Significance
The functional diversity of MAPKs is dependent on differential selection of substrates for phosphorylation. Nevertheless, the mechanisms assisting MAPKs to maintain substrate specificity in plants are not known. Our study shows that WRKY33, a pathogen-responsive MPK3/6 substrate, is SUMOylated in response to pathogen-associated molecular patterns and pathogen signals. SUMOylation mediates WRKY33 phosphorylation and its activation. This correlation is reliant on the selective identification of SUMOylated WRKY33 by MPK3/6 via SUMO interaction motif (SIM) sites. Therefore, WRKY33 SUMOylation is important to selectively interact with MAPKs via the SUMO–SIM module and undergo phosphorylation and thus activation. Our data reveal a mechanism by which MPK3/6 prioritize molecular pathways by differentially selecting substrates using the SUMO–SIM module.
Keywords: plants, immunity, SUMO, WRKY33, MAPKs
Abstract
The versatility of mitogen-activated protein kinases (MAPKs) in translating exogenous and endogenous stimuli into appropriate cellular responses depends on its substrate specificity. In animals, several mechanisms have been proposed about how MAPKs maintain specificity to regulate distinct functional pathways. However, little is known of mechanisms that enable substrate selectivity in plant MAPKs. Small ubiquitin-like modifier (SUMO), a posttranslational modification system, plays an important role in plant development and defense by rapid reprogramming of cellular events. In this study we identified a functional SUMO interaction motif (SIM) in Arabidopsis MPK3 and MPK6 that reveals a mechanism for selective interaction of MPK3/6 with SUMO-conjugated WRKY33, during defense. We show that WRKY33 is rapidly SUMOylated in response to Botrytis cinerea infection and flg22 elicitor treatment. SUMOylation mediates WRKY33 phosphorylation by MPKs and consequent transcription factor activity. Disruption of either WRKY33 SUMO or MPK3/6 SIM sites attenuates their interaction and inactivates WRKY33-mediated defense. However, MPK3/6 SIM mutants show normal interaction with a non-SUMOylated form of another transcription factor, SPEECHLESS, unraveling a role for SUMOylation in differential substrate selectivity by MPKs. We reveal that the SUMO proteases, SUMO PROTEASE RELATED TO FERTILITY1 (SPF1) and SPF2 control WRKY33 SUMOylation and demonstrate a role for these SUMO proteases in defense. Our data reveal a mechanism by which MPK3/6 prioritize molecular pathways by differentially selecting substrates using the SUMO–SIM module during defense responses.
One of the key aspects of plant development is the integration of environmental abiotic and biotic signals into ongoing growth processes for successful acclimatization of plants to their vicinal environment. Mitogen-activated protein kinase (MAPK) cascades are versatile signaling mechanisms in all eukaryotes, which translate exogenous and endogenous stimuli into appropriate cellular responses (1, 2). The MAPK cascade encompasses three kinase families activating each other by sequential phosphorylation (1–3). An exogenous stimulus or a developmental signal activates an MAPK kinase kinase (MAPKKK), which phosphorylates and activates an MAPKK, which in turn phosphorylates and activates an MAPK (3). Activated MAPKs phosphorylate specific substrates resulting in the activation of the corresponding cellular responses. The Arabidopsis thaliana genome codes for 20 MAPK, 10 MAPKK, and about 80 MAPKKK family members (4).
MAPK pathways function as central regulators of plant growth and development (3). They regulate stomatal patterning by phosphorylating the basic helix–loop–helix transcription factor (TF) SPEECHLESS (SPCH) (5, 6) and affect male gametogenesis by phosphorylating pollen-specific WRKY34 during early stages in pollen development (7). They also regulate zygotic asymmetry and embryo patterning by controlling WRKY2 phosphorylation (8).
MAPK cascades also play pivotal roles in plant immune responses. Activation of MAPKs is one of the earliest signaling events occurring after plants sense pathogen invasion (4). Recognition of pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) by pattern recognition receptors triggers early defense responses, including production of reactive oxygen species and activation of MAPKs (9). For example, MAPKs are activated by the pattern recognition receptor FLAGELLIN SENSING 2 (FLS2) upon perception of bacterial flagellin through a 22-amino acid sequence (flg22) as a PAMP. Activation of MAPKs results in multiple defense responses, including defense gene expression, the generation of reactive oxygen species, and stomatal closure leading to PAMP-triggered immunity (9). Moreover, the activation of key defense-responsive TFs, such as WRKY33 (10), ETHYLENE RESPONSE FACTOR 6 (ERF6) (11), and ERF104 (12) are also reliant on MAPKs-mediated phosphorylation. MAPKs also contribute to immune signaling by phosphorylating and stabilizing 1-AMINO-CYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE 2/6 (ACS2/ACS6) upon Botrytis cinerea infection (13). This ability of MAPKs to phosphorylate differentially expressed substrates in specific plant tissues represents the functional diversity of MAPKs. Nevertheless, it is intriguing how MAPKs maintain substrate specificity to regulate distinct functional pathways. In animals several mechanisms have been proposed to control the specificity determination of MAPKs (14, 15), however, not much is known about how this diversity is maintained in plants.
Small ubiquitin-like modifier (SUMO) is an emerging posttranslational modification (PTM) in plants with demonstrably important roles in plant development and defense pathways (16, 17) by rapid reprogramming of cellular events (18). SUMOylation entails three successive enzymatic reactions catalyzed by an E1 [a heterodimeric complex of SUMO activating enzymes 1a/b (SAE1a/b) and SAE2], an E2 or SUMO conjugating enzyme1 (SCE1), and E3s or SUMO ligases. Currently, there are four SUMO E3 ligases identified in Arabidopsis, SIZ1, MMS21, and PIAL1/2. The latter two are also known to promote SUMO chain elongation (17). Additionally, currently 16 SUMO proteases (19) have been identified that are responsible for deSUMOylation of target proteins. These proteases form an integral part of the SUMO machinery and provide flexibility and plasticity to the mechanism.
In this study, we show that SUMOylation of WRKY33 is an early plant response to stress perception and is essential for providing resistance against necrotrophs and regulating WRKY33 activation. Our data reveal that the SUMO proteases, SPF1 and SPF2 (20), have a specific role in immunity by governing WRKY33 SUMOylation. We further demonstrate that SUMOylation is essential for WRKY33 phosphorylation and WRKY33–MPK3/6 interaction. We also identified SUMO interacting motif (SIM) sites in both MPK3 and MPK6 that facilitate interaction with SUMOylated WRKY33. However, the SIM motifs in MPK3/6 do not play a role in modulating the interaction with SPCH, another TF substrate that is required for stomatal patterning. This emphasizes the important role of WRKY33 SUMOylation in mediating selective interaction and phosphorylation by MAPKs. Therefore, our study uncovers a PTM mechanism allowing MPK3/6 to prioritize molecular pathways to allow plants to adapt to their environment.
Results
WRKY33 SUMOylation Is an Early Plant Response to PAMPs and Regulates Immunity.
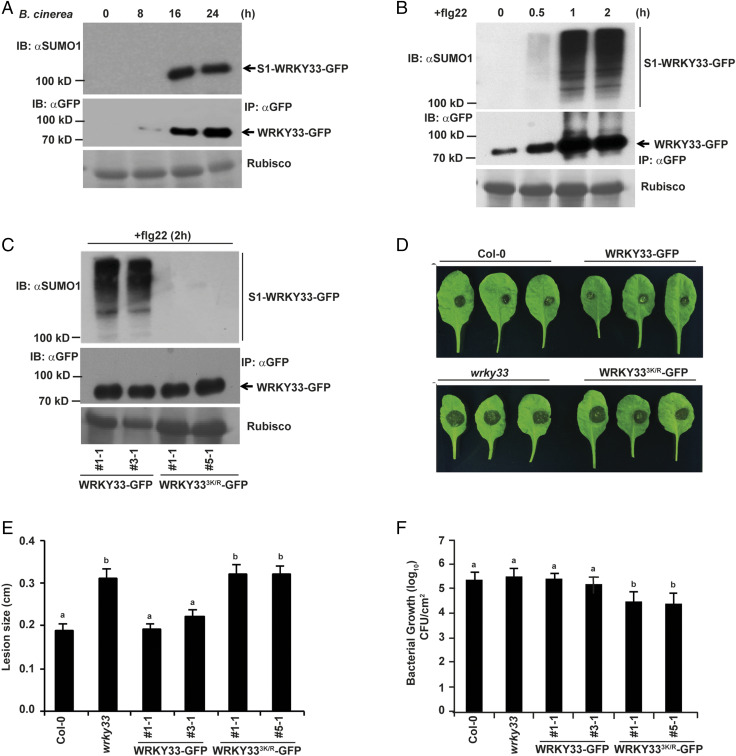
WRKY33 was identified as a potential SUMO target during stress conditions (21, 22), implicating SUMOylation of WRKY33 as a plant response to stress. However, the specific environmental conditions in which WRKY33 is conjugated to SUMO or its functional relevance is not known. Since WRKY33 is a PAMP/pathogen-responsive TF activated upon B. cinerea infection (23, 24) and flg22 treatment (25), we tested WRKY33 SUMOylation status under these conditions. This was done using a transgenic line expressing WRKY33 translationally fused to GFP driven by its own promoter (ProWRKY33::WRKY33WT-GFP) in the wrky33 knockout background (line 3-1). WRKY33WT-GFP immunopurified from leaves of the transgenic line spray-inoculated with B. cinerea was found to be SUMOylated at 16 and 24 h after B. cinerea infection (Fig. 1A). Similarly, WRKY33WT-GFP immunopurified after flg22 treatment exhibited several SUMO conjugated forms within 30 min of treatment that increased considerably after 1 h (Fig. 1B). Although the proportion of induced WRKY33-GFP protein undergoing SUMOylation is low, it is congruent with other reports (26) confirming that only a small fraction of the pool of target protein undergoes SUMOylation at any given point of time. Nevertheless, the data clearly indicated that SUMOylation of WRKY33 is an early plant response to pathogen infection.
