Significance
This work examines a fundamental question of how bacteria sense plant-released chemicals. We recently identified an effector of one member of a plant-responsive PipR family of transcription factors present in many plant-associated bacteria. This compound (abbreviated HEHEAA) requires a specific transporter for import into bacterial cells. We have solved crystal structures of one component of the transporter free and bound to HEHEAA. We discovered that a close homolog of the transporter protein cannot bind HEHEAA, implying there are other effector compound(s) for the widespread PipR signaling system family. Understanding the molecular details of these plant-responsive systems could identify a means of controlling plant colonization.
Keywords: Populus, Pseudomonas signaling, substrate-binding protein
Abstract
A number of plant-associated proteobacteria have LuxR family transcription factors that we refer to as PipR subfamily members. PipR proteins play roles in interactions between bacteria and their plant hosts, and some are important for bacterial virulence of plants. We identified an ethanolamine derivative, N-(2-hydroxyethyl)-2-(2-hydroxyethylamino) acetamide (HEHEAA), as a potent effector of PipR-mediated gene regulation in the plant endophyte Pseudomonas GM79. HEHEAA-dependent PipR activity requires an ATP-binding cassette-type active transport system, and the periplasmic substrate-binding protein (SBP) of that system binds HEHEAA. To begin to understand the molecular basis of PipR system responses to plant factors we crystallized a HEHEAA-responsive SBP in the free- and HEHEAA-bound forms. The SBP, which is similar to peptide-binding SBPs, was in a closed conformation. A narrow cavity at the interface of its two lobes is wide enough to bind HEHEAA, but it cannot accommodate peptides with side chains. The polar atoms of HEHEAA are recognized by hydrogen-bonding interactions, and additional SBP residues contribute to the binding site. This binding mode was confirmed by a structure-based mutational analysis. We also show that a closely related SBP from the plant pathogen Pseudomonas syringae pv tomato DC3000 does not recognize HEHEAA. However, a single amino acid substitution in the presumed effector-binding pocket of the P. syringae SBP converted it to a weak HEHEAA-binding protein. The P. syringae PipR depends on a plant effector for activity, and our findings imply that different PipR-associated SBPs bind different effectors.
Many plant-associated proteobacteria, including several plant pathogens, possess LuxR-type transcription factors (1–7), which we refer to as PipR transcriptional regulators. These widespread but understudied regulators are similar to acyl-homoserine lactone (AHL)-responsive quorum-sensing regulators (8), but PipR family members do not respond to AHLs and instead respond to effector chemicals that exist in plant tissues. Genes coding for PipR family members are located upstream of pipA, a gene coding for a proline iminopeptidase (Pip) (3). PipRs generally activate the expression of pipA and other genes in response to plant tissue or plant-derived chemicals (1, 3, 4, 7, 9). In several species, the pipR-type gene (1, 5, 7) is adjacent to and divergently transcribed from an operon encoding an ATP-binding cassette (ABC)-type transporter (10, 11). This is the case for the cottonwood tree (Populus) endophyte Pseudomonas sp. GM79 (Fig. 1A), and we have shown that the aapBCDEF transporter operon is required for the PipR response to plant effectors and activation of pipA expression.
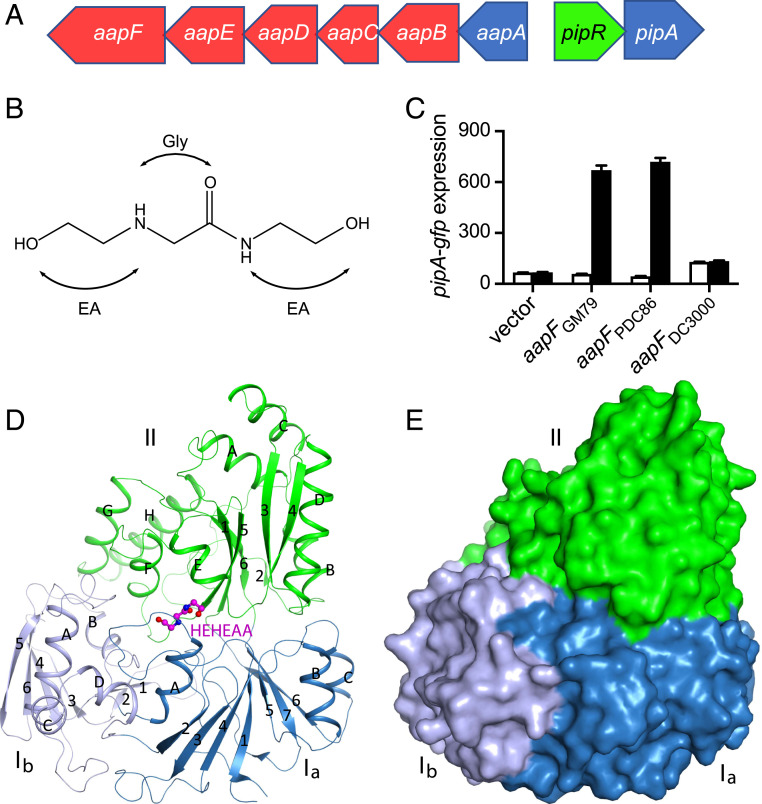
Fig. 1.
The Pseudomonas GM79 pipR genomic region, HEHEAA chemical structure, AapF activity analysis, and structure of SBPPDC86-HEHEAA complex. (A) The pipR (green) genomic region. Genes coding for the predicted ABC-type transporter are in red, and genes coding for peptidases are in blue. (B) Chemical structure of HEHEAA with the ethanolamine (EA) and glycine (Gly) substructures indicated. (C) SBPPDC86, but not SBPDC3000, can substitute for SBPGM79. Data are pipA-gfp fluorescence in a GM79 aapF mutant harboring plasmids with the indicated aapF gene and a pipA-gfp reporter. Cells were grown with (black bars) or without (white bars) 100 nM HEHEAA. The pipA-gfp expression is in relative fluorescence units (RFU) per optical density (OD600) of four biological replicates, and the error bars represent the SDs of the mean. (D) Ribbon diagram of the overall structure of SBPPDC86-HEHEAA complex. The three domains Ia, Ib, and II are labeled and colored in sky blue, gray blue, and green, respectively. β-strands and α-helices are labeled with numbers and letters, respectively. The ligand HEHEAA is shown as ball-and-stick model in magenta. (E) Surface representation of SBPPDC86 with the same color scheme as in D. The HEHEAA ligand is completely buried inside the structure. The structure figures were produced with PyMOL (www.pymol.org).
