Significance
Lipid droplets (LDs) are stored in the embryo throughout the preimplantation development. Yet, the role of LDs in the embryo remains unknown. Embryonic diapause (ED) is a temporary arrest of an embryo when it waits for the uterine receptivity signal to implant. We provide evidence that LDs play a crucial role in maintaining ED. Diapausing embryos exhibit increased release of exosomes reflecting elevated embryonic signaling to the mother. During ED, the decrease in lipid is caused by a switch from carbohydrate metabolism to lipid catabolism. We have also shown that presence of LDs in the oocytes of various mammals positively corelates with their species-specific length of diapause. Our results reveal the functional role of LDs in embryonic development.
Keywords: embryonic diapause, lipid droplets, blastocyst
Abstract
Embryonic diapause (ED) is a temporary arrest of an embryo at the blastocyst stage when it waits for the uterine receptivity signal to implant. ED used by over 100 species may also occur in normally “nondiapausing” mammals when the uterine receptivity signal is blocked or delayed. A large number of lipid droplets (LDs) are stored throughout the preimplantation embryo development, but the amount of lipids varies greatly across different mammalian species. Yet, the role of LDs in the mammalian egg and embryo remains unknown. Here, using a mouse model, we provide evidence that LDs play a crucial role in maintaining ED. By mechanical removal of LDs from zygotes, we demonstrated that delipidated embryos are unable to survive during ED. LDs are not essential for normal prompt implantation, without ED. We further demonstrated that with the progression of ED, the amount of intracellular lipid reduces, and composition changes. This decrease in lipid is caused by a switch from carbohydrate metabolism to lipid catabolism in diapausing blastocysts, which also exhibit increased release of exosomes reflecting elevated embryonic signaling to the mother. We have also shown that presence of LDs in the oocytes of various mammals positively corelates with their species-specific length of diapause. Our results reveal the functional role of LDs in embryonic development. These results can help to develop diagnostic techniques and treatment of recurrent implantation failure and will likely ignite further studies in developmental biology and reproductive medicine fields.
Lipid droplets (LDs) present in cells are used for energy production and signaling (1, 2). Persistent presence of LDs throughout the preimplantation period during embryonic development is found in a variety of mammalian species and suggests an important and as yet undiscovered function. Previous data showed that LDs stored in the mammalian egg are nonessential, as their removal from one cell-stage embryos did not influence further development (3). We propose that blastocysts use LDs to maintain their survival during embryonic diapause (ED). ED is a temporary arrest of the embryo while it waits for a signal to implant. As a result, the normal gestation time is extended to a species-specific time. This reproductive strategy is commonly used by over 100 mammalian species (4) and may also occur in normally “nondiapausing” mammals (5–7). Yet, the molecular mechanisms underpinning ED remain elusive.
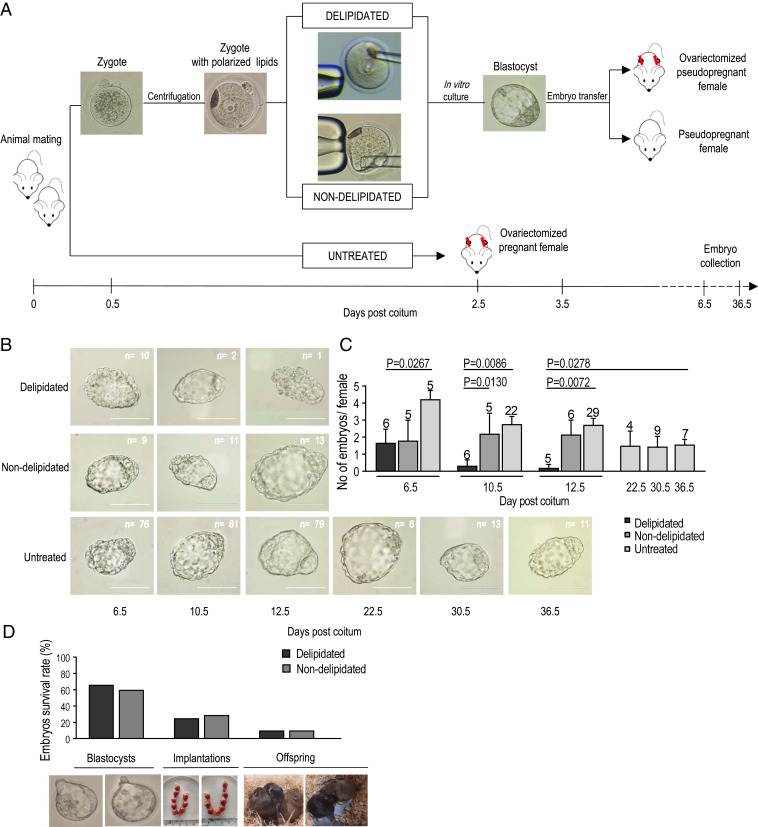
In mice, ovarian estradiol secretion initiates implantation. Ovariectomy of mated females performed before the ovarian estradiol surge (i.e., 4 d postcoitum [dpc]) induces ED and avoids embryo implantation (8). Therefore, to test our hypothesis that LDs are essential for maintenance of ED, LDs were mechanically removed from centrifuged one cell-stage embryos (Fig. 1A and Movie S1), and then, at the blastocyst stage, embryos were surgically transferred to ovariectomized pseudopregnant females to verify their survival during ED.
Fig. 1.
Delipidated embryos are unable to survive during the progression of ED. (A) Workflow of the generation of delipidated and nondelipidated embryos (control manipulation—removal of cytoplasm instead of lipids), their transfer to pseudopregnant ovariectomized females (number of transferred embryos per female = 16), and subsequent collection. Untreated embryos were developed entirely in vivo in ovariectomized females. (B) Representative images of blastocysts collected at progressive days of ED show temporal survival of delipidated and control (nondelipidated, untreated) embryos during ED. The proportions of collected delipidated and nondelipidated embryos were similar: 10 of 96 (10.42%) vs. 9 of 80 (11.25%) at 6.5 dpc. Then, they reduced drastically for delipidated embryos: 2 of 96 (2.08%) vs. 11 of 80 (13.75%) at 10.5 dpc and 1 of 80 (1.25%) vs. 13 of 96 (13.54%) at 12.5 dpc. (Scale bars, 100 μm.) (C) Histogram shows that only 3 (1.7%) delipidated embryos of 176 transferred were still present in the uterus of ovariectomized females at 10.5 and 12.5 dpc. At the same time, untreated embryos were collected from the uteri of ovariectomized mice without any significant decline in their survival until 36.5 dpc, which implies a crucial role of the lipid fraction in ED extension (demonstrated in the last part of this study) (Fig. 4). Numbers above the columns indicate the numbers of females carrying diapausing embryos. Values represent mean ± SEM; Mann–Whitney test. (D) Graph and representative images (Lower) demonstrate similar survival of delipidated and nondelipidated embryos at blastocyst stage (123 of 206 vs. 85 of 128) and following implantation (21 of 72 vs. 14 of 57) and delivery (3 of 30 vs. 2 of 20; χ2 test, no significant differences).