Fig. 1.
WRKY33 SUMOylation is essential for WRKY33-mediated defense responses against necrotrophs and biotrophs. (A) WRKY33 is SUMOylated in response to B. cinerea infection. Four-week-old plants of ProWRKY33::WRKY33-GFP (wrky33 kockout [k/o]) transgenic line 3-1 were spray-inoculated with B. cinerea spores (5 × 105 spores/mL) and leaf samples were collected at the indicated time points. Total protein from these samples were immunoprecipitated with anti-GFP beads (IP: αGFP) and immunoblotted with anti-GFP (IB: αGFP) for WRKY33-GFP and anti-SUMO1/2 (IB: αSUMO1) for SUMOylated WRKY33-GFP (S1-W33-GFP). Rubisco was used as the loading control. (B) WRKY33 is SUMOylated in response to flg22 treatment. Total protein from 12-d-old seedlings of WRKY33 transgenic line 3-1 treated with flg22 (1 µM) for indicated time points was immunoprecipitated with anti-GFP beads (IP: αGFP) and immunoblotted with anti-GFP (IB: αGFP) for WRKY33-GFP and anti-SUMO1/2 (IB: αSUMO1) for SUMOylated WRKY33-GFP (S1-W33-GFP). Rubisco was used as the loading control. (C) WRKY33 Lys182, Lys219 and Lys504 are essential for WRKY33 SUMOylation. Total protein from 12-d-old seedlings of ProWRKY33::WRKY33-GFP (wrky33 k/o) transgenic lines (1-1 and 3-1) and ProWRKY33::WRKY333K/R-GFP (wrky33 k/o) transgenic lines (1-1 and 5-1) treated with flg22 (1 µM) for 2 h was immunoprecipitated with anti-GFP beads (IP: αGFP) and immunoblotted with anti-GFP (IB: αGFP) for WRKY33-GFP and anti-SUMO1/2 (IB: αSUMO1) for SUMOylated WRKY33-GFP (S1-W33-GFP). Rubisco was used as the loading control. (D) WRKY33 SUMOylation is required for WRKY33-mediated resistance against B. cinerea. Fully expanded leaves from 4-wk-old plants of Col-0, wrky33 k/o and two independent transgenic lines each of WRKY33 and WRKY333K/R (mentioned above) were excised and drop inoculated with 5 µL B. cinerea spore suspension. Photographs were taken 72 h postinfection. (E) Quantification of the lesion size after B. cinerea infection in D. The data represent the means ± SE of lesion sizes from 20 to 25 individual leaves per genotype. A two-tailed Student’s t test was performed among means of lesion sizes. Genotypes with different letters were significantly different from others (P < 0.05). (F) WRKY33 SUMOylation promotes growth of P. syringae DC3000. Four-week-old leaves of the different genotypes indicated were infiltrated on the abaxial side of leaves with P. syringae pv. tomato culture and CFUs were counted 3 d postinfection. Data presented are means ± SE from two independent biological replicates with 11 to 12 plants per replicate (n = 2). Bars with different letters were significantly different from others; P < 0.05; two-tailed Student’s t test.
By comparing conserved amino acids surrounding established SUMO sites from other plant SUMO targets (17, 18, 26), we identified three lysine residues (Lys182, Lys219, and Lys504) in WRKY33 (conserved in WRKY33 homologs from other plant species) (SI Appendix, Fig. S1) as potential SUMO conjugation sites. Subsequently, all three potential SUMO sites were mutated to arginine residues and the resultant WRKY333K/R version was used to generate transgenic plants in the wrky33 knockout background (ProWRKY33::WRKY333K/R-GFP). A comparison of immunoprecipitates from WRKY33WT-GFP transgenic lines (1-1 and 3-1) and WRKY333K/R-GFP transgenic lines (1-1 and 5-1) after flg22 treatment clearly showed that WRKY333K/R-GFP failed to conjugate to SUMO1 (Fig. 1C). This indicated that one or more of these three lysines constitute all of the SUMO attachment sites on WRKY33. The SUMO site mutations did not affect the subcellular localization of the protein, since both GFP-WRKY33WT and GFP-WRKY333K/R showed fluorescent signals emanating from the nucleus when 35S::GFP-WRKY33WT/GFP-WRKY333K/R constructs were transiently expressed in Nicotiana benthamiana (25) (SI Appendix, Fig. S2).
Prompt activation of the transcription factor WRKY33 is essential for inducing downstream defense-related genes for resistance against the fungus B. cinerea (10, 23, 25). Therefore, to understand the role of WRKY33 SUMOylation in mediating resistance against B. cinerea infection, we subjected the above-mentioned WRKY33WT-GFP and WRKY333K/R-GFP complementation lines to B. cinerea infection. We found that in contrast to WRKY33WT-GFP, non-SUMO WRKY333K/R-GFP lines showed lesion sizes similar to wrky33 knockout mutant. These data indicated that the susceptibility of WRKY333K/R-GFP lines to B. cinerea were similar to that of wrky33 mutant plants (25) (Fig. 1 D and E and SI Appendix, Fig. S3).
Furthermore, to examine the impact of WRKY33 SUMOylation on bacterial infection we compared the response of WRKY33WT-GFP and WRKY333K/R-GFP Arabidopsis lines to Pseudomonas syringae pv. tomato DC3000 infection. Surprisingly, unlike B. cinerea infection, WRKY333K/R-GFP lines were more resistant to P. syringae pv. tomato infection as compared to wrky33 plants (Fig. 1F). The virulent bacterial growth in WRKY33WT-GFP lines, which was similar to Col-0 and wrky33 plants, was in accordance with earlier reports showing similar bacterial growth in these genetic backgrounds (25). Since WRKY33 prevents inappropriate induction of salicylic acid (SA) pathway (17), WRKY333K/R might have influenced this pathway resulting in increased activation of SA signaling and enhanced resistance to P. syringae pv. tomato. To test this hypothesis, we analyzed the expression levels of the PATHOGENESIS RELATED 1 (PR1) gene in WRKY33WT-GFP and WRKY333K/R-GFP transgenic lines after P. syringae pv. tomato infection. Interestingly, we observed higher levels of PR1 transcripts in WRKY333K/R-GFP transgenic lines as compared to WRKY33WT-GFP transgenic lines (SI Appendix, Fig. S4). This clearly indicated that WRKY333K/R positively impacts the SA pathway.
SUMOylation Regulates Transcriptional Activation of WRKY33.
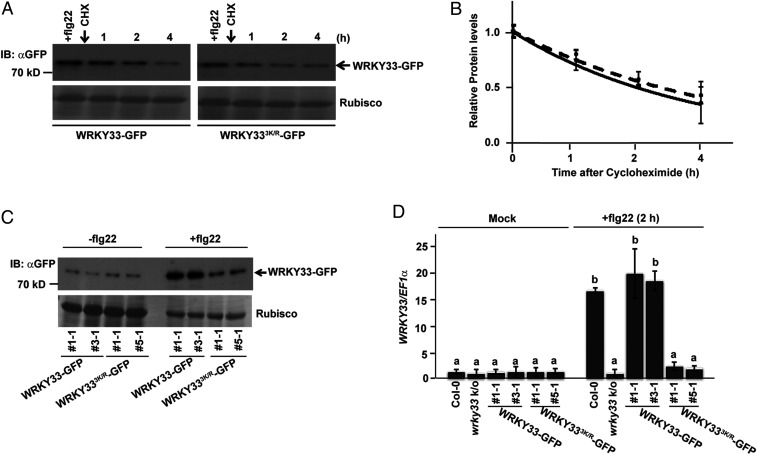
The altered response of Arabidopsis lines expressing non-SUMOylatable WRKY33 against fungal and bacterial pathogens prompted us to determine if SUMOylation affects the pool of WRKY33. A comparison of protein turnover rates of WRKY33WT-GFP and WRKY333K/R-GFP after flg22 treatment for 2 h followed by cycloheximide treatment showed comparable turnover for both (Fig. 2 A and B). However, we observed an interesting response when WRKY33-GFP protein levels in WRKY33WT-GFP transgenic lines were compared to WRKY333K/R-GFP levels in WRKY333K/R-GFP transgenic lines. In the absence of flg22, WRKY33WT-GFP and WRKY333K/R-GFP protein levels were similar in the two transgenic lines. However, in the presence of flg22 WRKY33WT-GFP, protein level was remarkably higher when compared to WRKY333K/R-GFP protein in the two transgenic lines (Fig. 2C). For this reason, for an unbiased comparison of their SUMOylation status a higher amount of WRKY333K/R-GFP protein was loaded in the gels in Fig. 1C to ensure equivalent protein levels to WRKY33WT-GFP (refer to the Rubisco levels in Fig. 1C). Similarly, although WRKY33 expression in WRKY33WT-GFP and WRKY333K/R-GFP transgenic lines were comparable before flg22 treatment, WRKY33 transcripts were higher in WRKY33WT-GFP transgenic lines as compared to WRKY333K/R-GFP transgenic lines after flg22 treatment (Fig. 2D). This clearly indicated that although SUMOylation does not impact WRKY33 protein stability, it does play a vital role in regulating WRKY33 gene induction and therefore affecting the cellular pool of WRKY33.
