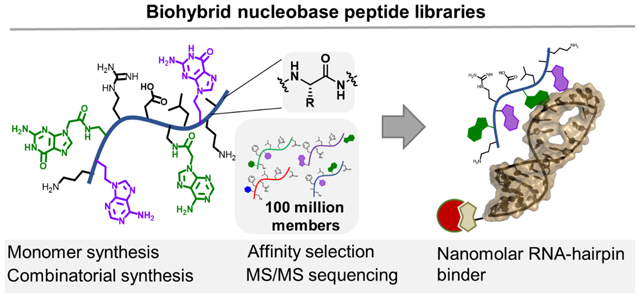
Abstract
Nature has three biopolymers: oligonucleotides, polypeptides, and oligosaccharides. Each biopolymer has independent functions, but when needed, they form mixed assemblies for higher-order purposes, as in the case of ribosomal protein synthesis. Rather than forming large complexes to coordinate the role of different biopolymers, we dovetail protein amino acids and nucleobases into a single low molecular weight precision polyamide polymer. We established efficient chemical synthesis and de novo sequencing procedures and prepared combinatorial libraries with up to 100 million biohybrid molecules. This biohybrid material has a higher bulk affinity to oligonucleotides than peptides composed exclusively of canonical amino acids. Using affinity selection mass spectrometry, we discovered variants with a high affinity for pre-microRNA hairpins. Our platform points toward the development of high throughput discovery of sequence defined polymers with designer properties, such as oligonucleotide binding.
Keywords: combinatorial libraries, nucleobase peptides, affinity selection, tandem mass spectrometry, nucleic acids, sequence defined polymers
Graphical Abstract
Introduction
Therapeutic targeting of nucleic acids (RNA and DNA) is of high interest. Targeting approaches span the full spectrum of molecules from small drugs to >100 kDa composite macromolecules (CRISPR-Cas).1–5 Nucleic acids are therapeutic targets because mutations of specific genes, dysregulation of transcription activity, and non-coding RNAs (ncRNA) are at the core of many genetic and acquired pathologies.6–8 CRISPR-Cas and antisense oligonucleotides (ASOs)4 restore these genetic abnormalities. Small molecule3,9–12 and peptide-based13–16 approaches are used for alternative targeting strategies.
Biohybrid abiotic polymers that resemble both peptides and nucleic acids are an intriguing modality to target nucleic acids. These variants should favor high affinity and selectivity for specific oligonucleotides. The amino acid side-chains allow for highly diverse structural and functional landscapes, while the nucleobases can interact with nucleic acids via base pairing. Over the years, scientists have aimed to generate two-in-one biomaterials. Previously, Mihara et al. designed peptides to bind RNA by rational insertion of nucleobase amino acids.17–19 Hecht et al. found that transcriptional DNA regulator proteins could be modified with nucleobases to improve their DNA-binding.20 Analogously, peptoids with nucleobase side chains were designed to targeted DNA.21–23 Peptide nucleic acids (PNAs) containing alanine and lysine side chains were found to assemble into supramolecular structures while maintaining sequence-specific RNA binding (Figure 1B).24 In an elegant approach, David Liu and coworkers built a platform for the in vitro evolution of nucleic acid polymers displaying amino acid side chains on nucleobases.25 The resulting greater structural diversity and hydrophobic side chains proved advantageous for the selection of potent aptamers (Figure 1A).26 Despite these efforts, no robust high-throughput selection/decoding strategies for mixed peptide biohybrids have been reported so far.
Figure 1. Recent examples of biohybrid sequence defined polymers.
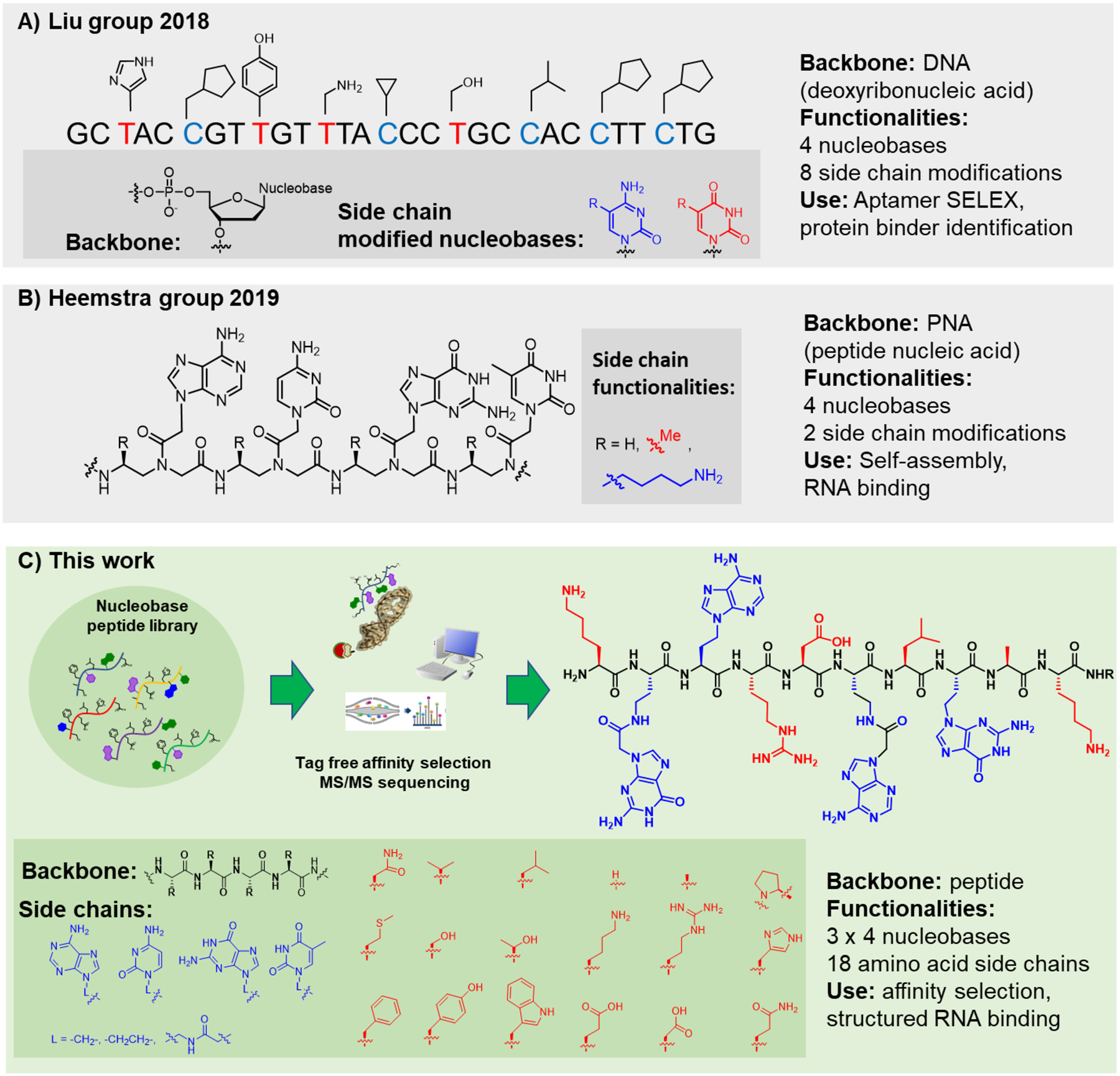
A) Highly functionalized nucleic acids developed in the Liu lab.25,26 These structures can be amplified by PCR and sequenced and therefor used for aptamer selections. B) Modified PNA developed in the Heemstra group.24 Hydrophobic and hydrophilic side chains drive PNA self-assembly to supramolecular structures. C) This work: efficient synthesis and sequencing strategies for nucleobase peptides were developed, enabling discovery experiment from combinatorial libraries.