We recently identified N-(2-hydroxyethyl)-2-(2-hydroxyethylamino) acetamide (HEHEAA), an ethanolamine derivative, as a potent PipR effector (Fig. 1B). Strain GM79 responds to picomolar concentrations of HEHEAA to activate expression of pipA (12). The first step in the response to HEHEAA is believed to be binding of this effector to the AapF periplasmic substrate binding protein (SBP). The current model for HEHEAA-dependent PipR activation of pipA expression is that the AapF protein binds HEHEAA and delivers it to the AapBCDE membrane transporter, and PipR then binds intracellular HEHEAA to activate pipA expression (4, 12).
To begin to understand the recognition of plant chemical effectors by PipR systems, we determined crystal structures of an AapF SBP. We report structures of both HEHEAA-bound and HEHEAA-free AapF and provide genetic evidence to support our findings about the HEHEAA-binding pocket. We report that the AapF homolog from the tomato pathogen Pseudomonas syringae pv tomato DC3000 does not bind HEHEAA but can be converted to a relatively inefficient HEHEAA-binding protein by introducing a single amino acid substitution in its predicted effector-binding pocket.
Results
Preparation and Structure Elucidation of an AapF in the Free and HEHEAA-Bound Forms.
Purified AapF from Pseudomonas GM79 binds HEHEAA, and an aapF-null mutant is impaired in HEHEAA-dependent activation of a pipA-gfp reporter (12) (Fig. 1C), presumably because HEHEAA is unable to enter cells. We wanted to determine the structure of the GM79 AapF in complex with HEHEAA to identify residues important for its binding. We purified a His-tagged cytoplasmic version of GM79 AapF, AapFGM79, from Escherichia coli by our previously described procedure (12) but were unable to obtain crystals. Thus, we examined two other Pseudomonas strains that had similar pipR, pipA, and ABC transporter (aapB–F) genes; Pseudomonas sp. PDC86, isolated from Populus roots (13), and the plant pathogen P. syringae pv tomato DC3000 (14). The AapF homologs of PDC86 and DC3000 have extensive amino acid sequence identity (PDC86, 81%; DC3000, 76%) and similarity (PDC86, 95%; DC3000, 93%) to GM79 AapF (SI Appendix, Fig. S1). The PDC86 aapF gene complemented the GM79 aapF mutant phenotype and activated pipA-gfp expression (Fig. 1C). From this, we conclude that the AapF from PDC86 can bind HEHEAA and deliver it to the GM79 AapBCDE active transport machinery. Interestingly, aapF from the plant pathogen DC3000 did not complement the GM79 aapF mutant phenotype (Fig. 1C), suggesting that the DC3000 PipR system may not respond to HEHEAA, or that the DC3000 AapF cannot interact properly with the GM79 Aap membrane transporter.
We expressed and purified a His-tagged cytoplasmic version of the Pseudomonas PDC86 AapF, also referred to as AapFPDC86 or SBPPDC86. In contrast to AapFGM79, the AapFPDC86 protein yielded high-resolution crystals (2.2 Å), from which we determined an HEHEAA-free structure. To obtain HEHEAA-bound AapFPDC86, we cocrystallized the purified protein with 25 mM chemically synthesized HEHEAA and successfully determined the structure of the complex at 1.9 Å resolution. The crystallographic information is summarized in SI Appendix, Table S1. Both structures have excellent agreement with the crystallographic data and the expected geometric parameters. We found 97% of the residues in the favored region of the Ramachandran plot, with no residues in the disallowed region.
Overall Structure of the HEHEAA-AapFPDC86 Complex and the Binding Mode of HEHEAA.
SBP family members bind a wide variety of ligands, including amino acids, peptides, saccharides, and inorganic and organic ions (15), all of which share a common three-dimensional structure. The superclass of SBPs has been divided into six clusters based on known ligand specificities and crystal structure data (16). Structurally, AapFPDC86 falls into cluster C. Like other cluster C SBPs (reviewed in refs. 10, 11), the AapFPDC86 HEHEAA-bound structure consists of two lobes connected by two loops and is in a closed conformation (Fig. 1 D and E). Lobe II has one α/β-type domain composed of a six-stranded β-sheet and eight α-helices flanking its two faces. Lobe I can be divided into two α/β-type domains, Ia and Ib, with the latter present only in C-type SBPs. Domain Ia is composed of a seven-stranded β-sheet surrounded by two α-helices on one face and one helix on the other face (11). Both the N and C termini of SBPPDC86 are located in this domain. Domain Ib contains a four-stranded β-sheet and is located at the periphery of the structure. Four α-helices and a β-hairpin are on one of its faces and have intimate contacts with domain Ia and lobe II (Fig. 1D). The other face of this β-sheet is exposed to the solvent. Residues on the surface of domain Ib, especially those in this β-sheet, have weaker electron density and higher Β-factors based on the crystallographic analysis, suggesting some flexibility for this region.
