane-binding assays and found that mutating the amphipathic helix, with residues 281 to 297 replaced by a linker (GGGSGGGSGGGS, AH-Mut), led to reduced binding of liposomes by SNX1 (Fig. 4 A and B). Next, performing liposome tubulation studies, we found that mutating the amphipathic helix also reduced the ability of SNX1 to induce membrane tubulation (Fig. 4C). To assess membrane tubulation in a more physiologic setting, we also performed cell-based studies. Overexpression of SNX1 has been shown previously to induce endosomal tubulation in cells (22). We found that the AH-Mut also impaired this ability of SNX1 (Fig. 4 D and E). Thus, the collective results supported that the linker region between the BAR and PX domains of SNX1 plays an important role in membrane binding and tubulation, which is predicted to involve an amphipathic helix.
Fig. 4.
Functional mutagenesis assessment of membrane binding and tabulation. (A) Binding of mutant forms of SNX1 (as indicated) to liposomes is assessed by the coprecipitation assay. Supernatant, S; Pellet, P. (B) Quantitation from three independent experiments is shown. All error bars represent SD from three independent experiments. Statistical analysis was performed comparing the wild-type and different mutants, with **P < 0.001 and *P < 0.01. (C) Negative-stain EM visualizing tubules induced by mutant forms of SNX1 (as indicated) (scale bar, 200 nm). (D) Confocal image of a HeLa cell expressing WT or mutant forms of GFP-tagged SNX1. Inset highlights endosomal tubulation seen in cells expressing WT, but not mutant, forms. (Scale bar, 10 μm.) (E) Quantitation of the above confocal analysis from three independent experiments. Error bars represent SD from three independent experiments.
Key Residues in SNX1 Acting in Membrane Binding and Tubulation.
The BAR domain has been suggested to bend the membrane through a scaffolding mechanism, which involves the BAR domain forming a banana-shaped dimer that binds to the negatively charged membrane surface through positively charged patches on the concave surface of the curved dimer (40, 42). On the concave surface of the SNX1 dimer that contains the BAR domain, we also observed several positively charged patches, R418/R458, K428/K429 and R435/K441/K444 (Fig. 3 E and F). Thus, we also performed functional mutagenesis studies to assess their role in membrane binding and bending.
When the R418/R458 residues were mutated to glutamates, we observed a relatively modest effect on membrane binding by SNX1 (Fig. 4 A and B). Mutating residues K428/K429 or R435/K441/K444, by substituting with alanines or glutamates, also had modest effects on membrane binding by SNX1 (Fig. 4 A and B). We also found that these mutations on the concave surface of the BAR domain had a modest impact on the ability of SNX1 to induce liposome tubulation (Fig. 4C). Thus, these results suggested that the positively charged patches at the ends of BAR domains are not the dominant elements in mediating membrane binding and bending by SNX1. In support of this conclusion, we found that high salt concentration did not affect membrane binding by SNX1 (SI Appendix, Fig. S8A).
We next considered that the curved structure of the SNX1 dimer extends to the PX domain and, notably, a positively charged patch exists on the concave side of this region. When residues (R185/K225) in this patch were mutated to alanines, we found that membrane binding by SNX1 was more dramatically reduced (Fig. 4 A and B). Moreover, as structure-based analysis predicted that R185/K225, along with F186, would participate in a PI3P binding pocket (SI Appendix, Fig. S8B), we next generated a more involved mutation (R185A/F186A/K225A) and found that membrane binding became even more reduced (Fig. 4 A and B). We also performed the liposome tubulation assay and found that mutating these residues affects membrane bending by SNX1 (Fig. 4C). This finding was further confirmed by the cell-based assay that examines the ability of overexpressed SNX1 to induce endosomal tubulation (Fig. 4 D and E).
Comparison of the SNX1 Lattice Structure with Other Structures of SNX1.