Fig. 2.
SUMOylation is essential for transcriptional activation of WRKY33. (A) SUMOylation does not affect WRKY33 protein turnover rates. Twelve-day-old seedlings of both WRKY33 and WRKY333K/R transgenic lines were first treated with flg22 for 2 h for induction of the respective proteins and then cycloheximide (200 μM) treatment was done for the indicated timepoints. The samples obtained were immunoblotted with αGFP for detecting WRKY33-GFP. Ponceau stained Rubisco was used as the loading control. (B) Quantification of the bands shown above were plotted. Relative protein abundances were normalized to the loading control (Rubisco) and to their respective levels at time 0 h. Results from two independent replicates were averaged and shown with error bars denoting SD. An exponential best-fit curve was fitted through the data points. (C) WRKY33 protein induction is attenuated in WRKY333K/R transgenics. Total protein from 12-d-old seedlings of WRKY33 and WRKY333K/R transgenics were treated with −/+flg22 (1 µM) and then samples were immunoprecipitated with anti-GFP (IP: αGFP) beads. The samples were immunoblotted with anti-GFP (IB: αGFP) antibodies. Rubisco was used as the loading control. (D) WRKY33 SUMOylation is essential for its transcriptional activation upon flg22 treatment. Twelve-day-old seedlings of the different genotypes indicated were treated with 1 μM flg22 for 2 h and WRKY33 expression was compared. The mock-treated seedling for each genotype was taken as controls. Bars represent means ± SD from two independent biological replicates. Col-0 was used as the reference and normalization was done using EF1α. Genotypes with different letters were significantly different from others (two-tailed Student’s t test; P < 0.05).
Furthermore, since chitin, rather than flg22, is the PAMP from B. cinerea, we analyzed WRKY33 regulation in response to chitin treatment to ascertain if chitin draws functional parallels with B. cinerea in regulating WRKY33. Similar to the induction upon B. cinerea infection, WRKY33 was also induced by chitin in a SUMOylation-dependent manner: That is, more induction in WRKY33-GFP lines than in WRKY333K/R-GFP lines (SI Appendix, Fig. S5). Moreover, the data were consistent with flg22-mediated WRKY33 regulation indicating that the two PAMPs show functional overlaps with respect to regulating WRKY33 gene expression.
SPF1 and SPF2 SUMO Proteases Control WRKY33 SUMOylation Levels.
SUMO proteases play an essential role in providing equilibrium to the SUMO system by deconjugating SUMO from target proteins (26). Moreover, unlike ubiquitination, which relies on a pool of E3 ligases for providing substrate specificity, in the SUMO system the current thinking is that SUMO proteases are entrusted with this responsibility (27), since it has only four E3 ligases (28). Therefore, to further corroborate the role of SUMOylation in regulating WRKY33 induction and identify the specific SUMO proteases responsible for WRKY33 deSUMOylation, we scanned the literature for the role of different SUMO proteases in defense response. The Arabidopsis genome is known to code for eight UBIQUITIN-LIKE PROTEASES (ULPs), which are the main deSUMOylating proteins in plants (29, 30). Of these, we judiciously selected SPF1 (also known as ULP2b) and SPF2 (also known as ULP2a) as the likely candidate SUMO proteases targeting WRKY33 for deSUMOylation. Several other functionally characterized SUMO proteases were not considered for the following reasons. OVER TOLERANT TO SALT1 (OTS1; ULP1d), OTS2 (ULP1c) (31), and EARLY IN SHORT DAYS4 (ESD4) (32) were omitted based on the observation that WRKY33 SUMOylation enhances resistance to B. cinerea (Fig. 1 D and E). Therefore, one would expect the corresponding SUMO protease mutant to be more resistant to B. cinerea. However, the ots1 ots2 double mutant is sensitive to B. cinerea infection (18) and similar to esd4 (32) mutant, has considerably higher levels of SA (33) that promote susceptibility (34) to B. cinerea. ESD4-LIKE SUMO PROTEASE1 (ELS1; ULP1a) was also ruled out because of its cytoplasmic localization (35), whereas WRKY33 is nuclear localized. ELS2 (ULP1b) and FOURTH ULP GENE CLASS 1 (FUG1; ULP1e or ULP3) were also left out as their role in immunity or development in plants has not been established (29). A recently identified SUMO protease from Arabidopsis, DESI3a, belonging to the DESI family was also omitted as the protease is membrane localized (26).
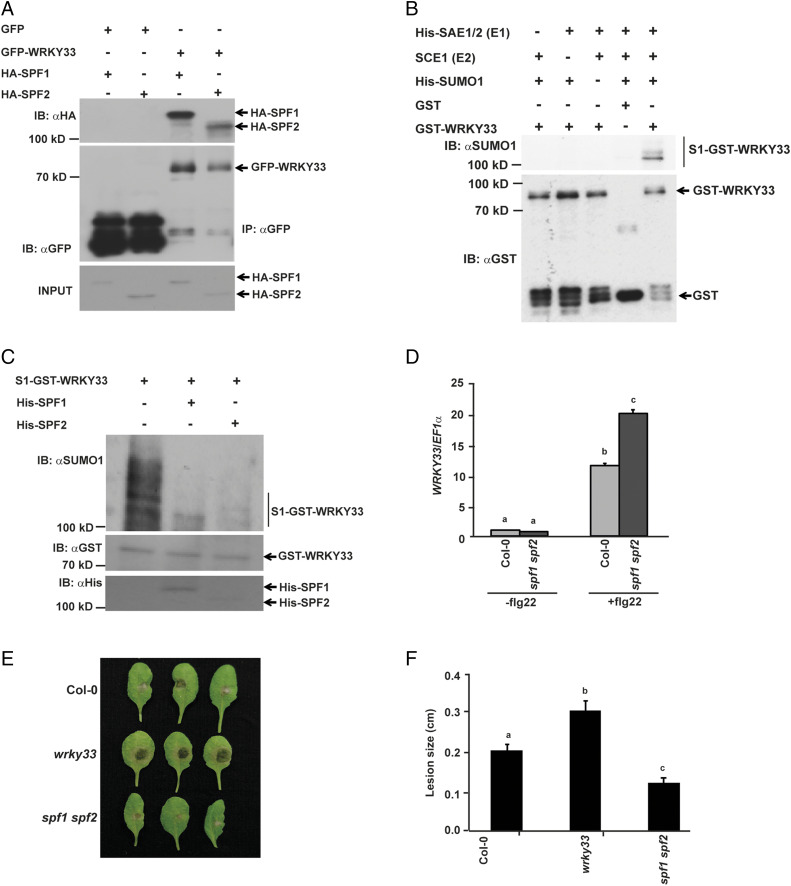
To validate whether SPF1/2 are the cognate SUMO proteases for WRKY33, we first immunopurified GFP-WRKY33 from N. benthamiana leaves coexpressing GFP-WRKY33 with HA-SPF1 or HA-SPF2 under the 35S promoter. Both HA-SPF1 and HA-SPF2 copurified with WRKY33 (Fig. 3A), suggesting a role for SPF1/2 in WRKY33 deSUMOylation. An in vitro assay was performed to examine if SPF1/2 can directly deSUMOylate WRKY33. For this, we first ascertained WRKY33 SUMOylation under in vitro conditions by incubating recombinant WRKY33 with affinity-purified His-tagged E1 heterodimer (SAE1/SAE2), SCE1, and His-SUMO1 (Fig. 3B and SI Appendix, Fig. S6). Subsequently, a deSUMOylation assay demonstrated a reduction in WRKY33 SUMOylation levels when SUMOylated GST-WRKY33 protein was incubated with recombinant His-SPF1 or His-SPF2 (Fig. 3C). This confirmed that SPF1/2 SUMO proteases target WRKY33 for deSUMOylation under in vitro conditions.
Fig. 3.