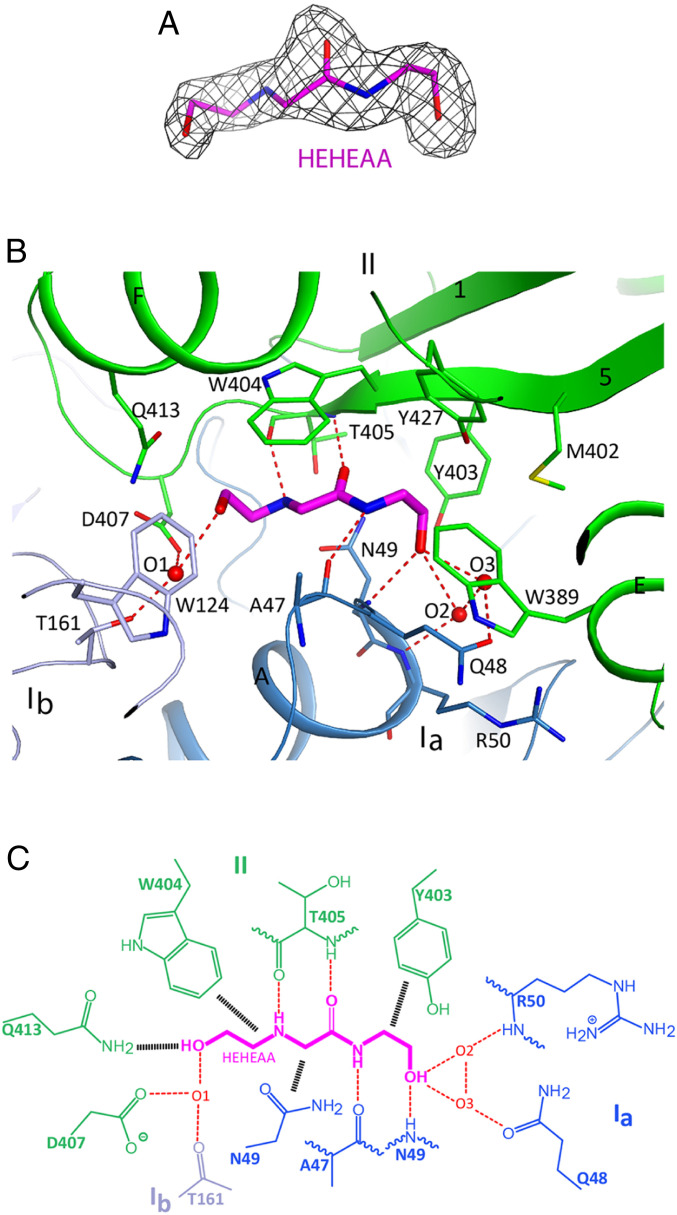
Strong electron density was observed for the HEHEAA ligand based on the crystallographic analysis at 1.9 Å resolution (Fig. 2A). The HEHEAA-binding site is in the protein center, at the interface between the two lobes, and the compound contacts all three domains (Fig. 1D). The ligand is buried deep in the structure and is inaccessible to the solvent (Fig. 1E). HEHEAA has extensive interactions with SBPPDC86, including hydrogen-bonding, π-stacking, and van der Waals interactions (Fig. 2 B and C). All the polar atoms of HEHEAA are hydrogen-bonded to SBPPDC86, including the central glycine moiety (Fig. 1B), which has an anti-parallel β interaction with the main chain of T405, while the amide nitrogen of HEHEAA is hydrogen-bonded to the main chain carbonyl of A47. The hydroxyl at one end of HEHEAA is hydrogen-bonded to the main chain amide of N49. Three water molecules mediate hydrogen-bonding interactions between the SBP and the hydroxyls at both ends of HEHEAA (Fig. 2C). The amide group of HEHEAA is π-stacked with the side chain of W404 on one side, while its other side is positioned against the N49 side chain (Fig. 2B). The side chains of several other amino acid residues, including W124, W389, Y403, D407, and Q413, are located near the HEHEAA ligand and exhibit polar and van der Waals interactions. All the HEHEAA-interacting residues are conserved between AapFPDC86 and AapFGM79 (SI Appendix, Fig. S2).
Fig. 2.
Binding mode of HEHEAA in SBPPDC86. (A) Simulated-annealing omit Fo–Fc electron density map for the HEHEAA ligand at 1.9 Å resolution, contoured at 2.5 σ. (B) Schematic drawing of the HEHEAA ligand-binding site showing detailed interactions with SBPPDC86. Residues in contact with HEHEAA are shown in sticks and labeled. Hydrogen bonds are indicated as red dashed lines. (C) Schematic drawing of the interactions between HEHEAA and SBPPDC86. Van der Waals and π-stacking interactions are indicated with black hatched lines. Three water molecules (O1 to O3) bridge the interactions between HEHEAA and SBPPDC86.
The structure of free SBPPDC86 is also in a closed conformation, with only a few side chains exhibiting conformational differences compared with the HEHEAA-bound complex, including D407, Q413, W124, and W404 (SI Appendix, Fig. S3). Although a closed-unliganded form of an SBP is somewhat unusual (10, 11, 17), there are reports of ligand-free closed forms for other SBPs, including glucose/galactose-binding protein (GGBP) (18), glutamine-binding protein (GlnBP) (17), and choline-binding protein (ChoX) (19), none of which, however, are members of cluster C (10, 11).
Comparison of SBPPDC86 with a Peptide-Binding SBP, OppA.