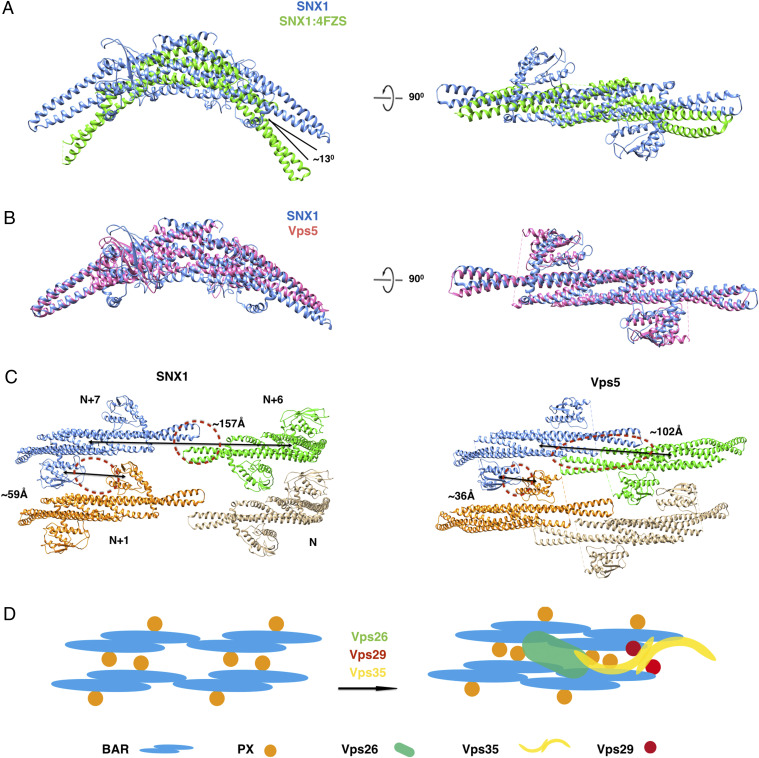
We next compared the SNX1 structure on the membrane to the structure of SNX1 previously solved in solution (PDB 4FZS) (22). This revealed that the positive curvature of the SNX1 dimer in the membrane-associated state is smaller to that seen in the solution state. Specifically, there is an ∼13° rotation of helix-α2 in SNX1 in the membrane-bound state as compared to that in the solution state (Fig. 5A), which predicts that a conformational change in the BAR domain occurs when SNX1 is recruited to the membrane, resulting in a more elongated SNX1 dimer along the long axis when it is assembled into the lattice structure. Our reconstructed SNX1 lattice also suggests that these changes likely involve flexibility in the kinks of the α2 and α3 helices of the BAR domain.
Fig. 5.
Structural comparisons between the SNX1 structure solved in this study versus those previously solved that contain SNX1 or SNX1-like protein. (A) Structural comparison of a SNX1 dimer solved in the context of the membrane versus the BAR domain of SNX1 solved previously in solution (PDB: 4FZS). The membrane structure of the SNX1 dimer is colored in blue while the solution structure of the BAR-domain dimer is colored in green. (B) Structural comparison of the SNX1 dimer solved in the current study versus the Vps5 dimer from Chaetomium thermophilum (PDB: 6H7W). The SNX1 dimer is colored in blue and the Vps5 dimer is colored in pink. (C) Structural comparison of the SNX1 assembly solved in the current study versus the Vps5 assembly solved previously in the context of a retromer-Vps5 assembly. Four dimers are shown in different colors which are labeled as N, N+1, N+6, and N+7. The distances between the centroids of adjacent dimers in the same helical row and also between the PX domains in different helical rows are labeled and indicated by black lines. Protein–protein interactions are highlighted by dashed red circles. (D) A cartoon representation of conformational changes in the SNX assembly upon the further binding by retromer.
We also compared our SNX1 structure to the previously solved structure of a retromer-SNX complex on the membrane, in which Vps5 (the yeast ortholog of the mammalian SNX1) was used (18). The diameters of tubules coated by the retromer-Vps5 complex were variable (SI Appendix, Fig. S9A), and thus the structure of retromer-Vps5 complex (PDB code 6H7W) could be only resolved by cryoelectron tomography and subtomogram averaging (18). To estimate the diameter of the retromer-Vps5–coated tubule, we superimposed the Vps5 dimer from one end of its structure to the other end and repeated this operation to generate a predicted helical model of Vps5 assembly. When this assembly was compared to the two classes of our SNX1 structures, similar diameters were observed (SI Appendix, Fig. S9B).