WRKY33 deSUMOylation is regulated by SPF1 and SPF2 SUMO proteases. (A) WRKY33 interacts with SPF1 and SPF2. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33 either with HA-SPF1 or HA-SPF2 was subjected to IP with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for SCE1-HA and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Total protein extracts were probed with anti-HA antibody (HA-SPF1/HA-SPF2 input) for equal HA-fusion proteins. (B) WRKY33 can be SUMOylated under in vitro conditions. Recombinantly expressed GST-WRKY33 was incubated with His-SAE1, SCE1, and His-SUMO1 at 30 °C for 2 h. Besides the GST-only control, independent reactions were set up as negative controls with one component missing to ensure that all components are essential for WRKY33 SUMOylation. Reactions were stopped with 4× SDS buffer and immunoblotted with anti-GST (IB: αGST) and anti-SUMO1/2 (IB: αSUMO1) antibodies. (C) About 10 μg of SUMOylated GST-WRKY33 was incubated either with His-SPF1 or His-SPF2 for 16 h and the reactions were stopped by adding 4× dye. The samples were immunoblotted with αGST for GST-WRKY33, αHis for His-SPF1 and SPF2, and with αSUMO1 for detecting SUMOylated WRKY33. (D) WRKY33 expression is higher in spf1 spf2 mutant compared to Col-0. WRKY33 expression was analyzed in 12-d-old seedlings of the different genotypes indicated before and after treatment with 1 µM flg22. Data presented are means ± SD from two independent biological replicates and normalized to EF1α expression. A two-tailed Student’s t test showed that WRKY33 expression was significantly different in genotypes with different letters (P < 0.05). (E) spf1 spf2 mutant is more resistant to B. cinerea infection. Fully expanded leaves from 4-wk-old plants of Col-0, wrky33 k/o and spf1 spf2 were excised and drop-inoculated with 5 µL B. cinerea spore suspension. Photographs were taken 72 h postinfection and quantified in F. (F) The data represent the means ± SE of lesion sizes from 20 to 25 individual leaves per genotype. A two-tailed Student’s t test was performed among means of lesion sizes. Genotypes with different letters were significantly different from others (P < 0.05).
Subsequently, to analyze the impact of SPF1/2 SUMO proteases on the regulation of WRKY33 induction we subjected the spf1 spf2 double gene-knockout mutant to B. cinerea infection and flg22 treatment. We found that WRKY33 transcript levels were increased in spf1 spf2 when compared to Col-0 upon B. cinerea infection (SI Appendix, Fig. S7) and flg22 treatment (Fig. 3D). This prompted us to assess the response of spf1 spf2 mutant against B. cinerea infection to understand the role of SPF1/2 in defense. The spf1 spf2 mutant showed enhanced resistance to B. cinerea as compared to Col-0 (Fig. 3 E and F and SI Appendix, Fig. S8), signifying a specific role for SPF1/2 in plant immunity. To further validate the in vitro data and confirm that SPF1 and SPF2 are indeed involved in deSUMOylating WRKY33, we generated ProWRKY33::WRKY33WT-GFP transgenic lines in the spf1 spf2 double-mutant background. We then subjected these transgenic lines and the transgenic lines in the wrky33 mutant background to flg22 treatment and compared the SUMOylation levels of WRKY33-GFP immunopurified from the two transgenic lines. We observed an increased abundance of the SUMOylated form of WRKY33 immunopurified from the spf1 spf2 mutant background (SI Appendix, Fig. S9), supporting the in vitro and infection data. Collectively, our data confirm that SPF1 and SPF2 target WRKY33 for deSUMOylation.
SUMOylation Affects WRKY33 Transcriptional Activity by Controlling Its Phosphorylation without Affecting Its DNA-Binding Activity.
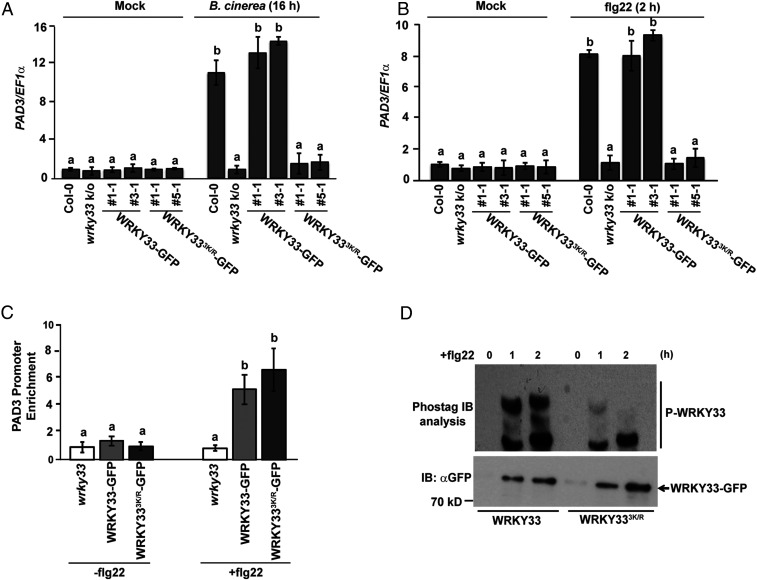
Intrigued by the role of SUMOylation in regulating WRKY33 transcription, we examined the impact of SUMOylation on WRKY33 target genes. WRKY33 is known to initiate the transcriptional activation of downstream genes, including its self-activation upon pathogen/PAMP signals (10). We observed enhanced expression of PAD3, one such downstream target of WRKY33 (10), in transgenic plants expressing WRKY33WT-GFP when compared to transgenic plants expressing WRKY333K/R-GFP upon B. cinerea infection (Fig. 4A) and flg22 treatment (Fig. 4B). This clearly suggested that SUMOylation is essential for transcriptional regulation by WRKY33. This evidence also confirmed that the attenuated induction of WRKY33 transcripts in transgenic plants expressing WRKY333K/R-GFP was due to reduced activity of WRKY333K/R. This further prompted us to compare the DNA-binding abilities of WRKY33WT-GFP and WRKY333K/R-GFP proteins. Surprisingly, no significant difference was observed (Fig. 4C) when the binding abilities of WRKY33WT-GFP and WRKY333K/R-GFP proteins onto PAD3 promoter were compared using a chromatin immunoprecipitation (ChIP) assay.
Fig. 4.
WRKY33 SUMOylation regulates phosphorylation of WRKY33 without affecting its DNA-binding activity. (A) B. cinerea induced WRKY33 SUMOylation is essential for its transcriptional activity. Four-week-old plants of the different genotypes indicated were either sprayed with water as mock or spray-inoculated with B. cinerea and PAD3 expression was compared 16 h postinfection in the different genotypes. Means from two independent biological replicates are denoted as bar graphs with error bars representing SD EF1α was used for normalization. Bars with different letters were significantly different from others (two-tailed Student’s t test; P < 0.05). (B) SUMOylation is required for transcriptional activity of WRKY33 after flg22 treatment. PAD3 expression was analyzed in 12-d-old seedlings of the different genotypes after treatment with 1 µM flg22. For mock-treatment the seedlings were left in water for the same duration. Data presented are means ± SD from two biological replicates and normalized to EF1α expression. Bars with different letters were significantly different from others (two-tailed Student’s t test; P < 0.05). (C) SUMOylation does not affect WRKY33 DNA-binding activity. ChIP assay showing WRKY33 DNA-binding activity in WRKY33 and WRKY333K/R ± flg22 (1 µM) by comparing the binding to PAD3 promoter. Fold enrichment was calculated relative to the last exon of At4g26410. Data presented are means ± SD from two biological replicates. A two-tailed Student’s t test showed that promoter enrichment was significantly different for genotypes with different letters (P < 0.05). (D) SUMOylation is required for efficient WRKY33 phosphorylation. Total protein from 12-d-old seedlings of WRKY33 and WRKY333K/R transgenics ± flg22 (1 µM) was immunoprecipitated with anti-GFP (IP: αGFP) beads and subjected to Phos-tag gel analysis to detect WRKY33 phosphorylation status. They were immunoblotted with anti-GFP (IB: αGFP) antibodies (Upper) and a regular immunoblot was also done to detect total WRKY33 protein (Lower).
It is known that the transcriptional activity of WRKY33 is reliant on its phosphorylation (10). This information led us to analyze the phosphorylation status of WRKY33 and WRKY333K/R proteins. A Phos-tag gel analysis of WRKY33-GFP protein immunopurified from the two independent transgenic lines (after flg22 treatment) showed that WRKY333K/R-GFP protein was noticeably less phosphorylated as compared to WRKY33-GFP protein (Fig. 4D). The data provide key evidence highlighting an essential role for WRKY33 SUMOylation in regulating its phosphorylation. To further validate this, we compared the phosphorylation status of WRKY33WT-GFP protein immunopurified from WRKY33WT-GFP transgenic lines in wrky33 and spf1 spf2 mutant backgrounds upon flg22 treatment. We observed a higher amount of phosphorylation of WRKY33WT-GFP protein immunopurified from spf1 spf2 mutant background (where more SUMOylated WRKY33 accumulates) as compared to WRKY33WT-GFP protein immunopurified from wrky33 background (SI Appendix, Fig. S10). This substantiated our earlier results that SUMOylation of WRKY33 is important for its phosphorylation.
SUMOylation Enables Interaction of WRKY33 with MPK3/6.
