Significance
Up to 30% of methicillin-resistant Staphylococcus aureus (MRSA) bloodstream infections fail to resolve despite appropriate therapy. Such infections are persistent and life threatening. Because persistent MRSA isolates are susceptible to antibiotics in vitro, unique interactions among the pathogen, patient, and antibiotic occur in the body that lead to persistent outcomes. To study host factors involved in persistence, we used state-of-the-art methods to explore how genes are modified by methylation in patients who experience persistent vs. resolving MRSA bacteremia. Findings established distinct methylation patterns in these patients, including differential methylation signatures associated with important immune system and related genes. These results may enable approaches to identify and treat patients prone to persistent infection to improve therapeutic outcomes and save lives.
Keywords: Staphylococcus aureus, MRSA, persistence, epigenetics, DNA methylation
Abstract
Persistent methicillin-resistant Staphylococcus aureus (MRSA) bacteremia is life threatening and occurs in up to 30% of MRSA bacteremia cases despite appropriate antimicrobial therapy. Isolates of MRSA that cause antibiotic-persistent methicillin-resistant S. aureus bacteremia (APMB) typically have in vitro antibiotic susceptibilities equivalent to those causing antibiotic-resolving methicillin-resistant S. aureus bacteremia (ARMB). Thus, persistence reflects host–pathogen interactions occurring uniquely in context of antibiotic therapy in vivo. However, host factors and mechanisms involved in APMB remain unclear. We compared DNA methylomes in circulating immune cells from patients experiencing APMB vs. ARMB. Overall, methylation signatures diverged in the distinct patient cohorts. Differentially methylated sites intensified proximate to transcription factor binding sites, primarily in enhancer regions. In APMB patients, significant hypomethylation was observed in binding sites for CCAAT enhancer binding protein-β (C/EBPβ) and signal transducer/activator of transcription 1 (STAT1). In contrast, hypomethylation in ARMB patients localized to glucocorticoid receptor and histone acetyltransferase p300 binding sites. These distinct methylation signatures were enriched in neutrophils and achieved a mean area under the curve of 0.85 when used to predict APMB using a classification model. These findings validated by targeted bisulfite sequencing (TBS-seq) differentiate epigenotypes in patients experiencing APMB vs. ARMB and suggest a risk stratification strategy for antibiotic persistence in patients treated for MRSA bacteremia.
Persistent Staphylococcus aureus bacteremia (SAB) is serious, common, and potentially lethal (1), particularly when involving methicillin-resistant S. aureus (MRSA). Persistent SAB occurs when the infecting isolate is not cleared from the bloodstream, despite appropriate treatment with an antibiotic to which it exhibits susceptibility in vitro. However, mechanism(s) responsible for antibiotic-persistent methicillin-resistant S. aureus bacteremia (APMB) are incompletely understood (2–5). Recent studies have begun to explore specific host–pathogen interactions in APMB (6, 7). Bacterial, viral, and parasitic pathogens can directly and indirectly use cellular methylation machinery to preserve their survival and as such, can evade clearance by suppressing an effective host immune system (8). Our discovery that MRSA infection encodes innate immune memory supports the essential concept of epigenetic regulation of host defense against this organism (9, 10). Moreover, we recently reported that a g.25498283A > C polymorphism in DNA methyltransferase-3A (DNMT3A) differentiates patients experiencing persistent vs. resolving MRSA bacteremia outcomes (11). These findings support our hypothesis that distinct DNA methylomes exist in patients infected by MRSA and correspond to immunologic functions potentially shaping APMB vs. antibiotic-resolving methicillin-resistant S. aureus bacteremia (ARMB) outcomes.
DNA methylation plays a critical role in host immune regulation by contributing to innate or adaptive gene expression shaping molecular and cellular plasticity in immune response (12). Mechanistically, the classical understanding is that DNA hypomethylation at gene promoters enables gene transcription (13), whereas hypermethylation suppresses gene expression. Moreover, recent discoveries emphasize how unique DNA methylation patterns may correlate with specific disease subtypes and function as clinical indicators (12, 14). Likewise, bacterial infections are known to modify DNA methylation in host cells (15–17). Integrating these concepts, in the present study we analyzed methylome patterns by reduced representation bisulfite sequencing (RRBS) in well-characterized, optimally matched APMB and ARMB patient cohorts to explore our hypothesis that distinct DNA methylation signatures are associated with clinical APMB vs. ARMB outcomes. We then assessed whether differentially methylated sites (DMSs) localized to specific immune functions relevant to host defense against S. aureus. Next, we applied innovative methods to resolve specific immune cell subsets enriched in DMS associated with APMB vs. ARMB. Finally, we utilized targeted bisulfite sequencing (TBS-seq) to validate the differentially methylation signatures associated with APMB/ARMB outcomes.
Results
APMB and ARMB Cohorts Exhibit Distinct Demographic and Clinical Characteristics.
The demographic and clinical variables of the study population are summarized in Table 1. APMB patients exhibited an increased incidence of death due to S. aureus infection (P = 0.04), a higher risk of metastatic infection (P < 0.001), and longer length of in-hospital stay (P < 0.001). In contrast, ARMB patients exhibited greater rates of cure (P = 0.01), neoplasm (P = 0.03), and recent surgeries (within 30 d prior to SAB; P = 0.02).
Table 1.
Characteristics of study population
| Characteristics | APMB, n = 70 | ARMB, n = 72 | P value* |
| Age, mean (median), y | 61 (62) | 61 (61) | 0.74 |
| Sex, male/female | 38/32 | 42/30 | 0.74 |
| Race, Caucasian/African American/unknown or others | 32/36/2 | 34/36/2 | 0.95 |
| Site of acquisition, hospital acquired/HCA community acquired/non-HCA community acquired | 8/56/6 | 8/56/8 | 0.91 |
| Source of bacteremia, endovascular infection/GI or GU infection/respiratory, lung or skin, soft tissue, joint/bone infection/unknown or none | 23/12/3/19/13 | 18/11/7/22/14 | 0.67 |
| Metastatic infection (abscess/arthritis/epidural abscess/meningitis/vertebral osteomyelitis/nonvertebral osteomyelitis/psoas abscess/septic emboli/septic thrombophlebitis/kidney abscess/endocarditis/others) | 54 (12/10/5/0/8/9/5/15/8/1/27/4) | 28 (9/2/2/0/1/7/2/3/4/2/5/3) | 4.14e-06 |
| APACHE II, mean (range) | 18 (6–37) | 16 (5–36) | 0.16 |
| LOS categories, <9/9–14/15–20/>20 d | 9/19/19/23 | 28/22/11/11 | 9.75e-04 |
| Type of procedures used to treat the infection | |||
| Surgical removal of foreign device | 33 | 16 | 2.55e-03 |
| Surgical debridement | 21 | 8 | 6.58e-03 |
| Surgical insertion of foreign device | 8 | 4 | 0.24 |
| Abscess drainage | 13 | 7 | 0.15 |
| Other | 33 | 34 | 1.00 |
| Outcome, 90 d | |||
| Cure | 46 | 62 | 0.01 |
| Recurrent SAB infection | 12 | 5 | 0.07 |
| Death due to SAB infection | 10 | 3 | 0.04 |
| Death due to other causes | 2 | 2 | 1.00 |
| Underlying comorbidity | |||
| Neoplasm | 7 | 18 | 0.03 |
| Diabetic | 37 | 42 | 0.61 |
| Hemodialysis dependent | 29 | 20 | 0.11 |
| HIV positive | 3 | 1 | 0.36 |
| Transplant recipient | 5 | 7 | 0.76 |
| Injection drug use | 1 | 1 | 1.00 |
| Corticosteroid use, 30 d | 16 | 16 | 1.00 |
| Surgery past 30 d | 11 | 24 | 0.02 |
For metastatic infection, some patients have more than one type of metastatic infection. APACHE II, Acute Physiology and Chronic Health Evaluation II; GI or GU, gastrointestinal or genitourinary; HCA, health care associated; LOS, length of stay.
The t test was used for continuous variables; the Fisher’s exact test was used for categorical variables.
RRBS DNA Methylation Signatures Differ in APMB vs. ARMB Patients’ Peripheral Blood.