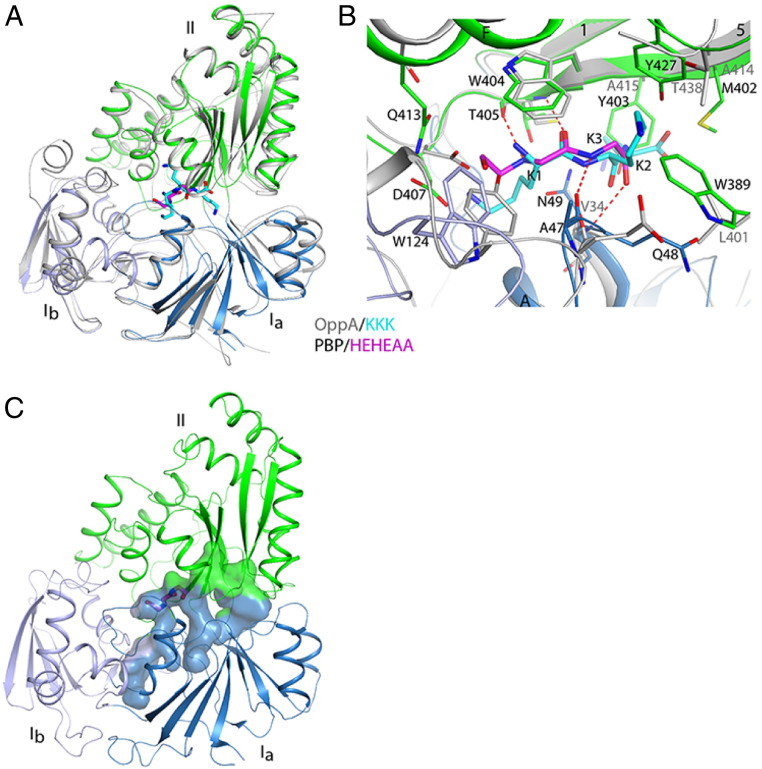
The SBPPDC86 structure shows extensive similarity to the cluster C SBPs that bind peptides, likely a reflection of the peptidomimetic nature of the HEHEAA chemical structure (Fig. 1B). A Dali search (20) of the Protein Data Bank identified >40 similar structures (Z-scores >20; average rms distance, 2.5 Å; sequence identity ∼22% for aligned Cα atoms). For a more detailed comparison with AapFPDC86, we selected the oligopeptide-binding protein OppA from Yersinia pestis (21), which has a Z-score of 42.7 and an rms distance of 2.2 Å to AppFPDC86, although OppA has a few extra helices in domain Ib (Fig. 3A). The binding mode for the tripeptide ligand (Lys-Lys-Lys) in OppA is similar to that of HEHEAA in SBPPDC86, especially in terms of the hydrogen-bonding interactions with the backbone carbonyls and amides (Fig. 3B). The glycine substructure in HEHEAA (Fig. 1B) overlaps the first residue of the Lys-Lys-Lys tripeptide (Fig. 3B). Although the W404 residue is conserved between SBPPDC86 and OppA, there are substantial differences between ligand-binding pocket residues (SBPPDC86 vs. OppA): N49 vs. V34; W389 vs. L401; M402 vs. A414; and Y403 vs. A415 (Fig. 3B). Each difference results in a comparatively smaller binding cavity for SBPPDC86, which allows for only small ligands lacking side chains (like HEHEAA) to fit. This is in contrast to the tripeptide-binding cavity in OppA, which is much wider (SI Appendix, Fig. S4). SBPPDC86 has a relatively long cavity, extending from either end of the HEHEAA ligand, suggesting that SBPPDC86 might be able to accommodate additional, similar ligands (Fig. 3C).
Fig. 3.
Comparison of SBPPDC86 with the structure of a peptide-binding SBP. (A) Superimposition of the SBPPDC86 (in color) structure in complex with HEHEAA (magenta) with the structure of Y. pestis OppA (gray) in complex with the Lys-Lys-Lys tripeptide (cyan; PDB ID code 2Z23). The Ib domain has higher variability than the other two domains. (B) Comparison of the binding mode of HEHEAA (magenta) in the complex with SBPPDC86 (colored) with that of Lys-Lys-Lys tripeptide (cyan) in the complex with OppA (gray). (C) Surface representation for the ligand-binding cavity in SBPPDC86. The HEHEAA ligand is shown as sticks.
Functional Studies Support the Crystal Structure Data.
To confirm our crystal structure data, we evaluated the effect of specific amino acid substitutions on HEHEAA binding in vivo. Because we have an established genetic system in Pseudomonas sp. GM79 (4), and because all the residues contained in the HEHEAA-binding pocket are conserved between SBPPDC86 and SBPGM79 (SI Appendix, Fig. S2), we created the desired SBP mutations in aapFGM79 and crossed the mutations into the GM79 aapF gene. We determined whether the mutant strains could still respond to HEHEAA using a pipA-gfp reporter plasmid (pJN-PpipA-gfp). We found that the substitutions N49P, W389A, W404A, and T405P abolished the response to HEHEAA, and that substitutions Y403A and D407A showed large reductions in the signal response (Table 1 and SI Appendix, Fig. S5). Only the gene encoding the T405A substitution remained capable of significant HEHEAA-dependent pipA-gfp activation, albeit to a lesser degree than wild-type SBPGM79 (Table 1).
Table 1.
Amino acid changes in the HEHEAA-binding pocket of AapFGM79 affect HEHEAA binding and protein thermal stability
| AapFGM79 seq* | Relative pipA-gfp† | ΔTm +/− HEHEAA‡ |
| None | 0.0 (0.4) | NA§ |
| Wild-type | 100.0 | 14 |
| N49P | 0.7 (0.4) | 0 |
| W389A | 1.4 (0.3) | 1 |
| Y403A | 9.7 (3.7) | 1 |
| W404A | 1.3 (0.7) | 1 |
| T405A | 64.0 (14.4) | 5 |
| T405P | 2.5 (1.4) | ND¶ |
| D407A | 16.3 (1.7) | 4 |
Amino acid sequence of the indicated Pseudomonas GM79 AapF polypeptides: wild-type, the aapF deletion mutant (none), or the indicated point mutants.
HEHEAA activation of the plasmid-borne pipA-gfp reporter in GM79 with the indicated AapF (aapF alleles and pipR are chromosomal). Data are normalized to the wild-type and are means of four biological replicates with the SDs in parentheses. Raw data and complementation data are presented in SI Appendix, Fig. S5.
Melting temperature changes (ΔTm) (Materials and Methods) of the indicated purified proteins in the presence (+) or absence (−) of 100 μM HEHEAA. Representative thermal shift data are shown in SI Appendix, Fig. S6.
Not applicable.
Not determined, no variant protein was obtained.
