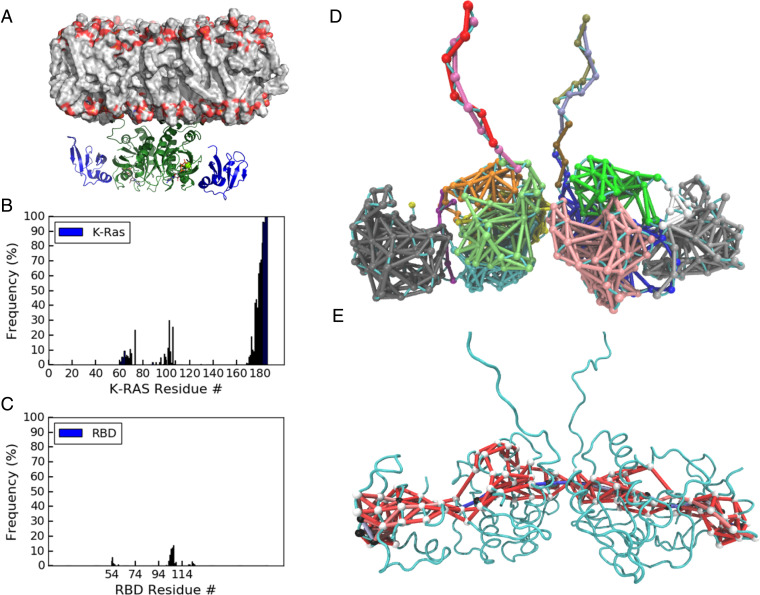
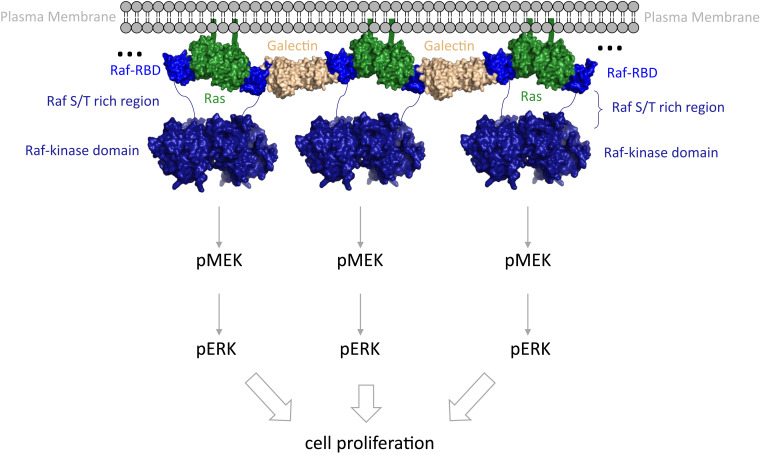
Significance
Ras GTPases, KRas, HRas, and NRas control cell proliferation via the Ras/Raf/MEK/ERK pathway through a process in which Ras dimerization is required but not mechanistically understood. Here, we show that Raf-RBD promotes Ras dimerization resulting in strong allosteric linkages between Galectin-binding residues on Raf-RBD at the two extreme ends of the dimer. This suggests that the dimer of the Ras/Raf–RBD complex couples to Galectin dimers to form a multivalent signaling complex for synchronized activation of Raf kinase, providing a new framework to be tested in the mechanistic understanding of Ras/Raf signaling.
Keywords: Ras, Raf, dimerization, allosteric connections
Abstract
Ras dimerization is critical for Raf activation. Here we show that the Ras binding domain of Raf (Raf-RBD) induces robust Ras dimerization at low surface densities on supported lipid bilayers and, to a lesser extent, in solution as observed by size exclusion chromatography and confirmed by SAXS. Community network analysis based on molecular dynamics simulations shows robust allosteric connections linking the two Raf-RBD D113 residues located in the Galectin scaffold protein binding site of each Raf-RBD molecule and 85 Å apart on opposite ends of the dimer complex. Our results suggest that Raf-RBD binding and Ras dimerization are concerted events that lead to a high-affinity signaling complex at the membrane that we propose is an essential unit in the macromolecular assembly of higher order Ras/Raf/Galectin complexes important for signaling through the Ras/Raf/MEK/ERK pathway.
