Significance
Metabolic disorders related to aberrant fat accumulation, including lipodystrophy and obesity, are a serious health concern. Inside cells, fat accumulates in organelles named lipid droplets (LDs). LDs form in the endoplasmic reticulum (ER), where triglycerides (TG), the most abundant form of fat, are produced. The Berardinelli–Seip congenital lipodystrophy type 2 protein, seipin, has been identified as a key regulator of LD formation, but its mechanism of action remains debated and its molecular details mostly obscure. Here, we use molecular dynamics simulations to investigate the mechanism of seipin action. We find that seipin clusters and traps both TG and its precursor, diacylglycerol. Our results suggest that seipin organizes the lipid composition of specific ER sites to prime them for LD biogenesis.
Keywords: lipid droplets, seipin, molecular dynamics, endoplasmic reticulum, lipid membranes
Abstract
Lipid droplets (LDs) are intracellular organelles responsible for lipid storage, and they emerge from the endoplasmic reticulum (ER) upon the accumulation of neutral lipids, mostly triglycerides (TG), between the two leaflets of the ER membrane. LD biogenesis takes place at ER sites that are marked by the protein seipin, which subsequently recruits additional proteins to catalyze LD formation. Deletion of seipin, however, does not abolish LD biogenesis, and its precise role in controlling LD assembly remains unclear. Here, we use molecular dynamics simulations to investigate the molecular mechanism through which seipin promotes LD formation. We find that seipin clusters TG, as well as its precursor diacylglycerol, inside its unconventional ring-like oligomeric structure and that both its luminal and transmembrane regions contribute to this process. This mechanism is abolished upon mutations of polar residues involved in protein–TG interactions into hydrophobic residues. Our results suggest that seipin remodels the membrane of specific ER sites to prime them for LD biogenesis.
Lipid droplets (LDs) are the intracellular organelles responsible for fat accumulation (1). As such, they play a central role in lipid and cellular metabolism (1–4), and they are crucially involved in metabolic diseases such as lipodystrophy and obesity (5–7).
Formation of LDs occurs in the endoplasmic reticulum (ER), where neutral lipids (NLs), namely triglycerides (TG) and cholesteryl esters, constituting the core of LDs are synthesized by acyltransferases that are essential for LD formation (8). The current model of LD formation posits that NLs are stored between the two leaflets of the ER bilayer, where they aggregate in nascent oblate lens-like structures with diameters of 40 to 60 nm (9) before complete maturation and budding toward the cytosol (10–13).
Recent experiments suggest that LDs form at specific ER sites marked by the protein seipin (14) upon arrival of its interaction partner protein promethin/LDAF1 (lipid droplet organization [LDO] in yeast) (15–19). These recent observations confirm previous works showing that seipin, in addition to modulating LD budding and growth (14, 19–21) and LD–ER contacts (22, 23), is also a major player in the early stages of LD formation, as deletion of seipin leads to TG accumulation in the ER and a delay in the formation of, possibly aberrant, LDs (20, 24).
The role of seipin in LD formation is potentially coupled to its function in regulating lipid metabolism (25, 26) and notably that of phosphatidic acid (PA) (27–31). Recently, seipin-positive ER loci have been shown to be part of a larger protein machinery that also includes membrane and lipid remodeling proteins of the TG synthesis pathway (32), most notably, Lipin (Pah1 in yeast) and FIT proteins (Yft2 and Scs3 in yeast), for which PA is either a known substrate (Lipin/Pah1) (33) or a likely one (FIT/Yft2/Scs3) (34).
Despite this thorough characterization of the cellular role of seipin in LD formation, the molecular details of its mechanism remain mostly unclear. Recently, the structure of the luminal part of the seipin oligomer has been solved at 3.7 to 4.0 Å resolution using electron microscopy (27, 35), paving the way for the investigation of the relationship between its three-dimensional structure and its mode of action. These studies revealed that the luminal domain of seipin consists of an eight-stranded beta sandwich, together with a hydrophobic helix (HH), positioned toward the ER bilayer. Notably, the seipin oligomer assembles into a ring-like architecture, an unconventional assembly in lipid bilayers that rather resembles the shape of microbial pore-forming assemblies (36) or GroEL-GroES chaperones (37, 38).
From a stochiometric point of view, both fluorescence and electron microscopy data are consistent with the presence of a single seipin oligomer per nascent LD (14, 15). Hence, the structure of the luminal part of seipin is consistent with two proposed modes of action: seipin could mark the sites of LD formation by controlling TG flow in and out of the nascent droplet (14), or, alternatively, seipin could help recognize and stabilize preexisting nascent droplets in the ER membrane (20, 21, 39). In both cases, however, the relationship between the role of seipin in LD formation and its ability to regulate lipid metabolism remains unclear.
Here, we use coarse-grain (CG) molecular dynamics (MD) simulations to investigate the mechanism of seipin in molecular detail. We find that seipin is able to cluster TG molecules inside its ring-like structure and that both the transmembrane (TM) helices and the luminal domain contribute to this process. Diacylglycerol (DG), the lipid intermediate between TG and PA in the Kennedy pathway, also accumulates around seipin, further promoting the accumulation of TG at very low TG-to-phospholipids ratios. Our data suggest that by accumulating DG and TG molecules, seipin generates ER sites with a specific lipid composition that in turn could promote the sequential recruitment of additional TG- and DG-sensing proteins involved in LD formation, including promethin/LDOs, FIT/Yft2/Scs3, and perilipins.
Results
Seipin Promotes the Accumulation of TG Molecules at Low TG Concentrations.
To investigate the role of seipin in LD formation in silico, we embedded a model of the seipin 11-mer, consisting of its luminal structure and of its TM helices (Fig. 1A), in lipid bilayers composed of di-oleoyl-phosphatidylcholine (DOPC) with varying concentrations of TG molecules. Our model derives from the luminal structure of human seipin recently solved using cryogenic electron microscopy (cryo-EM) (27), and our models for the protein and lipids are consistent with the CG Shinoda–Devane–Klein (SDK) and Surface Property fItting Coarse grAining (SPICA) force field (40–42).
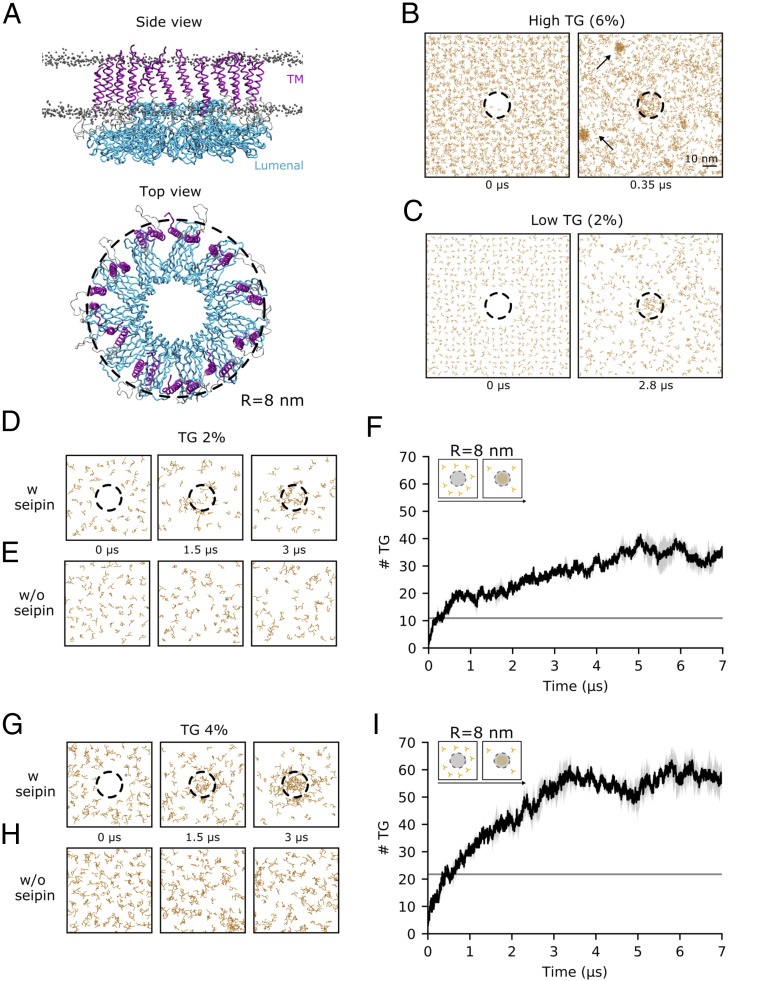
Fig. 1.