We found that diapausing blastocysts did not survive after the removal of LDs (Fig. 1 B and C). For further evidence that LDs play a major role in ED and to show that LDs are dispensable when direct implantation takes place (i.e., without ED), we show similar developmental progression of delipidated and nondelipidated embryos following their transfer to nonovariectomized pseudopregnant females (Fig. 1D). This demonstrates that LDs are dispensable when the embryo is not forced to diapause. Delipidated embryo survival remained unaffected during the first 4 d of development (9), for the blastocysts at 4.5 dpc, and subsequently, for blastocysts entering ED, at 6.5 dpc (Fig. 1C). Our finding confirms that embryos at early stages do not generally rely on LD reserves (10). Indeed, studies over decades have documented that pyruvate and subsequently (i.e., around embryo compaction), glucose are the predominant energy substrates used by mammalian embryos between the zygote and the blastocyst stage (11–14). Adenosine triphosphate (ATP) produced by oxidative phosphorylation of exogenous pyruvate is preferred by the embryo during its first stages of development in vitro (12, 13). In the absence of exogenous energy substrates, embryo development quickly arrests, usually within one to two cleavage divisions, which indicates that the cleaving embryo cannot substitute carbohydrates by using available stores of other substrates such as LDs (15).
The utilization of energy resources in early cleavage-stage embryos is limited because the embryo starts to grow and increase its protein content later, at the blastocyst stage (16). LDs may play a role in membrane biogenesis since a large increase in membrane surface area is observed in cleaving embryos. However, this activity is likely to account for only a small portion of the available lipid stores, and mechanical delipidation does not eliminate all available lipids from the cell (9). Altogether, our observation uncovers the essentiality of LDs for blastocyst viability maintenance during ED. In other developmental stages, either precedent or subsequent to ED, delipidated embryos develop normally.
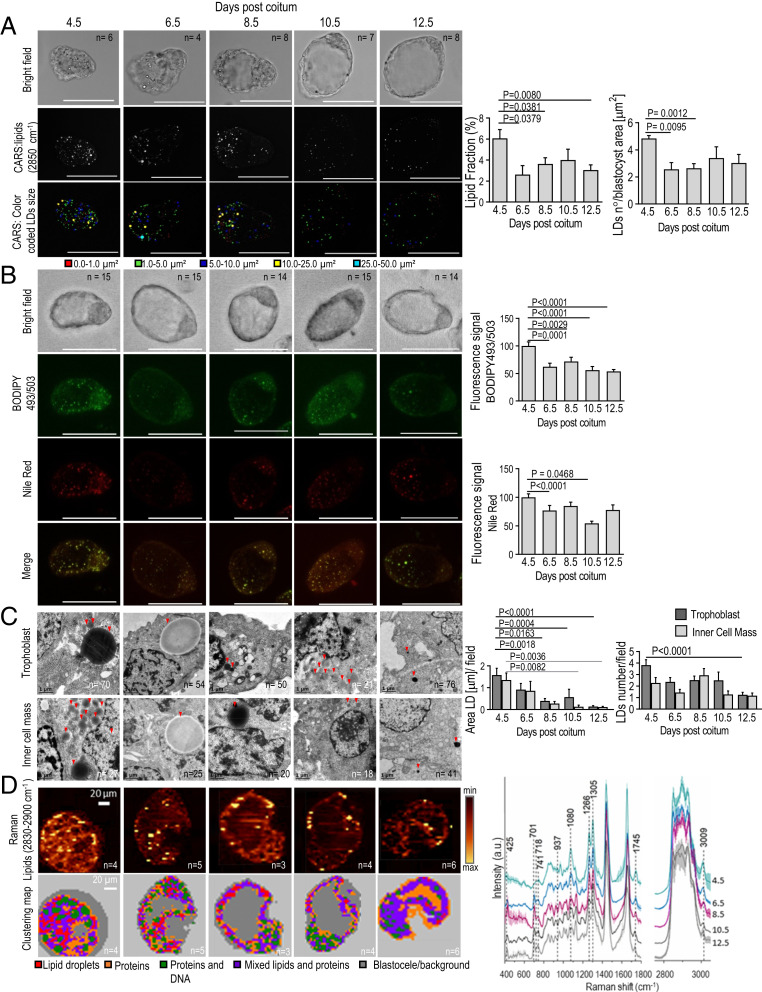
We then asked whether the LD fraction diminishes and/or changes its content throughout the progression of ED. To do this, blastocysts were collected from female mice at different dpc: 4.5 (day of implantation, nondiapausing control embryos), 6.5, 8.5, 10.5, and 12.5 (diapausing embryos from ovariectomized females). The blastocysts were then analyzed for LDs content. We found the reduction of LDs fraction (i.e., LDs area/blastocyst area) during the ED period by their direct visualization (via coherent anti-Stokes Raman scattering [CARS]) (Fig. 2A); by confocal fluorescence microscopy using two different markers: 4,4-difluoro1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503) and Nile Red (Fig. 2B); by transmission electron microscopy (TEM) (Fig. 2C); and by confocal Raman imaging (Fig. 2D). LDs number/blastocyst area also decreased during ED (Fig. 2 A and C).
Fig. 2.