The overall structure of Vps5, when assembled with retromer, was similar to that of our solved SNX1 structure, as the rmsd was 2.5 Å for 260 C-α when superimposing these two structures (Fig. 5B). However, some differences could be discerned in the packing lattice of these two structures. First, when superimposing four dimers (labeled as N, N+1, N+6, and N+7) that span across two helical rows, the distance between dimer N+6 and N+7 in the same helical row is decreased 55 Å, from 157Å in the SNX1 lattice to 102 Å in the retromer-SNX lattice (Fig. 5C). Second, the distance of the two PX domains between dimer N+1 and N+7 in adjacent helical rows is lowered to 36 Å from 59 Å, resulting in contacts formed between the helical rows through the PX domains, which involves loop1 between the first and second β-strands and the beginning of the second α-helix (Fig. 5C). These differences suggest that Vps5 exists in a more compact state when assembled into the retromer-SNX coat complex, for which the likely explanation is that contacts with retromer induces Vps5 to adopt this more compact state (schematized in Fig. 5D).
Discussion
We have advanced a molecular understanding of how an SNX deforms the membrane, a process that acts in generating endosomal transport carriers. SNX1 possesses both a BAR and a PX domain. Whereas the BAR domain in some proteins has been found to be sufficient in driving membrane deformation (39, 43, 44), SNX1 has been known to require not only its BAR domain but also its PX domain, as well as a linker region that connects these two domains (22, 23). Our structural reconstruction of the SNX1 protein lattice on the membrane now provides a molecular understanding of how all three parts of SNX1 contribute to its ability to deform the membrane.
Membrane Deformation by SNX1.
The ability of a BAR domain to bend the membrane has been explained through a scaffolding mechanism, which involves the concave surface of the curved BAR dimer possessing multiple patches of positively charged residues that interact electrostatically with the negatively charged membrane surface to induce membrane bending (40, 42). In the case of SNX1, although the concave surface of its curved dimer also contains positively charged clusters in the BAR domain, a previous study found that mutating them does not affect the ability of SNX1 to bend the membrane (22). However, an explanation remained elusive, as the study had only solved the structure of the SNX1 BAR dimer in solution (22). In the current study, by solving the structure of the protein lattice formed by SNX1 on the membrane, we find that the positively charged residues in the BAR domain of SNX1 do not form significant electrostatic interactions with the negatively charged membrane surface. Instead, the positively charged residues in the PX domain of the curved SNX1 dimer play a more significant role in mediating electrostatic interactions with the negatively charged membrane surface.
We have also elucidated how the BAR domain in SNX1, although not providing the main contacts with the membrane, is nevertheless important for the ability of SNX1 to deform the membrane. Our SNX1 structure reveals that the BAR domain participates in protein–protein interactions that are needed to maintain this lattice structure in two ways. First, the extension of a helical row involves the BAR domains of two adjoining SNX1 dimers in the same row interacting with each other. Second, the BAR domain participates in crosslinking adjacent helical rows, with the BAR domain of a SNX1 dimer in one helical row interacting with the PX domain of an SNX1 dimer in another row.
The cooperative roles of the BAR and PX domains in SNX1 bear some resemblance to that of another coat protein that we previously examined. Like SNX1, ACAP1 (Arfgap Coil-coil Ankryin-repeat Protein 1) also possesses a BAR domain in tandem with a domain that binds phosphoinositides, which in this case is known as a PH (pleckstrin homology) domain. Rather than the BAR domain, we found that the PH domain in ACAP1 plays the dominant role in membrane binding and bending (45). However, a notable difference is that membrane deformation by SNX1 has been known to involve not only the BAR and PX domains but also a linker region that connects these two domains (22). Thus, by solving the structure of SNX1 on the membrane, we have provided a detailed understanding of how all three parts of SNX1 cooperate to drive membrane deformation.
We further note that SNX6 contains an overall structural organization similar to SNX1, having also BAR and PX domains connected by a linker region, as well as a linker region predicted to possess an amphipathic helix. However, as SNX6 cannot induce membrane deformation by itself (22), this consideration further underscores the significance of our study that has provided a detailed molecular understanding of how SNX1 achieves membrane deformation.
Insight into the Assembly of the Retromer-SNX Coat Complex.