Seipin accumulates TG molecules in its ring-like structure at low TG concentrations. (A) Side and top view of the initial configuration of the MD simulations of the TM helices (purple) and luminal domain (cyan) of the human seipin undecamer embedded in a lipid bilayer. DOPC phosphate groups are shown as gray dots. (B and C) Top view of snapshots representing the time evolution of TG molecules in MD simulations at 6% (B) and 2% (C) TG concentrations. The black dotted ring represents the outermost radius of the seipin oligomer (R = 8 nm, the same radius used for the quantification of TG molecules in F and I) and indicates the presence of seipin in the simulations. TG molecules are colored in orange. (D and E) Top view of snapshots representing the time evolution of TG molecules in MD simulations at 2% TG concentration, with (D) or without (E) seipin. (F) Quantification of the number of TG molecules inside the seipin ring over time during the MD simulation depicted in D. The gray line indicates the amount of TG molecules that would be present in an area of equivalent size of that measured for seipin if the TG molecules were distributed uniformly in the bilayer. (G and H) Top view of snapshots representing the time evolution of TG molecules in MD simulations at 4% TG concentration, with (G) or without (H) seipin. (I) Quantification of the number of TG molecules inside the seipin ring over time during the MD simulation depicted in G. The gray line indicates the amount of TG molecules that would be present in an area of equivalent size of that measured for seipin if the TG molecules were distributed uniformly in the bilayer.
We first investigated whether seipin helps clustering TG molecules at low (2%) and high (6%) TG concentrations. We chose these two concentrations because previous works (43, 44) showed that at above 5% TG, spontaneous formation of TG blisters takes place in protein-free simulations, while this is not the case below this threshold. In agreement with those simulations, we observed that at higher concentrations (6% TG), spontaneous formation of TG blisters quickly (<300 ns) took place outside the seipin ring (Fig. 1B). At low concentrations (2% TG), however, we invariably observed that TG molecules only accumulated in the proximity of the protein (Fig. 1C).
To quantify the extent of seipin-induced TG accumulation at low TG concentrations (2%), we computed the amount of TG molecules per seipin oligomer over time, and we compared it with protein-free simulations (Fig. 1 D–F). We observed that TG accumulates in the presence of seipin (Fig. 1 D and F), reaching a plateau after a few microseconds at ∼35 TG molecules per seipin oligomer (Fig. 1F). On comparable time scales, no such accumulation was observed in the absence of seipin (Fig. 1E). To investigate whether the accumulation of TG molecules inside seipin is driven by specific protein–TG interactions or by demixing of TG molecules inside the lipid bilayer, we next estimated the amount of TG molecules inside the seipin ring that are in direct contact with the protein and compared it with those that do not interact with the protein but, rather, are in contact with other TG molecules (SI Appendix, Fig. S1). Notably, 90% of the TG molecules inside the seipin ring establish direct contacts with the protein, suggesting a protein-specific mechanism of TG accumulation.
To investigate whether the initial protein-bound TG molecules could act as seed for the growth of TG blisters via demixing of TG molecules from the bilayer, we repeated our simulations at a higher TG concentration (4%) (Fig. 1 G–I). At this concentration, spontaneous blister formation is not expected to occur in protein-free systems (43, 44), but the amount of TG molecules is substantially higher than that required for saturating seipin at 2% TG concentration according to Fig. 1F. Indeed, also at this TG concentration, we did not observe any spontaneous TG blister formation in the absence of seipin (Fig. 1H), whereas TG accumulated inside the seipin ring in the presence of the protein (Fig. 1G). Interestingly, the number of total TG molecules inside the seipin ring reached saturation at around 55 TG molecules (Fig. 1I), a value significantly higher than that observed at lower (2%) TG concentrations. An analysis of protein–TG contacts shows that while the amount of TG molecules in contact with the protein remains stable (SI Appendix, Fig. S1), the percentage of TG molecules inside the seipin ring that do not engage in interactions with the protein, but rather form TG–TG interactions, doubled (SI Appendix, Fig. S1).
Taken together, our results suggest that the seipin oligomer clusters TG molecules at low TG-to-phospholipid ratios. The finite size of the seipin oligomer sets a threshold on the amount of TG molecules that the protein can accumulate via direct protein–TG interactions, but those molecules can act as a seed for further growth of the TG blister inside the seipin ring. However, the observation that even in the presence of excess amounts of TG, blister growth is arrested at relatively low amounts of TG molecules [at least in comparison with protein-free systems, where TG blisters can accumulate up to hundreds of TG molecules (44)] indicates that seipin is likely insufficient to promote extensive LD growth, per se, in the absence of partner proteins (15, 17) or variations in membrane properties (45–47).
The Luminal Domain of Seipin Is Responsible for Accumulating and Retaining TG Molecules.
We next sought to investigate the molecular mechanism through which seipin accumulates TG molecules. To do so, we performed MD simulations, unlike otherwise stated, in pure DOPC bilayers after addition of TG molecules corresponding to 4% of total PC lipids, as in Fig. 1 G and I. We first investigated where TG molecules accumulate inside the seipin ring, and we specifically focused on the role of the individual components of seipin: the TM helices and the luminal region. These two domains are physically distinct in the seipin oligomer (Fig. 2 A and B), with the helices occupying the external ring of the complex and the luminal domain occupying the inner ring.
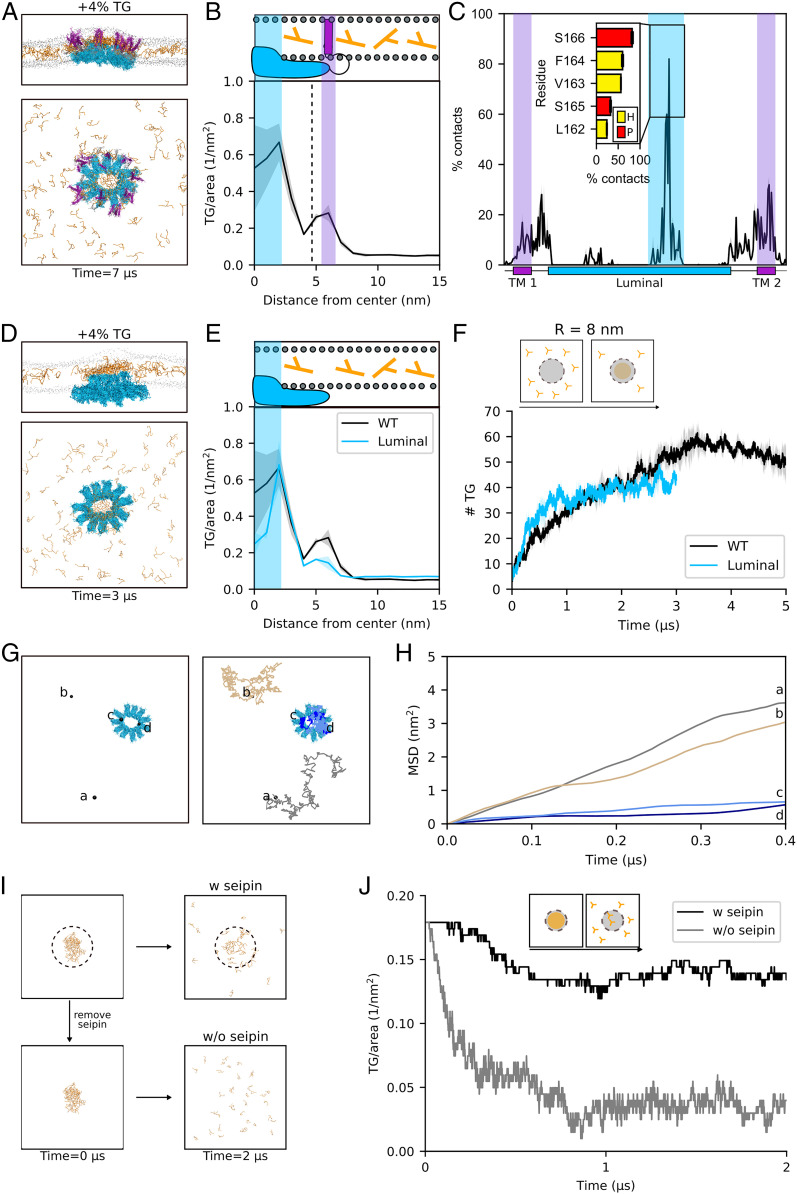
Fig. 2.