LDs diminish as ED progresses. (A) Reduction of LDs in the diapausing blastocyst demonstrated by CARS: the legend below the images indicates the color associated with each of the five LDs range/size. n = number of blastocysts. (Scale bars, 100 μm.) Values represent mean ± SEM; two-tailed unpaired Student’s t test performed after data normalization by arcsine transformation. (B) Blastocysts stained by BODIPY and Nile Red analyzed by confocal fluorescence microscopy. (Scale bars, 100 μm.) Histograms show the reduction of LDs. Values represent mean ± SEM; one-way ANOVA after Shapiro–Wilk normality test. (C) TEM analysis of blastocysts’ cells (i.e., trophoblast and inner cells mass). Red arrowheads indicate LDs. n = number of fields counted. (Scale bars, 1 μm.) Histograms show the progressive reduction of LDs area and number in diapaused blastocysts. Values represent mean ± SEM; Kruskal–Wallis test. (D) Confocal Raman imaging. Upper row: Integration Raman maps for lipid bands (range 2,830–2,900 cm−1). Lower row: KMC maps of blastocyst showing spectra grouping into five classes color coded as indicated by the legend below. (Scale bars, 20 μm.) Graph shows average LDs spectra of blastocysts obtained by KMC analysis. Bands at 425 cm−1, 701 cm−1, and 741 cm−1 relative to cholesterol were present at 6.5, 8.5, 10.5, and 12.5 dpc, but not at 4.5 dpc. Band at 1,745 cm−1, indicative for esterified form of the fatty acids, was temporally reduced in the diapausing blastocyst. Band at 3,009 cm−1 and the I1266/I1305 ratio, relative to unsaturated fatty acids, indicates that unsaturated lipids were temporally decreasing in the diapausing blastocyst. Band at 718 cm−1, present in blastocysts at 10.5 and 12.5 dpc, suggests increase in phospholipids.
In addition, important changes in the LDs composition occurred throughout the progression of ED as demonstrated by various spectroscopic techniques (Fig. 2D and SI Appendix, Fig. S1). Each time point of ED had its own characteristic “lipid fingerprint.” We were able to distinguish them using Raman spectroscopy and Matrix-Assisted Laser Desorption Ionization–tandem time of flight mass spectrometry. As ED progressed, the embryo contained more phospholipids and fewer fatty acids. Furthermore, the degree of lipid saturation increased during ED. The most prominent changes in the Raman spectra of LDs corresponded to a decrease in the esterified form of fatty acids (e.g., tripalmitolein and trilinolein) and to increased cholesterol (Fig. 2D). Analysis of microvesicles isolated from the embryo collection media, which can be considered a representation of embryonic content (bearing in mind nonactive uterus of ovariectomized mice), further demonstrated time-dependent variations in lipid fraction with regard to triglycerides, fatty acids, and cholesterol (SI Appendix, Fig. S1). Together, these data clearly show time-related consumption of lipids, which allowed us to distinguish progressive stages of ED.
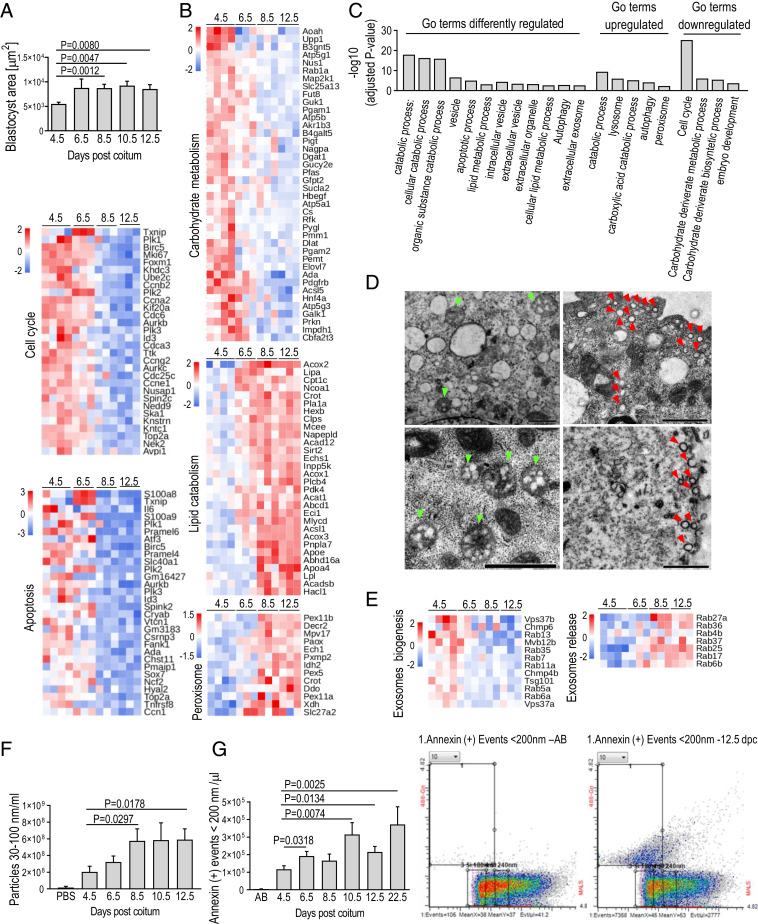
We next sought to understand the molecular pathways regulating utilization of LDs by the diapausing embryos. A transcriptional analysis of the diapausing mouse embryos demonstrated a temporal association between down-regulated carbohydrate metabolism pathways and up-regulated lipid metabolism pathways (Fig. 3 A and B). Carbohydrate metabolism was prevalent only in blastocysts at 4.5 dpc, which demonstrates that the embryo, when ready to implant, relies heavily on hydroxylic acid metabolism and that carbohydrates are the main ATP source. Here, we demonstrated that when diapause occurs, the situation changes, and the switch from carbohydrate to lipid metabolism occurs in the embryo between the beginning of diapause, at 6.5 dpc, and the subsequent evaluated time point, day 8.5, when the growth of the embryo had ceased (Fig. 3A). This change to lipid utilization occurs concomitantly with cessation of blastocyst growth, which is completed in mouse embryo by 8 dpc (17). With the cessation of blastocyst growth, fatty acid oxidation is the most pronounced cellular activity in the diapausing embryo (Fig. 3B), as evidenced by the two most highly up-regulated pathways: mitochondrial fatty acid oxidation and branched-chain amino acid catabolism (associated with increased fatty acid oxidation) (18). The diapausing blastocyst’s functional use of LDs is further evidenced by the up-regulation of genes characteristic of carboxylic acid utilization in peroxisomes (19) (Fig. 3A). Both autophagic and lysosomal genes, which play an important part in the early steps of lipid degradation (20), were heavily up-regulated during ED (Fig. 3). The switch to lipid utilization by the lysosomal (through autophagy) and further peroxisomal (Fig. 3 A and B) digestion of LDs in the diapausing embryo likely occurs because the carbohydrates are no longer available in the embryonic cells. Shifting the metabolism from reliance on carbohydrates to lipids as a consequence of starvation is a known adaptation of the cells (2). However, based on previous reports, so-called “reactivated” blastocysts (i.e., embryos that are answering a uterine receptivity signal following ED) (21, 22) switch back to carbohydrate metabolism. This may suggest that lipids are less important during implantation. However, studies focused on LDs involvement in embryo implantation were not performed. The role of LDs in cells constituting inner cell mass (ICM) and trophectoderm of those reactivated blastocysts is also not known. In diapausing blastocysts, similar LDs reduction in both lineages is observed (Fig. 2C). Having in mind that reactivated (e.g., implanting) blastocyst relies on carbohydrates (21, 22) and that its ICM displays relatively quiescent metabolism in comparison with that of trophectoderm (23), it would be interesting to know whenever LDs play any role in ICM maintenance during implantation. In the next step, we sought to determine for what purpose the diapausing blastocyst uses the energy generated by LDs catabolism. LDs are specialized energy reservoirs that take the form of triacylglycerol and sterol esters. The breakdown of LDs into free fatty acids may, therefore, both provide energy to the cell and activate cell signaling (18–20). Exosomes are small vesicles that upon release into the extracellular environment, play an important role in cell-to-cell communication (24). Here, we have shown that exosome production and release intensified substantially in the diapausing embryo (Fig. 3 C–G). Exosome biogenesis, multivesicular bodies transport, and exosome release as well as extracellular and intracellular vesicles are all processes most affected during ED, as documented by RNA sequencing analysis and by direct observations using TEM, nanotracking analysis, and flow cytometry (Fig. 3). Of relevance, the level of cholesterol, which plays a crucial role in signal transduction in mammalian cell (18), increased progressively during ED (Fig. 2D and SI Appendix, Fig. S1C). Strong overexpression of the genes involved in cholesterol transport and signaling pathways (i.e., APOA4 and APOE) as well as in peroxisomal genes regulating intracellular cholesterol transport (i.e., ABCD1, PEX5, PEX11) and in exosomes biogenesis and release further shows that communication via exosomes is strongly activated during ED (Fig. 3E).