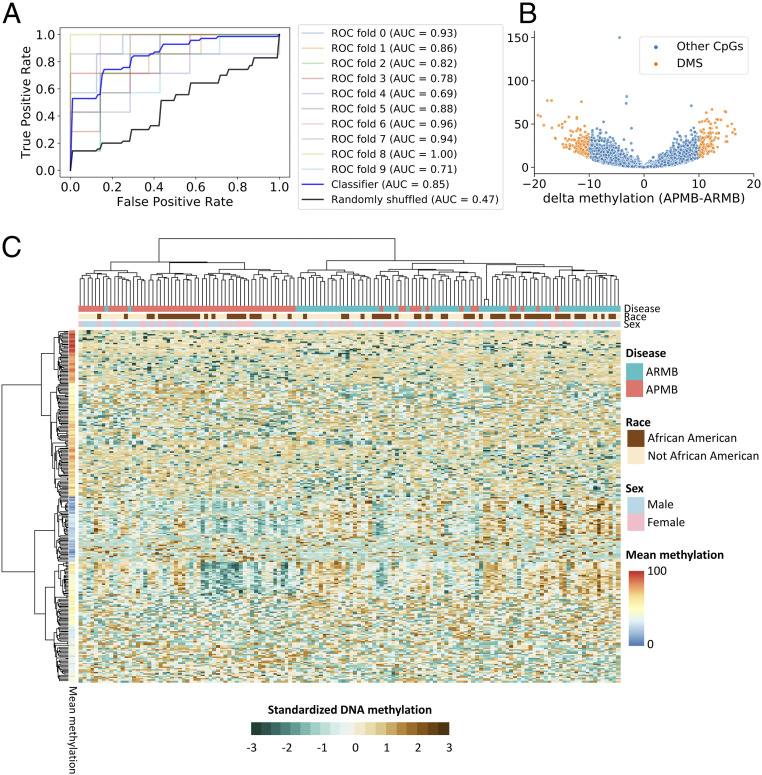
We utilized RRBS to determine if global methylation profiles distinguished APMB from ARMB. We then built a classification model based on the DNA methylation levels of all 749,212 5′-C-phosphate-G-3′ (CpG) sites using a logistic regression with elastic net regularization (18). The performance of the classification was evaluated by a 10-fold cross-validation of all the samples, and the average classification accuracy measured by a receiver operating characteristic (ROC) area under the curve (AUC) was 0.85 (Fig. 1A, blue bold line). This corresponds to a sensitivity of 75.7% and a specificity of 84.8% at the optimal cutoff, which maximized the sum of sensitivity and specificity. To contrast the classification performances with chance outcomes, we generated the ROC using randomly shuffled class labels (APMB and ARMB). As expected, AUC for the randomly shuffled class labels approximated 50% (Fig. 1A, black bold line). We then used methylkit (19) to identify methylation patterns associated specifically with APMB or ARMB. Methylation levels of 276 CpG DMSs were significantly associated with a persistent vs. resolving outcome (Fig. 1B and SI Appendix, Table S1). A total of 160 DMSs reflected relative hypomethylation and 116 DMSs exhibited hypermethylation in APMB compared with ARMB. Methylation patterns of the 276 DMSs encompassing 142 samples were then visualized (Fig. 1C and SI Appendix, Fig. S1). The hierarchical, agglomerative cluster analysis revealed an APMB cluster (left branch; containing 96.5% APMB samples) and an ARMB cluster (right branch; containing 82.4% ARMB samples). The algorithm also clustered DMSs into two major groups consisting of 131 DMSs in cluster 1 (comparatively hypermethylated; mean methylation levels, 46.8 to 87.3%) or 145 DMSs in cluster 2 (comparatively hypomethylated; mean methylation levels, 16 to 64.6%).
Fig. 1.
Peripheral blood leukocyte DNA methylation is altered in patients with APMB. (A) ROC curve summarizes the test set classification performance (estimated by 10-fold cross-validation) of a logistic regression model that uses the DNA methylation levels of 749,212 CpG sites to distinguish APMB and ARMB samples. (B) Volcano plot represents the methylation difference for 749,212 CpG by delta methylation (percentage) and –log10 adjusted P value (FDR) generated from methylkit. The cutoff to define DMS (orange dots) was set as FDR < 0.01 and |delta methylation| > 10%. (C) The heat map displays DNA methylation for DMS (row) across all SAB samples (column). The clusters were generated using the sex-, race-, and batch-corrected DNA methylation data with Ward.D2 method shown in SI Appendix, Fig. S1. Keeping the same clustering order shown in SI Appendix, Fig. S1, the standardized DNA methylation (z score) was applied to visualize the relative methylation differences in this figure.
To further investigate the differential methylation profiles focusing on sequence elements containing more than one CpG site (two or more CpG sites per region), CpG sites were clustered into 126,144 regions using the dynamic fragments method in CGmapTools (20). A classification model was then used to construct the 126,144 regions. The average AUC generated from 10-fold cross-validation using true class labeling was 0.88 (SI Appendix, Fig. S2A), with a sensitivity of 0.81 and a specificity of 0.85 at the optimal threshold. There were six CpG regions significantly associated with an APMB vs. ARMB outcome based on the methylkit analysis (SI Appendix, Fig. S2B and Table S3). All six of these differentially methylated regions (DMRs) were captured with at least one CpG site in the 276 identified DMSs. Together, these data reveal significant methylation signature differences in patients experiencing APMB vs. ARMB outcomes.
Specific DMSs Prioritize Differential APMB vs. ARMB Signatures in a Classification Model.
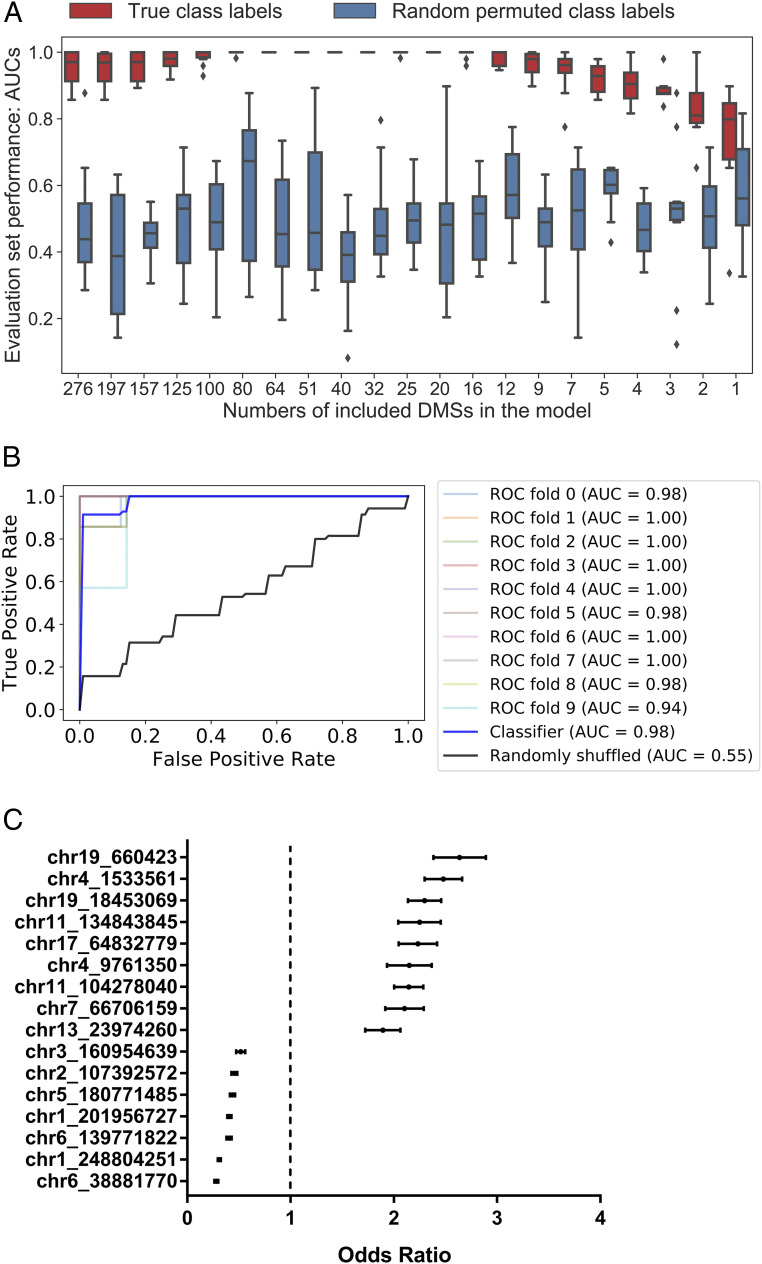
To evaluate and rank DMSs based on their discrimination power to identify APMB, we used the 276 DMSs to build a logistic regression model with elastic net regularization. A wrapper method (21) was used to iteratively concentrate the parsimonious subsets of DMSs with high absolute model coefficients and censor DMSs with low discriminating capability. Box plots summarize the results of this process by reporting AUC generated from each iteration (Fig. 2A). Average AUC with true labels systematically increased, as DMSs with high discriminating ability were concentrated, and peaked when including 16 specific DMSs. We contrasted the DMS performances with chance outcomes by calculating the AUC for randomly permuted class labels (APMB and ARMB), which approximated 50%, as expected. Next, we evaluated the classification performance of the top 16-ranked DMSs by 10-fold cross-validation. The average AUC for the classifiers built based on the DNA methylation levels of the top 16-ranked DMSs was 0.98 (Fig. 2B), corresponding to a sensitivity of 91.4% and a specificity of 98.9% at the optimal cutoff. By calculating the odds ratio using the model coefficients, nine DMSs (odds ratio > 1) were positively associated with APMB, while another seven DMSs (odds ratio < 1) were negatively associated with APMB (Fig. 2C). Thus, even with the degree of overfitting, these data suggest that DNA methylation profiles can aid in or even predict APMB classification, given methods of our unbiased classifier in which DMSs were selected based on all the samples.