The luminal domain of seipin is sufficient to promote TG accumulation and retention. (A) Side and top view of the accumulation of TG molecules in the presence of seipin. The color codes are the same as in Fig. 1. (B) Radial concentration of TG molecules in simulations of the entire seipin undecamer. (C) TG occupancy at seipin residues. The inset shows TG occupancy at the five protein residues that make the most contacts with TG molecules in the luminal domain during the MD simulations. Color code: hydrophobic, yellow; polar, red. (D) Side and top view of the accumulation of TG molecules in the presence of the luminal domain of seipin alone. (E) Radial concentration of TG molecules in simulations of seipin luminal domain alone. (F) Quantification of the number of TG molecules inside the seipin ring over time during the MD simulation of seipin luminal domain. (G) Top view of the diffusion path of four TG molecules representative of the free diffusion in the lipid bilayer (brown and gray) and of the confined diffusion in proximity of the luminal domain of seipin (dark and light blue) (H) Corresponding mean square displacements. (I) Top view of the initial and final snapshots of MD simulations of dissolution of the TG blister in the presence (Top) or absence (Bottom) of seipin. (J) Corresponding time evolution of the number of TG molecules inside the seipin ring when TG molecules are completely depleted from the surrounding lipid bilayer in the presence (black) or absence (gray) of seipin.
To do so, we first computed the distribution of TG molecules as a function of the radius from the center of the oligomer ring (Fig. 2B). We observed the presence of two clear peaks at which TG molecules accumulate (Fig. 2B). These two peaks appear at radius R = ∼2.5 nm and R = ∼6 nm, and they represent, respectively, the extent of TG accumulation by the luminal and TM domains. Notably, TG accumulation is significantly higher in the proximity of the luminal domain in comparison with the TM helices (Fig. 2B).
To further quantify the contribution of the two domains toward TG clustering, we next calculated the average residency time of TG molecules at specific protein interaction sites. To do so, we quantified the percentage of the trajectory during which the distance between a TG molecule and a specific residue was below 0.6 nm (Fig. 2C). Consistently with our previous observation (Fig. 2B), we found that TG molecules are in close contact with both TM helices and the luminal domain (Fig. 2C). In addition, we could identify that the binding of TG molecules to the luminal domain takes place mainly via its HH, with residues L162, V163, F164, S165, and S166 establishing the most contacts (Fig. 2 C, Inset).
The analyses of Fig. 2 B and C indicate that the luminal domain is primarily responsible for the accumulation of TG molecules inside seipin. To investigate whether the luminal domain is sufficient to promote this process, we performed analogous simulations in the presence of the luminal domain alone (Fig. 2D). Of note, in all simulations, the oligomeric structure of the protein was conserved during the simulations, a constraint which might not reflect physiological conditions in case some domains are also responsible for the oligomerization of the individual seipin monomers. We observed that in the absence of the TM helices, the luminal domain alone is able to promote TG accumulation comparably to that observed in the entire protein (Fig. 2E and SI Appendix, Fig. S2). Interestingly, the kinetics of TG accumulation is faster in the absence of the TM helices (Fig. 2F) and, specifically, in the inner part of the ring (R = 6 nm) (SI Appendix, Fig. S3).
Next, we investigated whether the TG molecules that are in contact with the luminal HH domain of seipin form stable contacts with the protein or rather exchange frequently with the surrounding lipid bilayer. To do so, we visually inspected the diffusion of TG molecules in contact with the protein and compared their diffusion with those that are free to diffuse in the bilayer (Fig. 2 G and H). A comparison of their mean square displacement clearly shows that molecules that engage in interactions with the luminal domain of seipin are restricted in their movement and that they establish long-lived interactions with the protein (Fig. 2H).
To further test the hypothesis of whether seipin, and specifically its luminal domain, is able to trap TG molecules in its proximity, we next investigated whether seipin is able to retain TG molecules when their concentration in the surrounding lipid bilayer is depleted. To do so, starting from the final state of our previous simulations in which TG molecules are clustered inside the seipin oligomer, we removed all free TG molecules that are not in direct contact with the luminal domain (Fig. 2I). In these conditions, if seipin is also removed, TG molecules dissolve in the bilayer, and no cluster is present (Fig. 2 I and J). In the presence of the protein, on the other hand, only few TG molecules dissolve in the bilayer, while the vast majority remain stably associated to the protein (Fig. 2J).
Taken together, our data suggest that the luminal domain of seipin, mostly through its HH region, is sufficient to accumulate TG molecules. The luminal domain is also responsible for the trapping of TG molecules inside the seipin ring by establishing long-lived interactions that retain them. These molecules infrequently escape the protein even when the surrounding bilayer is depleted of TG. This suggests that seipin-positive ER sites might be enriched with TG molecules even in the absence of contemporaneous TG synthesis. Thus, while seipin diffuses in the ER membrane before LD formation (15), it might sequester TG molecules from the ER and drag them along in its diffusion path.
The TM Helices of Seipin Control the Kinetics of TG Accumulation and Remodel the Lipid Composition at the Periphery of the Seipin Ring.
While the luminal domain of seipin appears to be sufficient to promote TG accumulation and to trap TG molecules (Fig. 2), the time-averaged analyses of Fig. 2 B and C indicate that both the luminal domain and the TM helices contribute to TG clustering inside the seipin ring.
To better characterize the role of seipin TM helices in the process of TG accumulation by seipin, we first computed the time evolution of the radial concentration of TG molecules (Fig. 3A). This analysis shows that TG molecules first accumulate around the TM helices before proceeding toward the core of the ring and engaging in interactions with the luminal domain (Fig. 3A). This suggests that the TM helices might play an important role in controlling the kinetics of TG accumulation.
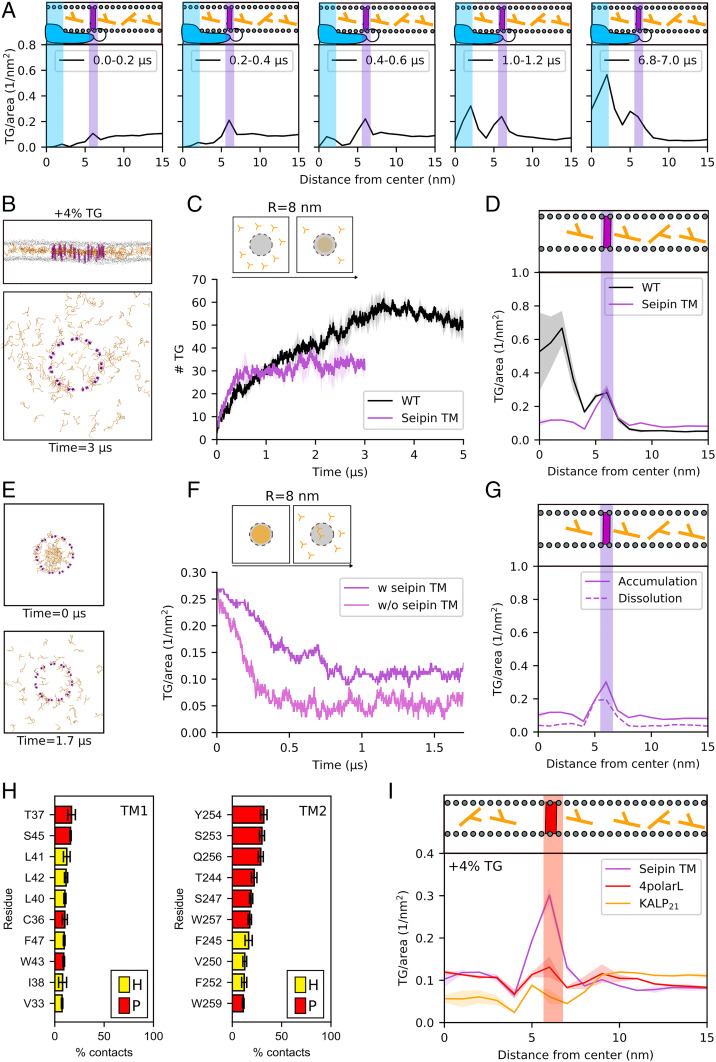
Fig. 3.