We also evaluated the ability of each variant polypeptide to bind HEHEAA in vitro by using a fluorescence-based thermal shift assay. In these experiments, the melting temperature (Tm) of wild-type and variant polypeptides (His-tagged, cytoplasmic versions; Materials and Methods) in the presence and absence of HEHEAA were compared. HEHEAA increased the Tm of wild-type AapFGM79 by 14 °C (Table 1 and SI Appendix, Fig. S6), consistent with the conclusion that AapFGM79 binds the ligand and renders it more resistant to thermal denaturation. In contrast, the variant polypeptides showed little or no Tm increase in response to HEHEAA (Table 1 and SI Appendix, Fig. S6). The two variants exhibiting moderate pipA-gfp reporter activity, T405A and D407A, showed some AapF-stability in the thermal shift assay, whereas the other four mutant proteins showed Tm shifts of 1 °C or less.
The P. syringae DC3000 AapF Homolog Does Not Bind HEHEAA, Indicating There May Be Additional Effectors for Other Plant- Responsive PipR Systems.
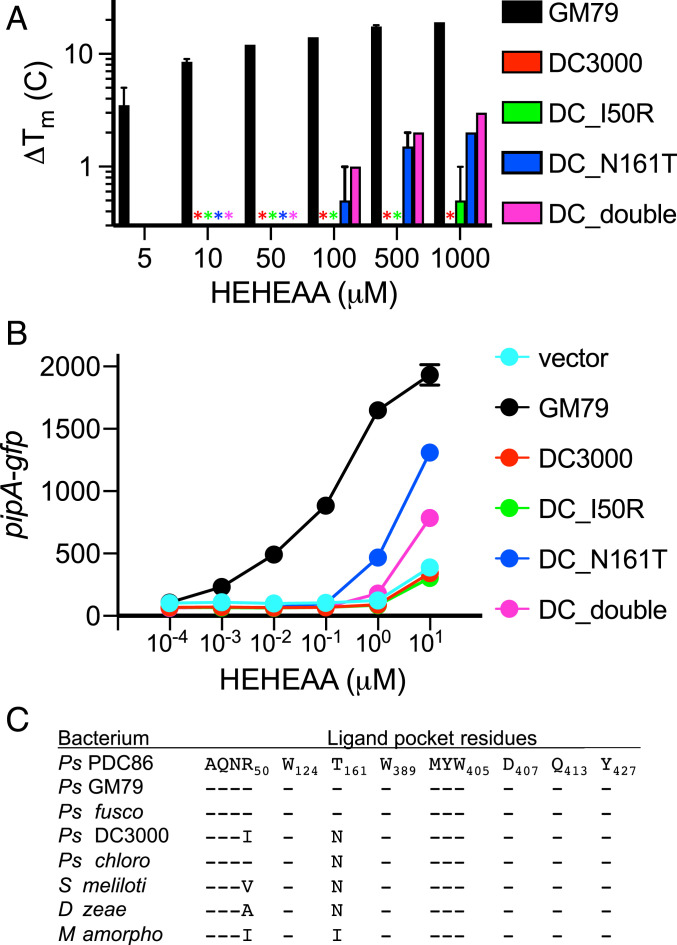
As discussed earlier, the AapF homolog from the plant pathogen P. syringae DC3000 (abbreviated as AapFDC3000 or SBPDC3000 hereinafter) was unresponsive to HEHEAA (Fig. 1C). We examined whether the 13 amino acid residues that compose the HEHEAA-binding pocket (SI Appendix, Fig. S1) were conserved and found two differences (SBPPDC86/GM79 vs. SBPDC3000), R50I and T161N, although neither of these residues interacts directly with HEHEAA in the SBPPDC86 structure (Fig. 2). To test their contribution to HEHEAA binding, we made appropriate changes to the aapFDC3000 gene so that amino acid residues in SBPDC3000 corresponded to the SBPGM79 residues. We then tested whether the altered SBPDC3000 proteins showed the capacity to bind HEHEAA using two different approaches. We first used the fluorescence-based thermal shift assay with purified DC3000 wild-type SBP and with purified DC3000 SBPs with an R50I substitution, a T161N substitution, or both substitutions (Fig. 4A and SI Appendix, Fig. S7). Over the range of HEHEAA concentrations tested (5 to 1,000 μM) the DC3000 wild-type protein showed no significant Tm change, and neither did the I50R substitution mutant. HEHEAA was able to induce a Tm shift for both the N161T and the I50R N161T substitution mutant proteins. These shifts were modest compared with that of purified GM79 AapF and required high concentrations of HEHEAA relative to those required to induce a shift of the GM79 SBP.
Fig. 4.
Not all AapF homologs bind HEHEAA. (A) Variant, but not wild-type, AapFDC3000 polypeptides bind HEHEAA in thermal shift experiments. The mean melting temperature changes (ΔTm) of three technical replicates for each of two biological replicates for each indicated AapF are shown and the error bars represent the range of values. The stars indicate there was no thermal shift. Thermal shift raw data are plotted in SI Appendix, Fig. S7. (B) Genes encoding a specific SBPDC3000 variant (N161T) can complement a GM79 aapF deletion mutant. Data are GFP fluorescence in a GM79 aapF mutant harboring the indicated plasmids. Plasmids used are pJN-PpipA-gfp (vector), pJN-aapFGM79PpipA-gfp (GM79), pJN-aapFDC3000PpipA-gfp (DC3000), pJN-aapFDC_I50RPpipA-gfp (DC_I50R), pJN-aapFDC_N161TPpipA-gfp (DC_N161T), and pJN-aapFDC_doublePpipA-gfp (DC_double). Data are mean RFU per optical density (OD600) of four biological replicates, and the error bars represent the SD of the mean. (C) The ligand-binding pocket residues in AapF homologs from eight bacterial species. The AapF homologs used for comparisons are provided in SI Appendix, Table S5. Residue comparisons are relative to the SBPPDC86 (Top), and a dash (-) indicates that the corresponding residue is the same as in SBPPDC86.