Fig. 3.
Lipids are catabolized and exosomes are intensively produced during ED. (A, Top) Histogram shows the cessation of diapausing blastocyst growth at 8.5 dpc. Mean ± SEM; two-tailed unpaired Student’s t test. (A, Middle and Bottom) The heat maps demonstrate down-regulation of genes controlling cell cycle and apoptosis activity in diapausing blastocysts at 8.5 and 12.5 dpc. (B) The heat maps indicate that diapausing blastocyst down-regulates carbohydrate metabolism while up-regulates lipid catabolism and peroxisomal activity. (C) Selected GO of differently regulated pathways in diapausing blastocysts at 12.5 vs. 4.5 dpc. (D) Representative TEM images show accumulation of spheroidal multivesicular bodies (green arrowheads) from 400 to 600 nm, containing numerous vesicles in the trophoblasts at 4.5 and 6.5 dpc, while the accumulation of numerous cup-shaped spherical vesicles sized <200 nm (red arrowheads) is present at 12.5 dpc. (Scale bars, Top Left, 0.5 µm; Top Right and Bottom, 1 µm.) (E) Sequential down- and up-regulation of genes involved in exosomes biogenesis, multivesicular bodies transport, and exosomes release. (F) Histogram shows an increased concentration of exosomes detected by NTA of embryo collection media. Mean ± SEM; two-tailed unpaired Student’s t test. (G) Histogram and representative cytograms of annexin-positive particles (<200 nm; exosomes marker) in embryo collection media at 12.5 dpc. Histogram shows an increased concentration of annexin-positive particles (<200 nm/μL) in uterine fluid collected at 6.5, 10.5, 12.5, and 22.5 dpc vs. 4.5 dpc. n samples/dpc ≥ 14. Mean ± SEM; two-tailed unpaired Student’s t test. AB, annexin in buffer.
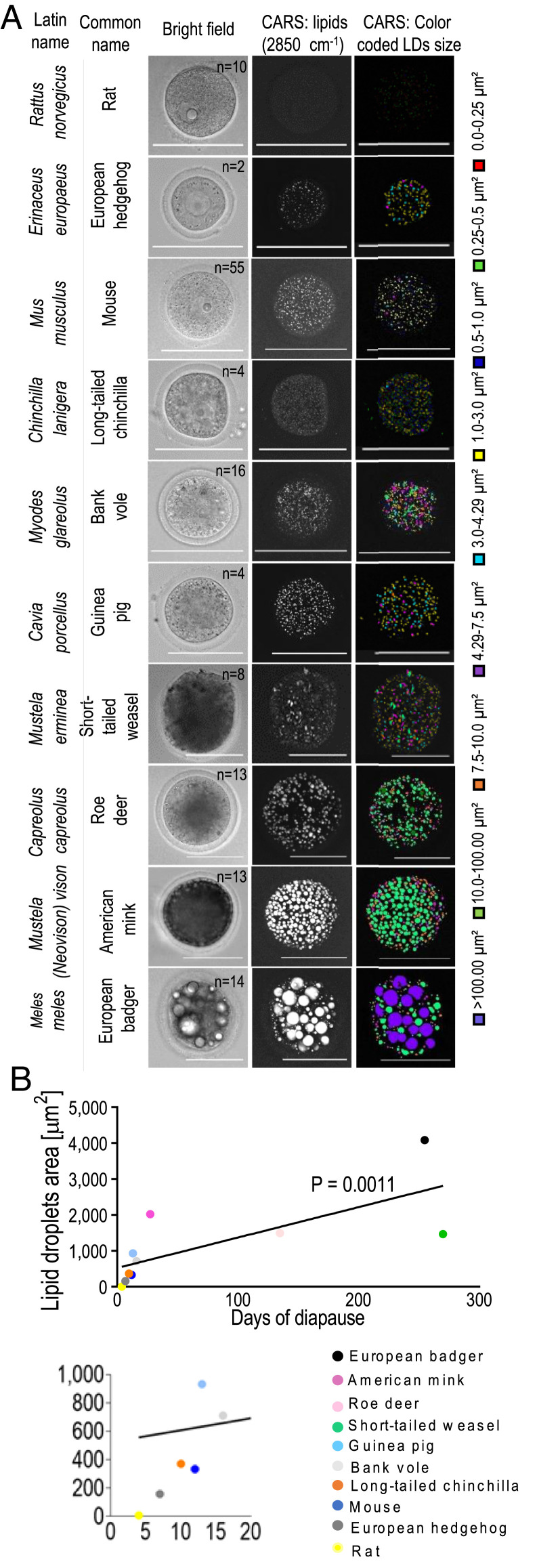
Finally, we sought to determine whether ED duration depends on the amount of lipids. To address this, we quantified the amount of intracellular lipids accumulated in the oocytes of several mammalian species. We found that oocytes of different mammalian species contain different amounts of lipids (Fig. 4). Species with very long ED (e.g., mustelids, with ED lasting up to 9 mo) were found to have a very high level of lipids in their oocytes. On the other hand, oocytes from species with ED lasting just few days (e.g., rodents) were characterized by a low quantity of lipids (Fig. 4 and Movie S2). Here, we showed that the total amount of lipids in the oocytes of various mammals positively correlates with their species-specific length of ED (Fig. 4).