Our reconstruction of the SNX1 lattice on the membrane also suggests insight into how the yeast retromer-Vps5 complex is assembled. In the initial stage that involves the recruitment of Vps5 from the cytosol to the membrane, a comparison of the previously solved structure of SNX1 in solution (22) and our currently solved SNX1 structure on the membrane suggests that the curvature of the Vps5 dimer would become reduced upon membrane association. As for the subsequent stage that involves the recruitment of retromer, a comparison of Vps5 in the previously solved retromer-Vps5 complex (18) to our currently solved SNX1 structure suggests that contact by the retromer results in the Vps5 dimer having a more compact state. As such, we propose the following sequential steps in the assembly of the yeast retromer-Vps5 complex on the membrane.
When Vps5 is initially recruited to the membrane, the curvature of the Vps5 dimer changes from a higher curved state to a lower one, resulting in a more extended state of the dimer. This change is likely due to Vps5 interacting with the membrane, for which we have identified molecular roles for the BAR and PX domains as well as the linker region that connects these two domains in the context of SNX1. Subsequently, when retromer is recruited to the Vps5 assembly, interactions between retromer and Vps5 induces the Vps5 dimer to achieve a more compressed state.
We further note that the elucidation of cargo binding by the mammalian SNX1-containing coats in recent years has suggested that retromer is not involved (19), in contrast to the situation in the yeast. However, as only exemplary cargoes have been examined, one cannot rule out that other cargoes, yet to be examined, may be found in the future to require retromer. As such, the prospect exists that our proposed model for how the retromer-Vps5 complex is assembled on the membrane may also be relevant to understanding how a potential mammalian equivalent of this yeast complex is assembled.
Materials and Methods
Protein Preparation.
Full-length mouse SNX1 (NP_062701.2) was subcloned into the plasmid pGEX-6P-1 (GE Healthcare) and then expressed as glutathione S-transferase (GST)-tagged fusion proteins in Escherichia coli BL21 (DE3) cells. Cells were grown in 2× yeast extract-tryptone (YT) medium at 37 °C until the optical density (OD) at 600 nm reached 1.2∼1.5 and then induced at 16 °C for 16 h with 0.1 mM isopropyl-β-d-thiogalactoside (IPTG). Cells were harvested by centrifugation, resuspended in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mMNa2HPO4, 1.8 mM KH2PO4, pH 7.4, and 10% Glycerol), and lysed by sonication. After centrifugation for 30 min at 15,000 rpm, the supernatant was collected and incubated with glutathione-Sepharose 4B at 4 °C and then washed using phosphate-buffered saline (PBS) buffer. After cleavage using Precision Protease (GE Healthcare) to remove the GST tag, the eluted proteins were further purified by size-exclusion chromatography using Superdex 200 10/300 glass (GL) column (GE Healthcare) with elution buffer (50 mM Hepes pH 7.4, 100 mM NaCl). The target protein was collected and stored at −80 °C for further experiments. Site-directed mutations of select residues were performed by overlap PCR, and mutant proteins were expressed and purified as described above for the wild-type protein.
Full-length mouse SNX6 (NP_081274.2) was subcloned into pET28a-His6-SUMO vector. Plasmids encoding SNX1 and SNX6 were cotransformed into Rosetta (DE3) cells. Heterodimer SNX1/SNX6 was expressed and purified firstly as described for SNX1 above. After cleavage using Precision Protease to remove the GST tag, the eluted fraction was loaded further onto Ni-NTA column, washed with the buffer of 20 mM Tris, pH 8.0, 300 mM NaCl, and 30 mM Imidazole to remove unbound samples, and eluted with the buffer containing 300 mM Imidazole. The elution was cleaved using Ubl-specific protease 1 (ULP1) and reloaded onto Ni-NTA column to remove SUMO tag. The target protein SNX1/SNX6 in the flow through was collected and stored at −80 °C for further experiments.
Liposome Production and Cosedimentation Assay.
All lipids were purchased from Avanti Polar lipids. Lipid mixtures, containing 40% phosphatidylcholine, 30% phosphatidylethanolamine, 20% phosphatidylserine, and 10% PI3P, were dried under gas nitrogen and then kept under vacuum for at least 3 h. Dry lipid mixtures were suspended in 50 mM Hepes, pH7.4, and 100 mM ⋅ NaCl for 30 min at 37 °C, frozen in liquid nitrogen, thawed at 37 °C for five cycles, and extruded through membrane filters of 0.05, 0.1, 0.2, and 0.4 μm for the production of 50-, 100-, 200-, and 400 nm diameter of liposomes, respectively. For cosedimentation assay, the 200 nm liposomes (1 mg/mL) and SNX1 protein (0.2 mg/mL) were incubated for 30 min at 20 °C before ultra-centrifugation at 250,000× g for 20 min and then the supernatants and pellets were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Negative-Stain EM.