A second way to test whether the DC3000 mutant proteins can bind HEHEAA is to ask whether genes coding for the wild-type or mutant DC3000 SBPs can complement a GM79 aapF mutation using our pipA transcription reporter assay (Fig. 4B). Compared with the melting temperature experiments, this approach involves some additional specificity. A response requires that an SBP binds HEHEAA and delivers it to the membrane-bound transporter. The effector then must be delivered to the cytoplasm, where it has to interact with and activate the GM79 PipR transcription factor. Results were consistent with the in vitro Tm shift results. The DC3000 wild-type or I50R-substituted polypeptide cellular responses were indistinguishable from the results with a vector control. The genes coding for the DC3000 N161T protein showed partial complementation of the GM79 aapF mutation at relatively high HEHEAA concentrations compared with cells with the native GM79 SBP.
Both the in vitro and in vivo experiments are consistent with the idea that the presence of an asparagine at position 161 could cause some steric hindrance in that area, which would be relieved when substituted with a threonine. The SBPDC3000 I50R, N161T mutant protein (SBPdouble) did not show enhanced activity over the single N161T variant protein (Fig. 4 A and B). That the N161T SBPDC3000 showed a significantly reduced sensitivity relative to SBPGM79 is consistent with the conclusion that amino acid residues outside of the HEHEAA-binding pocket affect the ability of these proteins to interact with their ligands. We assume that the P. syringae DC3000 AapF binds an effector other than, but possibly similar to, HEHEAA.
We identified aapF homologs in several sequenced genomes and compared them with one another (3) (Fig. 4C and SI Appendix, Table S5). The plant pathogen Pseudomonas fuscovaginae AapF shares all of the ligand-binding pocket residues identified in PDC86 and GM79, and thus this bacterium might respond to HEHEAA. However, AapF from the biocontrol strain Pseudomonas chlororaphis is like the DC3000 AapF protein and has an N rather than a T at position 161. We imagine that P. chlororaphis does not respond to HEHEAA. Beyond the genus Pseudomonas, the pathogen Dickeya zeae (22) and symbiont Sinorhiozbium meliloti (23) both possess PipR/PipA-type systems that influence their interactions with plants. AapF homologs from both of these strains have the N161T (relative to AapFGM79) difference, and thus we suspect that they do not respond to HEHEAA.
Discussion
Our studies have revealed molecular details of how HEHEAA is recognized by the AapF SBPs of Pseudomonas strains PDC86 and GM79. Our crystal structure analysis and associated functional analyses (Table 1) demonstrate that the two SBPs bind HEHEAA in a manner similar to how peptide-binding SBPs bind their ligands.
Although it is closely related to AapFPDC86 and AapFGM79, the P. syringae AapFDC3000 does not bind HEHEAA (Figs. 1C and 4). Analysis of the AapFPDC86 HEHEAA binding-pocket sequence revealed two residues that differed in AapFDC3000. Although AapFDC3000 could not substitute for AapFGM79, an AapFDC3000 variant with a single amino acid substitution (N161T) could serve as a poor substitute for AapFGM79 in Pseudomonas strain GM79, perhaps due to a relief of steric hindrance from the N residue in the area. Not surprisingly, the AapFDC3000 N161T mutant showed reduced activity relative to AapFGM79 in both in vitro and in vivo experiments (Fig. 4). The inability of the P. syringae DC3000 AapF to bind HEHEAA indicates that there is an alternative plant-derived effector for the P. syringae PipR system. Whether this effector can also bind the GM79/PDC86 AapFs is an interesting question, as our results suggest that the SBPPDC86 structure could accommodate additional similar, but longer, ligands (Fig. 3C). We found that the residue required for HEHEAA binding (T161) is present only in a subset of pseudomonads, and that most other AapF-containing bacteria possess N161. It may be that other ethanolamine derivatives serve as PipR signals in different bacteria, and because HEHEAA is a peptidomimic, there might even be dipeptides serving as PipR ligands for some systems. We are intrigued by the idea that fatty acyl-ethanolamine derivatives might be signals for some related systems. Such molecules are produced by plants and function as growth and development hormones (24).
Recognition of HEHEAA by AapFPDC86/GM79 and subsequent active transport by the AapBCDE transport apparatus is required for PipR-dependent plant–bacterial interactions (12), so understanding the details of how the ligand is recognized provides insight into how bacteria perceive an underexplored plant-derived compound. Our work indicates that there is specificity in PipR systems, and that different effectors will be discovered for different PipR systems. This specificity is at least in part dependent on the SBPs in the ABC transport systems. It remains to be determined if there are other layers of specificity to these systems. We can ask whether P. syringae DC3000 with its own SBP engineered to recognize HEHEAA can respond to HEHEAA. If not, we can then ask whether such a strain can transport HEHEAA, and whether the P. syringae PipR can recognize HEHEAA. Our identification of a diagnostic residue for HEHEAA binding (T161) could also aid future work focused on bioinformatic predictions of SBP ligands, an area of active interest (25–27). Building on this work, we hope to identify other plant inducers of Pip systems in other bacteria.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains and plasmids are described in SI Appendix, Tables S2 and S3. Pseudomonas sp. GM79 and its derived strains were grown in Lennox broth (LB) (28) or M9 minimal medium (29) with 10 mM succinate (succ-MM) at 30 °C with shaking unless indicated otherwise. E. coli strains were grown in LB or M9 minimal medium with 20 mM glucose, 1 μg/mL biotin, and 1 μg/mL thiamine (gluc-MM) at 37 °C with shaking. Antibiotics were used when appropriate at the following concentrations: 50 μg/mL (E. coli) or 25 μg/mL (Pseudomonas GM79) for kanamycin and 20 μg/mL (E. coli) or 50 μg/mL (Pseudomonas GM79) for gentamicin. Plating was done using media solidified with 1.5% (wt/vol) agar.
SBP Point Mutant Constructions, Activity Assays, and Complementation Assays.