Ras GTPases are at the hub of signal transduction cascades that control cell processes such as proliferation, migration, and survival, and their mutants appear in about a quarter of all human cancers with poor treatment prognosis (1). There are four main isoforms, each with specifically lipidated and highly divergent hypervariable regions (HVRs) that drive membrane localization and distinct biological outcomes (2). The Ras G-domain is similar between the isoforms, KRas (4A and 4B), HRas, and NRas with sequence identical effector lobes (residues 1 to 86) and small differences in the allosteric lobe (residues 87 to 166) (3). All four isoforms are generally observed as monomers in solution, although exceptions have been reported (4). The G-domain of HRas in particular has been shown by fluorescence anisotropy and nuclear magnetic resonance (NMR) to be strictly monomeric in solution (5). Several fluorescence-based studies reported dimerization on supported lipid bilayers (SLBs) (6, 7), but a subsequent study identified photo-oxidative crosslinking (including via dityrosine formation) to produce nonphysiological covalent Ras dimers when fluorescence is used in the absence of radical oxygen scavengers (8). A recent study that controlled for such artifacts determined the two-dimensional dissociation constant for dimerization for full-length K-Ras4B to be higher than the detection limit of 25,000 molecules/µm2 (loosely corresponding to a three-dimensional Kd minimum of 15 mM) (9). This work established that the higher-order organizations of Ras reported in live-cell contexts likely involve other molecules besides Ras. Indication that Ras functions through dimerization to activate Raf first appeared in the early days of Ras research (7), and Ras dimers have since been shown to be essential for signaling through the Ras/Raf/MEK/ERK pathway in cells and in mice (10, 11). Computational studies of G-domain dimerization have identified three low-affinity dimerization interfaces in the absence of the Raf Ras-Binding Domain (Raf-RBD): an extended β-sheet formed at the effector lobe coinciding with the effector Raf binding interface (4, 12) and two overlapping interfaces involving α3-α4 or α4-α5 in the allosteric lobe (6, 13, 14). Ras clustering on the membrane through these multiple interfaces may result in nanoclusters important for signaling, bringing together effector proteins and other components of the signaling machinery (15–17). While nanoclusters may form through multiple Ras interfaces (15), signaling through Ras/Raf occurs specifically through Ras dimers involving the α4-α5 interface (10, 18) which appears in several crystal forms of Ras and was first modeled to study the NRas dimer using molecular dynamics (MD) simulations (6). The dimer of the Ras/Raf complex by itself is most likely not the full signaling unit, as scaffold proteins such as Galectins interact with Raf-RBD and are required for activation of the Ras/Raf/MEK/ERK pathway (19, 20).
The monomeric nature of the Ras G-domain in solution has led to skepticism about its role in dimerization, obscured by the fact that interaction of the C-terminal HVR region with the lipid membrane is important for dimerization in cells (11, 21). However, membrane insertion is not sufficient for Ras dimerization as shown by experiments on supported membranes under conditions where reactive oxygen radicals are quenched (9). Here we use a combination of size exclusion chromatography (SEC) and small-angle X-ray scattering (SAXS) experiments to show that the presence of Raf-RBD is sufficient to promote Ras G-domain dimerization in solution at levels detected by SEC in the absence of the HVR and membrane. Single-molecule tracking (SMT) experiments and fluorescence correlation spectroscopy demonstrate that robust dimerization of Ras on supported membranes requires Raf-RBD. Starting from a crystallographic model of the HRas/CRaf-RBD dimer (2 Ras/Raf-RBD interacting through the Ras α4-α5 interface), we use MD simulations to show robust allosteric connections linking the Raf-RBD residues D113, located in the Raf-RBD binding site for Galectin (19) and at the two opposing ends of the dimer of the Ras/Raf-RBD complex. Two independent 1-μs simulations starting from a model of the KRas/CRaf-RBD dimer with farnesylated KRas at the membrane show a stable dimer with allosteric linkages similar to those we obtain for the HRas/CRaf-RBD dimer. Our results, combined with those from the literature, suggest a model in which the dimer of the Ras/Raf-RBD complex is a key feature of a signaling platform that also includes the Galectin dimer.
Results
Raf Promotes Ras G-Domain Dimerization in Solution and on Supported Lipid Membranes.
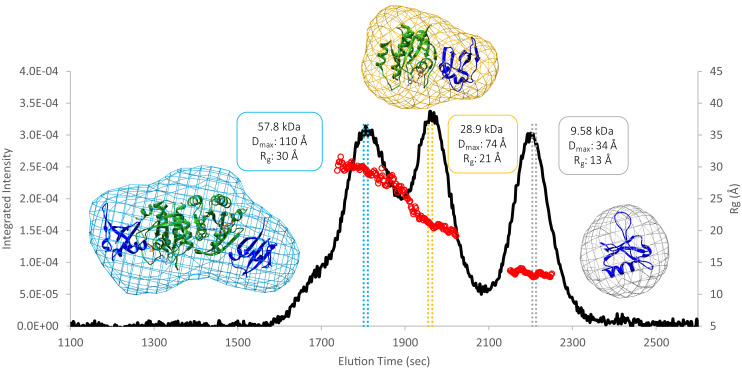