Fig. 4.
Storage of lipids in oocytes from various mammals is positively correlated with the duration of ED in a given species. (A) LD quantity and size vary in mammalian oocytes, as imaged by CARS microscopy. Columns from left to right are Latin names of species, common name of species, bright field, maximum intensity projection of CARS signal at 2,850 cm−1, LDs color-coded size mask, and relative legend. Legends indicate the color associated with each of the nine LDs range sizes. n = number of oocytes. (Scale bars, 100 µm.) (B, Upper) Graph shows a positive correlation between LDs amount and the length of ED as documented for the examined species (38–43). (B, Lower) The expansion of the graph is presented as the smaller graph evidencing an increased separation of the individual species. Spearman correlation coefficient.
It is interesting to note that rat oocytes contain less lipids than mice (Fig. 4B). It is not clear whenever those differences between oocytes of two rodent species are actually reflected by subtle dissimilarities in the extension of ED. One report from 1966 documented a similarly short extension of diapause (lasting 2 to 5 d) in lactating rats and mice (25). This previous report refers to natural occurrence of ED (25), where no more than a few days pregnancy extension was necessary for successful pregnancy completion, either for rats or mice. In our experimental settings, mouse embryos are actually able to extend diapause for more than 1 mo (Fig. 1C). It is not known whether rat embryos are similarly able to survive for such a long period.
One explanation of the differences between the quantity of lipids in rat and mice oocytes detected by CARS (Fig. 4) can rely on species-specific composition of LDs, although it is less likely that closely related species will differ significantly. It is possible that the technical limitation of CARS, in detection of particular types of lipids rather than their total amount, may account for such differences. Preferential detection of particular types of lipids in oocytes was reported by other microscopic techniques (26, 27).
Interestingly, oocytes from all mammals, not only those naturally diapausing, also possess LDs (10). Considering that the embryos of various naturally nondiapausing mammalian species may enter ED, if there is a lack of uterine receptivity signal (5–7, 28, 29), it is not surprising that in all mammals, ooplasmic LDs should be present. It is possible LDs act as a ballast that is not needed during prompt implantation and development but are indispensable during ED. As a result, the conservation of adequate content of lipids will be of relevance for all mammalian species. A similar strategy is also observed in different classes of animals, including insects, fish, and frogs, in which LDs provide a reservoir for generating energy for the developing embryo when the organism itself is not yet able to feed. In plant seeds, accumulated lipids also support the development of the embryonic plant before the onset of autonomous feeding (i.e., photosynthesis).
In conclusion, this study answers two fundamental developmental biology questions. What is the role of LDs in the preimplantation embryos? Why are there species-specific differences in the distribution of LDs? Attempts to answer both questions have been done before (3, 10, 30, 31). However, none of the previous studies considered ED as a stage when LDs are likely to be utilized. Our demonstration of the interplay between LDs and ED provides the long sought for answer regarding functional significance of lipids persisting in preimplantation mammalian embryos with implications for developmental biology and reproductive medicine fields. Our study shows that the diapausing blastocyst focuses its activity on signaling through exosomes. Persisting intensive signaling augments the chance of the embryo to be recognized by the mother and facilitates the establishment of pregnancy in the situations of disrupted maternal receptivity. In general, ED tends to increase embryo developmental potential and pregnancy rate (32–34). Thus, we propose that forcing the embryo to “wait” for uterine receptivity by the simulation of ED (for example, through asynchronous embryo transfer when assisted reproduction technologies are applied) can increase the success of implantation. Asynchronous transfer of frozen–thawed embryo can be an attractive solution in case of insufficient uterine receptivity and consequent recurrent implantation failure, which is considered to be one of the primary causes of low pregnancy rate (35, 36).
Materials and Methods
All chemicals, unless otherwise indicated, were obtained from Sigma-Aldrich.
Animal Studies.
All experimental procedures were conducted according to the guidelines of European Community Regulation 86/609 and conformed to the Polish Governmental Act for Animal Care. Animal procedures were conducted at the Malopolska Center of Biotechnology and the Institute of Zoology and Biomedical Research of Jagiellonian University (Kraków; permission no. 123/2018 approved by II Local Ethical Commission of Kraków). Animal experiments were performed on 11- to 14-wk-old C57BL/6xCBA mice. Animals were maintained in a temperature- and light-controlled room (22 °C and 12-h light–dark cycle) and were provided with food and water ad libitum. The experiments were not randomized, and the investigators were not blinded to experiment allocation or outcome assessment.
Embryo Manipulation.
Females were intraperitoneally (i.p.) injected with 5 international units (IU) pregnant mare serum gonadotropin followed 48 h later by an i.p. injection of 5 IU human chorionic gonadotrophin (hCG) to induce superovulation. Females were paired overnight with stud males; day of vaginal plug was considered as 0.5 dpc. The zygotes were isolated from ampullae 23 h after hCG administration in M2 medium. Lipid polarization in the zygotes was obtained by centrifugation, with a preincubation of 10 min, in modified M2 containing higher concentration of bovine albumin serum (1%) and 10 µg/mL cytochalasin B at 7,000 × g for 12 min at 37 °C. Centrifugated zygotes were allowed to recover for 1 h in KSOM (Millipore) and then manipulated. The zygotes were manipulated under a Leica inverted microscope equipped with Narishige micromanipulators (MO-108). The zygotes were manipulated based on a two-step protocol: the zona cut in KSOM and the removal of the lipids fraction or the same size fraction of cytoplasm in KSOM with 10 µg/mL cytochalasin B for the production of delipidated and nondelipidated embryos, respectively. The resulting embryos were in vitro cultured in KSOM at 37 °C at 7% O2 and 5% CO2 until 3.5 or 4.5 dpc.
Diapause Induction and Collection of Embryos.