Synthetic combinatorial chemistry and tandem mass spectrometry (MS/MS) sequencing can constitute a discovery platform for biohybrid nucleobase peptides. Combinatorial libraries give access to diverse chemical space for the discovery of compounds with novel properties. Our group recently reported affinity selection mass spectrometry (AS-MS) workflows, which enable high throughput screening of synthetic combinatorial peptide libraries;27–29 these strategies enable flexible incorporation of virtually any non-canonical amino acid. Success with high diversity AS-MS requires efficient synthesis and high fidelity MS/MS decoding. The AS-MS approach is tag-free as opposed to ‘nucleic acid-encoded’ libraries.30,31 The oligonucleotide tag might also bind to the target and interfere with the identification of real binders. By circumventing this limitation, ‘tag-free’ synthetic chemical libraries provide an additional advantage.
Here we describe the development of efficient synthesis and de novo sequencing procedures for biohybrid nucleobase peptides. We found that nucleobase containing peptides have a significantly higher affinity for nucleic acids, compared to canonical peptides. From a 100 million-membered nucleobase peptide library we enriched a nanomolar binder to RNA-hairpins, with selectivity over DNA. These results suggest a promising approach to target specific oligonucleotide strands.
Results & discussion
Design and synthesis of biohybrid nucleobase peptides
A polyamide peptide backbone with α-amino acid-based monomers was chosen for the synthesis of biohybrid nucleobase peptide libraries. We hypothesized that dovetailing amino acid side chains and nucleobases on a polyamide backbone would be an optimal choice for the following reasons: polyamides are chemically stable and can be assembled with high efficiency by Fmoc solid phase peptide synthesis (SPPS). Fmoc-protected amino acids are commercially available and several SPPS compatible nucleobase displaying amino acid monomers have been described in the literature.17,32–35 We decided to focus on building blocks based on α-amino acids. These monomers lead to sequence defined polymers having the potential to adopt secondary structure and are amenable to de novo sequencing with MS/MS methodologies and algorithms optimized for canonical peptides and proteomics.
Twelve nucleobase amino acid monomers were synthesized. To determine the synthetic accessibility and sequencing compatibility while covering a broad chemical space, we prepared three monomer sets with different linkers connecting each of the four nucleobases to the amino acid moiety (Scheme 1A). Serinolactone was transformed into alanyl nucleobase monomers by nucleophilic ring opening with the respective nucleobases.32,36–38 The resulting linker between the nucleobase and the alanyl scaffold is a methylene group. Bromohomoalanine was transformed into homoalanyl (hAla) nucleobase monomers by nucleophilic substitution of the bromine with the respective nucleobases.17–19,32 The resulting linker between the nucleobase and the hAla scaffold is an ethylene group. Diaminobutyric acid (Dab) was coupled to nucleobase acetic acid derivatives affording Dab-nucleobase monomers, with an ethyl amido methyl linker.33,34 Details regarding synthesis and protecting group strategies are discussed in the SI (SI schemes 1 and 2).
Scheme 1. Twelve nucleobase amino acid monomers were synthesized (A) and incorporated into peptides by SPPS (B).
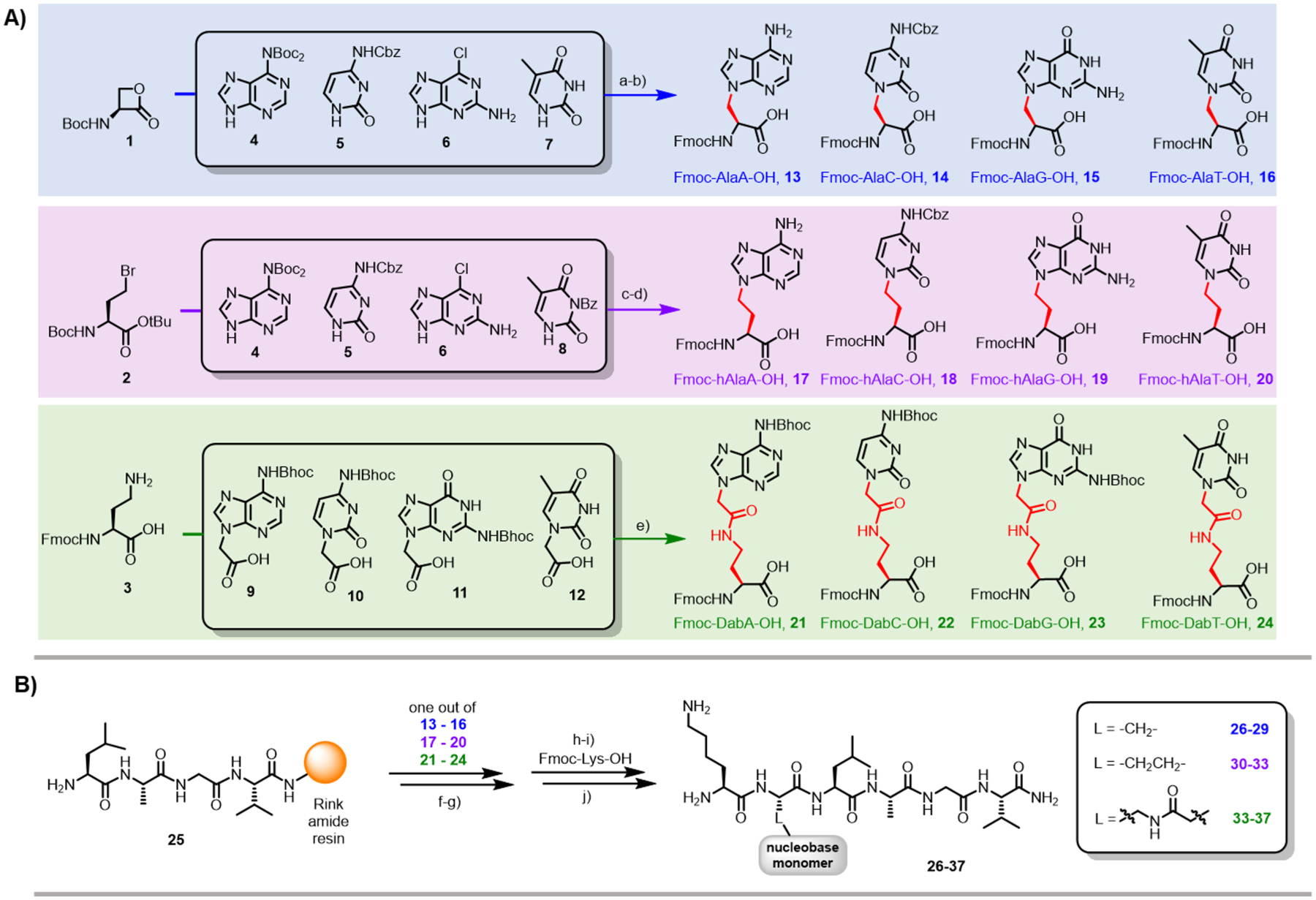
Reagents and conditions: a) DBU, DMSO, −78 °C, 2 h, 38% for 4, 39% for 5, 63% for 6, 29% for 7; b) TFA/CH2Cl2 (3:1), 90 min, r.t.; (for 6: TFA/H2O (3:1), 72 h, r.t.); then: Na2CO3, FmocOSu, H2O/1,4-dioxane, 16 h, 54% for 13, 61% for 14, 74% for 15, 63% for 16; c) K2CO3, Cs2CO3, DMF, 50 °C, 16 h, 58% for 4, 60% for 5, 70% for 6, 80% for 8; d) TFA/CH2Cl2 (3:1), 90 min, r.t.; (for 6: HCl (1 M), 2 h, 90 °C; for 8: HBr/CH3COOH, 1 h, r.t.); then: Na2CO3, FmocOSu, H2O/1,4-dioxane, 16 h, 73% for 17, 42% for 18, 42% for 19, 70% for 20; e) TSTU, DMF, DIEA, 2 h, 28% for 21, 22% for 22, 25% for 23, 28% for 24; f) PyAOP, DIEA, DMF, 20 min, r.t.; g) piperidine, DMF, 5 min, r.t.; h) HATU, DIEA, DMF, 10 min, r.t.; i) piperidine, DMF, 5 min, r.t.; j) TFA, TMSOTf, EDT, thioanisole, cresol, 1 h, r.t. Abbreviations: DBU = 1,8-Diazabicyclo[5.4.0]undec-7-ene, DMSO = dimethyl sulfoxide, TFA = trifluoroacetic acid, r.t. = room temperature, FmocOSu = N-(9H-fluoren-9-ylmethoxycarbonyloxy)succinimide, DMF = N,N-dimethylformamide, TSTU = N,N,N′,N′-tetramethyl-O-(N-succinimidyl)uronium tetrafluoroborate, DIEA = N,N-diisopropylethylamine, PyAOP = (7-azabenzotriazol-1-yloxy)trispyrrolidinophosphonium hexafluorophosphate, HATU = 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate, TMSOTf = trimethylsilyl trifluoromethanesulfonate, EDT = 1,2-ethanedithiol.