The TM helices of seipin control the kinetics of TG clustering and promote the accumulation of TG molecules at the periphery of the seipin ring. (A) Time evolution of the radial distribution of TG molecules inside seipin. (B) Side and top view of the accumulation of TG molecules in the presence of the TM helices of seipin alone (TM-only system). (C) Quantification of the number of TG molecules inside the seipin ring over time during MD simulation of the TM-only system. (D) The corresponding radial distribution of TG molecules. (E) Top view of initial and final snapshots of MD simulations of dissolution of the TG blister in the TM-only system. (F) Time evolution of the number of TG molecules inside the seipin ring when TG molecules are completely depleted from the surrounding lipid bilayer in the presence (purple) or absence (magenta) of seipin TMs. (G) Radial distribution of TG molecules around seipin in MD simulations of blister dissolution compared to the MD simulations of TG accumulation in TM-only systems. (H) TG occupancy of the 10 protein residues in the TM helices that make the most contacts with TG molecules during the MD simulations with the WT protein. Color code: hydrophobic, yellow; polar, red. (I) Radial distribution of TG molecules in the 4polarL (red) and KALP21 system (orange) in comparison with the TM-only system (purple).
To test this hypothesis, we investigated the process of TG accumulation in the presence of the TM helices alone (Fig. 3 B–D), and we compared its kinetics with that of the entire protein (Fig. 3C). We observed that the initial kinetics are similar in both cases before TG accumulation in the TM-only systems plateaus at a lower value in comparison with the entire protein (Fig. 3C and SI Appendix, Fig. S4). To further quantify this behavior, we computed the instances of TG molecules entering and exiting the seipin oligomer (SI Appendix, Fig. S5). We observed that the number of entry and exit events is comparable in the wild-type (WT) and in the TM-only system but remarkably different in the luminal-only system, further suggesting that the TM helices, which are located in the external part of the seipin ring, effectively act as gates for the entry of TG molecules.
Next, we investigated whether the TM-only system is able to retain TG molecules in its center once a preexisting TG blister is present (Fig. 3E). Indeed, the presence of the TM helices prevents the complete dissolution of the TG molecules in the bilayer, as is the case in the protein-free system (Fig. 3F). An analysis of the radial distribution of TG accumulation shows that this behavior originates from the fact that, similar to the protein-free system, the TM-only system fails to retain TG molecules in the inner region of the ring (Fig. 3G) but that a small, yet nonnegligible, TG accumulation can be observed around the TM helices (Fig. 3G). Of note, this TM-induced TG accumulation is identical with or without the luminal domain, as can be appreciated when comparing the peaks at 6 nm (Fig. 3 D and G). Consistent with the observed frequency of TG entries and exits in the TM-only system (SI Appendix, Fig. S5), the retention of TG molecules at the TM suggests that the ring-like structure alone is insufficient to retain a formed TG blister inside the ring but that TM helices play a role in regulating the flux of TG molecules inside and outside the ring.
Next, we analyzed which residues establish the most contacts between the TM helices and TG molecules (Fig. 3H). This analysis highlights the important role of multiple polar residues (T37, S45, T244, S247, S253, Y254, and Q256) that establish direct contacts with the slightly polar glycerol moiety of the TG molecules.
To further test whether the sequence of seipin TM helices, and in particular their polar residues, is indeed specific for TG recruitment, we performed MD simulations of different TM helices kept in a ring-like structure identical to that of the WT seipin helices. First, we randomly scrambled the residues of the seipin TM helices, thus keeping the overall ratio between polar and apolar residues constant but changing their distribution inside the helices. Next, we altered the polar/apolar ratio by mutating four polar residues into hydrophobic residues (S253L, Y254L, M255L, and Q256L, referred to subsequently as “4polarL”). Finally, we substituted the TM helices of seipin with artificial KALP21 peptides that lack polar residues in their sequence and are thus totally hydrophobic, being only composed of alanine and leucine residues (48) in their core. We observed that scrambling the order of residues has no impact on TG accumulation (SI Appendix, Fig. S6), while reducing the number of polar residues has a dramatic impact, with mutation of four polar residues decreasing TG accumulation (Fig. 3I) and total absence of polar residues completely abolishing TG accumulation (Fig. 3I).
On the one hand, these results further point to a specific role of polar residues in TG accumulation and indicate that the TM-induced TG accumulation at the periphery of the seipin oligomer might be important for its physiological role. On the other hand, the need for modeling of the TM helices, that are not explicitly resolved in the cryo-EM structure (27), suggests that these data must be interpreted with caution, as our models are likely unable to predict the existence of specific TG binding sites in the TM helices.
Seipin Is Able to Accumulate and Trap DG, and DG Enrichment Doesn’t Alter Seipin Ability to Cluster TG Molecules.
Our simulations show that the seipin oligomer clusters and traps TG molecules, and this activity is driven by its luminal domain. Even if the presence of the TM helices is not necessary toward this process, the TM helices of seipin distinctly promote local TG accumulation in their proximity by means of specific protein–TG interactions.
We thus reasoned that this local membrane remodeling could be of relevance to the mechanism of seipin, including, possibly, to establish protein–protein interactions with other membrane-embedded proteins, especially as the TM helices represent the outermost portion of the protein oligomer.
We thus investigated whether lipids other than TG could accumulate around seipin. To do so, we embedded the seipin oligomer in a model ER-like membrane containing PC, phosphatidyl-ethanolamine (PE), cholesterol, and DG. In this condition, we observed that TG molecules cluster inside the seipin ring with no discernible difference with respect to pure DOPC bilayers (Fig. 4 A and B). Next, we investigated whether specific ER lipids accumulate around seipin. We observed that PE lipids slightly accumulate around the seipin TM helices, while DG molecules significantly accumulate inside the seipin ring (Fig. 4C).
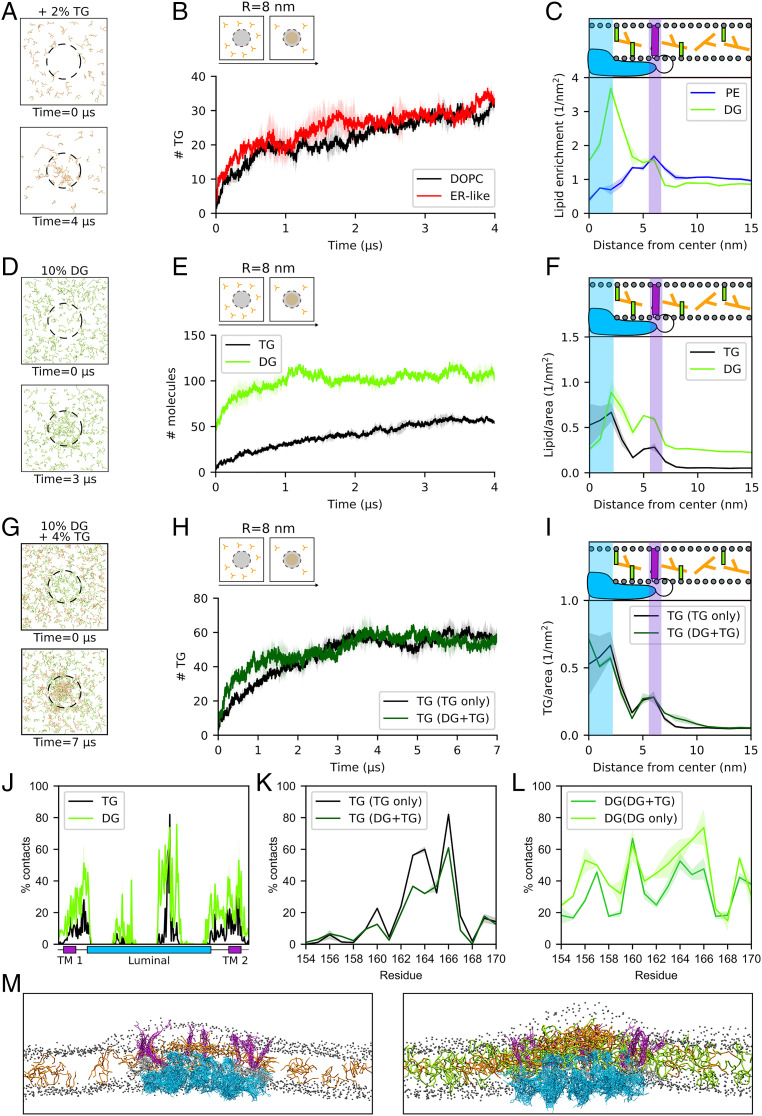
Fig. 4.