While our Ras purification protocol yields a single peak in the final gel filtration step, addition of Raf-RBD to form the complex is accompanied by a higher molecular weight peak. Recent discussion of Ras dimers in the literature prompted us to investigate the contents of this higher molecular weight species that appears in the presence of Raf-RBD, as we found that the peak increases at high protein concentrations. The SEC profile for a protein solution containing the truncated G-domain of wild-type KRas4B (referred to as KRas) bound to GppNHp in the presence of excess Raf-RBD shows three peaks that can be identified by Small Angle X-Ray Scattering (SEC-SAXS) data (22) as the dimer of the Ras/Raf-RBD complex, the monomer of the Ras/Raf-RBD complex and excess Raf-RBD (Fig. 1). These results are in agreement with the molecular weights for the species of each of the three elution volumes determined by SEC based on a standard curve (Fig. 1 and SI Appendix, Fig. S1). The SEC-SAXS results for HRas are very similar to those that we present here for KRas. In general, the relative intensities of the monomer and dimer peaks vary from one experiment to the next, depending on the Ras isoform, construct used (truncation at residue 166 or 173), protein concentration, the specific solution conditions, and experimental protocol. Raf-RBD is of paramount importance in these experiments, as Ras in the absence of Raf-RBD under similar experimental conditions does not form dimers (SI Appendix, Fig. S1). Due to the presence of solvent-exposed cysteine residues in the Ras/Raf-RBD complex (Raf-RBD C95 and C96 in particular), samples obtained from one of our SEC runs with the wild-type constructs in presence of 1 mM dithiothreitol (DTT) were submitted for mass spectrometry analysis. The results show that the observed Ras/Raf-RBD dimer peak does not contain disulfide bond crosslinks (SI Appendix, Fig. S2). In further support of this, a SEC run with the Raf-RBD C95S-C96S double mutant or with the wild-type Raf-RBD in the presence of 100 mM DTT shows that the dimer peak is still present (SI Appendix, Fig. S3). The molecular envelopes generated from the SAXS data unequivocally support a dimeric Ras/Raf-RBD structure for the contents of the first major peak eluted from the SEC column (Fig. 1 and SI Appendix, Fig. S4), with excellent χ2 fits to either the crystallographic dimer (Protein Data Bank [PDB] ID 4G0N) (23) or NMR dimer (PDB ID 6W4E) (18) (with Raf-RBD added), both containing the α4-α5 interface (SI Appendix, Fig. S5). Note that in Fig. 1, the Ras/Raf-RBD dimer and monomer peaks overlap and the Rg is sloping rather than flat in the peak regions, suggesting that the experimental profiles comprise scattering from mixed species. However, the homogeneity of the frames selected to calculate the molecular envelopes shown in Fig. 1 is demonstrated by an inability to optimize the χ2 values with a two-model system containing both the monomer and dimer of the Ras/Raf-RBD complexes. In contrast, near the midpoint between the monomer and dimer peaks, a χ2 of nearly 1 was obtained for a two-state model consisting of 63% dimer and 37% monomer of the Ras/Raf-RBD complex.
Fig. 1.
SEC-SAXS data collected for the three unique protein species that form in a solution containing the KRas-GppNHp G-domain and Raf-RBD. The black curve is the trace of the integrated X-ray scattering intensity (left axis), which correlates with protein concentration. The red data points correspond to the calculated radius of gyration (Rg, right axis) over elution time and indicate that the contents of each peak are monodisperse. The colored vertical bars through each peak indicate the data frames used to construct the corresponding molecular envelopes. Protein crystal structures were fit to the dimer (PDB ID 4G0N with dimer generated through a twofold crystallographic symmetry axis), monomer (PDB ID 4G0N asymmetric unity), and Raf-RBD (PDB ID 1RRB) envelopes using the volume fit function in Chimera.
Here we model the dimer based on our crystal structure of the HRas/CRaf-RBD complex generated by applying twofold crystallographic symmetry to the asymmetric unit (PDB ID 4G0N) (23). The resulting α4-α5 interface is stabilized by hydrogen bonding and salt bridge interactions across the two Ras molecules (Fig. 2) and is nearly identical to that identified previously for NRas (6). This is consistent with the α4-α5 dimer interface having been observed experimentally in crystals (6, 23, 24) and established as the active dimer that promotes signaling through the Ras/Raf/MEK/ERK pathway (10). The α4-α5 interface is also consistent with signal inhibition by a monobody shown to bind at this interface (24). In the recently published NMR data-driven model of KRas dimers on nanodiscs, the dimer affinity is low, and the interface is highly flexible in the absence of Raf-RBD (18). Although the average angle between helices across the dimer interface in the NMR model differs from that in the crystal structures, the key interacting residues are mostly the same.
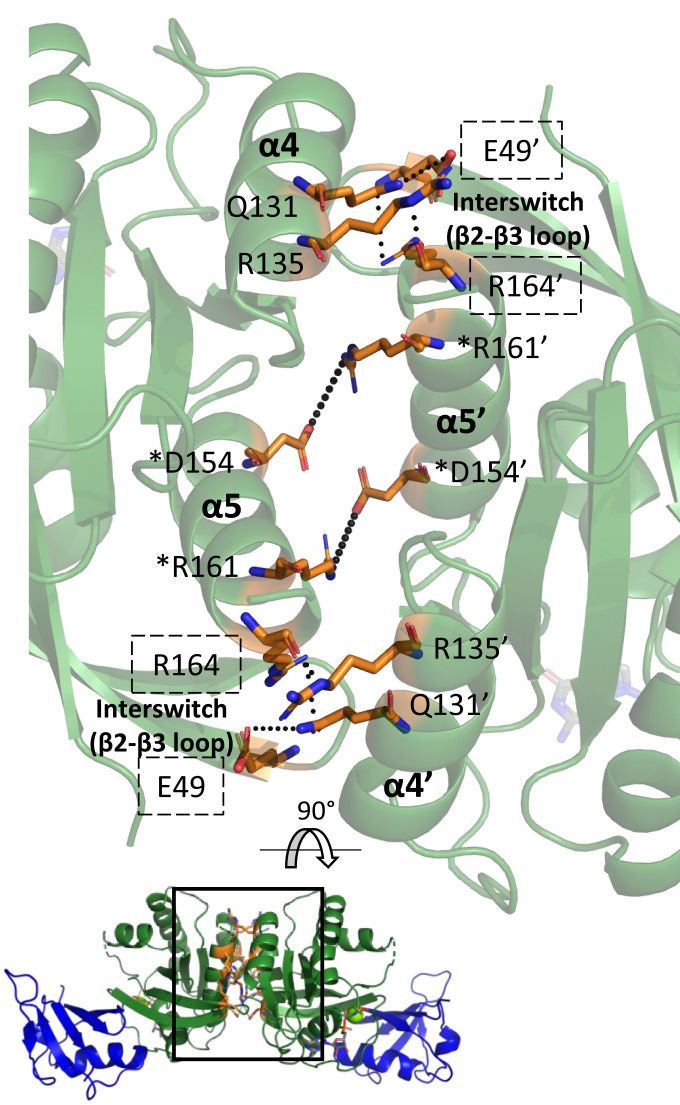
Fig. 2.