Combinatorial libraries require high efficiency for every chemical reaction. We confirmed the efficient incorporation of the twelve nucleobase monomers into short peptides with amino acid building blocks before and after incorporation of the nucleobase (K-Nucleobase-LAGV, Scheme 1B). In summary, the twelve crude nucleobase peptides were 70 to 99% pure, as determined by LC-MS (SI S38 – S43).
Tandem MS-based sequencing of biohybrid nucleobase peptides
High-fidelity de novo sequencing is a requirement for discovery experiments with non-natural polymers. We focused on combinatorial libraries with ~200 members to establish de novo sequencing procedures. Libraries with the three sets of nucleobase monomers were synthesized. We analyzed the alanyl, homoalanyl and Dab nucleobase monomers in three distinct libraries, to determine the influence of each linker on sequencing (Figure 2A). The library design was 12345678K, wherein 1, 3, 6 and 8 was one of two canonical amino acids, and 2, 4, 5 and 7 was either a canonical amino acid or one of the four nucleobase monomers. The total diversity of each library was 28 = 256. These libraries contain 16 possible sequences without any nucleobase and serve as positive controls. Each library contains 64 sequences with one nucleobase, 96 with two, 64 with three, and 16 with four nucleobases. Taken together, we answer how each non-natural monomer affects de novo sequencing efficiency.
Figure 2. De novo sequence determination of nucleobase peptides with up to four nucleobases per compound is possible using tandem MS/MS and sequencing software.
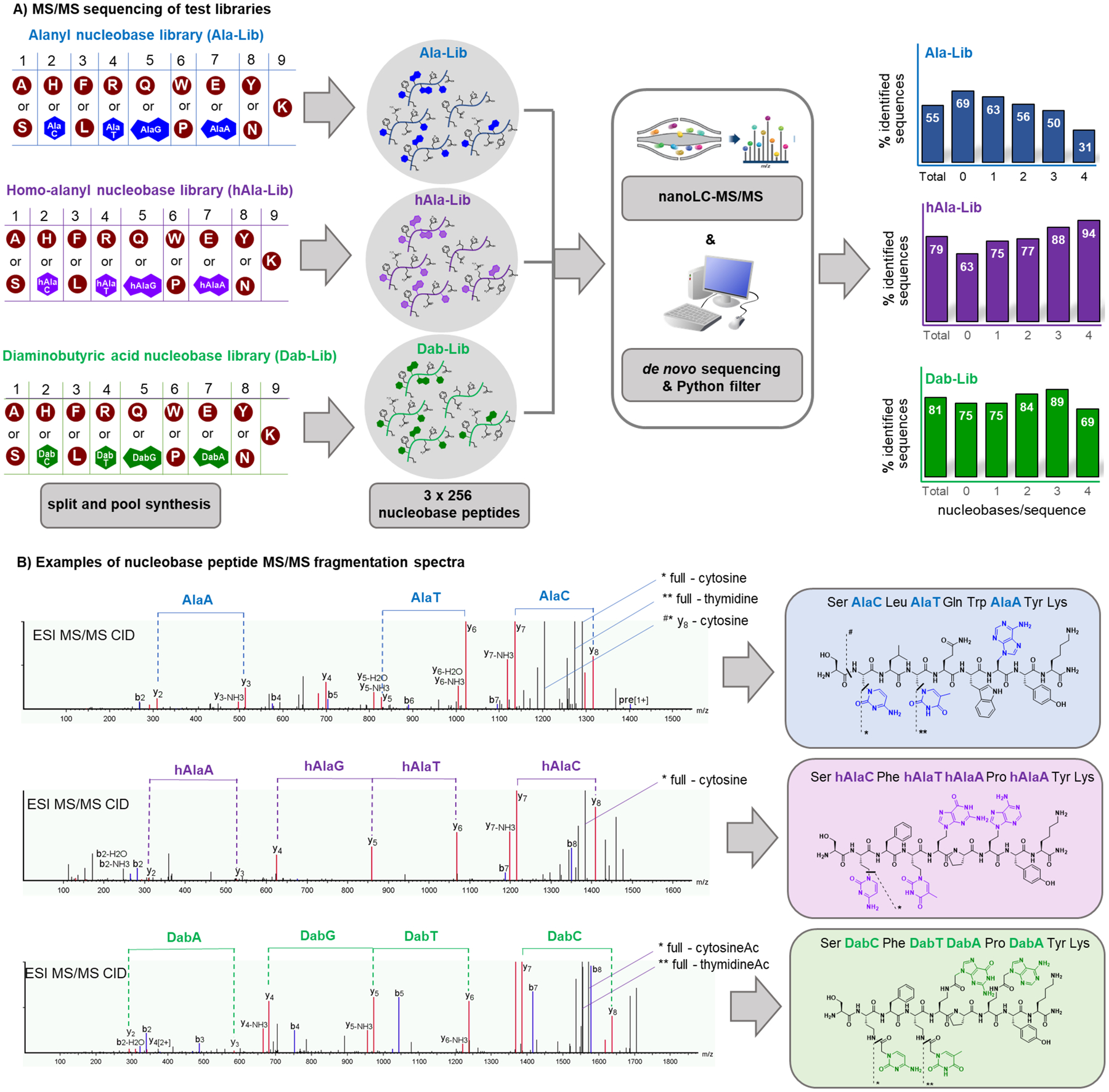
A) Three combinatorial libraries with the three nucleobase monomer sets (alanyl = blue; homoalanyl = purple; Dab = green) and canonical amino acid residues were synthesized and analyzed by nanoLC-MS/MS. De novo sequencing was performed with the software PEAKS 8.5 and the final list was filtered for sequences fitting the correct library design. The percentage of identified total sequences and sequences containing different numbers of nucleobases is reported on the graphs on the right. Expected total sequences with N nucleobases: 16 for N = 0, 64 for N = 1, 96 for N = 2, 64 for N = 3, 16 for N = 4. B) Exemplary MS/MS spectra of nucleobase peptides from the three libraries. Fragmentations of nucleobase side chains are shown.
Sequence identification by de novo MS/MS sequencing was possible for >70% of all nucleobase peptides. We analyzed each library by a single nanoLC-MS/MS run on an Orbitrap Fusion Lumos mass spectrometer. Three fragmentation modes were used in combination: collision induced dissociation (CID), higher energy collision induced dissociation (HCD) and electron transfer dissociation (ETD). The resultant MS spectra were analyzed with the sequencing software PEAKS 8.5. We used a Python script to filter candidate sequences provided from PEAKS according to library design criteria, including correct length and input monomers.39 The sequences were divided into categories. The percentage of sequences with zero, one, two, three or four nucleobases respectively, was determined. For the alanyl library we observed a decreased sequence recovery/identification with increasing nucleobases per sequence. For the homoalanyl and the Dab libraries, the presence of up to four nucleobase monomers per sequence did not adversely influence the sequence decoding. The MS/MS spectra show peptide backbone fragmentation and nucleobase and nucleobase-acetyl (Dab library) dissociations (exemplary spectra in Figure 2B). The most prominent nucleobase dissociations were observed in the alanyl library, possibly explaining the lower sequence identification rate. The MS/MS spectrum from a single fragmentation source did not enabled identification of the sequences, rather the combination of CID, HCD and ETD spectra was needed. The overall sequence identification rate (all three libraries taken together, 768 peptides) was 72%, comparable to the average identification of the control pool (69%).