Seipin accumulates and traps diacylglycerols (DG) without losing its ability to cluster TG molecules. (A) Top view of snapshots representing the evolution of TG in ER-like bilayers. (B) TG accumulation in an ER-like bilayer in comparison with a 100% DOPC bilayer. (C) Radial distribution of PE and DG enrichment around seipin. The two curves were normalized by the average number of lipid per square nanometer. (D) Top view of snapshots showing the time evolution of DG (green) in MD simulations in the presence of seipin. (E) Quantification of DG molecules inside the seipin ring over time in comparison with TG-only (4% TG) system. (F) Corresponding radial distribution of DG in comparison with TG-only (4% TG) system. (G) Top view of snapshots showing the time evolution of DG (green) and TG (orange) molecules in MD simulations in the presence of seipin when TG molecules are added to a lipid bilayer with preclustered DG molecules inside seipin. (H) Quantification of TG molecules inside seipin over time when DG is preclustered (dark green) in comparison with TG-only system (black). (I) Corresponding radial distribution of TG molecules in the DG+TG (dark green) and in the TG-only system (black). (J) TG occupancy at seipin in TG-only (4% TG) system in comparison with DG occupancy in DG-only systems. (K) TG occupancy at the HH of seipin in TG-only (4% TG) system in comparison with DG+TG systems. (L) DG occupancy at the HH of seipin in DG-only system in comparison with DG+TG systems. (M) Side view of snapshots representing the equilibrated conformation of seipin in the presence of TG-only (Left) or DG+TG (Right). Color code: seipin luminal domain, blue; seipin TM domains, purple; TG, orange; DG, green; and DOPC phosphate group, gray.
The observation that DG molecules accumulate inside the seipin ring is particularly intriguing. First, DG has been proposed to play multiple roles in LD formation, either directly (44, 45, 49) or possibly as a result of the activity of the PA-phosphatase Pah1 (29, 32, 50). Second, DG, while sharing some physicochemical properties with TG (42), is significantly more polar. Hence, its accumulation inside the seipin ring further supports the role of membrane-embedded polar residues in seipin activity.
To further investigate and quantify the observed mechanism of DG accumulation, we next prepared DOPC lipid bilayers containing 10% of DG or PA, the precursor of DG along the Kennedy pathway, as both DG and PA have been shown to accumulate at LD formation sites in various physiological (32) or nonphysiological conditions (28). Our data show that while PA does not accumulate around seipin (SI Appendix, Fig. S7), DG accumulation is remarkable and even faster than that of TG (Fig. 4 D and E). In particular, many more DG molecules (around 100) can be accommodated inside a single seipin oligomer (Fig. 4 D–F) when DG is present in the bilayer at 10 molar percentage (mol%). Correspondingly, DG accumulation around both TM helices and luminal domain is larger than that of TG (Fig. 4F). Also, as it is the case for TG molecules (Fig. 2 I and J), when excess DG molecules are depleted from the surrounding bilayer (SI Appendix, Fig. S8), DG molecules remain trapped inside the seipin ring, whereas they quickly dissolve in the bilayer if the protein is removed (SI Appendix, Fig. S8).
As DG is the precursor of TG, we next investigated whether the TG accumulation observed in the absence of DG is promoted or abolished by the preclustering of DG molecules inside the oligomer ring (Fig. 4G). We observed that in the presence of DG, TG molecules still accumulate inside seipin (Fig. 4H), and they do so with comparable kinetics, albeit slightly faster (Fig. 4H). Remarkably, the amount of TG molecules that accumulate inside the seipin ring is analogous to those observed in DG-free simulations (Fig. 4 H and I), despite the initial presence of DG molecules in contact with the protein. Finally, we tested whether seipin is able to retain both DG and TG molecules when they are depleted from the surrounding membrane (SI Appendix, Fig. S9). Also, in this case, both DG and TG molecules remain trapped inside the protein ring (SI Appendix, Fig. S9).
To better characterize the mechanism of TG accumulation in the presence of DG molecules, we first computed the occupancy of DG at protein residues in the absence of TG. We noticed that while DG interacts with similar residues as TG in the TM helices, the interaction of DG molecules with the HH is less specific for the 163 to 166 region and more diffuse along the entire HH (Fig. 4J). Next, we investigated what happens to the lipid–protein contacts in the HH when TG accumulates after preclustering of DG molecules (as in Fig. 4 G–I). We found that TG occupancy at HH residues is comparable with or without preclustering of DG molecules (Fig. 4K). DG, on the other hand, is displaced from residues S165 and S166 by the arrival of TG and redistributes more uniformly around the other residues of the HH (Fig. 4L).
Taken together, our data suggest that the seipin oligomer is able to significantly cluster DG molecules and that the presence of DG does not compete with the ability of seipin to cluster TG. Rather, even in the presence of a preexisting DG cluster inside seipin, TG molecules can still localize to their preferred binding site in the HH, and DG molecules can adapt to this specific TG localization by spreading around the remaining parts of the HH. In addition, the simultaneous presence of both DG and TG molecules in the seipin ring leads to a dramatic enhancement in overall blister size with respect to the presence of only TG molecules (Fig. 4M). As a consequence, the presence of seipin together with the associated TG and DG blister substantially remodel the surrounding bilayer in two distinct ways: first, it promotes a significant remodeling of the lipid composition in its surroundings, with a marked DG accumulation in its vicinity; second, it induces a nonnegligible positive curvature to the cytosolic leaflet of the lipid bilayer (Fig. 4M and SI Appendix, Fig. S10).
Interactions between Polar Residues in the HH of Seipin and TG Molecules Are Required for Its TG-Clustering Activity.
Our simulations indicate that seipin can trap and cluster TG and DG molecules inside its ring and that these interactions are promoted by a combination of hydrophobic and polar residues (Figs. 2C and 3H). In particular, the observations that DG can be trapped even more efficiently than TG (Fig. 4 E and F) and that removing polar residues from the TM helices abolishes TG clustering around them (Fig. 3I) suggest that polar residues might play a major role in this mechanism.