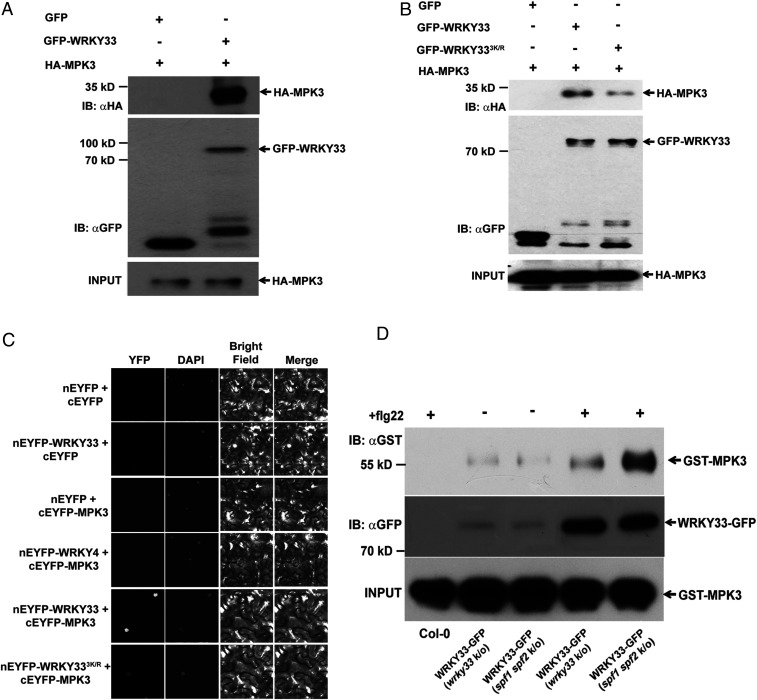
WRKY33 is known to be an MPK3/6 substrate (10), which activates the TF by phosphorylation in response to B. cinerea infection. Moreover, we noticed that WRKY33 SUMOylation occurs under similar conditions in which it is phosphorylated and activated by MPK3/6. In view of this and the above data, we hypothesized that SUMOylation facilitates WRKY33 phosphorylation by modulating its interaction with MPK3/6. To test this, we first determined if WRKY33 interacts with MPK3. Although WRKY33 was shown to be phosphorylated by MPK3/6 in an in vitro assay, evidence showing WRKY33 interaction with MPK3 or MPK6 is currently unavailable. Therefore, an IP assay was performed from N. benthamiana leaves coexpressing GFP-WRKY33 and HA-MPK3 proteins. We observed copurification of HA-MPK3 when GFP-WRKY33 was pulled down, confirming the interaction between the two proteins (Fig. 5A). Subsequently, to examine the impact of SUMOylation on WRKY33-MPK3 interaction, we analyzed the binding of MPK3 with non-SUMOylatable WRKY333K/R. Interestingly, in comparison to GFP-WRKY33WT, GFP-WRKY333K/R showed a diminished interaction with HA-MPK3 in a co-IP assay (Fig. 5B). This was further validated using bimolecular fluorescence complementation (BiFC) assays where WRKY33/WKY333K/R and MPK3 were expressed as fusion proteins with N- and C-terminal halves of the YFP protein, respectively, in N. benthamiana leaves. A reconstituted YFP signal was observed for nEYFP-WRKY33WT and cEYFP-MPK3 but no signal was detected for nEYFP-WRKY333K/R and cEYFP-MPK3 infiltrations, further illustrating the clear interaction between WRKY33WT and MPK3 (Fig. 5C). As a negative control, we also tested the interaction of cEYFP-MPK3 with nEYFP-WRKY4, another group I WRKY family member. Lack of any YFP signal confirmed that WRKY33 specifically interacts with MPK3 and the signal was not an artifact.
Fig. 5.
SUMOylation facilitates WRKY33 interaction with MPK3. (A) WRKY33 interacts with MPK3 in the transient expression system. Total protein extracted from N. benthamiana leaves transiently expressing GFP-WRKY33 with HA-MPK3 was subjected to IP with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for HA-MPK3 and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Total protein extracts were probed with anti-HA antibody (HA-MPK3 input) for equal HA-fusion protein. (B) SUMOylation is important for facilitating WRKY33-MPK3 interaction. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33 or GFP-WRKY33 with HA-MPK3 was subjected to IP with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for HA-MPK3 and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Total protein extracts were probed with anti-HA antibody (HA-MPK3 input) for equal HA-fusion protein. (C) BiFC analysis to show that WRKY333K/R interacted weakly with MPK3 in N. benthamiana leaves. Confocal images of N. benthamiana leaves coexpressing the N-terminal domain of enhanced yellow fluorescent protein (nEYFP):WRKY33 or nEYFP:WRKY333K/R with the C-terminal domain of YFP (cEYFP):MPK3. nEYFP with cEYFP and cEYFP-MPK3 and cEYFP with nEYFP-WRKY33 were used as negative controls. An additional control was used by testing the interaction of MPK3 with WRKY4, another group I WRKY member. YFP channel: reconstituted EYFP; DAPI channel: DAPI; Bright field: transmitted; Merge: superimposition of all the three channels; Magnification: 40×. (D) GFP-WRKY33 immunoprecipitated from spf1 spf2 mutant background interacts more strongly with MPK3. GFP-WRKY33 immunoprecipitated from spf1 spf2 mutant was mixed with recombinantly expressed GST-MPK3 protein followed by pulling down GFP-WRKY33. The sample was immunoblotted with anti-GST antibody (IB: αGST) to probe for GST-MPK3 and anti-GFP antibody (IB: αGFP) for probing WRKY33-GFP. Equal amount of GST-MPK3 was used for all samples and is represented as an input lane. Col-0 was used as negative control.
To further establish this correlation between WRKY33 SUMOylation and its interaction with MPK3, we compared the amount of MPK3 pulled down with WRKY33WT-GFP protein immunoprecipitated from transgenic lines expressing WRKY33WT-GFP in wrky33 and spf1 spf2 mutant backgrounds after flg22 treatment. This was performed by mixing WRKY33WT-GFP immunoprecipitate with recombinantly expressed GST-MPK3 protein followed by pulling down WRKY33WT-GFP. We observed more GST-MPK3 pulled down with WRKY33WT-GFP immunoprecipitated from spf1 spf2 mutant background as compared to the protein immunoprecipitated from wrky33 mutant background (Fig. 5D). These results clearly indicated that SUMOylation plays an important role in facilitating the interaction of WRKY33 with MPK3. Moreover, this further implied that SUMOylation of WRKY33 in response to pathogen and PAMP signals is important for WRKY33 to be selected by the two MAPKs for phosphorylation and, hence, activation of defense-related down-stream genes, including self-activation of WRKY33-gene expression.
MPK3/6 SIM Sites Are Important for Selective Interaction with WRKY33.
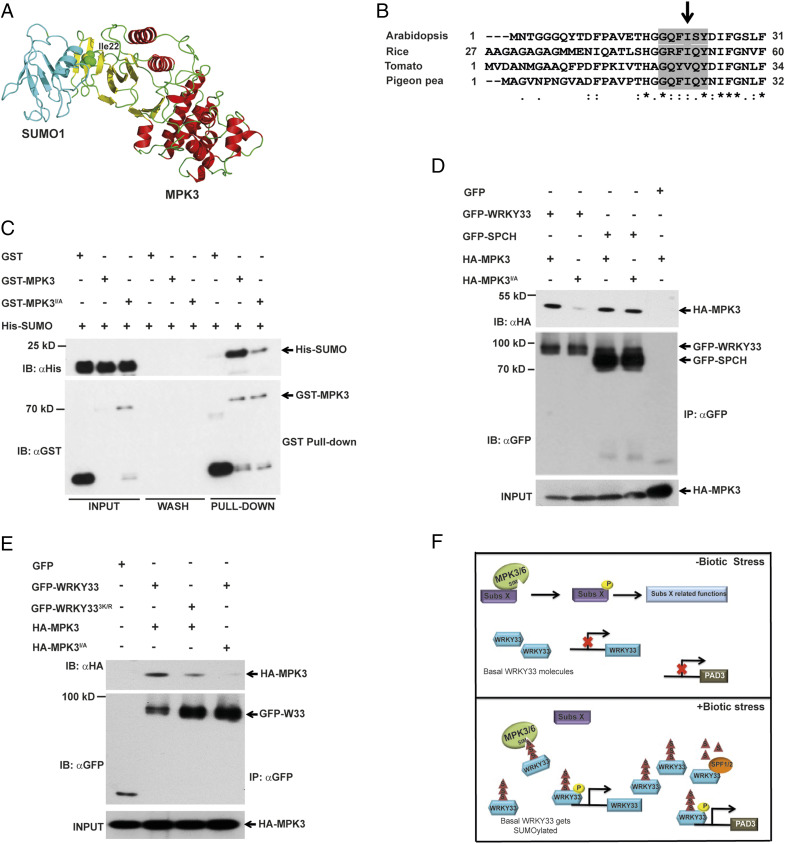
An essential role of SUMO in modulating WRKY33 interaction with MPK3 prompted us to postulate that WRKY33–MPK3 interaction is dependent on the canonical SUMO–SIM interaction module, where SUMO on WRKY33 interacts with the SIM motif on MPK3/6. SIMs are generally characterized by a short stretch of hydrophobic amino acids flanked by acidic residues and arranged in a parallel or antiparallel β-strand to interact with a hydrophobic pocket on the SUMO surface (36). This structural arrangement helps SIM sites in providing specificity to interactions of SUMO-conjugated proteins (WRKY33 in this case) with their SIM-containing binding partners (37, 38) (MPK3 and MPK6 in this case). A thorough scan of MPK3 and MPK6 primary, secondary, and three-dimensional structures (MPK3 structure modeled from MPK6) identified the amino acid sequences GQFISY and GRFIQY as potential SIM motifs in MPK3 and MPK6, respectively. To assess the structural feasibility, we modeled SUMO1 structure on the MPK3 homology model and found that SUMO1 covers the SIM motif at the N-terminal β-sheet–rich region of MPK3 (Fig. 6A).