Fig. 2.
Specific DMSs prioritize differential APMB vs. ARMB signatures in a classification model. (A) Evaluation set performance of the logistic regression classifiers, in order of decreasing number of DMSs used in model construction, from left to right (x axis). Red box–whisker plots show the quartiles of 10 AUC values generated from 10-fold of leave one class out cross-validations, and blue box–whisker plots depict the performance based on permuted class labels to represent the random background distribution. (B) ROC curve of the logistic regression model that uses the DNA methylation levels of the top 16-ranked CpG sites to distinguish APMB and ARMB samples. (C) Plots depicting the mean with 95% CI of odds ratio for the top 16 DMSs in the model shown in B.
DMSs Are Enriched in Introns and Intergenic Regions.
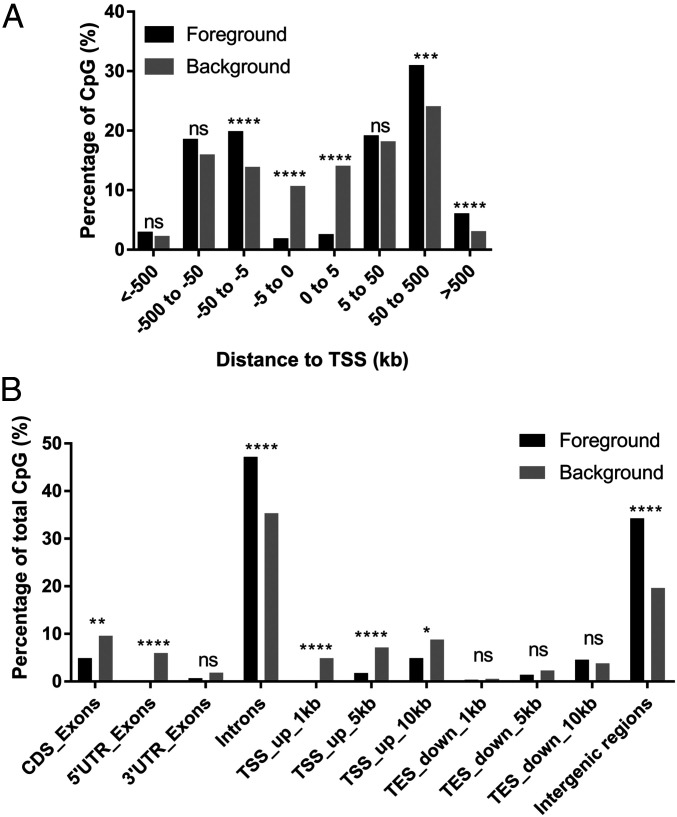
Next, we investigated the immunological relevance of the principal 276 DMSs identified. First, we explored whether the DMSs localized to promoter regions with potential to directly regulate gene transcription. Here, distances between a target DMS and the transcription start sites of its proximal genes were determined (SI Appendix, Table S2). We found that DMSs are significantly enriched 5 to 50 kb upstream and >50 kb downstream relative to transcriptional start sites (TSSs) and largely depleted between 5 kb upstream and 5 kb downstream to the TSS (Fig. 3A). Another view of the genomic distribution for DMSs revealed enrichment of DMSs at introns and intergenic regions, with significant depletion 1 to 10 kb upstream of TSS, 5′ untranslated region (UTR), and coding sequence exon regions (Fig. 3B). Together, these results indicate that the differential DMSs in APMB vs. ARMB are enriched at regions distal to promoters.
Fig. 3.
DMSs are enriched in introns and intergenic regions. (A) Distance of DMSs (foreground) to the TSSs of the proximal genes. Background was defined by total detected CpG sites excluding DMS sites. The statistics were calculated using χ2 test with Yates’ correction. (B) DMS locations in genomic regions. (CDS, coding sequence; UTR, untranslated region; TSS, transcriptional start site; TES, transcriptional end site.) The statistics were calculated using Fisher’s exact test. (ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.)
DMSs Localize to Regulatory Regions Containing Relevant Transcription Factor Binding Sites.
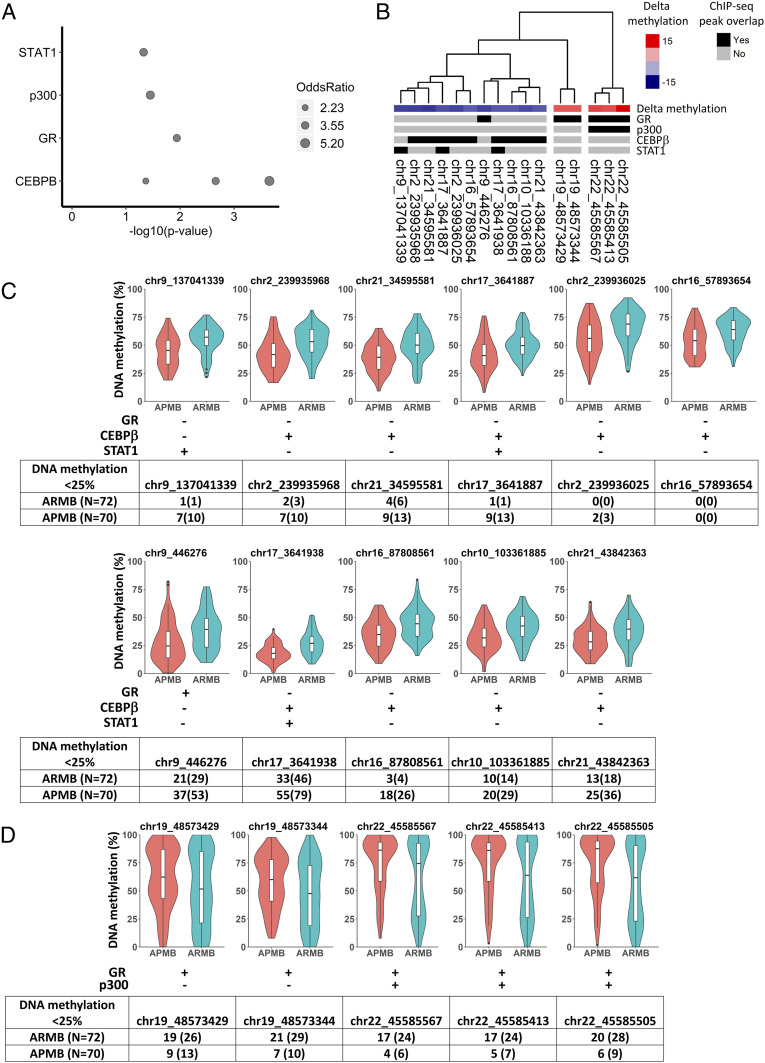
To further explore the link between differential methylation and regulation of gene expression, we analyzed DMS enrichment in transcription factor binding sites. To this end, we used Locus Overlap Analysis (LOLA) (22) to comparatively overlay the DMS locations with experimentally validated chromatin immunoprecipitation sequencing (ChIP-seq) peaks (SI Appendix, Table S4). Significant overlap (P < 0.05) was observed between the DMS and specific transcription factor binding sites, including the signal transducer/activator of transcription 1 (STAT1), histone acetyltransferase p300 (p300), CCAAT enhancer binding protein-β (C/EBPβ), and glucocorticoid receptor (GR) (Fig. 4A).