Plasmids and primers are described in SI Appendix, Tables S3 and S4. We generated aapFGM79 point mutant plasmids by PCR sequence overlap extension (30). We assembled the construct on the suicide vector pEX19Gm by E. coli DH5α-mediated assembly (31). The mutant aapFGM79 genes were crossed into their native location on the GM79 chromosome as described previously (32). We used pipA-gfp to monitor HEHEAA-dependent SBP activity. Overnight cultures of GM79 wild-type or SBPGM79 point mutant strains harboring pJN-PpipA-gfp (SI Appendix, Tables S2 and S3) were used to inoculate succ-MM (5% vol/vol). Cultures were dispensed into 384-well microtiter wells (70 μL) containing either 100 nM final concentration HEHEAA (purchased from Chiron AS) or water as indicated. After 8 h at room temperature, we measured GFP fluorescence (excitation, 485 nm; emission, 535 nm) and growth (OD600) using a Synergy H1 plate reader (BioTek). To test whether the aapF homologs from other Pseudomonas strains could complement a Pseudomonas GM79 ΔaapF mutation, the GM79 aapF gene encoded by the pJN-AapFGM79PpipA-gfp plasmid was replaced with the indicated aapF homolog by E. coli DH5α-mediated assembly (31). To assess HEHEAA-dependent AapF activity, we followed GFP fluorescence in GM79 containing the indicated pJN-AapFPpipA-gfp (SI Appendix, Table S3) as described previously (12) but modified by adding 0.5% l-arabinose to the medium to induce AapF production for all experiments except those shown in Fig. 4B. To evaluate the influence of amino acid substitutions in AapFDC3000, we generated the indicated point mutants using sequence overlap extension PCR (30). Single mutations utilized pJN-AapFDC3000PpipA-gfp for the DNA template. For the double mutant aapFDC_double, we used pJN-AapFDC_I50RPpipA-gfp as the DNA template. The resulting PCR products were digested with HindIII and NheI and then ligated to similarly digested pJN-AapFDC3000PpipA-gfp. All constructs were verified by Sanger sequencing and used to transform E. coli S17-1, which was used for conjugal transfer into strain GM79ΔaapF. Data are reported either as relative fluorescence units per OD600 (Figs. 1C and 4B and SI Appendix, Fig. S5) or normalized to the wild-type after subtraction of background GFP fluorescence levels, as determined in the aapFGM79 null mutant (Table 1).
SBPPDC86 Purification.
We created an E. coli that produced cytoplasmic AapF by cloning a version of the PDC86 aapF gene (locus tag Ga0115494_13757) that omitted the 5′ nucleotides coding for the AapF secretion signal (nucleotides 1 to 78 of the aapF ORF) into the N-terminal hexahistidine-tagged (His6) expression vector pET-28a (EMD Millipore) to create pETaapFPDC86. To obtain purified His6-AapFPDC86, E. coli BL21 Star (DE3) containing pETaapFPDC86 was grown at 37 °C in gluc-MM. When cultures reached mid-logarithmic phase (OD600 = 0.6), we added 0.4 mM isopropyl β-d-1-thiogalactopyranoside and 10 mM ethanolamine. Cells were then incubated overnight at 18 °C with shaking and then harvested by centrifugation. The cell pellet was suspended in buffer (50 mM NaH2PO4 pH 7.6, 500 mM NaCl, 20 mM imidazole, and 5% [vol/vol] glycerol), cells were lysed by sonication, and the lysate was centrifuged to remove cell debris. Lysate was incubated with Ni-nitrilotriacetic acid beads, and the protein was eluted with 500 mM imidazole, followed by size-exclusion chromatography on a HiLoad Superdex 200 16/600 (GE Healthcare) in a buffer containing 10 mM Tris (pH 7.6) and 150 mM NaCl. Protein was followed by UV absorbance (λ = 280 nm), and peak fractions were collected and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis to verify the molecular weight and purity. SBPPDC86-containing fractions were combined and concentrated to ∼100 mg/mL, flash-frozen in liquid nitrogen, and stored at –80 °C.
SBPPDC86 Crystallization and Structure Determination.
Crystals were grown by the sitting-drop vapor-diffusion method at 20 °C by mixing 20 mg/mL protein with crystallization buffer containing 0.1 M Bis-Tris (pH 6.5) and 2.6 M ammonium sulfate. Crystals grew as thin-plate clusters typically appearing within 2 d and were flash-frozen in liquid nitrogen in mother liquor supplemented with 20 to 30% (vol/vol) glycerol. X-ray diffraction data were collected at the National Synchrotron Light Source II Frontier Microfocusing Macromolecular Crystallography (NSLSII FMX) beamline and processed using XDS software (33). The structure was solved by the molecular replacement method with Phaser (34), using a homology model of SBP generated by SwissModel using E. coli YliB (Protein Data Bank ID code 1UQW). The initial search using the entire model directly was not successful, and a solution was found when the model was split into two parts. The structure was refined with PHENIX (35).
We were concerned that there might be some ethanolamine-derived HEHEAA in the purified SBP preparations from E. coli grown with ethanolamine. To estimate the HEHEAA occupancy, we boiled an aliquot of SBPPDC86 to release any bound ligand, measured HEHEAA levels using the bioassay reporter GM79 pPpipA-gfp (12), and calculated the HEHEAA occupancy to be ∼3 mol HEHEAA per 100 mol AapFPDC86. To obtain HEHEAA-bound SBPPDC86, we incubated SBPPDC86 (20 mg/mL) in the presence of 25 mM HEHEAA in the protein buffer (10 mM Tris pH 7.6 and 150 mM NaCl) for 30 min before crystallization. This sample crystallized in the same condition, but the space group symmetry was reduced from P21 to P1, with similar unit cell parameters. The crystallographic information is summarized in SI Appendix, Table S1.
Fluorescence-Based Thermal Shift Assays.
His-tagged, cytoplasmic versions of each AapFGM79 variant were created in the vector pQE30, and aapFDC3000 variants were created in pET-16b (SI Appendix, Table S3). The variant polypeptides were purified in ligand-free form from E. coli grown in gluc-MM as described previously (12), except AapFT405P, for which no soluble protein was obtained. The purified proteins were used in thermal shift assays as described elsewhere (36). In brief, the denaturation temperature of a given polypeptide (10 μM) was determined in the presence or absence of HEHEAA and 5X SYPRO orange (Thermo Fisher Scientific) with the Bio-Rad CFX96 real-time PCR system. Temperatures were increased by 1 °C at each 1-min cycle over a temperature range of 20 to 90 °C. All thermal shift reactions were performed in triplicate in a 25-µL volume of 100 mM ammonium acetate buffer (pH 7.5), and the reproducibility of the results was confirmed by performing two separate protein purifications for the AapFDC3000 variants and AapFDC3000 wild-type polypeptides. Data were analyzed as the derivative of the fluorescence signal as a function of temperature, and the Tm was defined as the temperature at which the derivative is the lowest (SI Appendix, Figs. S6 and S7).