Fig. 6.
SUMO–SIM module facilitates WRKY33-MAPK Interaction. (A) A structural snapshot of MPK3–SUMO1 protein complex model highlighting the docking of SUMO1 onto MPK3 N-terminal region containing the SIM site (Ile22). MPK6 structure (PDB ID: 5CI6) was used as a template for MPK3 model (red and yellow). SUMO1 (PDB ID: 4WJO) is in turquoise. MPK3 Ile22 as green spheres. (B) Arabidopsis MPK3 Ile22 is conserved among its homologs from rice, tomato and pigeon pea. SIM is highlighted in gray. (C) MPK3 SIM site is required for interaction with SUMO. An in vitro pull-down assay exhibited that MPK3 SIM mutant (MPK3I22A) interacted weakly with SUMO. Purified GST-MPK3 and GST-MPK3I22A were incubated with His-SUMO for 1 h and eluted after unbound proteins were washed. Input, wash, and elution (pull-down) were immunoblotted with anti-GST (IB: αGST) and anti-His (IB: αHis) antibodies. GST was used as a negative control. (D) MPK3 SIM site is required for WRKY33 but not SPCH. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33/GFP-SPCH with HA-MPK3/HA-MPK3I22A was immunoprecipitated with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for HA-MPK3/HA-MPK3I22A and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Extracts were probed with anti-HA antibody (HA-MPK3 input) for equal MPK3/MPK3I22A. (E) WRKY33-MPK3 interaction is reliant on SUMO–SIM module. Total protein from N. benthamiana leaves transiently expressing GFP-WRKY33/GFP-WRKY333K/R with either HA-MPK3/HA-MPK3I22A was subjected to IP with anti-GFP beads (IP: αGFP) and immunoblotted with anti-HA (IB: αHA) for HA-fusion proteins and anti-GFP (IB: αGFP) for GFP/GFP-fusion proteins. Extracts were probed with anti-HA antibody (HA-MPK3/HA-MPK3I22A input) for equal HA-fusion proteins. (F) A simplistic model showing that WRKY33 SUMOylation is an early plant response upon pathogen infection and allows WRKY33 to be quickly and specifically identified by MAPKs SIM site via the SUMO–SIM module. In absence of stress, basal-level WRKY33 is unperturbed and MAPKs interact and phosphorylate substrate X (Subs X), resulting in regulation of its functions. However, during biotic stress, basal-level WRKY33 is rapidly SUMOylated to be selectively and quickly identified by MPK3/6 via the SUMO–SIM module for phosphorylation and activation, resulting in activation of downstream genes (PAD3). SPF1/2 regulates WRKY33 SUMO levels. Yellow circles with “P” are phosphorylation; triangles with “S” are SUMOylation.
To validate if the identified motif is a potential SIM site, we mutated isoleucine at position 22 of MPK3 and isoleucine at position 47 of MPK6 to alanines to disrupt the SIM site. The respective amino acid residues were selected for mutation as they are highly conserved in other Arabidopsis MPKs, as well as with MPK3 homologs from other plant species (Fig. 6B and SI Appendix, Fig. S11). The recombinantly expressed mutated MPKs were then used to examine the interaction with purified His-SUMO1. Both GST-MPK3I22A and GST-MPK6I47A exhibited a remarkably diminished interaction with His-SUMO1 as compared to the respective wild-type proteins (Fig. 6C and SI Appendix, Fig. S12), confirming that the identified motif is a SIM site in MPK3/6. Furthermore, we ascertained if the SIM site has specificity for SUMO-conjugated WRKY33 by analyzing the interaction of HA-MPK3 and HA-MPK3I22A with GFP-WRKY33 and SPCH (GFP-SPCH) (5), another well-characterized TF substrate of MAPKs fused to GFP, in N. benthamiana. Interestingly, SPCH did not differentiate between the two MPK3 versions, whereas WRKY33 showed a reduced interaction with MPK3I22A as compared to MPK3 (Fig. 6D). This evidence clearly suggested that the SIM site is essential for interacting with WRKY33 but not with SPCH, hence playing an important role in MPK3 substrate selection.
As a definitive test of our hypothesis that the MPK3/6-WRKY33 interaction is dependent on the SUMO–SIM module, we examined the interaction of WRKY33WT/WRKY333K/R with MPK3/6 and of WRKY33WT with MPK3I22A/MPK6I47A (Fig. 6E and SI Appendix, Fig. S13). As expected, although WRKY33 showed a strong interaction with MPK3/6, disruption of either WRKY33 SUMO or MPK3/6 SIM sites strongly affected their interaction, thereby confirming an important role for the SUMO–SIM module in WRKY33–MPK3/6 interaction.
Discussion
MAPKs are functionally diverse proteins regulating a wide range of plant developmental processes from growth to defense (3). However, it is unclear how they maintain substrate selectivity for regulating such functional diversity in plants. SUMOylation is known to regulate cellular responses by covalently conjugating to target proteins and affecting their functions, partly by modulating protein–protein interaction. Besides, noncovalent interaction to SUMO-conjugated protein SIM sites also plays a vital role in determining interaction specificity of SUMO-conjugated proteins with their SIM-containing binding partners. This intricate involvement of the SUMO–SIM module in controlling specificity of protein–protein interaction and the functional overlap between regulatory roles of MAPKs and SUMO suggested that SUMO modification might have an important role in determining how MAPKs show selectivity toward different substrates. SUMOylation of WRKY33, an MPK3/6 substrate, under similar conditions (Fig. 1) in which it is phosphorylated by MAPKs (10), as well as SUMOylation-mediated control of WRKY33–MPK3/6 interaction (Figs. 5 and 6), strongly substantiated the view that SUMO is important for substrate selectivity of MPK3 and MPK6.
Mao et al. (10) highlighted that induction of WRKY33 during pathogen- or PAMP-triggered gene activation is independent of de novo protein synthesis and suggested the involvement of PTM of basal-level WRKY33 protein for activation of WRKY33. They speculated phosphorylation by MAPKs as the PTM governing activation of basal-level WRKY33. However, our data showed SUMOylation of WRKY33 upon pathogen infection as the PTM controlling WRKY33 transcriptional activation (Fig. 4). Enhanced WRKY33 transcripts in the spf1 spf2 mutant corroborated a tight correlation between WRKY33 induction and SUMOylation (Fig. 3).
Moreover, the enhanced SUMOylation (SI Appendix, Fig. S9) and phosphorylation of WRKY33 (SI Appendix, Fig. S10) in the spf1 spf2 mutant background resulting from increased selective interaction with MPK3 in that background (Fig. 5) together highlight how WRKY33 SUMOylation is pivotal for its induction. The stronger defense response in spf1 spf2 mutant against B. cinerea (Fig. 3) further supports this finding. Moreover, the activation of basal-level WRKY33 protein demands a quick adaptive PTM response for rapid elicitation of defense signaling. Our data fulfill this criterion, since WRKY33 SUMOylation was an early plant response (Fig. 1) to PAMP and pathogen signals. This also implies that WRKY33 should first be SUMOylated for it to be selectively identified by MPK3/6 for phosphorylation. Taken together, this evidence indicates that the PTM regulating basal-level WRKY33 protein is SUMOylation.
The finding that WRKY33 SUMOylation is a prompt adaptive response to pathogen infection compels one to think that B. cinerea must have devised strategies to deSUMOylate WRKY33 to impede the associated defense response. In light of this, it is noteworthy that WRKY33 is mono-SUMOylated upon B. cinerea infection, whereas it is poly-SUMOylated in response to flg22 treatment (Fig. 1), connoting that B. cinerea might have injected effectors targeting higher-order SUMO conjugates of WRKY33 for deSUMOylation. This would lower the magnitude of WRKY33 phosphorylation and activation resulting in a weakened defense response against B. cinerea. In congruence with this, we observed poly-SUMOylated forms of WRKY33 upon flg22 treatment, since flg22 is a PAMP and not a live pathogen (Fig. 1). Furthermore, the in vitro SUMOylation assay also depicted that WRKY33 is poly-SUMOylated (Fig. 3), thereby supporting our hypothesis that higher-order SUMO conjugates can only be detected in the absence of the fungal pathogen. A similar SUMOylation pattern observed for the pathogen recognition receptor FLS2 further substantiates our viewpoint. FLS2 was mono-SUMOylated upon P. syringae pv. tomato infection, but poly-SUMOylated under in vitro conditions (26).