Fig. 4.
DMSs localize to regulatory regions containing transcription factor binding sites. (A) LOLA region set enrichment analysis displays the significant associations (P < 0.05) between transcription factor binding sites (y axis) and DMS. The area of points represents the magnitude of odds ratio (a value of zero is mapped to a size of zero). (B) The heat map shows the overlap of DMS with transcription factor binding sites that LOLA analysis of the ChIP-seq datasets identified as enriched. Columns were arranged by hierarchical clustering with Ward.D2 method based on the methylation profiles of the 16 transcription factor binding DMSs across all SAB patients. (C and D) Violin plots of methylation levels for (C) DMS with reduced methylation in APMB and (D) DMS with reduced methylation in ARMB. Overlap with transcription factor binding site shown as + for yes and − for no. Frequency of patients with hypomethylated DMS (<25% methylation level) in APMB and ARMB shown as number (percentage).
Since DNA hypomethylation at a transcription factor binding motif is often associated with increased transcription factor binding and gene expression, we investigated methylation levels of DMS with potential transcription factor binding sites. We calculated the delta methylation levels of DMS with at least one overlap with transcription factor binding site (n = 16) and annotated this mapping with transcription factor binding enrichment (Fig. 4B). This analysis revealed 11 DMSs with hypomethylation in APMB, and 5 DMSs having hypomethylation in ARMB. Nine of the 11 DMSs hypomethylated in APMB overlapped with transcription factor binding sites C/EBPβ, and 3 of 11 overlapped with those for STAT1. These genes are well-established regulators of immune and antimicrobial host defense responses (23, 24). Similarly, all five DMSs with hypomethylation in ARMB overlapped with transcription factor binding sites for GR, known to regulate stress responses to sepsis (25, 26). p300 functions as a coactivator by interacting with several transcription factors including GR to increase gene expression (27). The distributions of DNA methylation in each cohort of patients are summarized in Fig. 4 C and D, segregated by directionality of change in methylation levels relative to APMB vs. ARMB.
For the 11 DMSs with reduced methylation levels in APMB, we observed a populational shift, suggesting overall increased immunologic functions orchestrated by the binding of C/EBPβ and STAT1 in APMB patients (Fig. 4C). For the five DMSs having hypomethylation in ARMB, the individual DNA methylation levels ranged from 0 to 100% among SAB patients, demonstrating high methylation variability at the GR and p300 transcription factor binding sites. Specifically, we observed a shift toward M-shaped distributions in ARMB, resulting from a larger subgroup (24 to 29%) of ARMB patients with hypomethylated DMS (Fig. 4D). This result suggested that there was a larger subgroup of ARMB patients with immune cells accessible for GR and p300 transcription factor binding. Collectively, these data demonstrate two distinct DNA methylation patterns at transcription factor binding motifs differentiating APMB from ARMB.
Differential Methylation Is Enriched in Myeloid Lineages of APMB vs. ARMB Patients.
To investigate whether the 16 transcription factor binding DMSs are located at imputed enhancer regions, we searched for enhancers reported in the GeneHancer database (28). We confirmed a total of six imputed enhancers reported in GeneHancer associated with nine transcription factor binding DMSs (Table 2). Four of 11 DMSs exhibiting hypomethylation in APMB localized at imputed enhancer regions known to regulate SETD6, ADGRG1, and SLC7A5 proteins involved in innate immune cell signal transduction. All five DMSs having hypomethylation in ARMB (two DMSs located at chromosome 19; three DMSs located at chromosome 22) were associated with two enhancers, which regulate lysosome-membrane protein (CTNS) or extracellular matrix protein (FBLN1), respectively.
Table 2.
DMSs localizing to enhancers as identified from the GeneHancer databank
| DMS | GH identifier | Enhancer score | GH sources | No. of putative transcription factor binding sites | Putative gene targets |
| chr16_87808561 | GH16J087805 | 2 | FANTOM5, Ensembl, ENCODE, dbSUPER | 171 | 8 genes (ENSG00000260177, SLC7A5, LOC101928659, CTU2, ZC3H18, ZCCHC14, RNF166, MIR6775) |
| chr16_57893654 | GH16J057891 | 1.5 | FANTOM5, ENCODE, dbSUPER | 142 | 13 genes (RSPRY1, ADGRG3, CSNK2A2, SETD6, KIFC3, LOC388282, CNGB1, ADGRG1, CNOT1, CFAP20, ZNF319, GC16P057890, GC16P057897) |
| chr19_48573344, chr19_48573429 | GH19J048572 | 1.4 | Ensembl, ENCODE, dbSUPER | 121 | 3 genes (SULT2B1, DKKL1, SPACA4) |
| chr17_3641938 | GH17J003639 | 1 | ENCODE | 108 | 3 genes (CTNS, TAX1BP3, LOC105371493) |
| chr21_34595581 | GH21J034589 | 0.9 | ENCODE, dbSUPER | 20 | 7 genes (RCAN1, KCNE1, FAM243A, MRPS6, GC21M034577, GC21M034614, GC21M034615) |
| chr22_45585413, chr22_45585505, chr22_45585567 | GH22J045584 | 0.8 | ENCODE, dbSUPER | 9 | 6 genes (LINC01589, PIR61892, GC22M045121, GC22M045550, FBLN1, GC22P045074) |
GH, GeneHancer; FANTOM5, Funational annotation of the mammalian genome project; Ensembl, Ensembl gene annotation; dbSUPER, Database of super-enhancers,
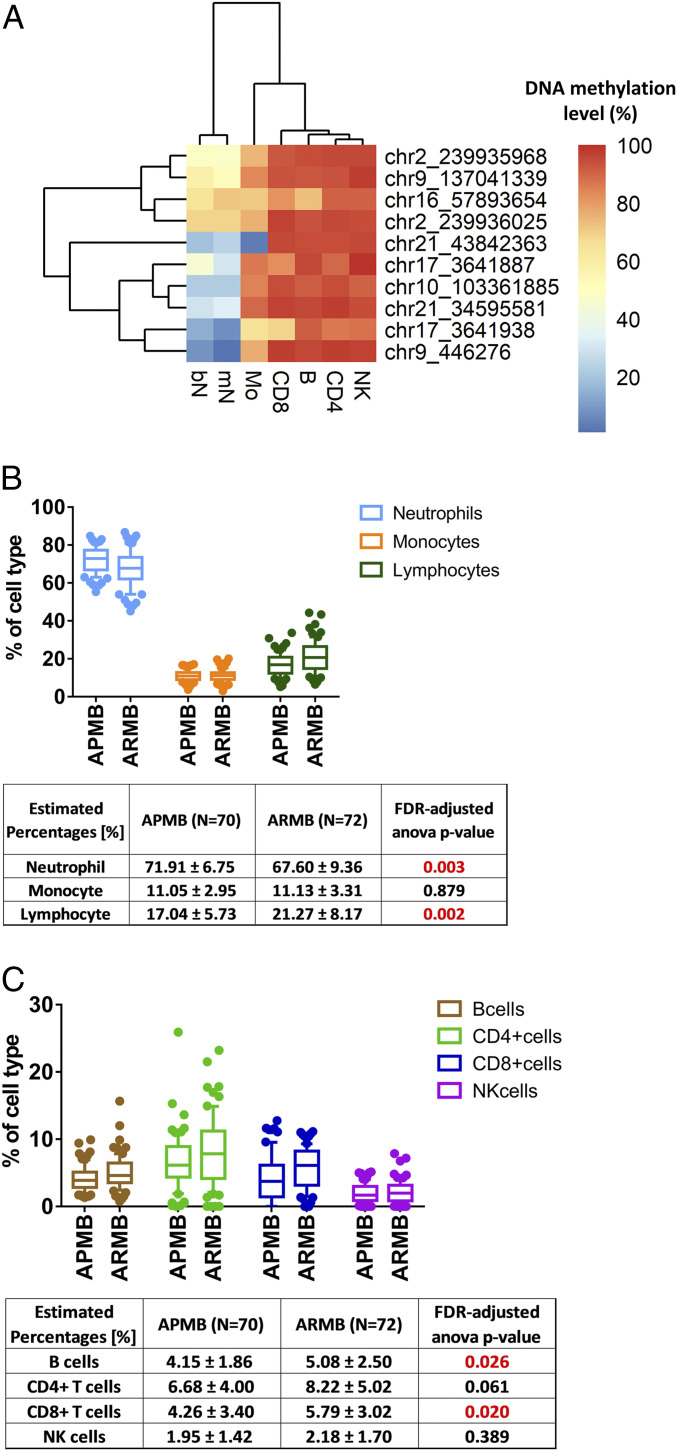
As transcription factor binding patterns are typically cell-type specific (29, 30), we analyzed whether the 16 transcription factor binding DMSs are enriched in specific cell populations. Methylation data from healthy human adult blood cells were obtained through the Blueprint Human Blood Cell Epigenome project (31). To ensure high-confidence leukocyte-specific DNA methylation data, we included CpG sites with at least 10-read coverage across all 37 leukocyte samples (SI Appendix, Table S5) in this analysis (10 of 16 DMSs). All 10 DMSs with validated data quality exhibited hypomethylation in the myeloid lineages as compared with cell types from lymphoid lineage (Fig. 5A).