SNX1 protein (0.9 mg/mL) was incubated with liposomes (0.5 mg/mL) for 30 min, and then the mixture was applied onto a glow-discharged carbon-coated EM grid and stained with uranyl acetate. The EM grids were examined with a transmission EM (FEI Spirit 120), and the micrographs were recorded with an EMSIS VELETA 2K*2K CCD camera under the nominate magnification of 49,000×.
Cryo-EM and Helical Reconstruction.
SNX1 protein (5 mg/mL) was incubated with 200 nm diameter liposomes (1.7 mg/mL) at room temperature for 60 min. A drop (3.5 µl) of the mixture was then applied onto a quantifoil 300-mesh R2/1 holy carbon grid that was pretreated in plasma cleaner (PDC-32G, Harrick Plasma). The grid was then blotted for 3.5 s with a blot force 2 at 100% humidity, using FEI Vitrobot (Mark IV), before it was quickly frozen in liquid ethane that was cooled by liquid nitrogen.
The SNX1 coating membrane tubules were imaged with a FEI Titan Krios cryoelectron microscope that was operated under 300 kV and equipped with a direct electron-detector device Falcon II camera (Thermo Fisher Scientific Incorporated). Low-dose images (25 e-/Å2) were collected manually. The nominal magnification was set to 59,000×, which corresponds to a pixel size of 1.42 Å. The defocus range was set to 0.5 to ∼2.5 µm. A total of 500 cryo-EM micrograph movie stacks were collected. Motion correction and defocus estimation for all these micrographs were performed using MotionCorr2 (46) and Gctf (GPU-accelerated contrast transfer function program) (47), respectively. Those raw micrographs with bad qualities such as ice contamination, poor Thon rings, or too large defocus values (greater than 3 μm) were sorted out and not included for further image processing. About 300 remaining micrographs were then multiplied by their theoretical contrast transfer function (CTF) for initial correction of CTF. A total of 476 SNX1 tubes were boxed using e2helixboxer.py in the package of EMAN2 (48) with a box width of 384 pixels. All tubes were classified according to their diameters. Each tube was projected along the z-axis of the tube. The minimum peak values of intensities at both sides of the center for one-dimensional projection array were searched. The distance between the two peaks was calculated which approximately equaled to the diameter of the tube. All tubes were then classified according to their diameter values. This diameter measurement and classifying process was performed by MATLAB code. Two main classes with diameters of 39 and 43 nm were obtained, and we performed helical reconstruction for these two classes, defined as class I and class II, respectively. An initial segment stack was generated from the two classes’ tubes with an overlap of 90%. Class I generated 11,795 segments, while 11,677 segments were generated for class II. The diffraction patterns for each class were then calculated. The diffraction patterns in each class were found to be homogeneous although they were classified from tubes with diverse diameters. Cylinders with 39- and 43 nm diameters, respectively, were generated by SPIDER (49), and they were set as the initial models for following reconstructions. Initial helical parameters were calculated by indexing the layer lines in the power spectrums of the boxed tubes in each class. An initial helical rise of 8.76Å and twist of 60.13° were obtained for the class I, while the corresponding values were 7.51Å and 51.51° for class II. All these values were used as initial parameters for helical reconstruction through an iterative helical real space reconstruction (IHRSR) algorithm (31, 32). Different out-of-plane tilt ranges were inspected during the projection matching procedure. Finally, (−15° to 15°) range can have a normal azimuth angle distribution and was used for IHRSR helical reconstruction of both classes. For class I, the helical parameters finally converged to 8.74 Å for the helical rise, and 60.11° for the helical twist, while these parameters converged to 7.53 Å and 51.53° for class II. We multiplied CTF for raw micrographs according to their estimated defocus values, with the two reconstructed maps for the two classes being divided by corresponding CTF2 for amplitude correction. SPIDER was used for negative B-factor sharpening. Resolutions of the final maps were estimated based on the gold standard FSC 0.143 criterion, which revealed ∼9.0 Å resolution for class I map and 10.0 Å resolution for class II map.