Interactions at the α4–α5 interface revealed by crystal structures and NMR models. Interactions are shown for the interface found in our crystal structure of the Ras/Raf-RBD complex (PDB ID 4G0N with dimer generated through a twofold symmetry axis). Ras is in green, and Raf-RBD is in blue. Residues at the dimer interface are in orange. The boxed interface area is rotated and magnified for clarity. Ras forms a symmetric dimer with corresponding residues in one of the monomers denoted by a prime (‘). Residues present within the crystallographic interface but not the interface observed by NMR have their names boxed by dashed lines. Residues denoted by a star (*) have been validated in vivo. Only the key residue interactions are depicted for clarity.
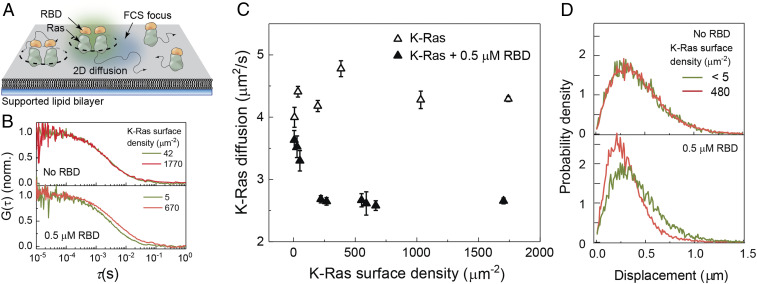
On SLBs, the addition of Raf-RBD to GppNHp-bound Ras leads to a robust formation of complexes containing two Ras molecules under conditions similar to those in which Ras alone does not dimerize (9) (Fig. 3 A and B). We detect this complex formation by the protein surface density-dependent decrease in diffusion, which can be used as an indicator for dimerization on a homogeneous membrane environment such as the one provided by SLBs (8, 9, 25). Fig. 3 shows full-length farnesylated and carboxy-methylated KRas (KRas-FMe) (26) density-dependent changes in KRas diffusion with or without Raf-RBD measured by fluorescence correlation spectroscopy (FCS) and SMT. In the FCS measurements, the diffusion coefficient of KRas is unchanged across all observed surface densities but is reduced from 4.5 to 2.3 µm2/s in the presence of Raf-RBD (Fig. 3C). A similar change can be seen in the SMT step size distributions (Fig. 3D). The change in the diffusion coefficient is similar to that due to dimerization by a crosslinker, suggesting that this complex contains two Ras molecules (9). The two-dimensional dissociation constant (Kd) of Ras dimerization is ∼62 molecules/μm2 by fitting the binding curve obtained by diffusion measurement shown in Fig. 3C (9). This two-dimensional Kd may be translated into a three-dimensional dissociation constant of 37 µM using the percent occupancy formalism (9, 25). This corresponds to a high-affinity interaction likely due to allosteric effects leading to concerted binding of Raf-RBD and dimerization in the presence of the membrane. HRas shows a similar diffusion behavior (SI Appendix, Fig. S6A). The type and location of the fluorescent tag on Raf-RBD, necessary for the SLB experiments (27), was found to be capable of disrupting the complex formation (SI Appendix, Fig. S6B). While the addition of the SNAP-tag (19.4 kDa) at the N terminus removed the density-dependent decrease in diffusion, mCherry (28.8 kDa) on the C terminus had no effect on complex formation. This is consistent with our model of the dimer, in which the N terminus of Raf-RBD is located close to the Ras/Raf-RBD interface and the C terminus is in a position remote from the interfaces where an added tag would not be expected to interfere with either Ras binding or dimerization. The enhanced green fluorescent protein (eGFP) fluorescent tag used to detect Ras in the absence of Raf-RBD does not affect dimerization (9). Monomeric behavior is also observed for KRas4B labeled with Atto488-GppNHp and tracked with Alexa647-GppNHp in oxygen scavenger buffer (SI Appendix, Fig. S7). Overall, our experiments show robust dimerization of the Ras/Raf-RBD complex in supported membranes for both KRas and HRas.
Fig. 3.
Ras diffusion measurements on SLBs. (A) The experimental setup on SLB. Farnesylated and methylated full-length KRas is allowed to spontaneously insert to the SLB, and Raf-RBD is introduced. In FCS, the surface density and the diffusion coefficient of fluorescent species diffusing together are measured. (B) FCS autocorrelation functions for KRas without (Top) and with (Bottom) 0.5 µM Raf-RBD for multiple KRas membrane surface densities. Only in the presence of RBD is there a Ras density-dependent change. (C) The apparent diffusion coefficients for KRas measured by FCS; the surface density-dependent decrease in diffusion indicates RBD-dependent oligomerization of Ras. (D) Single-molecule step size distributions without (Top) and with (Bottom) 0.5 µM Raf-RBD for KRas at low and high surface densities. SLBs were composed of 20% DOPS and 80% DOPC.