Fig. 5.
Cell-type proportions estimated from DNA methylation differentiating APMB from ARMB. (A) Cell type-specific methylation levels obtained from Blueprint for the transcription factor binding DMS. B, B cell; bN, band neutrophil; mN, mature neutrophil; Mo, monocyte; NK, natural killer cell. (B and C) Box plots of proportions of each cell type within APMB/ARMB groups. The FDR-corrected ANOVA test within each cell type was used, and the adjusted P values smaller than 0.05 are highlighted in red. (B) The proportions of estimated neutrophils, lymphocytes (combining the estimated proportions of B, CD4+ T, CD8+ T, and NK cells), and Mos within APMB/ARMB groups. (C) The proportions of estimated lymphocytes (including B, CD4+ T, CD8+ T, and NK cells).
Cell-Type Proportions Estimated from DNA Methylation Differentiate APMB from ARMB.
To address the possibility that DNA methylation levels could vary due to changes in cell-type abundance across individuals, analyses were performed to assess cellular composition in blood samples in APMB and ARMB patients. A cell-type deconvolution method was used to estimate cell-type proportions (32–34). This approach obviated potential bias with respect to missing complete blood count data in the study cohort (up to 57%). The DNA methylation-based cell-type decomposition analysis results confirm that estimated cell-type compositions in APMB and ARMB groups had significant differences in relevant cell subsets (multivariate ANOVA, P = 0.004). We found significantly higher neutrophils (adjusted P value = 0.003), lower B cells (adjusted P value = 0.026), and lower CD8+ T cells (adjusted P value = 0.02) associated with APMB outcome (Fig. 5 B and C). Incorporating estimated cell-type composition into the logistic regression model, we obtained average AUC equal to 0.87 (SI Appendix, Fig. S3), suggesting that cell-type composition had only a minor effect on improving classification.
TBS-seq Confirmed the Disease-Associated Differential DNA Methylation Signatures.
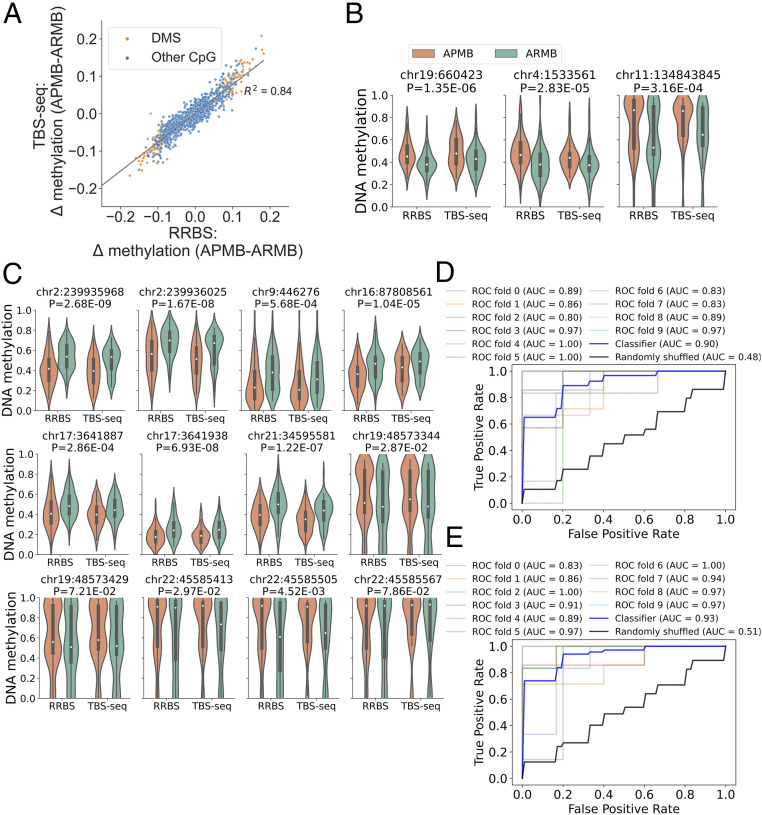
To further validate the DMS identified by RRBS, we carried out TBS-seq (35). A panel of probes was designed to target the DMS and DMR identified by RRBS (SI Appendix, Table S6). Both DNA methylation and genetic data were computed from the bisulfite converted reads aligned to the genome. The intersecting CpG methylation data generated using TBS-seq showed significant correlation between the two techniques (SI Appendix, Fig. S5B) and with the MRSA disease-associated DMS detected by RRBS (Fig. 6A). Overall, there was excellent concordance between outcomes of the two methods. As an example, all three of the DMSs positively associated with APMB by RRBS (Fig. 2C) for which probes could be designed were confirmed by TBS-seq (Fig. 6B). Furthermore, all seven hypomethylated DMSs (Fig. 4C) and five hypermethylated DMSs associated with transcription factor binding for GR, C/EBPβ, and/or STAT1 (Fig. 4D) were confirmed in the TBS-seq data (Fig. 6C). The performance of the classification of the 121 DMSs overlapping between TBS-seq and RRBS was evaluated by a 10-fold cross-validation. The average classification accuracy resulted in an AUC of 0.90 for TBS-seq (Fig. 6D) and 0.93 for RRBS (Fig. 6E), confirming that these DNA methylation signatures differentiate APMB from ARMB.
Fig. 6.
TBS-seq confirmed the disease-associated differential DNA methylation signatures. (A) Scatterplots of the delta methylation (APMB–ARMB) in RRBS (x axis) and in TBS-seq (y axis). (B and C) Violin plots of methylation levels in RRBS and TBS-seq for (B) top-ranked DMSs reported in Fig. 2 and (C) transcription factor binding DMSs reported in Fig. 4. Two-way ANOVA was used to determine the disease associated (P value shown). (D and E) ROC curves of the logistic regression model that uses the DNA methylation levels of 121 detected DMSs from (D) TBS-seq and (E) RRBS.
To determine if the disease-associated DMRs overlap with genetic polymorphisms that may be attributable to disease phenotype, we analyzed the sequence of the AA, AG, and GG alleles based on reads mapping to the forward strand and CC, CT, and TT alleles based on reads mapping to the reverse strand. Genotypes were determined for the majority of alleles in our target regions by correlating the observed counts with the expected counts for each allele combination (Materials and Methods). Fisher’s exact test was computed for each single-nucleotide polymorphism (SNP) genotype, and the adjusted P values were calculated based on the false discovery rate (FDR) methods. Of the 774 SNP genotypes that were identified within the targeted regions, no significant statistical associations were found between APMB/ARMB outcomes (SI Appendix, Table S7). This pattern of results signifies that epigenetic, not genetic, determinants accounted for these distinct outcomes of MRSA bacteremia.