Model Building and MDFF.
Since the crystal structure of the SNX1 BAR domain (PDB code 4FSZ) is highly curved and cannot be docked into the BAR densities in our reconstruction, we generated the initial model of the full-length SNX1 by i-TASSER online service, which started from structure of Vps5 (PDB code 6H7W). For each class, the model was rigid fitted onto the map using Chimera (50). Then the model was adjusted manually in Coot (51). The region not fitted well with the map was truncated and remodeled as a helix corresponding to residues 279 to 302 that fits into the density between BAR and PX domain. Then a full-length model was generated by combining different segments. Finally, using this full-length model, we performed MDFF simulation using NAMD2.10 (52) with CHARMM 36 force field (53). The temperature was kept at 300 K with a 5 ps−1 damping coefficient. The time step was 1fs, and cutoff distance was 10 Å for nonbonded interactions. Restraints for secondary structures were introduced to the system. Finally, the model generated after MDFF simulation was further refined by Phenix (54).
Immunofluorescence Microscopy.
SNX1 forms were subcloned into the plasmid pEGFP-C1 (Clontech) to generate GFP-tagged forms and then transfected into HeLa cells using Fugene 6 reagent (Roche). After 24 h of expression, cells were fixed with 2% paraformaldehyde in PBS for 15 min and then coverslips were mounted onto slides by Fluoromount G (Thermo Fisher). Cells were examined using a Zeiss LSM-800 confocal microscope with Plan-Apochromat 63× objective and 488 nm laser line. The images were acquired using the Zen 2.6 Blue edition software (Zeiss) and processed by NIH Image J 1.54e. A total of 50 cells were examined in each condition. The percentage of cells with tubules labeled with GFP-tagged SNX1 were quantified.
Supplementary Material
Acknowledgments
We thank Prof. Edward H. Egelman from the University of Virginia for his generous help on image processing of helical reconstruction. We are grateful to Prof. Jizhong Lou and Dr. Yi Shi from the Institute of Biophysics, Chinese Academy of Sciences, for their help on model building. We would also like to thank Ping Shan, Ruigang Su, and Mengyue Lou (F.S.’s group) for assistance in laboratory management. This work was supported by grants from the National Natural Science Foundation of China (31670744, 31961160723, 31770794, and 31925026) and from the NIH to V.W.H. (GM058615 and GM115683). All EM data were collected at the Center for Biological Imaging (CBI, cbi.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences. We are grateful to Dr. Xiaojun Huang and Boling Zhu (CBI) for their assistance in EM data collection. Immunofluorescence images were collected at the Confocal Microscopy Core Facility of the Brigham and Women’s Hospital.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022614118/-/DCSupplemental.
Data Availability
The EM maps of helical tubules have been deposited in the EM Data Bank with the accession codes EMD-30592 (55) for class I and EMD-30593 (56) for class II, respectively. The corresponding fitted coordinates of SNX1 helical array on the tubule have been deposited in the PDB with the accession codes 7D6D (57) and 7D6E (58), respectively. Uncropped gel scans are shown in SI Appendix, Fig. S10.