The ED was induced as previously described (7, 8). To induce diapause of naturally conceived and developed embryos, pregnant females at 2.5 dpc were ovariectomized and daily subcutaneously injected with Depo-Provera (1 mg medroxyprogesterone acetate/0.1 mL sterile saline solution; Pfizer). Similarly, to induce diapause on manipulated embryos, pseudopregnant females were ovariectomized and injected with Depo-Provera after asynchronous transfer of manipulated embryos in vitro cultured until 3.5 dpc. According to the addressed aim and analysis, the resulting diapausing embryos were collected by uterine flushing with phosphate-buffered saline solution and 0.4% polyvinylpyrrolidone (0.4% PBS-PVP), 1 mL for each horn, and then, embryos were processed for further analysis. The embryo collection media of each horn were centrifuged at 3,500 × g for 10 min at 4 °C to remove cellular fraction and blood contamination. The supernatant was frozen at −80 °C until analysis.
Induction of the Implantation of Manipulated Embryos.
To induce the implantation, in vitro cultured manipulated embryos at 3.5 dpc were transferred into asynchronous pseudopregnant females. Recipient females were killed at 12.5 dpc for the evaluation of the implantation sites or allowed to deliver spontaneously for assessment of the offspring rate.
Isolation of Extracellular Vesicles.
Extracellular vesicles (EVs) were obtained from the supernatant of embryo collection medium by differential centrifugation, according to previous protocols (37). The same protocol of EV isolation was used for all samples considered in this study. To remove debris, macroparticles, and apoptotic bodies, the supernatant was thawed and centrifugated at 20,000 × g for 30 min at 4 °C. The resulting pellet enriched in EVs was resuspended in 15 µL of phosphate-buffered saline (PBS) and stored at −80 °C. Then, the supernatant was transferred to 11 × 34-mm polycarbonate centrifuge tubes (Beckman Coulter Inc.) to perform the first ultracentrifugation step at 100,000 × g for 2 h at 4 °C in an MLA-130 (Beckman Coulter Inc.) rotor using an Optima MAX-XP ultracentrifuge (Beckman Coulter Inc.); the supernatant was removed completely, and the obtained pellet was washed in 1 mL of PBS and centrifuged at 100,000 × g for 1 h at 4 °C. The final pellet enriched in small EVs (exosomes) was resuspended in 15 µL of PBS. The pellet of exosomes was added to the pellet of EVs and stored at −80 °C for nanoparticle tracking analysis (NTA), flow cytometry, and Fourier transform–Raman analysis.
Collection of Oocytes from Various Species.
Ovarian oocytes were collected postmortem from mammalian species with known durations of ED (38–43). None of the animals were euthanized specifically for the purpose of the paper: mice, rats, and bank voles were available from the animal facilities of Jagiellonian University; American minks were from farms; long-tailed chinchillas, guinea pigs, and short-tailed weasels were from veterinary clinics; and roe deer and European badgers were from the Polish Hunting Association. The ovaries were kept in PBS with antibiotics at 4 °C during transport. The oocytes were collected by follicular aspiration and then, by slicing of the ovaries; then, the oocytes were washed in 0.4% PBS-PVP and fixed in 4% paraformaldehyde for 20 min. After fixation, the oocytes were washed twice in 0.4% PBS-PVP for 5 min and stored in 0.4% PBS-PVP at 4 °C until analysis.
CARS Microscopy.
The multimodal nonlinear microscope used in this study consisted of a Leica DMi8 inverted microscope equipped with the Leica TCS SP8 CARS module and the Leica SP8 confocal module (Leica Microsystems). The Leica TCS SP8 CARS uses a tunable pump laser with a tuning range of 780 to 940 nm combined with a Stokes laser at 1,064 nm provided by Laser picoEmerald and integrated with a 750-mW power optical parametric oscillator. At the time of analysis, the sample was placed on a Lab-Tek chamber (Thermo Scientific) after fixation in 0.4% paraformaldehyde for 20 min. To detect the LD signal, the optical parametric oscillator’s wavelength was tuned to 816.7 nm to serve as the pump beam, in combination with the 1,064-nm Stokes beam to probe the CH2 stretch vibration. All signals were detected using a photomultiplier tube by collecting the photons in a forward direction through a 40× objective. Each sample was scanned across all of its sizes in the z axis using a z step of 1.5 µm. For the analysis, all stacks from a single sample were processed to obtain a maximum projection image. The threshold was adjusted specifically for all samples to collect all positive signals. When necessary, LDs were separated manually in cases of obvious pixel overlay. Statistics regarding the LDs area and number were generated automatically by LAS X software (Leica Microsystems). The LAS X software was used to apply color-coded LD size masks to the maximum projection images for graphical observation of the LDs’ size distribution of the analyzed samples. The blastocyst area was calculated using ImageJ (NIH) due to limitations of the LAS X software.
LD Staining by BODIPY 493/503 and Nile Red.
For LD staining, the fixed blastocysts were incubated with 1 µg/mL BODIPY 493/503 (Invitrogen) and 10 µg/mL Nile Red in 0.4% PBS-PVP for 1 h and then washed three times in 0.4% PBS-PVP. Mounted specimens were analyzed with a confocal ZEISS LSM 880 Confocal Laser Scanning Microscope using a 20× Zeiss Plan-Apochromat Infinitive corrected objective with a numerical aperture of 0.8. Relative fluorescence signal intensities were analyzed using ImageJ and normalized to control (4.5 dpc).
Confocal Raman Imaging.
Raman imaging was performed with a WITec alpha300 Raman spectrometer. At the time of analysis, the embryos were thawed, fixed in 4% paraformaldehyde for 20 min, washed first in 0.4% PBS-PVP, and then, placed on a calcium fluoride slide in drops of 300 µL of 0.4% PBS-PVP. The blastocysts were measured with a laser line of 532 nm, an immersive objective (60×), and 600-grooves∙mm−1 grating. Step sizes equal to 3 µm and integration time of 0.5 s were applied. Raman images were generated for lipids (2,830 to 2,900 cm−1). Data preprocessing and analysis were performed with WITec Project Plus software. Preprocessing included cosmic ray spike removal and background subtraction (third-order polynomial). K-Means Clustering (KMC) analysis enabled the spectra to be grouped into classes, each of which represented the main blastocyst biochemical composition. Additionally, averaged spectra of each class were generated showing their characteristic spectral profile.
TEM.