Supplementary Material
Acknowledgments
We thank W. Shi and M. Fuchs for their help with data collection at the NSLS II FMX beamline. This research was supported by NIH Grants R35 GM118093 and S10 OD012018 (to L.T.) and R35 GM136218 (to E.P.G.), and by the Genomic Science Program, US Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area (pmi.ornl.gov) (C.S.H. and E.P.G.). The Oak Ridge National Laboratory is managed by UT-Battelle LLC for the US Department of Energy under Contract DE-AC05-00OR227525. J.R. was supported by the Fund for the Shanxi 1331 Project.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019462118/-/DCSupplemental.
Data Availability
Coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 7KZ8 for HEHEAA-free AapFPDC86 and 7KZ9 for HEHEAA-bound AapFPDC86).
References
- 1.Chatnaparat T., Prathuangwong S., Ionescu M., Lindow S. E., Xag R., XagR, a LuxR homolog, contributes to the virulence of Xanthomonas axonopodis pv. glycines to soybean. Mol. Plant Microbe Interact. 25, 1104–1117 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Ferluga S., Bigirimana J., Höfte M., Venturi V., A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 8, 529–538 (2007). [DOI] [PubMed] [Google Scholar]
- 3.González J. F., Venturi V., A novel widespread interkingdom signaling circuit. Trends Plant Sci. 18, 167–174 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Schaefer A. L., et al., A LuxR homolog in a cottonwood tree endophyte that activates gene expression in response to a plant signal or specific peptides. MBio 7, e01101-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramoni S., et al., Bacterial subfamily of LuxR regulators that respond to plant compounds. Appl. Environ. Microbiol. 77, 4579–4588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H., Zhao Y., Qian G., Liu F., Xoc R., XocR, a LuxR solo required for virulence in Xanthomonas oryzae pv. oryzicola. Front. Cell. Infect. Microbiol. 5, 37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Jia Y., Wang L., Fang R., A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris. Mol. Microbiol. 65, 121–136 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Schuster M., Sexton D. J., Diggle S. P., Greenberg E. P., Acyl-homoserine lactone quorum sensing: From evolution to application. Annu. Rev. Microbiol. 67, 43–63 (2013). [DOI] [PubMed] [Google Scholar]
- 9.González J. F., Myers M. P., Venturi V., The inter-kingdom solo OryR regulator of Xanthomonas oryzae is important for motility. Mol. Plant Pathol. 14, 211–221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson A. L., Dassa E., Orelle C., Chen J., Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quiocho F. A., Ledvina P. S., Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 20, 17–25 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Coutinho B. G., et al., A plant-responsive bacterial-signaling system senses an ethanolamine derivative. Proc. Natl. Acad. Sci. U.S.A. 115, 9785–9790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blair P. M., et al., Exploration of the biosynthetic potential of the Populus microbiome. mSystems 3, e00045-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin X. F., He S. Y., Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Doeven M. K., Kok J., Poolman B., Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Mol. Microbiol. 57, 640–649 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Berntsson R. P., Smits S. H., Schmitt L., Slotboom D. J., Poolman B., A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Bermejo G. A., Strub M. P., Ho C., Tjandra N., Ligand-free open-closed transitions of periplasmic binding proteins: The case of glutamine-binding protein. Biochemistry 49, 1893–1902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flocco M. M., Mowbray S. L., The 1.9-Å X-ray structure of a closed unliganded form of the periplasmic glucose/galactose receptor from Salmonella typhimurium. J. Biol. Chem. 269, 8931–8936 (1994). [PubMed] [Google Scholar]
- 19.Oswald C., et al., Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J. Biol. Chem. 283, 32848–32859 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Holm L., Kääriäinen S., Rosenström P., Schenkel A., Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanabe M., et al., Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains. Acta Crystallogr. D Biol. Crystallogr. 63, 1185–1193 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Feng L., et al., Virulence factor identification in the banana pathogen Dickeya zeae MS2. Appl. Environ. Microbiol. 85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patankar A. V., González J. E., An orphan LuxR homolog of Sinorhizobium meliloti affects stress adaptation and competition for nodulation. Appl. Environ. Microbiol. 75, 946–955 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blancaflor E. B., et al., N-acylethanolamines: Lipid metabolites with functions in plant growth and development. Plant J. 79, 568–583 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Elbourne L. D., Tetu S. G., Hassan K. A., Paulsen I. T., TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandre G., Greer-Phillips S., Zhulin I. B., Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28, 113–126 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Fernández M., et al., Determination of ligand profiles for Pseudomonas aeruginosa solute-binding proteins. Int. J. Mol. Sci. 20, 5156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J., Russel R., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- 29.Miller J., Experiments in Molecular Genetics (Cold Spring Harbor Laboratory Press, 1972). [Google Scholar]
- 30.Horton R. M., Cai Z. L., Ho S. N., Pease L. R., Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques 8, 528–535 (1990). [PubMed] [Google Scholar]
- 31.Kostylev M., Otwell A. E., Richardson R. E., Suzuki Y., Cloning should be simple: Escherichia coli DH5alpha-mediated assembly of multiple DNA fragments with short end homologies. PLoS One 10, e0137466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P., A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy A. J., et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams P. D., et al., PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Giuliani S. E., Frank A. M., Collart F. R., Functional assignment of solute-binding proteins of ABC transporters using a fluorescence-based thermal shift assay. Biochemistry 47, 13974–13984 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 7KZ8 for HEHEAA-free AapFPDC86 and 7KZ9 for HEHEAA-bound AapFPDC86).