Synthesis of a nucleobase peptide library
A nucleobase peptide library with 100 million members was synthesized. The outcome of the sequencing experiments indicated that all three sets of nucleobase monomers can be used for library synthesis and sequencing. We prepared a combinatorial library with the format X9K, using twelve nucleobase monomers and 18 canonical amino acids for each variable position. A fixed Lys residue was placed at the C-terminus to facilitate the MS/MS sequencing with a constant charge on all y-type ions. One-bead-one-compound (OBOC)40,41 split-and-pool synthesis was used to prepare a library with ~108 members (Figure 3A), a suitable size for magnetic bead-based affinity selections.29 This library size is a compromise between high diversity, which enhances the probability of identifying binders, and a sufficient concentration of each library member to ensure enough material for affinity selection and subsequent MS/MS sequencing (~10 fmol per compound). To control the number of nucleobases per sequence, the split-and-pool process was biased: in each coupling step 20% of the resin was used for the 12 nucleobase peptides and 80% for the canonical residues. Based on this, the library distribution indicates that 86% of sequences would have one to four nucleobases (which we showed being sequenced with high confidence), 11% no nucleobases and 3% five or more nucleobases (Figure 3B).
Figure 3. A 100-million membered nucleobase peptide library was synthesized.
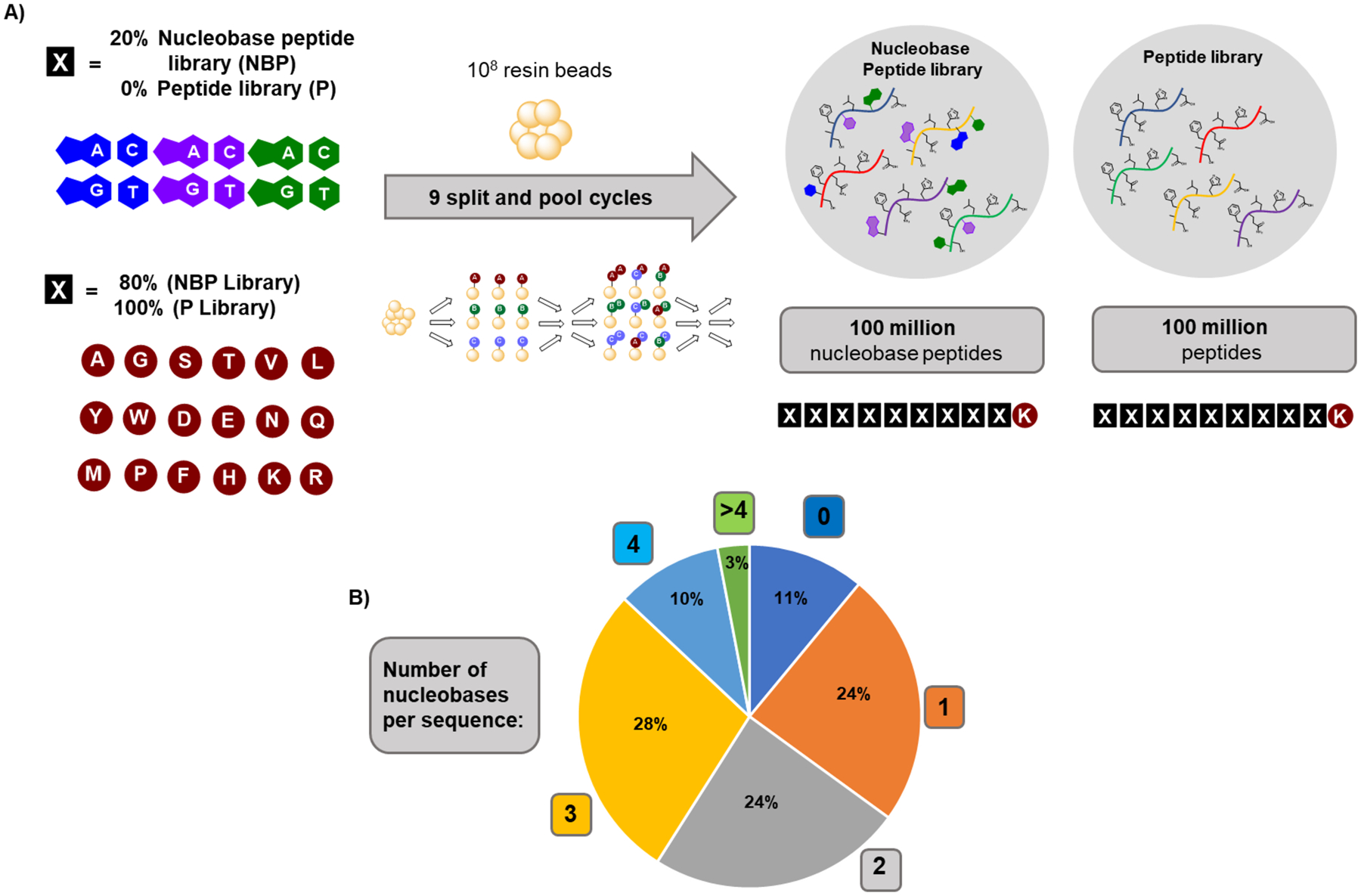
A) Schematic representation of the monomer set and the synthesis of a 100 million-membered library of biohybrid nucleobase peptides or canonical peptides, respectively. B) Calculated number of nucleobases per sequence in the library based on the percentage of monomers used.
Bulk affinity of biohybrid nucleobase peptides towards nucleic acids
Nucleobase peptides have higher bulk affinity to oligonucleotides than canonical peptides. The nucleobase peptide library and a canonical peptide library with the same format (X9K, 100 million member) were immobilized on an ELISA (enzyme linked immunosorbent assay) plate and incubated with a biotinylated DNA library (Biotin-N20, with N = adenine, cytosine, guanidine or thymidine) (Figure 4A). Also, a positively charged peptide 38 (LKKLLKLLKKWLKLK)14 was immobilized, as to probe for electrostatic interactions. Association was determined after exposure to horseradish peroxidase (HRP) conjugated to streptavidin (SA) and tetramethylbenzidine (TMB). The nucleobase peptide library showed significantly higher signal when compared to both the canonical peptide library and the positively charged peptide (Figure 4B). These results indicate that nucleobase peptide libraries are a preferred pool for the identification of binders to nucleic acids.
Figure 4. Hybrid nucleobase peptides have higher bulk affinity to oligonucleotides than canonical peptides.