Discussion
The current study compared DNA methylomes in whole-blood leukocytes from patients with APMB and ARMB to assess if differential methylation signatures exist and if so, whether such epigenetic signatures associate with relevant immune functions. Key findings revealed significant differences of DMSs in APMB vs. ARMB patients and localized predominant signals proximate to transcription factor binding sites of genes known to influence immune functions. These data strongly support our hypothesis that differential methylation in circulating immune cells may shape protective vs. nonprotective responses during early stages of MRSA bacteremia that affect ensuing APMB vs. ARMB outcomes. These insights serve as a proof of concept that DNA methylation profiles within host immune cells may aid in APMB classification or predict therapeutic outcomes.
Our findings emphasize that epigenetic regulation correlates with protective immunity in response to S. aureus infection. Importantly, DMSs identified were enriched at gene enhancers known to regulate transcription factor binding sites and control cell type-specific gene activation or repression (36, 37). DNA hypomethylation at gene enhancers can switch such enhancers to an active state (38), resulting in enhanced transcription factor binding and gene expression. Our results demonstrated two groups of transcription factor binding patterns associated with significant differences in leukocyte methylation associated with APMB. In turn, these patterns offer plausible mechanistic roles of these transcription factors in the development of APMB.
First, our data support the concept of emergency granulopoiesis in APMB. We observed 11 DMSs exhibiting hypomethylation in APMB, 9 of which were bound by C/EBPβ. The C/EBPβ protein is a key transcription factor in emergency granulopoiesis, rapidly mobilizing bone marrow granulocyte progenitors during severe systemic infection (23, 39). Additionally, C/EBPβ amplifies emergency granulopoiesis by inducing FancC expression in infection (39). In support of this finding, up-regulation of C/EBPβ transcripts is observed in granulocyte progenitors, and C/EBPβ-deficient progenitors have impaired pathogen-induced granulopoiesis in vitro and in vivo (23). Because immature neutrophils (e.g., band cells) may be ineffective at killing S. aureus or may promote harmful nonspecific inflammation, ostensibly they may be adverse in resolving SAB, facilitating persistence. Further studies are needed to determine if APMB patients have increased release of early granulocyte precursors during MRSA bacteremia.
Second, the polarization of CD4+ T helper (Th) cells during early stages of SAB may be critical in determining subsequent APMB from ARMB outcomes. For example, Th17 responses during S. aureus infection are important for protective immunity, as the cytokines interleukin-1 (IL-1) and IL-17 mediate neutrophil recruitment and activation necessary for bacterial clearance (40, 41). However, the present results in APMB patients reveal hypomethylation at transcription binding sites for STAT1 (Th1 promoting), not STAT3 (Th17 promoting). Importantly, STAT1 induces gamma interferon signaling during infections (42, 43) but counterregulates Th17 polarization (44). Incorporating these findings, we posit that APMB may result in part from a profusive epigenetic activation of STAT1, which in turn, suppresses protective antistaphylococcal immune response mechanisms induced via Th17 pathways. This phenomenon is also consistent with inadequate neutrophil-mediated protection associated with emergency granulopoiesis.
Third, we identified five GR transcription factor binding sites to be hypomethylated in the ARMB patient cohort. Glucocorticoid-mediated homeostasis is important in the stress response to sepsis by modulating excessive proinflammatory activities through inactive activator protein-1 (AP-1) or nuclear factor kappa B (NF-κB) (25). In this respect, activated GR can directly engage its coactivator p300 to regulate GR signaling. Thus, our data suggest a mechanistic link between GR–p300 signaling and ARMB, in which modulated immune responses facilitate bacteremia resolution. While glucocorticoids are often used in therapy of infections involving the central nervous system, uncertainty remains about their use as adjuvant therapy for bacteremia and sepsis (45). Previous studies suggest that distinct GR expression levels in septic patients may contribute to treatment responsiveness (46, 47). Our results showing diverse DNA methylation levels in GR transcriptional binding sites among SAB patients may offer further insights into patient responses to glucocorticoid therapy through epigenetic regulation of glucocorticoid response.
Finally, current data reveal important insights into diversity of biological responsiveness to S. aureus infection based on interindividual variance of methylomic signatures. We recently identified a genetic polymorphism located in the DNMT3A gene (g.25498283A > C genotype), which correlates with resolving MRSA bacteremia outcomes, potentially via hypermethylation gene regulation resulting in reduced IL-10 levels (11). Furthermore, circulating immune cell-type profiles are dynamic and may also influence differences in methylation patterns of immune cells in APMB vs. ARMB. For example, DNA methylation in human hematopoietic development is affected by adaptive changes in lineage of specific immune cell subsets (29, 30). Our current results highlighted that the majority of transcription factor binding DMSs were demethylated in neutrophils. Therefore, this observation suggests that overall differences in methylation result from a higher proportion of DMSs in circulating neutrophils or band neutrophils in APMB compared with ARMB. This conclusion is also highly consistent with our present findings regarding C/EBPβ and STAT1 methylation signatures with respect to neutrophils.
It is important to note that differential methylation using RRBS analysis can potentially be confounded by the presence of single-nucleotide variants in cis. To validate our initial RRBS findings, TBS-seq was performed. Findings from TBS-seq strongly supported the interpretation that disease-associated interindividual differences were only driven by changes in DNA methylation levels and not SNPs found in our target regions. Notwithstanding this key finding, it should also be understood that variation in gene sequence can influence persistent vs. resolving MRSA bacteremia outcomes (11). Thus, it is plausible that both epigenetic and genetic signatures are associated with APMB/ARMB outcomes in MRSA bacteremia.
Initial results raised the possibility that cellular composition profiles could be influencing APMB classification. Importantly, methylation variability may result from several potential factors, including 1) direct changes of methylation levels at a regulatory element in one or more specific cell type(s) or 2) altered cell-type compositions indirectly impacting methylation levels at a cell type-specific regulatory element (29, 30). To explicitly segregate these two potential sources of variability, obtaining cell composition information is required. Given the limitation that complete blood counts were not available from every patient studied, we employed a method to correct methylation profile data based on potential cell-type composition. We found that the cell-type abundances were similar between the persistent and resolving groups. However, some of these differences achieved statistical significance for relevant immune cell subsets. Moreover, we demonstrated that our classification model, which has an AUC = 0.85 without accounting for cell-type composition, is only modestly improved by explicitly including inferred cell-type abundances as covariates. These results support the conclusion that methylation differences, comprising both disease- and cell type-associated features, may be useful to develop an agnostic algorithm to inform or predict clinical outcomes in MRSA bacteremia patients.
Whether the host, pathogen, or a combination of the two drives differential methylation is the key question that remains to be determined. Pathogens may drive alterations of DNA methylation in host immune cells, supporting the emerging concept of innate immune memory (48–50). For example, Mycobacterium tuberculosis (15) and Helicobacter pylori (51) have been reported to subvert host DNA epigenetics through ten-eleven translocation (TET) enzyme family and inducing de novo DNA methylation. One recent observation further supported this concept that DNA demethylation was induced after M. tuberculosis infection (52). Thus, it is possible that the host epigenetic changes identified in the current study were driven by prior episodes of infection, S. aureus or otherwise.