References
- 1.Teasdale R. D., Loci D., Houghton F., Karlsson L., Gleeson P. A., A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J. 358, 7–16 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen P. J., Endosomal sorting and signalling: An emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 9, 574–582 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Cullen P. J., Steinberg F., To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Weeratunga S., Paul B., Collins B. M., Recognising the signals for endosomal trafficking. Curr. Opin. Cell Biol. 65, 17–27 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., et al., The retromer complex and sorting nexins in neurodegenerative diseases. Front. Aging Neurosci. 10, 79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C., Shah S. Z. A., Zhao D., Yang L., Role of the retromer complex in neurodegenerative diseases. Front. Aging Neurosci. 8, 42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonifacino J. S., Glick B. S., The mechanisms of vesicle budding and fusion. Cell 116, 153–166 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Seaman M. N., McCaffery J. M., Emr S. D., A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665–681 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seaman M. N., Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arighi C. N., Hartnell L. M., Aguilar R. C., Haft C. R., Bonifacino J. S., Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123–133 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strochlic T. I., Setty T. G., Sitaram A., Burd C. G., Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177, 115–125 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harterink M., et al., A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13, 914–923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temkin P., et al., SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13, 715–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg F., et al., A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15, 461–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison M. S., et al., A mechanism for retromer endosomal coat complex assembly with cargo. Proc. Natl. Acad. Sci. U.S.A. 111, 267–272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clairfeuille T., et al., A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat. Struct. Mol. Biol. 23, 921–932 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Lucas M., et al., Structural mechanism for cargo recognition by the retromer complex. Cell 167, 1623–1635.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovtun O., et al., Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonetti B., et al., Molecular identification of a BAR domain-containing coat complex for endosomal recycling of transmembrane proteins. Nat. Cell Biol. 21, 1219–1233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost A., Unger V. M., De Camilli P., The BAR domain superfamily: Membrane-molding macromolecules. Cell 137, 191–196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daumke O., Roux A., Haucke V., BAR domain scaffolds in dynamin-mediated membrane fission. Cell 156, 882–892 (2014). [DOI] [PubMed] [Google Scholar]
- 22.van Weering J. R., et al., Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 31, 4466–4480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlton J., et al., Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 14, 1791–1800 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Hurley J. H., Boura E., Carlson L. A., Różycki B., Membrane budding. Cell 143, 875–887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hierro A., et al., Functional architecture of the retromer cargo-recognition complex. Nature 449, 1063–1067 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendall A. K., et al., Mammalian retromer is an adaptable scaffold for cargo sorting from endosomes. Structure 28, 393–405.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D., et al., The cryo-EM structure of the SNX-BAR Mvp1 tetramer. Nat. Commun. 11, 1506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cozier G. E., et al., The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 277, 48730–48736 (2002). [DOI] [PubMed] [Google Scholar]
- 29.van Weering J. R., Verkade P., Cullen P. J., SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin. Cell Dev. Biol. 21, 371–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada A., et al., Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129, 761–772 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Egelman E. H., A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Egelman E. H., The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., et al., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGreevy R., Teo I., Singharoy A., Schulten K., Advances in the molecular dynamics flexible fitting method for cryo-EM modeling. Methods 100, 50–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhatt V. S., Ashley R., Sundborger-Lunna A., Amphipathic motifs regulate N-BAR protein endophilin B1 auto-inhibition and drive membrane remodeling. Structure 29, 61–69.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Mim C., et al., Structural basis of membrane bending by the N-BAR protein endophilin. Cell 149, 137–145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frost A., et al., Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pylypenko O., Lundmark R., Rasmuson E., Carlsson S. R., Rak A., The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 26, 4788–4800 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H., Schulten K., Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput. Biol. 9, e1002892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peter B. J., et al., BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303, 495–499 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Mizuno N., Jao C. C., Langen R., Steven A. C., Multiple modes of endophilin-mediated conversion of lipid vesicles into coated tubes: Implications for synaptic endocytosis. J. Biol. Chem. 285, 23351–23358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang X., et al., A PH domain in ACAP1 possesses key features of the BAR domain in promoting membrane curvature. Dev. Cell 31, 73–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang G., et al., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Shaikh T. R., et al., SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallographica 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Phillips J. C., et al., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Best R. B., et al., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liebschner D., et al., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., Pang X., Sun F., Structural insights into membrane remodeling by SNX1. Electron Microscopy Data Bank. https://www.emdataresource.org/EMD-30592. Deposited 30 September 2020.
- 56.Zhang Y., Pang X., Sun F., Structural insights into membrane remodeling by SNX1. Electron Microscopy Data Bank. https://www.emdataresource.org/EMD-30593. Deposited 30 September 2020.
- 57.Zhang Y., Pang X., Sun F., Structural insights into membrane remodeling by SNX1. Protein Data Bank. https://www.rcsb.org/structure/unreleased/7D6D. Deposited 30 September 2020.
- 58.Zhang Y., Pang X., Sun F., Structural insights into membrane remodeling by SNX1. Protein Data Bank. https://www.rcsb.org/structure/unreleased/7D6E. Deposited 30 September 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EM maps of helical tubules have been deposited in the EM Data Bank with the accession codes EMD-30592 (55) for class I and EMD-30593 (56) for class II, respectively. The corresponding fitted coordinates of SNX1 helical array on the tubule have been deposited in the PDB with the accession codes 7D6D (57) and 7D6E (58), respectively. Uncropped gel scans are shown in SI Appendix, Fig. S10.