Diapaused blastocysts were fixed in 2.5% glutaraldehyde overnight at 4 °C, washed in 0.1 M cacodylate buffer, and treated in 1% OsO4 for 1 h. Samples were dehydrated through a graded series of ethanol and then infiltrated in pure resin (Poly/Bed 812; Polysciences, Inc.) through growing concentrations of resin in 100% propylene oxide, propylene oxide:resin 3:1 for 1 h and 1:1 overnight at room temperature. The samples were transferred into pure resin, and polymerization was carried out at 60 °C for 3 d. Sections of ∼70 nm were cut by an ultramicrotome (Leica) contrasting with uranyl acetate and lead citrate. Images were captured by Tecnai Osiris 200 kV (FEI). LDs area was quantified using the ImageJ segmentation tool (NIH).
Messenger RNA (mRNA) Sequencing.
Poly (A)+ RNA was isolated from individual blastocysts using the Dynabeads mRNA DIRECT Kit (Invitrogen). mRNA integrity was assessed by PCR, and complementary DNA (cDNA) was prepared from the mRNA samples using a SMARTer Ultra Low Input RNA Kit for Illumina Sequencing (Takara). The cDNA was used to generate the libraries for sequencing utilizing Nextera XT DNA Library Preparation Kits (Illumina). The sequencing was performed on a NextSeq 500 system (Illumina). The quantity and quality of the RNA were evaluated with an Agilent 2100 Bioanalyser (Agilent) system, and only samples with an RNA integrity number (RIN) > 7.5 were selected for further library preparation and sequencing experiments. A library was prepared from each sample using an Ion Total RNA-Seq Kit v2 (Thermo Fisher).
RNA Sequencing (RNA-Seq) Data Analysis.
Raw reads were checked for quality using FastQC software (Babraham Bioinformatics) filtered to remove accidental adapter sequences and low-quality reads and mapped against the Mus musculus GRCm38 genome assembly using TopHat2 (44) software set for paired-end reads. The reads mapped to separate genes were counted using HTSeq software [with “Union” mode (45)], and for additional verification, pseudoalignment with the use of Kallisto was performed as well (46). In all cases, differential expression analysis was performed with DESeq2 (47). Only genes with an adjusted P value < 0.05 (after false discovery rate [FDR] correction using the Benjamini–Hochberg procedure, q value) were considered differentially expressed. Comparisons were made between control embryos (4.5 dpc) and embryos at subsequent time points of diapause (6.5, 8.5, and 12.5 dpc). Differentially expressed genes (adjusted P < 0.05) were listed, functionally annotated, and analyzed in terms of overrepresented biological processes/pathways using g:Profiler—a web-based toolset (48). For this purpose, the information obtained from the Gene Ontology (GO), Wikipathways, Panther Pathways, and Reactome Pathways databases was used. All overrepresentation tests were performed with respect to all annotated M. musculus genes (genome) with multiple testing correction (FDR).
NTA.
NTA was performed using a NanoSight LM10 microscope (Malvern Instruments) equipped with an SCMOS camera (Hamamatsu Photonics). For the analysis, 10-µL uterine fluid pellet was diluted in 1 mL of PBS to fit the optimal working range (particles per frame) of the instrument. Five videos were recorded for each sample, with the camera level set at 14 and the detection threshold set at five. The camera level and overall settings were selected as optimal for the accurate detection of EVs, following the manufacturer’s recommendations and as confirmed during instrument setting. Videos were analyzed with NTA software version 3.1 build 3.1.45 to determinate the concentration of measured particles.
Flow Cytometry Analysis.
The annexin A5/fluorescein isothiocyanate (FITC) and the CD63/PE Cyanine7 assay were performed using the A50 Micro (Apogee) flow cytometer. The Annexin A5/FITC (Beckman Coulter) was used to label the phospatidylserine in the outer membrane leaflet of EVs. A 2-µL uterine fluid pellet was incubated with Annexin A5/FITC and 100 µL of staining buffer for 15 min on ice. After incubation, another 100 µL of staining buffer was added. A dot plot cytogram with green fluorescence signal for Annexin A5/FITC vs. medium-angle light scatter (MALS) was used to collect the data. A calibration bead mix (Apogee) containing beads of known sizes was used to scale the MALS signal first. Populations of Annexin A5-positive events were divided on the dot plot between three ranges: 1) <200 nm, 2) 200 to 500 nm, and 3) 500 to 650 nm. The absolute number of Annexin A5-positive events per microliter for each size range was calculated by the cytometer’s software after the acquisition of 2.5 to 3 µL of sample.
Supplementary Material
Acknowledgments
We thank Profs. M. Ward, M. Żylicz, J. Heddle, and D. Mossakowska Earnshaw for critical reading of the manuscript. We also thank Profs. H. Okarma, T. Motyl, and K. Zub and Drs. P. Niedbała and A. Zoń for the gift of ovaries from the nondomestic mammals. This research was funded by National Science Centre of Poland Grants 2018/29/N/NZ3/02177 (to R.A.), 2016/22/M/ST4/00150 (to M. Barańska), and 2016/21/B/NZ3/03631 and 2019/35/B/NZ4/03547 (to G.E.P.); by Leading National Research Centre Grant KNOW/IGHZ/RMK/PhD/2016/07 (to G.E.P.); and by the European Union’s Horizon 2020 Research and Innovation Programme Grant 692185 (to G.E.P.). F.Z. is supported by Maria Skłodowska-Curie Action Individual European Fellowship Society and Enterprise Grant 834621. K.F. is a Diamond Grant fellow (Ministerstwo Nauki i Szkolnictwa Wyższego; Grant 0175/DIA/2019/28).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018362118/-/DCSupplemental.
Data Availability
The FASTQ file for RNA-Seq data has been deposited in the National Center for Biotechnology Information Sequence Read Archive database under BioProject (accession no. PRJNA63160).