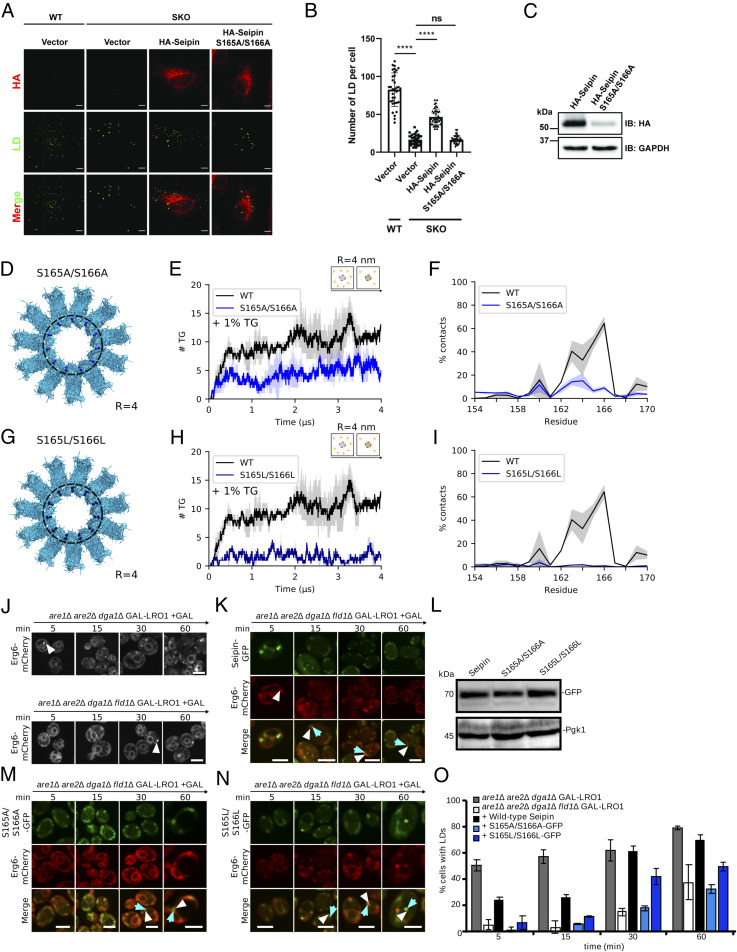
As the luminal domain is the one mainly responsible for TG accumulation (Fig. 2) via its HH (Fig. 2C), we next focused on the two polar residues of the HH that establish the most contacts with TG molecules: S165 and S166 (Fig. 2C). Of note, when these two serine residues were mutated into alanine (S165A/S166A) in SUM159 cells, this resulted in a seipin loss of function for what pertains to LD formation (15). However, this phenotype was attributed to a loss of interaction between seipin and its interaction partner in LD formation LDAF1 (15).
To test this hypothesis, as well as the ability of our computational model to reproduce and interpret cellular observations, we first confirmed that the double mutant S165A/S166A abolishes the ability of seipin to rescue the normal LD biogenesis phenotype in seipin knockout (KO) HeLa cells at early time points (15 min after oleate loading) (Fig. 5 A–C). Indeed, we observed that expressing the S165A/S166A mutant in seipin-KO cells does not change the number of LD per cell with respect to the seipin-KO, unlike the expression of WT seipin that instead significantly increases the number of LD per cell (Fig. 5B).
Fig. 5.
Polar residues in the hydrophobic helix (HH) modulate seipin function. (A) Immunofluorescence of WT and seipin-KO (SKO) HeLa cells transiently transfected with HA-tagged seipin or S165A/S166A mutant. Cells were lipid starved with 1% LPDS for 16 h before incubation with 400 µM OA for 15 min. LDs were stained with BODIPY493/503. (Scale bar, 5 µm.) (B) Quantification of the number of LDs per cell shown in A. ****P < 0.0001. (C) Western blot of HeLa cells transiently transfected with HA-tagged seipin or S165A/S166A. (D) Top view of seipin and localization of the Ser to Ala mutations. (E) Quantification of TG molecules inside seipin (R = 4 nm) over time for WT (black) and S165A/S166A (light blue) at low (1%) TG. (F) Contacts between TG and the HH of WT (black) and S165A/S166A (light blue). (G) Top view of seipin and localization of the Ser to Leu mutations. (H) Quantification of TG inside seipin (R = 4 nm) over time for WT (black) and S165L/S166L (dark blue) at low (1%) TG. (I) Contacts between TG and the HH of WT (black) and S165L/S166L (dark blue). (J) LD-inducible strains expressing WT seipin (Upper) or lacking seipin (Lower) expressing the LD marker Erg6-mCherry were transferred to galactose-containing media for the indicated time. White arrow heads designate LDs. (K) Fluorescence microscopy images of LD-inducible strains GAL-LRO1are1∆are2∆dga1∆fld1∆ expressing the LD marker Erg6-mCherry and WT human seipin tagged with GFP. (L) Western blot showing expression of seipin-GFP, S165A/S166A-GFP, and S165L/S166L-GFP and that of Pgk1. (M and N) S165A/S166A and S165L/S166L mutants do not rescue the LD biogenesis defect in seipin-deficient yeast cells. White arrowheads designate LDs, blue arrows designate seipin dots. (Scale bar, 5 µm.) (O) Quantification of cells containing LDs at the indicated time points.
We next performed MD simulations of two double mutants, in which the two serines were mutated into apolar resides: alanine (S165A/S166A) and leucine (S165L/S166L). In both cases, the mutations significantly decrease the ability of the seipin luminal domain to cluster TG molecules in its inner region (R = 4 nm), even at very low TG concentrations (1%) (Fig. 5 D–I). This mechanism is strongly hindered even in the presence of the entire protein (luminal + TM domains) and at higher TG concentrations (SI Appendix, Fig. S11).
To further test our hypothesis in vivo, and to partially remove potential effects arising from protein–protein interactions, including with its interaction partner LDAF1, we took advantage of a previously characterized protocol to express human seipin in yeast cells (27). In detail, we expressed human seipin and the two mutants S165A/S166A and S165L/S166L in yeast cells devoid of the yeast seipin fld1 in the presence of an inducible promoter of TG synthesis (are1Δare2Δdga1Δfld1ΔGAL-Lro1) (Fig. 5J). In this system, expression of human seipin rescues normal LD formation (Fig. 5K), but expression of the two mutants does not (Fig. 5 L–N). An analysis of the time evolution of LD formation (Fig. 5O) shows almost negligible LD formation after 5 and 15 min for the two mutants, while significant LD formation can be observed after only 5 min in WT yeast seipin or when human seipin is expressed in yeast. Of note, while the S165A/S166A mutant behaves similarly to seipin-null cells up to late time points (60 min after TG synthesis induction), in agreement with what is observed in human cells (Fig. 5A and ref. 15), this is not the case for the S165L/S166L mutant where LD formation is almost entirely recovered after 60 min. However, we noticed that the ER localization of both mutants is markedly different from that of human seipin after 60 min from TG induction (Fig. 5 K, M, and N). Hence, care must be taken when interpreting observations at late time points (e.g., after 15 min in our assay) as part of the bona fide LD formation process, as additional factors, including protein degradation or recruitment of multiple proteins to the nascent LD blister, are likely to contribute to this process at late time stages.
Taken together, the agreement between our in silico results and the in vivo quantification of the early stages of LD formation provides further validation of our observation that the presence of polar residues in the HH helix of seipin is required for its TG-clustering activity.
Discussion
Identified as a crucial protein in LD biogenesis and homeostasis many years ago (22), the molecular mechanism of seipin has remained enigmatic and debated (14–17, 20–22, 24, 28, 29, 34, 48), in large part because of its role in multiple steps of LD biology, including LD localization (14, 32), stabilization of ER-LD contacts (14, 21, 39), control of TG flow between the ER and LDs (14), LD budding (20, 51), and PA metabolism (27–30). Here, we took advantage of the recently solved cryo-EM structures of the luminal domain of seipin (27, 35) to perform MD simulations of the seipin oligomer embedded in explicit lipid bilayers to investigate the role of seipin in the early stages of LD formation.
Our simulations suggest that seipin can cluster TG molecules even when they are present at low concentrations in the surrounding membrane and that it does so by first binding to them through several polar residues located in its TM helices. These interactions are short lived, and TG molecules can subsequently diffuse toward the interior of the ring and engage in stable interactions with polar residues of the HH of the luminal domain, from which they hardly detach. These strongly bound TG molecules can act as nucleation seeds for newly arriving TG molecules, further promoting TG accumulation. In addition, we found that a similar mechanism is true for the TG precursor, DG, and that accumulation of DG inside seipin does not prevent the subsequent accumulation of TG.