Raf Binding and Dimerization of the Ras G-Domain Increase Allosteric Connections.
Given our previous work showing that the dynamics of the Ras G-domain are affected by Raf-RBD through allosteric effects (23), we performed 90-ns MD simulations on HRas, on the HRas/CRaf-RBD complex, and on the dimer of the HRas/CRaf-RBD complex to determine possible differences in allosteric communication between the Raf-RBD binding site and the dimer interface on Ras. HRas was used so that the simulations could be started directly from our crystal structures of HRas alone (PDB ID 3K8Y) and in complex with Raf-RBD (PDB ID 4G0N monomer and dimer). Dynamic network analysis (28) of the protein trajectories using the CARMA software package (29) identifies correlated motion between protein residues. Visualizing these correlations reveals edges connecting the alpha carbon of residues, represented by spherical nodes, whose atoms are within 4.5 Å of one another throughout at least 75% of the simulation time, highlighting areas of correlated movements within a protein system (28). These networks can be subdivided into communities of nodes that interact more with one another than with nodes of other communities, helping to identify allosteric networks. The communities calculated from the three simulations are shown in Fig. 4. The simulation of Ras by itself shows six distinct communities, indicating six regions of the protein with relatively independent dynamics (Fig. 4 A, Left). Note that neither of the helices involved in Ras dimerization, helices 4 (yellow) and 5 (blue), are connected to each other or to the effector lobe of Ras, and the Ras active site is divided into three communities. In addition to determining communities of connected residues, it is possible to trace the edges connecting any two residues using optimal and suboptimal path calculations in which the optimal path shows the smallest number of nodes, or shortest path, between “source” and “sink” residues connected through long-distance interactions, and the suboptimal paths show all other connections differing by no more than 20 edges from the optimal path (28). Taking our simulation of Ras-GTP by itself, we checked our previously identified allosteric network connecting the allosteric site residue R97 to the active site residue Q61 on switch II (30) and found a total of 10 optimal and suboptimal paths connecting the two residues (Fig. 4 A, Right). Raf-RBD by itself also has six communities and, as with Ras, this indicates various regions with relatively independent dynamics (Fig. 4B).
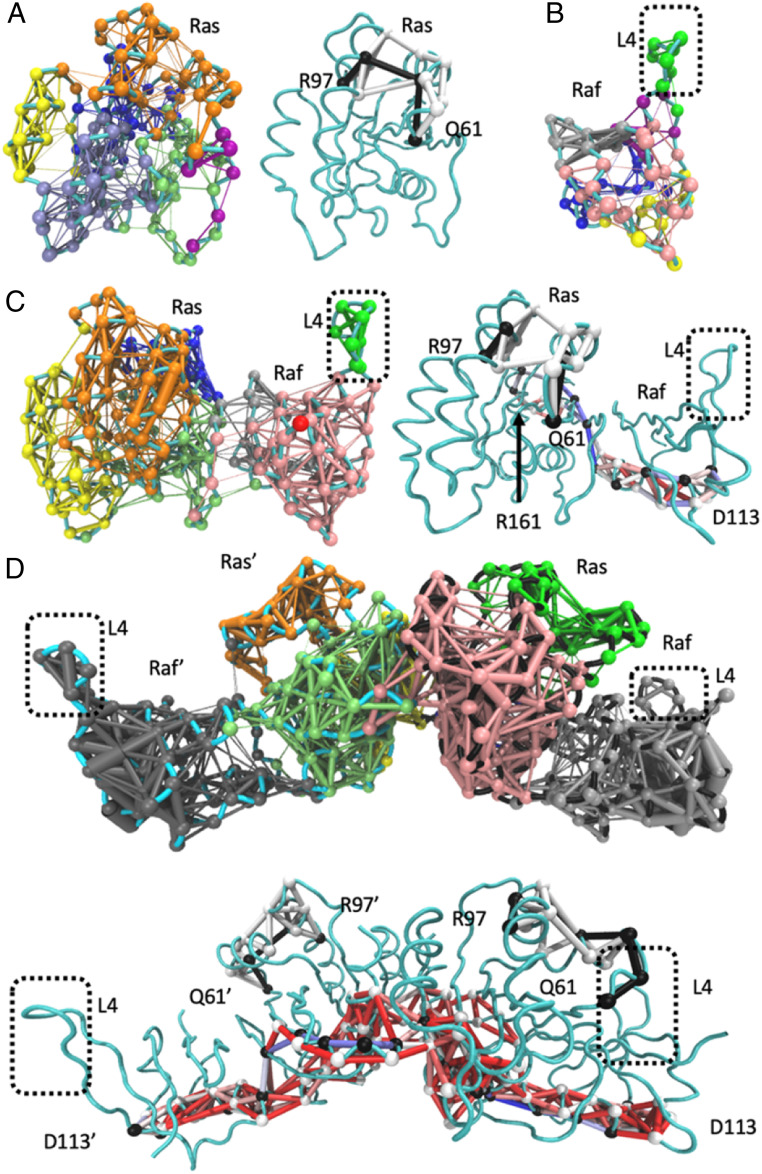
Fig. 4.