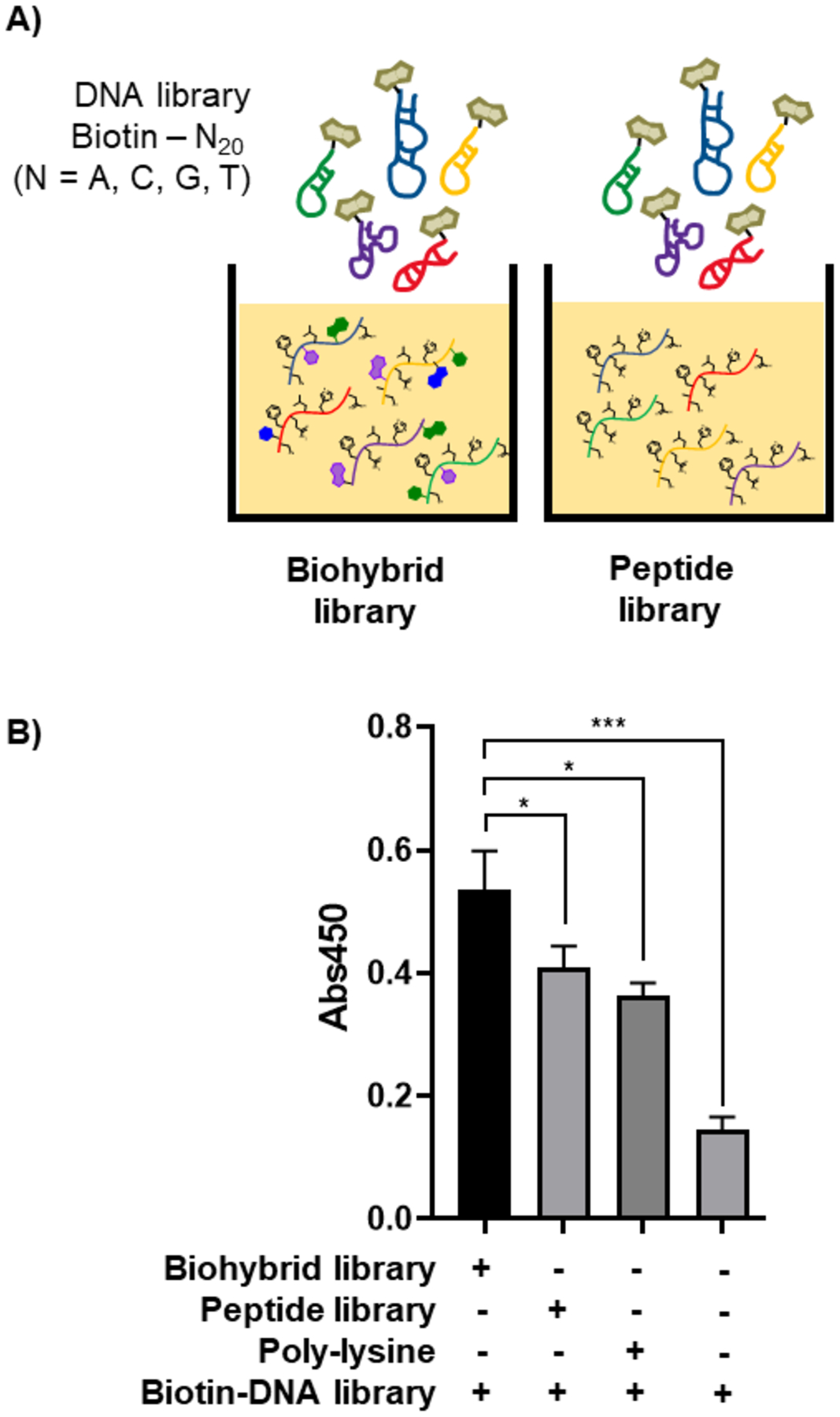
A) Schematic representation of the ELISA assay: immobilized nucleobase peptide or canonical peptide library, respectively, is incubated with a biotinylated DNA library. B) ELISA readout diagram: nucleobase peptides have higher affinity towards DNA than canonical peptides.
Identification of an RNA-hairpin-binding nucleobase peptide by affinity selection – mass spectrometry
The targeting of pre-miRNA hairpins is of high interest: gene expression in cells is controlled by miRNAs and miRNAs are correlated with disease.42,43 Binders to structured hairpin precursor miRNA can potentially inhibit the processing to the mature and active miRNA.44 Two pre-miRNAs involved in disease are pre-miRNA-2145 and pre-miRNA-155.46
We envisioned our nucleobase peptide library could be used to discover high affinity pre-miRNA-hairpin binders. Biotinylated pre-miRNAs (21 and 155) were immobilized on magnetic streptavidin beads and incubated with the 100-million membered nucleobase peptide library ([library] = 1 mM; [individual library member] = 10 pM; Figure 5A). Yeast RNA was added to the selection conditions to scavenge for non-specific RNA binders. After washing to remove unbound material, potential hits were eluted and subjected to nanoLC-MS/MS. As a negative control, the same workflow was done with unmodified streptavidin beads. The resultant MS spectra were visualized with the sequencing software PEAKS 8.5 and further refined with a Python script that nominated variants matching the library design. Finally, an extracted ion chromatogram was created for each variant (based on the precursor ion used by PEAKS for sequencing): only compounds resulting in a clear peak in the chromatogram were considered ‘hits’ (Figure 5D). Cross-analysis of the same ion in the control chromatograms (beads only) allowed to exclude potential unspecific bead or streptavidin binders. After these steps, for enrichments with the pre-miRNA21, only one nucleobase peptide was identified: Lys-DabG-hAlaA-Arg-Asp-DabA-Leu-hAlaG-Ala-Lys (Fig 5B–C). The variant ions were also identified in the LCMS chromatograms resulting from enrichments with pre-miRNA-155: due to their low intensity, however, no sequencing data were obtained from these samples. It was absent in the unmodified streptavidin bead control sample (Figure 5D). No specific nucleobase peptide variants were identified from the pre-miRNA-155 enrichments.The biohybrid nucleobase peptide binds to RNA-hairpins with nanomolar affinity. To validate the binding of the hit sequence to the pre-miRNA hairpins, we synthesized the compound with a C-terminal biotin handle (39-biotin). 39-biotin was immobilized on biolayer interferometry (BLI) streptavidin tips and the association/dissociation to pre-miRNA21 in solution at different concentrations was determined: the dissociation constant (Kd) was found to be 9 nM for pre-miRNA21 (Figure 4E) and 12 nM for pre-miRNA-155 (Figure 4F). The affinity towards the pre-miRNA-21 stem, missing the hairpin loop, was reduced by over 50-fold, indicating the importance of the hairpin loop for the binding of 39 (SI Figure1). Indeed, 39 was found to bind with high affinity to an unrelated hairpin derived from HIV RNA (SI Figure 1). We also determined melting temperatures of pre-miRNA-21 alone and in the presence of 39 using circular dichroism: no significant shift in melting temperature was observed between the two samples (SI Figure 3); in presence of 39, however, a more defined melting transition was found.
Figure 5. A nucleobase peptide with nanomolar affinity for RNA-hairpins was identified by affinity selection.