It is also important to note the limitations of the current investigation. First, this study was not designed to explicitly determine APMB vs. ARMB outcome-associated SNPs, and it is challenging to accurately identify the heterozygous single-nucleotide variants (SNVs) from partially C to T converted sequences in the bisulfite conversion conditions. Second, our annotation analyses were limited by the scope of data available in the University of California Santa Cruz (UCSC) genome browser, the Encyclopedia of DNA Elements (ENCODE), and the CODEX databases, which are largely generated from uninfected human cell lines. Third, the current SAB classification model has not been evaluated using an external validation cohort. However, a principal goal of this study was to compare RRBS sequencing data and methylation characteristics in SAB patients experiencing APMB vs. ARMB outcomes. Thus, we demonstrate findings not available from any prior investigation to our knowledge, namely genome-wide methylation signatures associated with persistent outcomes in MRSA bacteremia. Fourth, precise impact of the methylation patterns uncovered in this study requires future investigation. While hypomethylation is generally felt to promote gene expression, recent studies have challenged this view. For example, Pacis et al. (52) suggest that gain or loss of methylation may be associated with increased transcriptional activity, particularly at nonpromoter, enhancer regions. Finally, the current study focused on patients with clinically confirmed MRSA bacteremia receiving appropriate vancomycin therapy over a 5-d course. Whether the specific epigenetic pattern differences are the same in such patients treated with other antibiotic regimens remains to be determined. In this respect, the current study establishes an important reference point. Thus, our ongoing efforts are focused on determining the impact of methylation status on expression of key immune system genes and their expression in MRSA infection and beyond.
In summary, the current findings support a proof of principle that DNA methylation signatures in host immune effectors can differentiate persistent vs. resolving outcomes in MRSA infection. Moreover, specific DMSs identified correspond to immune cell regulatory functions of direct and established relevance to host defense against S. aureus. The fact that targeted methods validated general methods suggests that targeted methods may be readily developed into much more expedient and practical assays for rapid assessment of likelihood for persistent MRSA bacteremia outcomes. In turn, such knowledge could guide interventions that minimize likelihood for such adverse outcomes. These concepts could also logically extend to many other types of infections. The implications for future studies include testing the predictive value of targeted CpG sites for APMB detection, as well as studying in vivo how DNA methylation correlates with gene expression in specific cell subsets and in context of antiinfective therapy.
Materials and Methods
Case Selection.
All human studies were conducted in accordance with Good Clinical Practice and Human Subjects Research as approved by the Duke University Medical Center (DUMC) Institutional Review Board. SAB cases were evaluated and consented for enrollment in the S. aureus Bacteremia Group (SABG) biorepository at DUMC. Cases for the current study were carefully selected based on the following inclusion criteria: laboratory-confirmed MRSA bacteremia, received appropriate vancomycin therapy, and enrolled in the SABG study between 2007 and 2017 (to ensure contemporary medical practices). APMB was defined in patients who had continuous MRSA-positive blood cultures for at least 5 d after vancomycin antibiotic treatment (11); ARMB patients had initial blood cultures that were positive for MRSA, but the subsequent blood cultures became negative. Thus, epigenetic methylome patterns in this study focused on circulating host immune cells proximate to the time of SAB diagnosis. Stratified propensity scoring was used to match the demographic and clinical variables for selection of APMB vs. ARMB cases, including race, sex, age, dialysis, diabetes diagnosis, and/or presence of indwelling device (artificial heart valve, peritoneal dialysis port, central venous/arterial catheter, or left ventricular assist device) and vancomycin treatment. This case-controlled study was based on a total of 142 SAB samples (70 APMBs and 72 ARMBs) with matched criteria and qualified DNA quality control. Details of clinical characteristics of the study cohort are presented in Table 1.
RRBS.
RRBS libraries were prepared as described previously (53) and sequenced by University of California, Los Angeles, Broad Stem Cell Research Center (BSCRC) sequencing core. The data were subjected to a quality control step using FastQC (version 0.11.7) (54). Sequencing data were trimmed to remove the adaptors and the terminal CG nucleotides using the trimmomatic software (version 0.38) (55). The reads were aligned to the human genome GRCh38/hg38 (release 93, downloaded from Ensembl), and DNA methylation levels were determined using the BS-Seeker2 pipeline (56). Data were filtered to include the autosomal CpG sites with at least 15-read coverage across all samples, and the potential confounding factors, including batch, race, and sex, were accounted for by using an empirical Bayesian framework, as implemented in the ComBat function of R package SVA as in previous reports (30, 57). Details are in SI Appendix.
TBS-seq.
A total of 1,117 probes were designed by Integrated DNA Technologies based on the coordinates of the DMSs we discovered using RRBS data. The libraries were prepared as described in SI Appendix. Data were preprocessed with FastQC (version 0.11.8) and cutadapt (version 2.10) to remove the adapter sequences. Trimmed reads were aligned against the GRCh38 genome using BSBolt (58) according to the pipeline described in ref. 35. Methylation was called on aligned reads after PCR duplicates removal, and the DNA methylation matrix was assembled using the common CpG sites covered by at least 20 reads across all samples. A total of 3,614 intersecting CpGs detected from both RRBS and TBS-seq were collected, which included the 121 DMSs and five DMRs reported from RRBS. The correlation between data generated from the two methods was computed, and the two-factor ANOVA was used to determine the statistical difference among factors including the disease outcomes and techniques. The genotype data were generated as described in SI Appendix. Fisher’s exact test was used to determine the association between SNP genotypes and APMB/ARMB outcomes.
Classification of Patients.
Logistic regression was used to build a classifier using the Python package, scikit-learn version 0.21.2. A regularization method elastic net (18) was incorporated in the classification model to build a generalizable classifier and reduce model overfitting. All RRBS-detected 749,212 CpGs were included in the optimization phase using both precision and recall score for the model evaluation. Then, the classifiers were trained and tested using a 10-fold cross-validation strategy. ROCs were used to estimate the sensitivity and specificity of the APMB classification method. The AUC was calculated for each ROC to evaluate the accuracy of APMB classification.
CpG Clustering and Identification of DMSs and Regions.
The clustered CpG regions were generated based on the dynamic fragmentation strategy in CGmapTools (20) (with the following parameters: maximal distance between two adjacent CpGs, s = 100 bp; maximal fragment size, S = 1,000 bp; minimal CpG number in one fragment, n = 2) The matrix of the single CpG site or the clustered CpG region was then used to calculate the differential methylation sites using the methylkit package in R (19). The cutoff of q value < 0.01 and methylation difference larger than 10% were used to define DMSs.
Genomic Landscape of DMS.
The DMS distances to TSSs were determined using the Genomic Regions Enrichment of Annotations Tool (http://great.stanford.edu/public/html/) (details are in SI Appendix), and the genomic distribution was determined by using the script of read_distribution.py file with the reference file (hg38_RefSeq.bed) in the software package, RSeQC (version 2.6.4). LOLA (22) was to identify significant overlaps of DMS regions with transcription factor binding sites based on ChIP-seq datasets obtained from ENCODE (59) and CODEX (60) databases. The Fisher’s exact test was used with a significance threshold of 0.05 on FDR-adjusted P values. The whole-enrichment results together with their original and curated annotations are presented in SI Appendix, Table S4.
Cell Type-Specific DNA Methylation.
The DNA methylation data (data type: methylation signal) for specific leukocytes were downloaded from Blueprint Epigenome Project DCC portal (31). The analysis was performed using the selected 37 Blueprint Bisulfite-Seq experiments (SI Appendix, Table S5). The low-coverage CpG sites (<10-read coverage) in each sample were first excluded, and intersection against the genome-wide locations of autosomal CpG sites was performed, resulting in a total of 4,861,556 CpG sites. The details are in SI Appendix.
Supplementary Material
Acknowledgments
These studies were supported in part by NIH Grants U01-AI124319 (to M.R.Y.), R01-AI068804 (to V.G.F.), R33-AI111661 (to M.R.Y.), U01-AI124319 (to E.F.R.), and U19AI128913 (to E.F.R.).
Footnotes
Competing interest statement: V.G.F. reports grant/research support from MedImmune, Cerexa/Forest/Actavis/Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Cubist/Merck, Medical Biosurfaces, Locus, Affinergy, Contrafect, Karius, Genentech, Regeneron, and Basilea; paid consultant for Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy, Janssen, xBiotech, Contrafect, Regeneron, Basilea, and Destiny; membership with Merck as cochair for V710 vaccine; educational fees from Green Cross, Cubist, Cerexa, Durata, Theravance, and Debiopharm; and royalties from UpToDate. M.R.Y. is a founder and shareholder of NovaDigm Therapeutics, Inc., which develops vaccines and immunotherapeutics targeting multidrug-resistant pathogens, including S. aureus.
This article is a PNAS Direct Submission.
3A complete list of the MRSA Systems Immunobiology Group can be found in SI Appendix.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000663118/-/DCSupplemental.