Community network and path analyses performed by Carma and visualized using NetworkView in VMD. (A) Ras alone, (B) Raf-RBD alone, (C) the monomer of the Ras/Raf complex, and (D) the dimer of the Ras/Raf complex. Left panels in A and C show community network analyses with each community in a distinct color, and right panels show optimal (black nodes and edges) and suboptimal (white nodes and edges) path analyses between R97 and Q61. Paths between R161 and D113 in the Ras/Raf-RBD complex or D113 and D113’ in the dimer (black nodes, optimal; white nodes, suboptimal) are shown with all edges colored by the number of paths that cross them from the highest colored red to the lowest colored blue. In D, the analogous panels are on top and bottom. Spheres are nodes centered on the alpha carbons of amino acid residues, and sticks are edges calculated by Carma.
In the monomer of the Ras/Raf-RBD complex, Ras coalesces into four communities with residues within the Ras active site establishing connections to helix 5 (green), Raf-RBD (pink), and the allosteric site (orange) (Fig. 4 C, Left). Helix 4 strengthens its connections across the allosteric lobe toward the Ras active site (yellow). Upon complex formation, Raf-RBD also experiences an increase in allosteric connections with three major communities (Fig. 4C). For calculation of the optimal and suboptimal paths across the monomeric complex, we chose R161 in helix 5 at one end and D113 on Raf-RBD at the opposite end of the complex. Ras R161 is in a critical position in helix 5 at the dimer interface (Fig. 2). Raf-RBD D113 is situated near loop 4 and has been previously identified for its long-distance contribution to the Ras and Raf-RBD interaction (31, 32). D113 is also in the binding site between Raf-RBD and the scaffold protein Galectin (19). In the Ras/Raf-RBD monomer, communication develops between Ras helix 5 across the complex interface to residues near Raf-RBD loop 4, which is in its own dynamic community as it is in Raf-RBD alone (Fig. 4C) (23). There are over 50 paths linking Ras R161 to Raf-RBD D113 in the monomer of the complex, while the number of paths linking allosteric site R97 and active site Q61 on Ras remains small.
Simulations of the dimer of the Ras/Raf-RBD complex show a robust increase in connectivity, with each Ras molecule having only three communities and Raf-RBD linked in a single allosteric network from the interface with Ras all the way to loop 4, near D113 on the opposite side of the molecule (Fig. 4 D, Top). The increase in allosteric connections upon dimerization of the complex links the entire β-sheet core of Ras to both of the dimerization helices (pink and light green) and unifies helix 5 into a single community with the entire Ras active site (light green). Despite the increase in allosteric connection from one end of the dimer to the other, the portion of Ras that undergoes a conformational shift upon ligand binding at the allosteric site remains in an isolated community in each protomer (orange and bright green). The two Ras/Raf-RBD complexes in the dimer are approximately symmetric, although some asymmetry is observed, perhaps due to stochastic motions over the course of the simulations. In the highly connected dimer complex, about 20 paths link the allosteric site and active site residues, and over 300 paths exist between helix 5 residue 161 at the dimer interface and the opposite end of the complex at Raf residue D113 (Fig. 4 D, Top). Residue D154, another key residue at the dimer interface, also has over 300 paths to the Raf residue D113. However, when the D113 residues in the two Raf-RBD molecules, at a distance of 85 Å apart on opposite ends of the Ras/Raf-RBD dimer, are used as “source” and “sink” nodes for allosteric connectivity, over 25,000 paths are identified, reflecting a strong allosteric linkage between the two ends of the dimer through the dimer interface with R161 included in about 40% of the paths. The most traversed inter-Ras edge among the possible D113-D113’ paths involves residue E143 of one Ras molecule and residue D47 of the other, consistent with residue D47 of each Ras molecule belonging to the same community as helix 4 in the opposite Ras protomer (Fig. 4D, pink and green). Interestingly, E143 is part of the active site ExSAK motif in which it forms a salt bridge with loop 8 residue R123 immediately following the nucleotide-binding NKxD motif (3). Thus, the Ras/Ras interface is strongly connected to the Ras/Raf-RBD interface and the active site at switch I in the dimer of the complex, providing a venue through which Raf-RBD binding may modulate Ras helices 4 and 5 to form a high-affinity dimer. From Ras loop 8, the allosteric paths go through Ras helix 1 near the active site across the Ras/Raf-RBD interface to Raf-RBD helix 1 toward D113 near loop 4, spanning the entire base of the complex (Fig. 4 D, Bottom). Overall, it is clear that allosteric connectivity and information transfer are significanly strengthened in the dimer of the Ras/Raf-RBD complex as supported both by community network analysis and allosteric paths between connected nodes that span the entire complex (Fig. 4D). Interestingly, the connection between the active and allosteric sites remains relatively weak and outside of the strong allosteric network that links the two Raf-RBD ends in the dimer of the Ras/Raf-RBD complex.
MD Simulations of the KRas/CRaf-RBD Dimer on the Membrane.