Despite such a vital role of the SUMO–SIM module in mediating WRKY33–MPK3/6 interaction, we did not observe a complete abolition of interaction after mutating the SUMO or SIM sites (Figs. 5 and 6). This could be because SUMOylation might have a fundamental role in structurally orientating the two proteins for a tighter interaction rather than just facilitating a physical contact. The inability of WRKY333K/R to complement wrky33 mutant (Fig. 1), despite its (weak) interactions with MPK3 and MPK6 (Fig. 6), supports this point.
WRKY33 overexpressing plants are more susceptible to P. syringae pv. tomato, whereas wrky33 mutant responds normally to the pathogen, probably due to functional redundancy among different WRKY proteins (25). It was, therefore, surprising to see enhanced resistance of WRKY333K/R transgenic lines in comparison to Col-0, wrky33 knockout, and WRKY33WT transgenic lines against P. syringae pv. tomato infection (Fig. 1F). Conceptually, being unable to complement wrky33, the response of WRKY333K/R transgenic plants to P. syringae pv. tomato should be comparable to wrky33 mutant. However, unlike wrky33 that lacks functional WRKY33 protein, WRKY333K/R transgenic plants possess a basal-level of non-SUMOylatable (and nonphosphorylatable) WRKY33 protein (Fig. 2C). This would exhibit a dominant-negative effect on the activation of downstream target genes by occupying their promoter-binding sites (Fig. 4C) and preventing other WRKYs from binding. Since WRKY33 is known to prevent inappropriate induction of the SA pathway (23), a dominant-negative effect by WRKY333K/R would have resulted in enhanced resistance against P. syringae pv. tomato via increased activation of the SA pathway.
Therefore, we propose that during normal growth conditions, basal-level WRKY33 is unperturbed and MPK3/6 interact with other substrates, for example, SPCH, resulting in the regulation of SPCH-related functions (Fig. 6F). However, during biotic stress conditions, WRKY33 rapidly undergoes SUMOylation so that it can be selectively preferred by the two MAPKs via the SUMO–SIM module to undergo phosphorylation and activation resulting in the up-regulation of downstream genes (Fig. 6F). SPF1/2 SUMO proteases modulate WRKY33 SUMO levels to prevent uncontrolled activation. In conclusion, the study reveals a mechanism that allows MPK3/6 to prioritize molecular pathways, in this case immune responses through WRKY33, enabling plants to rapidly respond to changing environmental conditions.
Methods
Plant Materials and Growth Conditions.
A. thaliana ecotype Columbia-0 (Col-0) was used as the wild-type control for in planta experiments. Plants used in the study were grown in environmentally controlled growth chambers at 21 °C under long-day conditions (16:8 h, light:dark) for generating and growing transgenic plants and under short-day (8:16 h, light:dark) conditions for B. cinerea and P. syringae pv. tomato DC3000 infection assays. For seedling growth, seeds were surface-sterilized and sown on Murashige and Skoog (MS) (Duchefa) plates supplemented with 0.8% Phytoagar. The seeds of wrky33-1 (SALK-006603) used in the study were obtained from Nottingham Arabidopsis Stock Centre. It is noteworthy that wrky33-1 lines, in addition to T-DNA insertion in the WRKY33 gene, has another linked WRKY33 copy with no T-DNA insertion but with a 33-bp deletion followed by a 23-bp duplication abolishing proper splicing after transcription (25). The second copy of WRKY33 results in the synthesis of a larger transcript with an unspliced intron translating into a truncated protein of 209 amino acids with no WRKY zinc-finger motif.
Plasmid Construction, Site-Directed Mutagenesis, and Plant Transformation.
To generate the ProWRKY33::WRKY33-GFP construct, full-length WRKY33 CDS (without stop codon) with a 1.5-kb region of genomic DNA upstream of WRKY33 ATG were cloned into a pGreen binary vector (HY105 backbone). All other constructs, including GFP-WRKY33, GFP-SPCH, HA-MPK3, HA-MPK6, HA-SCE1, HA-SPF1, HA-SPF2, GST-WRKY33, GST-MPK3, and His-SUMO were generated using the GATEWAY cloning method –pEarleyGate 103 for GFP, pEarleyGate 201 for HA, pDEST 15 for GST and pDEST 17 for His tag clones. All the constructs were confirmed by sequencing.
WRKY333K/R, MPK3I22A, and MPK6I47A were generated using a site-directed mutagenesis approach. For WRKY333K/R, the mutations were introduced sequentially by first mutating Lys-504, followed by Lys-182 and Lys-219 and confirmed by sequencing.
For generating transgenic plants, ProWRKY33::WRKY33-GFP and ProWRKY33::WRKY333K/R-GFP constructs were introduced into Agrobacterium tumefaciens strain GV3101:pMP90 and used for transforming wrky33 and spf1 spf2 mutant plants via a floral-dip method. The transgenics were selected using BASTA and progressed to T3 generation before they were used for analysis.
Elicitors Used.
Flg22 peptide used in the study was obtained from Peptron and the lyophilized powder was solubilized in water to make stock solutions, aliquoted, and stored at −80 °C for further use.
The other elicitor used in the study was chitin. It was derived from crab-shell (a chitin mixture; Sigma-Aldrich) and the stock was prepared in water containing 1 M acetic acid under overnight continuous stirring, as described previously (39).
B. cinerea Infection Assay.
B. cinerea infection was performed as described previously with minor modifications (33). B. cinerea isolate 2100 was cultivated on potato dextrose agar plates at 22 °C for 12 to 14 d and the spores were counted using a hematocytometer and diluted with potato dextrose broth to a final concentration of 5 × 105 spores/mL. For droplet inoculations, about 20 to 25 leaves from 4-wk-old plants grown under short-day conditions were infected per genotype by placing 5 μL inoculum on them. The mock leaves were inoculated with 5 μL drops of potato dextrose broth. Leaf images were taken 3 d postinfection and lesion sizes were quantified using ImageJ software (rsb.info.nih.gov/ij). The data presented are mean ± SE from 20 leaves (n = 20).
For SUMO blots and real-time analysis, 4-wk-old plants were spray-inoculated with the same B. cinerea inoculum with the addition of 0.02% (vol/vol) Silwet L-77 and leaf samples were collected at the indicated time points and stored at −80 °C until being processed. The experiments were repeated two more times with consistent results.
P. syringae Infection Assay.
P. syringae pv. tomato DC3000 used for bacterial infection assays was grown onto a Kings B Agar medium (20.0 g Bacto Peptone, 0.75 g K2HPO4⋅7H2O, 10 mL glycerol/L) plate with kanamycin and rifampicin antibiotics (50 mg/L) in the dark for 2 d at 28 °C. For inoculation, P. syringae pv. tomato DC3000 was grown overnight in Kings B broth at 28 °C and then pelleted and washed with sterile MgCl2. Subsequently, the suspension was adjusted to 1 × 105 colony forming units (CFU)/mL in MgCl2 and used for infection by infiltrating the lower surface of Arabidopsis leaves. The disease rate was scored 72 h postinfection by counting the CFUs on Kings B Agar plates. The experiment was performed twice with each biological replicate consisting of leaf samples from 12 individual plants.
RNA Extraction and Quantitative Real-Time PCR Analysis.
Total RNA was extracted either from 12-d-old seedlings in case of flg22 (1 μM) and chitin (100 μg/mL) treatments or from 4-wk-old plant after B. cinerea infection using Spectrum Plant Total RNA kit (Sigma-Aldrich) as per the manufacturer’s protocol. cDNA synthesis was done using SuperScript-II Reverse Transcriptase (Invitrogen) following the manufacturer’s guidelines. Quantitative real-time PCR was performed using Brilliant III Ultra-Fast SYBR QPCR master mix (Agilent) by using Rotor-Gene Q (Qiagen) instrument. The analysis was done with the software provided using comparative quantification methods. EF1α (At1g07920) was used as internal control for normalization. The experiments were repeated two times. For quantification of the B. cinerea infection via real-time PCR, the expression of B. cinerea Cutinase A gene was analyzed.
IP and Co-IP Assays.
For detecting WRKY33 SUMOylation, total protein was extracted either from 12-d-old seedlings upon flg22 treatment or from 4-wk-old plant leaves after B. cinerea infection using SUMO extraction buffer, as mentioned in Orosa et al. (26). The extract was then incubated with 50 μL anti-GFP beads (Chromotek) for 30 min at 4 °C, washed three times, and finally the immuno-complex was eluted with 50 μL of preheated (95 °C) 1× SDS-loading buffer. For other IP experiments—such as Phos-tag analysis, WRKY33 protein levels, and protein turnover rates—total protein was isolated using 150 mM NaCl, 100 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 0.5% Trition X-100, 1 mM DTT and 5% (vol/vol) glycerol followed by an anti-GFP IP, as mentioned above.
For co-IP assays, overnight-grown Agrobacterium cultures containing the respective constructs were resuspended in MgCl2 to a final OD600 of 0.2. The cultures of constructs to be tested for interaction were mixed and incubated with acetosyringone for 60 to 90 min and subsequently infiltrated on the abaxial side of N. benthamiana leaves. The plants were grown further for 3 d before the leaf samples were used for co-IP, as mentioned above. All IP and co-IP experiments were performed twice with consistent results.