While these observations suggest a mechanistic role for seipin in the early stages of LD formation, the presence of seipin alone seems to be insufficient to promote TG accumulation beyond a certain size threshold, as in our simulations, TG blister growth reaches a plateau even in the presence of excess TG molecules in the lipid bilayer. Rather, the initial TG clustering promoted by seipin could be functional to promote the subsequent recruitment of additional protein factors that are required for normal LD biogenesis, such as LDAF1/promethin in human (15) or LDO16/45 in yeast (17). Our data could also explain why TG was not identified to coprecipitate with seipin in the absence of its partner protein LDAF1 (15) given that a limited accumulation of TG molecules paired to minor loss during the purification process could produce a signal below the detection limit.
Interestingly, the main binding site we identified for TG binding overlaps with the proposed seipin-LDAF1 interface based on low-resolution cryo-EM density maps (15). Our yeast data, however, indicate that mutations to this binding site result in delayed LD formation even in the absence of LDAF1. Even if we can’t exclude potential direct interactions between heterologous-expressed human seipin and LDO16/45 in yeast, the absence of the HH in the yeast version of seipin (fld1/sei1) suggests that the observed delay in LD formation is likely to result from the action of seipin alone. Hence, our data are consistent with a model in which seipin-driven preexisting TG accumulation promotes subsequent LDAF1 recruitment, possibly via a combination of direct seipin–LDAF1 interactions as well as preference of LDAF1 for TG-rich membranes.
Our simulations suggest that the membrane-remodeling activity of seipin might play a major role in the subsequent recruitment of additional proteins that are required for the completion of LD biogenesis (15–18, 52) and its regulation (11, 34, 49). To this extent, the unexpected observation that seipin also accumulates DG, and that it does so even more efficiently than TG, is extremely intriguing. Unlike TG, DG is a constitutive ER lipid that is generally present at nonnegligible concentrations, and it has been shown to promote the recruitment of several LD-associated proteins, including perilipins (53), FIT proteins (54), and the TG-synthesizing acyltransferase Lro1 (32). Notably, our results are in agreement with the recent observation that seipin-positive ER sites are also enriched in DG, even in the absence of LD formation (32), and they suggest that seipin could be directly responsible for the observed DG enrichment via direct accumulation of DG molecules in its structure. Also, our observation that seipin, upon TG and DG accumulation, induces a positive curvature to the lipid bilayer, and specifically to the cytosolic leaflet, not only agrees with the experimental observation that seipin preferentially localizes in curved regions of the ER (55) but also potentially suggests that the generation of local curvature might help in the recruiting of downstream protein partners (e.g., LDAF1, perilipins, etc.).
The mechanism through which DG accumulation could facilitate subsequent TG accumulations is also intriguing due to the unique physical chemistry properties of this lipid. First, DG accumulates more efficiently than TG close to the TM helices (Fig. 4 F and J). This corresponds to the region with the largest negative curvature in the budding LD, and, hence, DG could help stabilize those sites by specifically partitioning there. Second, the presence of DG in lipid bilayers favors the spontaneous nucleation of TG blisters (44). Hence, seipin-driven DG clustering could deplete TG from the ER membrane in the promixity of seipin, leading to TG demixing in the seipin ring. Third, DG molecules can behave as a pure oil when present in the bilayer at high concentrations, spontaneously undergoing phase separation from the bilayer and forming pure DG blisters (42). Under these circumstances, TG molecules would likely incorporate into preexisting DG blisters because of favorable entropic DG–TG interactions as opposed to entropy-frustrated interactions with the acyl chains of the bilayer lipids.
Interestingly, DG has also been implicated in later stages of LD formation, namely, LD budding from the ER, where the concentration of DG has to decrease in order to promote directional budding (49). Thus, our data are consistent with a role of DG as a master regulator of LD biogenesis, whereby a high initial concentration promotes TG depletion from the ER and its accumulation around seipin-positive ER sites, while subsequent conversion of DG into TG, and corresponding DG depletion, promotes LD growth and budding.
Quite unexpectedly, we did not observe specific accumulation of PA around seipin in our simulations. However, PA binding to seipin has been attributed to the activity of its NPC1-like luminal domains (27) rather than to direct interactions with the membrane-embedded regions, and these interactions would take place on time scales beyond those explored in our simulations. Additionally, a role of the N-terminal domain of seipin has been proposed in relation to PA homeostasis (28), and this region of the protein is absent from our molecular model.
In addition to its role in LD biogenesis, our simulations hint at an intriguing possible role of seipin in maintaining ER homeostasis; as it diffuses through the ER bilayer (15), seipin would be able to remove potentially lipotoxic TG and DG molecules from the ER by accumulating them in its structure, where they could subsequently promote protein recruitment and act as a seed for the biogenesis of LDs. Interestingly, our data are consistent with recent experiments suggesting that deletion of seipin leads to an accumulation of PA in the ER and nuclear membrane (30), as, in the absence of this DG-scavenging activity of seipin, an excess of DG in the ER membrane would lead to an excess of PA because the two lipids are homeostatically linked (56). We expect that future studies will further address the role of seipin in ER homeostasis and whether the seipin mechanism we have identified is specific to DG and TG accumulation, or if it is also valid for other NLs, such as steryl ester.
Finally, we want to highlight the limitations of our modeling approach. First, our seipin model lacks the N-terminal domain (residues 1 through 20). We opted not to model this domain ab initio, as it has been shown that while deletion of the first 14 amino acids at the N terminus of seipin results in a delay in LD formation, this doesn’t affect LD morphology (24). In addition, in our model, only the luminal part of seipin was built using experimentally derived coordinates, while the TM helices were modeled ab initio as straight helices based on the coordinates of the cryo-EM structure of the luminal domains. While this leaves room for improving the accuracy of the TM helix region model with respect to experiment, the observation that the luminal domain alone is sufficient for the accumulation and trapping of TG molecules indicates that the bias introduced in any potentially erroneous modeling of the TM helices is likely insufficient to alter the main conclusions of our work. Second, the use of CG modeling introduces inherent quantitative shortcomings in the accurate reproduction of molecular interactions, most notably in the case of charged or polar residues where electrostatic effects or hydrogen bonding might play a major role. In our case, this might be particularly important pertaining to the observation that seipin interacts with TG molecules via multiple polar residues. However, a specific role of the polar moiety of TG in establishing interactions with proteins is supported by recent experimental observations (57) and could be the subject of future validation by atomistic simulations.
Overall, our simulations provide a high-resolution description of the mechanism through which seipin promotes LD formation, and they indicate that seipin ER remodeling activity is a crucial component of its molecular mode of action. We foresee that future studies including additional proteins that are known to interact with seipin at sites of LD biogenesis will shed further light on the entire process of LD formation and growth with unprecedented molecular detail.
Materials and Methods
All MD simulations were performed using the software LAMMPS (58) in combination with the SDK (41, 42, 59–61) and the SPICA force field (62) for proteins (https://www.spica-ff.org). The MD simulations are based on the structure of the luminal portion of the human seipin (ID: 6DS5) (27). Our model includes the TM domains of the protein (two per monomer) and the loops connecting them with the luminal domains. The modeling strategy used to build these domains is described in SI Appendix. The structures of the WT protein, luminal-only, and TM-only domain as well as single-point mutants were then embedded in lipid bilayers using CHARMM-GUI (23). In certain systems a varying amount of TG was added. Details of all systems are described in SI Appendix.
Site-directed mutagenesis in human cells was performed using high-fidelity Thermococcus kodakaraensis (KOD) HotStart DNA polymerase (Merck Millipore), and imaging was performed using immunofluorescence of HA-tagged seipin, after transfection with vector or seipin constructs for 48 h. Images were collected after 15 min of oleic acid (OA) treatment following fixation and permeabilization. Additional details are described in SI Appendix.