Because KRas is a major Ras isoform of interest in Ras mutant cancers, we modeled the KRas/CRaf-RBD dimer complex based on our HRas/CRaf-RBD dimer (PDB ID 4G0N) and performed two independent 1-μs MD simulations of the model dimer of the KRas/CRaf-RBD complex on an 80%:20% POPC:POPS membrane. The average structure of the complex from the simulations is shown in Fig. 5A. The part of the dimer containing the G-domains bound to Raf-RBD has an excellent fit to our KRas/Raf-RBD SAXS data obtained from the dimer SEC peak, with a χ2 of 1.04 (SI Appendix, Fig. S8A). The fit for the average model from our 90-ns simulations of the HRas/Raf-RBD dimer to the KRas/Raf-RBD SAXS data has a χ2 of 1.21 (SI Appendix, Fig. S8B). Overall, the dimer of the KRas/CRaf-RBD complex is well maintained on the model membrane, consistent with the experimental finding on SLBs (Fig. 3). The rmsd of the KRas dimer from the starting structure shows variations less than 6 Å (SI Appendix, Fig. S9A). The fluctuations of the KRas/CRaf-RBD dimer relative to the model membrane are limited as reflected by its orientation angle (SI Appendix, Fig. S9B) with the Ras helices 3, 4, and 5 remaining roughly perpendicular to the membrane as was previously observed for the NRas dimer simulation on the membrane in the absence of Raf (6). The dimer of the KRas/CRaf-RBD complex lifts away from the membrane in comparison to previous simulations of a model of the KRas/CRaf-RBD–CRD monomer (33). While there are extensive membrane contacts for the HVR, the KRas G-domain as well as Raf-RBD are mostly away from the membrane throughout the simulations (SI Appendix, Fig. S9 C and D), as major membrane interaction regions in KRas helices 4 and 5 observed for monomeric KRas are now occupied by the dimerization interface. Membrane interactions are occasionally seen for R73 in switch II and R102 and D105 in loop 7 at the allosteric site (Fig. 5B). These residues are predicted to be exposed in the dimer to membrane phospholipid headgroups for possible allosteric modulation of GTP hydrolysis in the presence of Raf (23, 30). There is a significant increase in fluctuation of the angle between the α4-α5 helices across the Ras dimer interface in the simulations of the KRas/Raf-RBD dimer in the presence of the membrane compared to what we observe for the HRas/Raf-RBD dimer in the absence of the membrane, although the average angle differs by less than 10° (SI Appendix, Fig. S9E). The most notable change is weakening of interactions at the bottom half of the dimer interface on the KRas dimer complex as Raf-RBD is drawn toward the membrane to form transient electrostatic interactions involving residues 101 to 109 in loop 4 (Fig. 5C). In vivo, the interactions between Raf-RBD and the membrane in the context of the dimer could be shielded with the binding of the scaffold protein Galectin at the site including loop 4 and extending to D113 (19), perhaps resulting in a more stable Ras dimer interface on the membrane.
Fig. 5.
Dimer of the KRas/CRaf-RBD complex simulations at the membrane. (A) Average structure of the complex at a model membrane. The proteins are shown as ribbon diagram, with Ras in green and Raf-RBD in blue. The phospholipids on the membrane are colored by atoms with carbon in gray, phosphorous in orange, and oxygen in red. (B, C) Frequency of KRas and Raf-RBD residues in the dimer coming within 4 Å of the membrane surface. (D) Community network analysis for the KRas/Raf-RBD dimer with communities colored as in Fig. 4 D, Top. (E) Optimal and suboptimal paths connecting Raf-RBD D113 residues on opposite ends of the complex. Nodes are white in the optimal path and black in the suboptimal paths, with all edges colored by the number of paths that cross through them as described in Fig. 4.
Despite the differences noted above, community network analysis and suboptimal path calculations on simulations of the KRas/Raf-RBD dimer complex show similar patterns of allosteric connectivity as described for the HRas/Raf-RBD dimer (Fig. 5D). Once again incorporation of the Ras active site into the dimer interface communities was observed with Raf coalescing into a single community of synchronous residues. There are minor communities observed in the switches of the KRas dimer complex not present in the HRas simulations, consistent with empirical data showing greater dynamics for KRas than for HRas in this region (34). Importantly, the community containing the allosteric site and switch II (Fig. 5D orange and bright green) is segregated from the communities linking the two ends of Raf-RBD, as observed in the HRas/Raf-RBD simulations. Strong connections across the length of the Ras dimer linking residues D113 on the opposing Raf molecules are still present, although the optimal and suboptimal paths that cross the dimer interface traverse further up in the complex on KRas (Fig. 5E), closer to the membrane and further from loop 8 than observed for HRas (Fig. 4D). In the KRas dimer simulations, the edge between residues I139 and isoform-specific residue K165 at the top of helices 4 and 5 respectively provides the most prominent connection across the dimer interface. Although interactions between residues I139 and Q165 are present in the dimer of the HRas/Raf-RBD complex, they are not part of a major allosteric pathway. Conversely, the interswitch loop 3 residues D47 and E49, which in the HRas/Raf-RBD dimer are at the center of allosteric pathways across the interface, interact with isoform specific helix 4 residue K128 (R128 in HRas) and with R135 but are not part of the calculated optimal and suboptimal paths in the KRas/Raf-RBD dimer. These differences between the KRas and HRas simulations could stem from isoform-specific residues at the dimer interface (K/R128, D/E153, and K/Q165 KRas/HRas residues) or elsewhere from the presence of the longer helix 5 leading to the HVR in the KRas simulations or from the fact that the membrane is present for the KRas simulations but not for the HRas simulations. Furthermore, these details may change in the presence of Galectin. Importantly, regardless of whether the simulations include KRas or HRas, the community network analyses described above (Figs. 4 and 5) show strong allosteric connections that link the Galectin-binding D113 residue near loop 4 in one Raf-RBD molecule to D113 on the other across the two Ras molecules.