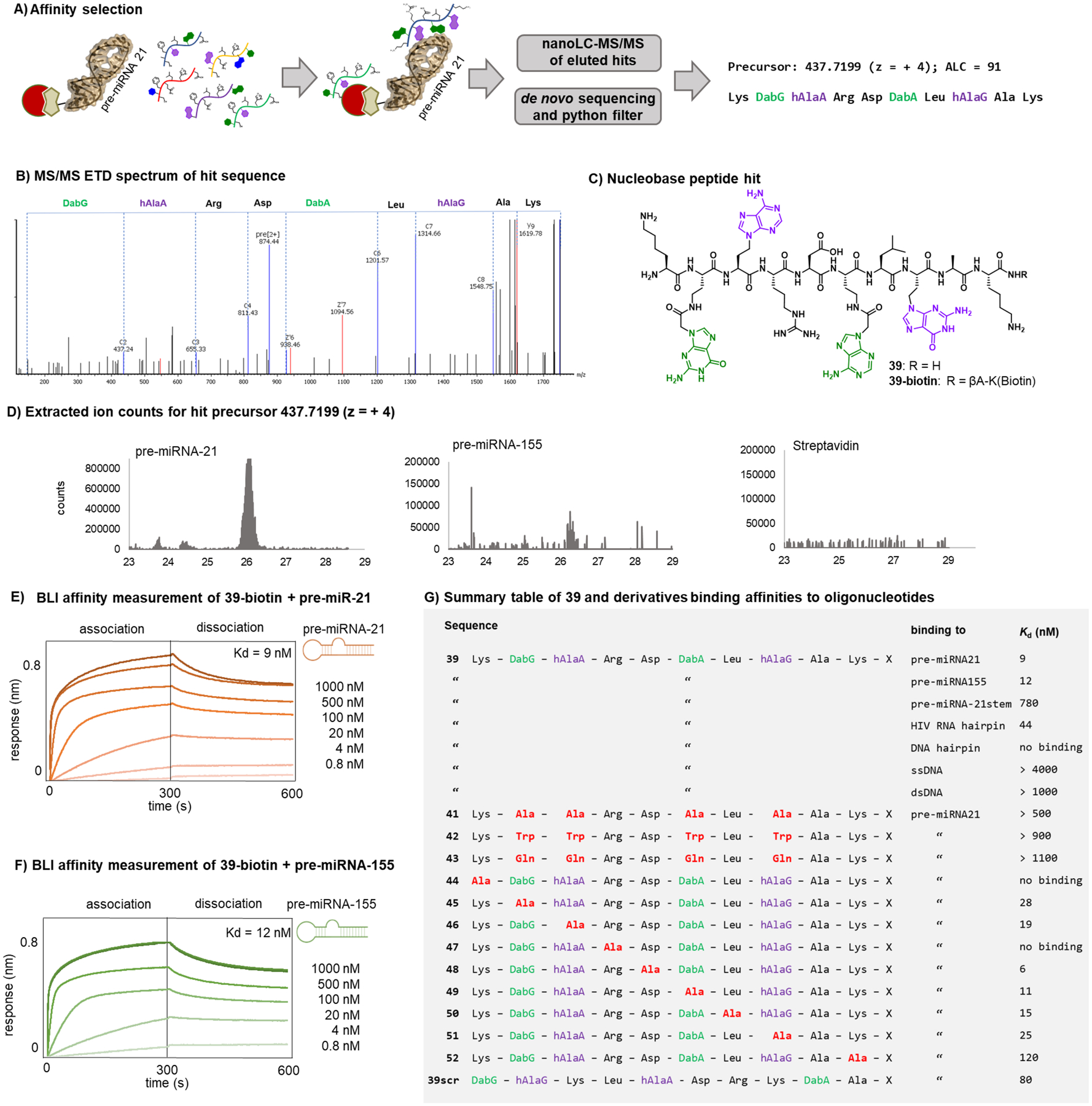
A) Schematic representation of the magnetic bead-based affinity selection with nucleobase peptide library (streptavidin coated magnetic beads = red; biotinylated pre-miRNA = brown). Total immobilized RNA: 100 pmol; [library] = 1 mM; [individual library member] = 10 pM; incubation buffer: Tris 20 mM, pH = 7.5, NaCl 300 mM, MgCl2 20 mM, Tween 0.05 %, Yeast RNA 0.2 mg/mL, Biotin 2 mM; B) ETD spectrum of hit sequence identified by affinity selection. C) structure of hit sequence. D) extracted ion counts for the precursor mass (437.72; z = + 4) in the MS chromatograms of the affinity selection with pre-miRNA-21 (Biotin- UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUCUGACA), pre-miRNA-155 (Biotin-CUGUUAAUGCUAAUCGUGAUAGGGGUUUUUGCCUCCAACUGACUCCUACAUAUUAGCAUUAACAG) and the negative control streptavidin beads only. E) BLI association/dissociation of pre-miRNA-21 to immobilized 39-biotin. F) BLI association/dissociation of pre-miRNA-155 to immobilized 39-biotin. G) Summary table of 39 and derivatives binding to RNA and DNA oligonucleotides. The table lists also the affinities towards pre-miRNA-21 of alanine scan mutants and nucleobase mutants of 39. X = -βAla-Lys(Biotin)-CONH2
The binding of the nucleobase peptide hit to RNA hairpins is selective over DNA hairpins, single stranded DNA (ssDNA) and double stranded DNA (dsDNA). We wanted to determine whether the binding of 39-biotin to pre-miRNA-hairpins was based solely on nonspecific electrostatic interactions due to its 4 positively charged residues. Other negatively charged oligonucleotides, such as a DNA sequence with the same primary sequence as pre-miR-21, ssDNA or dsDNA, in that case, would be expected to bind to the biohybrid compound with similarly high affinity. BLI measurements indicated that 39-biotin has >400-fold selectivity for pre-miRNA-hairpins relative to all DNA samples tested. (SI Figure 1, Figure 5G).
Single-point nucleobase substitutions in 39 to alanine were tolerated, as mutation of all four nucleobases caused up to 100-fold decrease in affinity for RNA. To investigate the role of the nucleobase monomers and individual residues in the hit sequence, we synthesized several analogues. First, the 4 nucleobase monomers were mutated to either alanine, tryptophan, or glutamine. These variants showed reduced binding affinity for pre-miRNA21 (> 50–100-fold decrease; Figure 5G). The need for a combination of side chains might provide clues regarding why one sequence was identified in the affinity selection and no other consensus compounds as observed in other target based AS-MS approaches.29 Next, we performed an alanine scan at each position. Mutation of the N-terminal lysine and the arginine in position 4 depleted the binding. Single mutations all other positions were tolerated and did not significantly affect the binding to pre-miR21. A scrambled variant of 39 showed ~10-fold decreased binding affinity (Figure 5G).
Discussion
Abiotic sequence defined polymers might be new modalities for the development of functional materials or therapeutics.47–50 In this work, we showed the design and synthesis of SPPS-ready nucleobase monomers. The robust synthesis procedures allowed for preparation of multimillion membered combinatorial libraries and this class of materials had enhanced affinity for nucleic acids, compared to canonical peptides. Several classes of sequence defined polymers (beyond canonical peptides and nucleic acids) can be synthesized and sequenced by MS/MS.48 No robust approaches for high throughput discovery have been described so far. Our platform shows the possibility of high throughput discovery of sequence defined polymers with favorable properties for oligonucleotide binding.
We established procedures for the synthesis and sequencing of complex mixtures of biohybrid nucleobase peptides with up to ten residues and four nucleobases per sequence. Several reports on the synthesis of individual nucleobase peptides with varying length and nucleobase content have been published.32–36,51 We designed our libraries with sequence lengths of 9–10 residues, based on previous results in our laboratory which showed the sequencing outcome for peptide libraries with a length of over 12 residues being less accurate compared to shorter sequences.27 Using model libraries, we showed efficient sequencing of biohybrids with up to four nucleobases per compound (Fig 2A). In the quality control samples of the 100-million membered library we identified sequences with up to five nucleobases. Higher nucleobase content might be possible, but not likely frequent in our library, due to the probabilistic residue distribution (Figure 3B).
We identified a single binder upon screening a 100-million membered library in presence of pre-miRNA-21, and none for pre-miR-155. Several reasons might be at the basis of this outcome: AS-MS can achieve high enrichment for peptides sequenced, typically identifying only binders with low nanomolar affinities. Weaker binders might be present but not enriched enough for the mass spectrometry threshold. The excess of competitor yeast RNA, present in the selection buffer, may successfully scavenge nonspecific oligonucleotide binders. Overall, the library size, monomer composition, and enrichment conditions affect the likelihood of discovering novel binders.29,52 Libraries beyond 108 members and iterative designs might enable the identification of additional hits.
Often, hot-spot residues within peptides or proteins drive the binding to their partner (usually ~3 residues in a 10-mer).53,54 For DNA or RNA duplexes, the binding affinity does not depend on a single nucleobase ‘hotspot’, but on synergistic base-pairing along the whole oligonucleotide. Compound 39 exhibited hybrid recognition: the Lys1 and Arg4 behave as hotspot residues and this observation is in line with the general finding that basic residues are important for protein-RNA interactions.54 The nucleobases within 39, on the other hand, tolerate single substitutions without significant effects on the binding affinity. However, after mutating all four nucleobases to canonical residues, the binding decreased by 50-fold. Open questions about the specific contributions of the nucleobases remain at this stage and will be subject of future investigations. Also, scrambled 39 with 4 positive charges and 4 nucleobases maintains binding to the miRNA-hairpin, although with 10-fold reduced affinity. The main driving force for the interaction of nucleobase peptide 39 and scrambled variant might be the overall content of positively charged residues and nucleobases and not the order of the primary sequence.
While peptide nucleic acids (PNAs) can target specific oligonucleotide sequences by direct hybridization or strand invasion, they rarely recognize the defined secondary or tertiary structures formed by RNA, beyond triple helix binding.55,56 Our results suggest a mixed two-in-one molecule might lead to classes of variants that are not only selective toward RNA over DNA, but also for specific structural RNA elements.