Subcellular Localization Studies.
For transient expression, 4-wk-old N. benthamiana plants were infiltrated on the abaxial side of the leaf with Agrobacterium cultures, as indicated above. Seventy-two hours postinfiltration, sections of N. benthamiana leaves transiently expressing GFP only, WRKY33-GFP, or WRKY333K/R-GFP were observed using a Zeiss LSM 880 laser scanning confocal microscope. An excitation wavelength of 488 nm and an emission wavelength of 505 to 530 nm was used for GFP. Image analysis was done using a ZEN microscope imaging software and converted to JPEG format.
In Vitro SUMOylation, deSUMOylation, and Pull-Down Assays.
For examining WRKY33 SUMOylation under in vitro conditions, recombinantly expressed GST-WRKY33 (5 μg) was incubated with His-tagged E1 heterodimer (His-SAE1/SAE2) (2 μg), SCE1 (2 μg), and His-SUMO1 (1 μg) at 30 °C for 2 h. GST was used as a negative control. For additional controls, five independent reactions were set up with one component missing in each. The reactions were stopped by adding 4× SDS sample buffer and heating to 98 °C for 3 min.
For deSUMOylation assay, about 10 μg of SUMOylated GST-WRKY33 obtained above was subjected to a quick purification by centrifugation for removal of SUMO machinery components and the purified SUMOylated protein was incubated either with 5 μg of His-SPF1 or His-SPF2 in deSUMOylation buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1 mM DTT and 0.2% Igepal) at 30 °C for 16 h. The reactions were stopped by adding 4× SDS sample buffer and heating to 98 °C for 3 min.
For in vitro pull-down assay, purified GST-MPK3/MPK3I22A (5 μg) protein was immobilized to GST resin in an Eppendorf tube and unbound protein was removed by centrifugation. Subsequently, purified His-SUMO1 (1 μg) was mixed with GST-tagged protein in interaction buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1 mM DTT, 5% glycerol) and incubated at room temperature for 1 h. The mix was thoroughly washed with the interaction buffer and the putative protein complex was eluted in 4× SDS sample buffer and heated to 98 °C for 3 min.
Western Blot Analysis.
For Western blotting analysis, the samples were first subjected to 10% SDS/PAGE and subsequently, transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane is then blocked with 5% semiskimmed milk powder at room temperature and probed with the respective primary antibodies. Anti-GFP (Abcam; dilution 1:6,000), anti-HA (Roche; dilution 1:4,000), and anti-SUMO1/2 (generated against AtSUMO1 in rabbit; dilution 1:5,000) antibodies were used for immunoblotting the IPs and co-IPs. Anti-GST (Merck; dilution 1:6,000) and anti-His antibodies were used for in vitro experiments. Secondary horseradish peroxidase-conjugated antibodies (anti-rabbit for anti-GFP and anti-SUMO1/2 antibodies, anti-rat for anti-HA and anti-GST antibodies, and anti-mouse for anti-His antibody) were applied before developing the blots with X-ray film using an automated developer.
To detect phosphorylation state of WRKY33, Phos-tag analysis was done as per the manufacturer’s protocol. Briefly, 5 μM phos-tag (Wako) and 10 μM MnCl2 were included in the resolving gel mix during preparation. The SDS/PAGE was run at 50 mV at 4 °C and the gel was incubated with EDTA (1 mM) containing transfer buffer and then washed three times with transfer buffer without EDTA before subjecting for overnight transfer. Western blotting was done as indicated above.
ChIP Assay.
ChIP was conducted as described in Chromotek protocol for A. thaliana with the following modifications: 1 g (fresh weight) of 12-d-old Arabidopsis seedlings, treated with 1 μM flg22 for 2 h or mock-treated (control) were harvested and cross-linked for 15 min under vacuum in 1% formaldehyde on ice. For processing, samples were sonicated using Covaris M220 (peak power = 75.0, duty factor = 5.0, cycles/burst = 200, at 7 °C) for 6 min. Five percent volume was taken as input, and the remaining sample volume was incubated with 25 µL anti-GFP beads (Chromotek) for 2 h at 4 °C. The samples were then washed and sample elution was carried out according to manufacturer’s protocol. One microliter of input or immunoprecipitated sample was used for quantitative real-time PCR as mentioned above. Primer pairs used were: WRKY33 ChIP FP GATGATGAAAGCATTGAGCCG; WRKY33 ChIP RP TATTGCACGTCTGATTGGTGAA; PAD3 ChIP FP GAAGTTTACTGACGGCTTCC; PAD3 ChIP RP CCATTTCTAATATGAGACACTACC; At4g26410 ChIP FP GAGCTGAAGTGGCTTCCATGAC; At4g26410 ChIP RP; GGTCCGACATACCCATGATCC.
Statistical Analyses.
Statistical differences between samples were determined by Student’s t test two-tailed, unpaired with equal variance (computed with Microsoft Excel). Sample differences with P < 0.05 were considered to be statistically significant and is indicated in the figure legends as *P < 0.05.
Supplementary Material
Acknowledgments
We thank Tim Hawkins and Joanne Robson for help with confocal microscopy. This work was supported by a Ramalingaswami Re-entry grant, Department of Biotechnology, India (to V.V.), and European Research Council and Biotechnology and Biological Sciences Research Council, United Kingdom grants (to A.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021351118/-/DCSupplemental.
Data Availability
All study data are included in the article and supporting information.
References
- 1.Krysan P. J., Colcombet J., Cellular complexity in MAPK signaling in plants: Questions and emerging tools to answer them. Front. Plant Sci. 9, 1674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigeard J., Hirt H., Nuclear signaling of plant MAPKs. Front. Plant Sci. 9, 469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taj G., Agarwal P., Grant M., Kumar A., MAPK machinery in plants: Recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 5, 1370–1378 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng X., Zhang S., MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Lampard G. R., Macalister C. A., Bergmann D. C., Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 (2008). [DOI] [PubMed] [Google Scholar]
- 6.MacAlister C. A., Ohashi-Ito K., Bergmann D. C., Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Guan Y., et al., Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10, e1004384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueda M., et al., Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 31, 617–627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mithoe S. C., et al., Attenuation of pattern recognition receptor signaling is mediated by a MAP kinase kinase kinase. EMBO Rep. 17, 441–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao G., et al., Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng X., et al., Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bethke G., et al., Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 8067–8072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han L., et al., Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64, 114–127 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Bardwell A. J., Frankson E., Bardwell L., Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 284, 13165–13173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheridan D. L., Kong Y., Parker S. A., Dalby K. N., Turk B. E., Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J. Biol. Chem. 283, 19511–19520 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu D., et al., SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol. Plant 12, 215–228 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Orosa-Puente B., et al., Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362, 1407–1410 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Srivastava A. K., et al., SUMO suppresses the activity of the jasmonic acid receptor CORONATINE INSENSITIVE1. Plant Cell 30, 2099–2115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yates G., Srivastava A. K., Sadanandom A., SUMO proteases: Uncovering the roles of deSUMOylation in plants. J. Exp. Bot. 67, 2541–2548 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Liu L., et al., Two SUMO proteases SUMO PROTEASE RELATED TO FERTILITY1 and 2 are required for fertility in Arabidopsis. Plant Physiol. 175, 1703–1719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller M. J., Barrett-Wilt G. A., Hua Z., Vierstra R. D., Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 16512–16517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rytz T. C., et al., SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 30, 1077–1099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birkenbihl R. P., Diezel C., Somssich I. E., Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birkenbihl R. P., Kracher B., Roccaro M., Somssich I. E., Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29, 20–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Z., Qamar S. A., Chen Z., Mengiste T., Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Orosa B., et al., SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 9, 5185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava A. K., Zhang C., Caine R. S., Gray J., Sadanandom A., Rice SUMO protease Overly Tolerant to Salt 1 targets the transcription factor, OsbZIP23 to promote drought tolerance in rice. Plant J. 92, 1031–1043 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Verma V., Croley F., Sadanandom A., Fifty shades of SUMO: Its role in immunity and at the fulcrum of the growth-defence balance. Mol. Plant Pathol. 19, 1537–1544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castro P. H., et al., Revised nomenclature and functional overview of the ULP gene family of plant deSUMOylating proteases. J. Exp. Bot. 69, 4505–4509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrido E., Srivastava A. K., Sadanandom A., Exploiting protein modification systems to boost crop productivity: SUMO proteases in focus. J. Exp. Bot. 69, 4625–4632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conti L., et al., Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20, 2894–2908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villajuana-Bonequi M., et al., Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J. 79, 206–219 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bailey M., et al., Stability of small ubiquitin-like modifier (SUMO) proteases OVERLY TOLERANT TO SALT1 and -2 modulates salicylic acid signalling and SUMO1/2 conjugation in Arabidopsis thaliana. J. Exp. Bot. 67, 353–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van der Does D., et al., Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25, 744–761 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermkes R., et al., Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233, 63–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gareau J. R., Lima C. D., The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolesar P., Sarangi P., Altmannova V., Zhao X., Krejci L., Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 40, 7831–7843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y., Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan J., Zhang S., Stacey G., Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 5, 125–135 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.