References
- 1.Rambold A. S., Cohen S., Lippincott-Schwartz J., Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa A. D., Savage D. B., Siniossoglou S., Lipid droplet-organelle interactions: Emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 35, 91–97 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Nagashima H., Kashiwazaki N., Ashman R. J., Grupen C. G., Nottle M. B., Cryopreservation of porcine embryos. Nature 374, 416 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Renfree M. B., Fenelon J. C., The enigma of embryonic diapause. Development 144, 3199–3210 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Chang M. C., Reciprocal insemination and egg transfer between ferrets and mink. J. Exp. Zool. 168, 49–59 (1968). [DOI] [PubMed] [Google Scholar]
- 6.Murphy B. D., The role of prolactin in implantation and luteal maintenance in the ferret. Biol. Reprod. 21, 517–521 (1979). [DOI] [PubMed] [Google Scholar]
- 7.Ptak G. E., et al., Embryonic diapause is conserved across mammals. PLoS One 7, e33027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamatani T., et al., Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. U.S.A. 101, 10326–10331 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisogno S., Arena R., Fic K., Gąsior Ł., Ptak G. E., Lipid droplets utilization by mouse embryo. Bioscientifica Proceedings 10, ISEDISED9 (2020). [Google Scholar]
- 10.Leese H. J., Metabolism of the preimplantation embryo: 40 years on. Reproduction 143, 417–427 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Leese H. J., Barton A. M., Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J. Reprod. Fertil. 72, 9–13 (1984). [DOI] [PubMed] [Google Scholar]
- 12.Gardner D. K., Leese H. J., Concentrations of nutrients in mouse oviduct fluid and their effects on embryo development and metabolism in vitro. J. Reprod. Fertil. 88, 361–368 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Thompson J. G., Partridge R. J., Houghton F. D., Cox C. I., Leese H. J., Oxygen uptake and carbohydrate metabolism by in vitro derived bovine embryos. J. Reprod. Fertil. 106, 299–306 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Gopichandran N., Leese H. J., Metabolic characterization of the bovine blastocyst, inner cell mass, trophectoderm and blastocoel fluid. Reproduction 126, 299–308 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Kane M. T., Minimal nutrient requirements for culture of one-cell rabbit embryos. Biol. Reprod. 37, 775–778 (1987). [DOI] [PubMed] [Google Scholar]
- 16.Sellens M. H., Stein S., Sherman M. I., Protein and free amino acid content in preimplantation mouse embryos and in blastocysts under various culture conditions. J. Reprod. Fertil. 61, 307–315 (1981). [DOI] [PubMed] [Google Scholar]
- 17.McLaren A., A study of blastocysts during delay and subsequent implantation in lactating mice. J. Endocrinol. 42, 453–463 (1968). [DOI] [PubMed] [Google Scholar]
- 18.Cingolani F., Czaja M. J., Regulation and functions of autophagic lipolysis. Trends Endocrinol. Metab. 27, 696–705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu B. B., et al., Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 161, 291–306 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Kaushik S., Cuervo A. M., Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Z., et al., Integral proteomic analysis of blastocysts reveals key molecular machinery governing embryonic diapause and reactivation for implantation in mice. Biol. Reprod. 90, 52 (2014). [DOI] [PubMed] [Google Scholar]
- 22.He B., et al., Blastocyst activation engenders transcriptome reprogram affecting X-chromosome reactivation and inflammatory trigger of implantation. Proc. Natl. Acad. Sci. U.S.A. 116, 16621–16630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houghton F. D., Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation 74, 11–18 (2006). [DOI] [PubMed] [Google Scholar]
- 24.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Mantalenakis S. J., Ketchel M. M., Frequency and extent of delayed implantation in lactating rats and mice. J. Reprod. Fertil. 12, 391–394 (1966). [DOI] [PubMed] [Google Scholar]
- 26.Angermüller S., Fahimi H. D., Imidazole-buffered osmium tetroxide: An excellent stain for visualization of lipids in transmission electron microscopy. Histochem. J. 14, 823–835 (1982). [DOI] [PubMed] [Google Scholar]
- 27.Aardema H., et al., Oleic acid prevents detrimental effects of saturated fatty acids on bovine oocyte developmental competence. Biol. Reprod. 85, 62–69 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Tarín J. J., Cano A., Do human concepti have the potential to enter into diapause? Hum. Reprod. 14, 2434–2436 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Nicholls H., Mammals put embryo development on hold. Nature News, 15 March 2012. https://www.nature.com/news/mammals-put-embryo-development-on-hold-1.10228. Accessed 24 November 2018.
- 30.Tatsumi T., et al., Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development 145, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Bradley J., et al., Quantitative imaging of lipids in live mouse oocytes and early embryos using CARS microscopy. Development 143, 2238–2247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaren A., Michie D., Studies on the transfer of fertilized mouse eggs to uterine foster-mothers. I. Factors affecting the implantation and survival of native and transferred eggs. J. Exp. Biol. 33, 394–416 (1956). [Google Scholar]
- 33.Kaufman M. H., Barton S. C., Surani M. A., Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature 265, 53–55 (1977). [DOI] [PubMed] [Google Scholar]
- 34.Nagasawa H., Furukoshi K., Effects of concurrent pregnancy and lactation on reproduction in four strains of mice. Lab. Anim. Sci. 35, 142–145 (1985). [PubMed] [Google Scholar]
- 35.Paria B. C., Reese J., Das S. K., Dey S. K., Deciphering the cross-talk of implantation: Advances and challenges. Science 296, 2185–2188 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Cha J., Dey S. K., Cadence of procreation: Orchestrating embryo-uterine interactions. Semin. Cell Dev. Biol. 34, 56–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witwer K. W., et al., Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orr T. J., Zuk M., Reproductive delays in mammals: An unexplored avenue for post-copulatory sexual selection. Biol. Rev. Camb. Philos. Soc. 89, 889–912 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Gustafsson T. O., Andersson C. B., Westlin L. M., Reproduction in laboratory colonies of bank vole, Clethrionomys glareolus, originating from populations with different degrees of cyclicity. Oikos 40, 182 (1983). [Google Scholar]
- 40.Pritchett-Corning K. R., Clifford C. B., Festing M. F. W., The effects of shipping on early pregnancy in laboratory rats. Birth Defects Res. B Dev. Reprod. Toxicol. 98, 200–205 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Ranson R. M., New laboratory animals from wild species: Breeding a laboratory stock of hedgehogs (Erinaceus europaeus L.). J. Hyg. (Lond.) 41, 131–138 (1941). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrell M. I., et al. , Exploring the pregnant guinea pig as a model for group Streptococcus intrauterine infection. J. Infect. Dis. Med. 2, 109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikkelsen E., et al., The chinchilla as a novel animal model of pregnancy. R. Soc. Open Sci. 4, 161098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S., Pyl P. T., Huber W., HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C., Zhang B., Lin L. L., Zhao S., Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 18, 583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimand J., Kull M., Peterson H., Hansen J., Vilo J., g: Profiler–A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The FASTQ file for RNA-Seq data has been deposited in the National Center for Biotechnology Information Sequence Read Archive database under BioProject (accession no. PRJNA63160).