Taken together, our results highlight the potential of synthetic combinatorial libraries combined with modern mass spectrometry for the exploration of new chemical space and ligand discovery. We envision that continued access to areas of chemical space beyond natural building blocks will create opportunities for the discovery of materials with novel functions, properties, or even enzymatic activity.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Dr. Andrei Loas for proofreading the manuscript. S.P. thanks the Deutsche Forschungsgemeinschaft for a postdoctoral fellowship (DFG, PO 2413/1–1). We thank the Biophysical Instrumentation Facility at MIT for providing access to the Octet Red96 Bio-Layer Interferometry System (NIH S10OD016326).
Funding Sources
This research was funded by the National Institutes of Health (NIH, grant R01 GM110535 to B.L.P.) and a Bristol-Myers Squibb Unrestricted Grant in Synthetic Organic Chemistry (to B.L.P.).
B.L.P. is a co‐founder of Amide Technologies and Resolute Bio. Both companies focus on the development of protein and peptide therapeutics.
Footnotes
Supporting information containing synthesis and experimental procedures as well as characterization of all compounds and libraries is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E A Programmable Dual-RNA – Guided. 2012, 337 (August), 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sander JD; Joung JK CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol 2014, 32 (4), 347–350. 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Velagapudi SP; Gallo SM; Disney MD Sequence-Based Design of Bioactive Small Molecules That Target Precursor MicroRNAs. Nat. Chem. Biol 2014, 10 (4), 291–297. 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bennett CF Therapeutic Antisense Oligonucleotides Are Coming of Age. Annu. Rev. Med 2019, 70 (1), 307–321. 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- (5).Wen D; Danquah M; Chaudhary AK; Mahato RI Small Molecules Targeting MicroRNA for Cancer Therapy: Promises and Obstacles. J. Control. Release 2015, 219, 237–247. 10.1016/j.jconrel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jackson M; Marks L; May GHW; Wilson JB The Genetic Basis of Disease. Essays Biochem. 2018, 62 (5), 643–723. 10.1042/EBC20170053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lee TI; Young RA Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152 (6), 1237–1251. 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Matsui M; Corey DR Non-Coding RNAs as Drug Targets. Nat. Rev. Drug Discov 2017, 16 (3), 167–179. 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Disney MD Targeting RNA with Small Molecules To Capture Opportunities at the Intersection of Chemistry, Biology, and Medicine. J. Am. Chem. Soc 2019, 141 (17), 6776–6790. 10.1021/jacs.8b13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Velagapudi SP; Cameron MD; Haga CL; Rosenberg LH; Lafitte M; Duckett DR; Phinney DG; Disney MD Design of a Small Molecule against an Oncogenic Noncoding RNA. Proc. Natl. Acad. Sci 2016, 113 (21), 5898–5903. 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Warner KD; Hajdin CE; Weeks KM Principles for Targeting RNA with Drug-like Small Molecules. Nat. Rev. Drug Discov 2018, 17 (8), 547–558. 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Van Meter EN; Onyango JA; Teske KA A Review of Currently Identified Small Molecule Modulators of MicroRNA Function. Eur. J. Med. Chem 2020, 188. 10.1016/j.ejmech.2019.112008. [DOI] [PubMed] [Google Scholar]
- (13).Bose D; Nahar S; Rai MK; Ray A; Chakraborty K; Maiti S Selective Inhibition of MiR-21 by Phage Display Screened Peptide. Nucleic Acids Res. 2015, 43 (8), 4342–4352. 10.1093/nar/gkv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pai J; Hyun S; Hyun JY; Park SH; Kim WJ; Bae SH; Kim NK; Yu J; Shin I Screening of Pre-MiRNA-155 Binding Peptides for Apoptosis Inducing Activity Using Peptide Microarrays. J. Am. Chem. Soc 2016, 138 (3), 857–867. 10.1021/jacs.5b09216. [DOI] [PubMed] [Google Scholar]
- (15).Pai J; Yoon T; Kim ND; Lee IS; Yu J; Shin I High-Throughput Profiling of Peptide-RNA Interactions Using Peptide Microarrays. J. Am. Chem. Soc 2012, 134 (46), 19287–19296. 10.1021/ja309760g. [DOI] [PubMed] [Google Scholar]
- (16).Shortridge MD; Walker MJ; Pavelitz T; Chen Y; Yang W; Varani G A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing. ACS Chem. Biol 2017, 12 (6), 1611–1620. 10.1021/acschembio.7b00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Takahashi T; Hamasaki K; Kumagai I; Ueno A; Mihara H Design of a Nucleobase-Conjugated Peptide That Recognizes HIV-1 RRE IIB RNA with High Affinity and Specificity. Chem. Commun 2000, 1 (5), 349–350. 10.1039/a909579i. [DOI] [Google Scholar]
- (18).Miyanishi H; Takahashi T; Mihara H De Novo Design of Peptides with L-α-Nucleobase Amino Acids and Their Binding Properties to the P22 BoxB RNA and Its Mutants. Bioconjug. Chem 2004, 15 (4), 694–698. 10.1021/bc034210n. [DOI] [PubMed] [Google Scholar]
- (19).Takahashi T; Hamasaki K; Ueno A; Mihara H Construction of Peptides with Nucleobase Amino Acids: Design and Synthesis of the Nucleobase-Conjugated Peptides Derived from HIV-1 Rev and Their Binding Properties to HIV-1 RRE RNA. Bioorganic Med. Chem 2001, 9 (4), 991–1000. 10.1016/S0968-0896(00)00324-2. [DOI] [PubMed] [Google Scholar]
- (20).Zhang C; Chen S; Bai X; Dedkova LM; Hecht SM Alteration of Transcriptional Regulator Rob In Vivo: Enhancement of Promoter DNA Binding and Antibiotic Resistance in the Presence of Nucleobase Amino Acids. Biochemistry 2020, 59 (12), 1217–1220. 10.1021/acs.biochem.0c00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rozenski J; Lescrinier T; Rosemeyer H; Chaltin P; Lescrinier E; Busson R; Van Aerschot A; Hendrix C; Herdewijn P Selection of New Sequence-Selective Unnatural Peptides Binding to Double-Stranded Deoxyribonucleic Acids (DsDNA) by Means of a Gel-Retardation Experiment for Library Analysis. Helv. Chim. Acta 2002, 85, 2258–2283. 10.1002/chin.200249170. [DOI] [Google Scholar]
- (22).Huang Y; Dey S; Zhang X; Sönnichsen F; Garner P The α-Helical Peptide Nucleic Acid Concept: Merger of Peptide Secondary Structure and Codified Nucleic Acid Recognition. J. Am. Chem. Soc 2004, 126 (14), 4626–4640. 10.1021/ja038434s. [DOI] [PubMed] [Google Scholar]
- (23).He Y; Zheng K; Wen C; Li X; Gong J; Hao Y; Zhao Y; Tan Z Selective Targeting of Guanine-Vacancy-Bearing G-Quadruplexes by G-Quartet Complementation and Stabilization with a Guanine–Peptide Conjugate. J. Am. Chem. Soc 2020. 10.1021/jacs.0c00774. [DOI] [PubMed] [Google Scholar]
- (24).Swenson CS; Velusamy A; Argueta-Gonzalez HS; Heemstra JM Bilingual Peptide Nucleic Acids: Encoding the Languages of Nucleic Acids and Proteins in a Single Self-Assembling Biopolymer. J. Am. Chem. Soc 2019, 141 (48), 19038–19047. 10.1021/jacs.9b09146. [DOI] [PubMed] [Google Scholar]