Experiments in yeast were performed after expression of WT and mutant versions of human seipin, fused to GFP, in LD-inducible strains expressing WT seipin (GAL-LRO1 are1∆ are2∆ dga1∆) or lacking seipin (GAL-LRO1 are1∆ are2∆ dga1∆ fld1∆) expressing the LD marker Erg6-mCherry. Images were acquired after 5, 15, 30, and 60 min of LD induction by resuspension in selective galactose medium containing 0.67% yeast nitrogen base without amino acids (USBiological), 0.73 g/L amino acids, 2% galactose. Microscope experiments were performed on three different clones with essentially similar results. The counting of LDs was performed manually in 100 cells for each time point. Additional details are described in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (Grant 163966). This work was supported by a grant from the Swiss National Supercomputing Centre (CSCS) under Project ID s980. We acknowledge Partnership for Advanced Computing in Europe for awarding us access to Piz Daint, ETH Zurich/CSCS. V.Z. acknowledges the Japan Society for the Promotion of Science/ETH young researchers’ exchange program (Fellowship GR19107). R.S. acknowledges support from Swiss National Science Foundation (Grant 31003A_173003). We thank Pablo Campomanes for providing the script for the mean square displacement calculations.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017205118/-/DCSupplemental.
Data Availability
Data collected and analyzed in this paper are available on Zenodo (https://doi.org/10.5281/zenodo.4544256), and supplementary information about computational methods and experimental data analyses are included in the SI Appendix.
References
- 1.Walther T. C., Farese R. V. Jr, Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Q., Goodman J. M., The lipid droplet-a well-connected organelle. Front. Cell Dev. Biol. 3, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pol A., Gross S. P., Parton R. G., Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olzmann J. A., Carvalho P., Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu S., Zhang X., Liu P., Lipid droplet proteins and metabolic diseases. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1968–1983 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Dalhaimer P., Lipid droplets in disease. Cells 8, 974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gluchowski N. L., Becuwe M., Walther T. C., R. V. J. Farese, Jr, Lipid droplets and liver disease: From basic biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 14, 343–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandager L., et al., Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277, 6478–6482 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Choudhary V., Ojha N., Golden A., Prinz W. A., A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211, 261–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Goodman J. M., The collaborative work of droplet assembly. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1205–1211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nettebrock N. T., Bohnert M., Born this way – Biogenesis of lipid droplets from specialized ER subdomains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158448 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Walther T. C., Chung J., Farese R. V. Jr, Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33, 1–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiam A. R., Forêt L., The physics of lipid droplet nucleation, growth and budding. Biochim. Biophys. Acta 1861, 715–722 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Salo V. T., et al., Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev. Cell 50, 478–493.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Chung J., et al., LDAF1 and seipin form a lipid droplet assembly complex. Dev. Cell 51, 551–563.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teixeira V., et al., Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217, 127–138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohnert M., New friends for seipin–Implications of seipin partner proteins in the life cycle of lipid droplets. Semin. Cell Dev. Biol. 108, 24–32 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg-Bord M., et al., Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217, 269–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagac M., et al., SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17, 1546–1559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., et al., Seipin is required for converting nascent to mature lipid droplets. eLife 5, 1–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salo V. T., et al., Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szymanski K. M., et al., The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U.S.A. 104, 20890–20895 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fei W., et al., Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–482 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cartwright B. R., et al., Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell 26, 726–739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutet E., et al., Seipin deficiency alters fatty acid Delta9 desaturation and lipid droplet formation in Berardinelli-Seip congenital lipodystrophy. Biochimie 91, 796–803 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Wang M., et al., Dysfunction of lipid metabolism in lipodystrophic Seipin-deficient mice. Biochem. Biophys. Res. Commun. 461, 206–210 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Yan R., et al., Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248–256.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Han S., Binns D. D., Chang Y.-F., Goodman J. M., Dissecting seipin function: The localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol. 16, 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolinski H., et al., Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta 1851, 1450–1464 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Sołtysik K., et al., Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol. 220, e202005026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fei W., et al., A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7, e1002201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choudhary V., El Atab O., Mizzon G., Prinz W. A., Schneiter R., Seipin and Nem1 establish discrete ER subdomains to initiate yeast lipid droplet biogenesis. J. Cell Biol. 219, e201910177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith S. W., Weiss S. B., Kennedy E. P., The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 (1957). [PubMed] [Google Scholar]
- 34.Hayes M., et al., Fat storage-inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes. Microb. Cell 5, 88–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sui X., et al., Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. J. Cell Biol. 217, 4080–4091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacovache I., Bischofberger M., van der Goot F. G., Structure and assembly of pore-forming proteins. Curr. Opin. Struct. Biol. 20, 241–246 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Hunt J. F., Weaver A. J., Landry S. J., Gierasch L., Deisenhofer J., The crystal structure of the GroES co-chaperonin at 2.8 A resolution. Nature 379, 37–45 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Braig K., et al., The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 371, 578–586 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Grippa A., et al., The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinoda W., DeVane R., Klein M. L., Zwitterionic lipid assemblies: Molecular dynamics studies of monolayers, bilayers, and vesicles using a new coarse grain force field. J. Phys. Chem. B 114, 6836–6849 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacle A., Gautier R., Jackson C. L., Fuchs P. F. J., Vanni S., Interdigitation between triglycerides and lipids modulates surface properties of lipid droplets. Biophys. J. 112, 1417–1430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campomanes P., Zoni V., Vanni S., Local accumulation of diacylglycerol alters membrane properties nonlinearly due to its transbilayer activity. Commun. Chem. 2, 72 (2019). [Google Scholar]
- 43.Khandelia H., Duelund L., Pakkanen K. I., Ipsen J. H., Triglyceride blisters in lipid bilayers: Implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS One 5, e12811 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zoni V., et al., Pre-existing bilayer stresses modulate triglyceride accumulation in the ER versus lipid droplets. eLife, 10.7554/eLife.62886 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben M’barek K., et al., ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell 41, 591–604.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Zoni V., et al., To bud or not to bud: A perspective on molecular simulations of lipid droplet budding. Front. Mol. Biosci. 6, 124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chorlay A., et al., Membrane asymmetry imposes directionality on lipid droplet emergence from the ER. Dev. Cell 50, 25–42.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y.-P., Lewis R. N. A. H., Hodges R. S., McElhaney R. N., Peptide models of helical hydrophobic transmembrane segments of membrane proteins. 2. Differential scanning calorimetric and FTIR spectroscopic studies of the interaction of Ac-K2-(LA)12-K2-amide with phosphatidylcholine bilayers. Biochemistry 34, 2362–2371 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Choudhary V., et al., Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curvature. Curr. Biol. 28, 915–926.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adeyo O., et al., The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S., et al., Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9, 2939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castro I. G., et al., Promethin is a conserved seipin partner protein. Cells 8, 268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skinner J. R., et al., Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284, 30941–30948 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gross D. A., Zhan C., Silver D. L., Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. U.S.A. 108, 19581–19586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santinho A., et al., Membrane curvature catalyzes lipid droplet assembly. Curr. Biol. 30, 2481–2494.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Barbosa A. D., et al., Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 26, 3641–3657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olarte M.-J., et al., Determinants of endoplasmic reticulum-to-lipid droplet protein targeting. Dev. Cell 54, 471–487.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plimpton S., Fast parallel algorithms for short–range molecular dynamics. J. Comput. Phys. 117, 1–19 (1995). [Google Scholar]
- 59.Shinoda W., DeVane R., Klein M. L., Multi-property fitting and parameterization of a coarse grained model for aqueous surfactants. Mol. Simul. 33, 27–36 (2007). [Google Scholar]
- 60.MacDermaid C. M., et al., Molecular dynamics simulations of cholesterol-rich membranes using a coarse-grained force field for cyclic alkanes. J. Chem. Phys. 143, 243144 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Shinoda W., DeVane R., Klein M. L., Computer simulation studies of self-assembling macromolecules. Curr. Opin. Struct. Biol. 22, 175–186 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Seo S., Shinoda W., SPICA force field for lipid membranes: Domain formation induced by cholesterol. J. Chem. Theory Comput. 15, 762–774 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data collected and analyzed in this paper are available on Zenodo (https://doi.org/10.5281/zenodo.4544256), and supplementary information about computational methods and experimental data analyses are included in the SI Appendix.