Contributor Information
Collaborators: Richard Ahn, Arnold S. Bayer, Liana C. Chan, Yu-Ling Chang, Scott G. Filler, Vance G. Fowler, Jr, David Gjertson, Alexander Hoffmann, Felix Mba Medie, Tsuyoshi Mikkaichi, Simon Mitchell, Yan Qin, Elaine F. Reed, Maura Rossetti, Felicia Ruffin, Batu K. Sharma-Kuinkel, Katherine Sheu, Joshua Thaden, Alan J. Waring, Yan Q. Xiong, Ying Zheng, and Michael R. Yeaman
Data Availability
The DNA methylation sequence data reported in this paper have been deposited into the Gene Expression Omnibus (accession no. GSE150144). All other data, associated protocols, and materials used in this work are described in the paper.
References
- 1.Tong S. Y., Davis J. S., Eichenberger E., Holland T. L., Fowler V. G. Jr, Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowler V. G. Jr, et al., Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190, 1140–1149 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Sakoulas G., et al., Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49, 2687–2692 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y. Q., et al., Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J. Infect. Dis. 199, 201–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner N. A., et al., Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harms A., Maisonneuve E., Gerdes K., Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354, aaf4268 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jiang J. H., et al., Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. U.S.A. 116, 3722–3727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Pietro A., Good-Jacobson K. L., Disrupting the code: Epigenetic dysregulation of lymphocyte function during infectious disease and lymphoma development. J. Immunol. 201, 1109–1118 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Chan L. C., et al., Innate immune memory contributes to host defense against recurrent skin and skin structure infections caused by methicillin-resistant Staphylococcus aureus. Infect. Immun. 85, e00876-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan L. C.et al.; MRSA Systems Immunobiology Group , Protective immunity in recurrent Staphylococcus aureus infection reflects localized immune signatures and macrophage-conferred memory. Proc. Natl. Acad. Sci. U.S.A. 115, E11111–E11119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mba Medie F.et al.; MRSA Systems Immunobiology Group , Genetic variation of DNA methyltransferase-3A contributes to protection against persistent MRSA bacteremia in patients. Proc. Natl. Acad. Sci. U.S.A. 116, 20087–20096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suarez-Alvarez B., Rodriguez R. M., Fraga M. F., López-Larrea C., DNA methylation: A promising landscape for immune system-related diseases. Trends Genet. 28, 506–514 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Bhutani N., Burns D. M., Blau H. M., DNA demethylation dynamics. Cell 146, 866–872 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moinova H. R., et al., Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Sci. Transl. Med. 10, eaao5848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacis A., et al., Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 25, 1801–1811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinclair S. H., Yegnasubramanian S., Dumler J. S., Global DNA methylation changes and differential gene expression in Anaplasma phagocytophilum-infected human neutrophils. Clin. Epigenetics 7, 77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michigami Y., et al., Long-term effects of H. pylori eradication on epigenetic alterations related to gastric carcinogenesis. Sci. Rep. 8, 14369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou H., Hastie T., Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67, 301–320 (2005). [Google Scholar]
- 19.Akalin A., et al., methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W., et al., CGmapTools improves the precision of heterozygous SNV calls and supports allele-specific methylation detection and visualization in bisulfite-sequencing data. Bioinformatics 34, 381–387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyon I., Weston J., Barnhill S., Vapnik V., Gene selection for cancer classification using support vector machines. Mach. Learn. 46, 389–422 (2002). [Google Scholar]
- 22.Sheffield N. C., Bock C., LOLA: Enrichment analysis for genomic region sets and regulatory elements in R and bioconductor. Bioinformatics 32, 587–589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirai H., et al., C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7, 732–739 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Sow F. B., et al., Role of STAT1, NF-kappaB, and C/EBPbeta in the macrophage transcriptional regulation of hepcidin by mycobacterial infection and IFN-gamma. J. Leukoc. Biol. 86, 1247–1258 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Adcock I. M., Caramori G., Cross-talk between pro-inflammatory transcription factors and glucocorticoids. Immunol. Cell Biol. 79, 376–384 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Adcock I. M., Ito K., Barnes P. J., Glucocorticoids: Effects on gene transcription. Proc. Am. Thorac. Soc. 1, 247–254 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Dendoncker K., et al., TNF-α inhibits glucocorticoid receptor-induced gene expression by reshaping the GR nuclear cofactor profile. Proc. Natl. Acad. Sci. U.S.A. 116, 12942–12951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fishilevich S., et al., GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017, bax028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farlik M., et al., DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 19, 808–822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L., et al., Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martens J. H., Stunnenberg H. G., BLUEPRINT: Mapping human blood cell epigenomes. Haematologica 98, 1487–1489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orozco L. D., et al., Epigenome-wide association in adipose tissue from the METSIM cohort. Hum. Mol. Genet. 27, 2586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen P. Y., et al., Prenatal growth patterns and birthweight are associated with differential DNA methylation and gene expression of cardiometabolic risk genes in human placentas: A discovery-based approach. Reprod. Sci. 25, 523–539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam L., et al., Epigenetic changes in T-cell and monocyte signatures and production of neurotoxic cytokines in ALS patients. FASEB J. 30, 3461–3473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morselli M., et al., Targeted bisulfite sequencing for biomarker discovery. Methods, 10.1016/j.ymeth.2020.07.006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz S., Romanoski C. E., Benner C., Glass C. K., The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clermont P. L., Parolia A., Liu H. H., Helgason C. D., DNA methylation at enhancer regions: Novel avenues for epigenetic biomarker development. Front. Biosci. 21, 430–446 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Sharifi-Zarchi A., et al., DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1-H3K4me3 seesaw mechanism. BMC Genomics 18, 964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah C. A., et al., Stat3 and CCAAT enhancer-binding protein β (C/ebpβ) activate Fanconi C gene transcription during emergency granulopoiesis. J. Biol. Chem. 293, 3937–3948 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller L. S., Cho J. S., Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 11, 505–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bröker B. M., Mrochen D., Péton V., The T cell response to Staphylococcus aureus. Pathogens 5, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapgier A., et al., A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 119, 1502–1514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toubiana J.et al.; International STAT1 Gain-of-Function Study Group , Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood 127, 3154–3164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villarino A. V., Gallo E., Abbas A. K., STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J. Immunol. 185, 6461–6471 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkatesh B.et al.; ADRENAL Trial Investigators and the Australian–New Zealand Intensive Care Society Clinical Trials Group , Adjunctive glucocorticoid therapy in patients with septic shock. N. Engl. J. Med. 378, 797–808 (2018). [DOI] [PubMed] [Google Scholar]
- 46.van den Akker E. L., et al., Glucocorticoid receptor mRNA levels are selectively decreased in neutrophils of children with sepsis. Intensive Care Med. 35, 1247–1254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vardas K., et al., Increased glucocorticoid receptor expression in sepsis is related to heat shock proteins, cytokines, and cortisol and is associated with increased mortality. Intensive Care Med. Exp. 5, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostuni R., et al., Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Quintin J., Cheng S. C., van der Meer J. W., Netea M. G., Innate immune memory: Towards a better understanding of host defense mechanisms. Curr. Opin. Immunol. 29, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Saeed S., et al., Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maekita T., et al., High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin. Cancer Res. 12, 989–995 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Pacis A., et al., Gene activation precedes DNA demethylation in response to infection in human dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 116, 6938–6943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu K., et al., A temporal transcriptome and methylome in human embryonic stem cell-derived cardiomyocytes identifies novel regulators of early cardiac development. Epigenetics 13, 1013–1026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leggett R. M., Ramirez-Gonzalez R. H., Clavijo B. J., Waite D., Davey R. P., Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front. Genet. 4, 288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo W., et al., BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14, 774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ecker S.et al.; BLUEPRINT Consortium , Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types. Genome Biol. 18, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrell C., Thompson M., Tosevska A., Oyetunde A., Pellegrini M., BiSulfite bolt: A BiSulfite sequencing analysis platform. 10.1101/2020.10.06.328559 (8 October 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harrow J., et al., GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sánchez-Castillo M., et al., CODEX: A next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 43, D1117–D1123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA methylation sequence data reported in this paper have been deposited into the Gene Expression Omnibus (accession no. GSE150144). All other data, associated protocols, and materials used in this work are described in the paper.