- (25).Chen Z; Lichtor PA; Berliner AP; Chen JC; Liu DR Evolution of Sequence-Defined Highly Functionalized Nucleic Acid Polymers. Nat. Chem 2018, 10 (4), 420–427. 10.1038/s41557-018-0008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lichtor PA; Chen Z; Elowe NH; Chen JC; Liu DR Side Chain Determinants of Biopolymer Function during Selection and Replication. Nat. Chem. Biol 2019, 1. 10.1038/s41589-019-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gates ZP; Vinogradov AA; Quartararo AJ; Bandyopadhyay A; Choo ZN; Evans ED; Halloran KH; Mijalis AJ; Mong SK; Simon MD; Standley EA; Styduhar ED; Tasker SZ; Touti F; Weber JM; Wilson JL; Jamison TF; Pentelute BL Xenoprotein Engineering via Synthetic Libraries. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (23), E5298–E5306. 10.1073/pnas.1722633115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Touti F; Gates ZP; Bandyopdhyay A; Lautrette G In-Solution Enrichment Identifies Peptide Inhibitors of Protein – Protein Interactions Protein-Protein Interactions. Nat. Chem. Biol 2019, 318–339. 10.1038/s41589-019-0245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Quartararo AJ; Gates ZP; Somsen BA; Hartrampf N; Ye X; Shimada A; Kajihara Y; Ottman C; Pentelute BL Ultra-Large Chemical Libraries for the Discovery of High-Affinity Peptide Binders. Nat. Commun 2020, 11 (3183). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Galán A; Comor L; Horvatić A; Kuleš J; Guillemin N; Mrljak V; Bhide M Library-Based Display Technologies: Where Do We Stand? Mol. Biosyst 2016, 12 (8), 2342–2358. 10.1039/c6mb00219f. [DOI] [PubMed] [Google Scholar]
- (31).Passioura T; Suga H A RaPID Way to Discover Nonstandard Macrocyclic Peptide Modulators of Drug Targets. Chem. Commun 2017, 53 (12), 1931–1940. 10.1039/c6cc06951g. [DOI] [PubMed] [Google Scholar]
- (32).Diederichsen U; Weicherding D; Diezemann N Side Chain Homologation of Alanyl Peptide Nucleic Acids: Pairing Selectivity and Stacking. Org. Biomol. Chem 2005, 3 (6), 1058–1066. 10.1039/b411545g. [DOI] [PubMed] [Google Scholar]
- (33).Roviello GN; Musumeci D Synthetic Approaches to Nucleopeptides Containing All Four Nucleobases, and Nucleic Acid-Binding Studies on a Mixed-Sequence Nucleo-Oligolysine. RSC Adv. 2016, 6 (68), 63578–63585. 10.1039/c6ra08765e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Roviello GN; Musumeci D; D’Alessandro C; Pedone C Synthesis of a Thymine-Functionalized Nucleoamino Acid for the Solid Phase Assembly of Cationic Nucleopeptides. Amino Acids 2013, 45 (4), 779–784. 10.1007/s00726-013-1520-2. [DOI] [PubMed] [Google Scholar]
- (35).Roviello GN; Benedetti E; Pedone C; Bucci EM Nucleobase-Containing Peptides: An Overview of Their Characteristic Features and Applications. Amino Acids 2010, 39 (1), 45–57. 10.1007/s00726-010-0567-6. [DOI] [PubMed] [Google Scholar]
- (36).Diederichsen U Pairing Properties of Alanyl Peptide Nucleic Acids Containing an Amino Acid Backbone with Alternating Configuration. Angew. Chemie (International Ed. English) 1996, 35 (4), 445–448. 10.1002/anie.199604451. [DOI] [Google Scholar]
- (37).Talukder P; Dedkova LM; Ellington AD; Yakovchuk P; Lim J; Anslyn EV; Hecht SM Synthesis of Alanyl Nucleobase Amino Acids and Their Incorporation into Proteins. Bioorganic Med. Chem 2016, 24 (18), 4177–4187. 10.1016/j.bmc.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Geotti-Bianchini P; Beyrath J; Chaloin O; Formaggio F; Bianco A Design and Synthesis of Intrinsically Cell-Penetrating Nucleopeptides. Org. Biomol. Chem 2008, 6 (20), 3661–3663. 10.1039/b811639c. [DOI] [PubMed] [Google Scholar]
- (39).Vinogradov A; Gates ZP; Zhang C; Quartararo AJ; Halloran KH; Pentelute BL Library Design-Facilitated High-Throughput Sequencing of Synthetic Peptide Libraries. ACS Comb. Sci 2017, 19 (11), 694–701. 10.1021/acscombsci.7b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lam KS; Salmon SE; Hersh EM; Hruby VJ; Kazmierski WM; Knapp RJ A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity [Published Errata Appear in Nature 1992 Jul 30;358(6385):434 and 1992 Dec 24–31;360(6406):768]. Nature 1991, 354 (6348), 82–84. [DOI] [PubMed] [Google Scholar]
- (41).FURKA Á; SEBESTYÉN F; ASGEDOM M; DIBÓ G General Method for Rapid Synthesis of Multicomponent Peptide Mixtures. Int. J. Pept. Protein Res 1991, 37 (6), 487–493. 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- (42).Bartel DP MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136 (2), 215–233. 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Calin GA; Croce CM MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6 (11), 857–866. 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- (44).Shah MY; Calin GA MicroRNAs as Therapeutic Targets in Human Cancers. Wiley Interdiscip. Rev. RNA 2014, 5 (4), 537–548. 10.1002/wrna.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Feng Y-H; Tsao C-J Emerging Role of MicroRNA-21 in Cancer. Biomed. Reports 2016, 5 (4), 395–402. 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Esquela-Kerscher A; Slack FJ Oncomirs - MicroRNAs with a Role in Cancer. Nat. Rev. Cancer 2006, 6 (4), 259–269. 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- (47).Porel M; Thornlow DN; Phan NN; Alabi CA Sequence-Defined Bioactive Macrocycles via an Acid-Catalysed Cascade Reaction. Nat. Chem 2016, 8 (6), 590–596. 10.1038/nchem.2508. [DOI] [PubMed] [Google Scholar]
- (48).Lutz JF Defining the Field of Sequence-Controlled Polymers. Macromol. Rapid Commun 2017, 38 (24), 1–12. 10.1002/marc.201700582. [DOI] [PubMed] [Google Scholar]
- (49).Dong R; Liu R; Gaffney PRJ; Schaepertoens M; Marchetti P; Williams CM; Chen R; Livingston AG Sequence-Defined Multifunctional Polyethers via Liquid-Phase Synthesis with Molecular Sieving. Nat. Chem 2019, 11 (2), 136–145. 10.1038/s41557-018-0169-6. [DOI] [PubMed] [Google Scholar]
- (50).Dahlhauser SD; Escamilla PR; Vandewalle AN; York JT; Rapagnani RM; Shei JS; Glass SA; Coronado JN; Moor SR; Saunders DP; Anslyn EV Sequencing of Sequence-Defined Oligourethanes via Controlled Self-Immolation. J. Am. Chem. Soc 2020, 142 (6), 2744–2749. 10.1021/jacs.9b12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Diederichsen U; Weicherding D Interactions of Amino Acid Side Chains with Nucleobases in Alanyl-PNA. Synlett 2017, 1999 (Sup. 1), 917–920. 10.1055/s-1999-3117. [DOI] [Google Scholar]
- (52).Liu R; Li X; Lam KS Combinatorial Chemistry in Drug Discovery. Curr. Opin. Chem. Biol 2017, 38, 117–126. 10.1016/j.cbpa.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).London N; Movshovitz-Attias D; Schueler-Furman O The Structural Basis of Peptide-Protein Binding Strategies. Structure 2010, 18 (2), 188–199. 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- (54).Krüger DM; Neubacher S; Grossmann TN Protein–RNA Interactions: Structural Characteristics and Hotspot Amino Acids. Rna 2018, 24 (11), 1457–1465. 10.1261/rna.066464.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Nielsen PE; Egholm M; Berg RH; Buchardt O Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science (80-.) 1991, 254 (5037), 1497–1500. 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- (56).Kesy J; Patil KM; Kumar SR; Shu Z; Yong HY; Zimmermann L; Ong AAL; Toh DFK; Krishna MS; Yang L; Decout JL; Luo D; Prabakaran M; Chen G; Kierzek E A Short Chemically Modified DsRNA-Binding PNA (DbPNA) Inhibits Influenza Viral Replication by Targeting Viral RNA Panhandle Structure. Bioconjug. Chem 2019, 30 (3), 931–943. 10.1021/acs.bioconjchem.9b00039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