Discussion
Signaling through Raf is one of the major pathways by which Ras promotes cell proliferation and its mutants result in oncogenic phenotypes. Yet, the molecular mechanisms associated with Ras activation of Raf remain obscure. Here we show that dimerization of Ras is promoted by both Raf-RBD and the membrane as dimerization in the presence of Raf-RBD in solution is only partial. While our SEC experiments show a clear role of the G-domain, only in the SLB experiments, in which both the membrane and Raf-RBD are present, is robust dimerization observed with no detectable diffusion associated with the monomer. Thus, we advance mechanistic understanding of Raf activation by Ras with evidence that Raf and the membrane act in concert to promote dimerization of the Ras/Raf complex. Furthermore, we show that dimerization dramatically increases allosteric connections linking the two Raf-RBD molecules at opposite ends of the dimer formed by the α4-α5 interface across the two Ras protomers, leading to the proposal of a signaling model that also includes Galectin. Having previously shown that HRas and KRas have distinct conformational preferences (3, 34), here we compare dynamic network analysis from simulations of the HRas/CRaf-RBD dimer started directly from the crystal structure of the complex with longer simulations on the membrane, starting from a model of the KRas/CRaf-RBD dimer generated from the HRas/CRaf-RBD structure. Despite the different setups, we found similar patterns of allosteric connections in our simulations (Figs. 4 and 5). Furthermore, SEC-SAXS experiments yielded similar molecular envelopes for dimers containing KRas/CRaf-RBD and HRas/CRaf-RBD, the SLB experiments gave similar results for both isoforms (Fig. 3 and SI Appendix, Fig. S6), and the dimerization interface for the KRas/CRaf-RBD dimer modeled on HRas/CRaf-RBD is stable throughout the 1-μs simulations, yielding a structure with excellent agreement to both KRas/CRaf-RBD (χ2 of 1.04, SI Appendix, Fig. S7A) and HRas/CRaf-RBD (χ2 of 1.26) SEC-SAXS data sets. Together, these data point to a similar mechanism between the two Ras isoforms for signaling through the Ras/Raf/MEK/ERK pathway which we expect would also extend to the NRas isoform.
The question of whether Ras forms dimers on the membrane in the absence of other factors has been met with paradoxical answers in which several studies based on fluorescence or light scattering experiments detected dimers (4, 6, 25) while others found Ras to be monomeric (5, 9, 35, 36). The detection of Ras dimers using biophysical methods based on photoexcitation was eventually explained by the formation of dityrosine crosslinks resulting from a photosensitization reaction that produces reactive radial oxygen species (8). As the authors point out, the distribution of tyrosine and histidine residues on the surface of Ras renders these proteins particularly susceptible to this crosslinking reaction (SI Appendix, Fig. S10). Here, we reproduce published results that Ras is monomeric by itself on SLBs in the presence of radical oxygen scavenger in the buffer (9) and show that Raf-RBD is the additional factor that promotes dimerization of Ras (Fig. 3).
Computational studies have predicted various dimer interfaces and estimated the binding free energy associated with each one (13). This work has identified weak dimer interfaces involving the α3-α4 or α4-α5 Ras allosteric lobe helices. Which of these interfaces is involved in signaling through Raf has until recently remained an open question, primarily because mutations in both helices 3 (37) and 5 (10) result in attenuated signaling through Ras/Raf. However, we have previously shown that a single mutation can have global allosteric effects on the structure and dynamics of the HRas/CRaf-RBD complex (23). Additionally, given the long-range allosteric connections that we have demonstrated here through our community network analysis based on MD simulations, it is likely that mutations can affect dimerization without being at the interface. In general, given the highly allosteric impact of the Ras/Raf-RBD complex formation on both Ras and Raf-RBD (23, 38), great caution is needed in correlating the effects of specific mutations, or chemical shift perturbations obtained by NMR between bound and unbound species, to the locations of binding sites on Ras. The recent NMR structure of the KRas dimer on a nanodisc (PDB ID 6W4E) unequivocally shows that the dimer forms through the α4-α5 helical interface (18), consistent with its prominent appearance in crystal structures of Ras (6, 24) and the stability of this interface in our simulations. Although captured in the NMR model, the Ras dimer interaction is weak in nanodiscs, with a Kd of 530 μM in the absence of Raf-RBD (18), and is most likely observed due to the small membrane area on the nanodisc, leading to a crowded environment for the two Ras molecules. An analogous crowding situation occurs in Ras crystals, explaining the appearance of the dimer as previously tabulated (24). In contrast, no dimerization is observed for Ras in the absence of Raf-RBD in SLBs (9) which provide a larger area through which Ras molecules can diffuse.
It has been previously observed that uncomplexed Ras has several regions of correlated motion such as those between the C-terminal end of helix 3 and the switch regions (39, 40) and that the binding of Raf-RBD affects the dynamics of the entire monomer of the complex (23). The present analysis identifies links between the dimer interface and the Raf-RBD loop 4 region in complexes with both HRas and KRas in addition to further validating the connection between R97 in the allosteric site and Q61 in switch II, which we have previously studied (23, 30). The community on Ras that includes the calcium-binding allosteric site involving loop 7, which in our simulations transiently interacts with the membrane (Fig. 5B), remains relatively unchanged with binding of Raf-RBD and dimerization of the complex. However, while the allosteric connections linking the dimer interface in Ras to loop 4 in Raf are modest in the monomer of the complex, a remarkable enhancement of allosteric connection occurs in the dimer of the Ras/Raf-RBD complex, linking residue D113 near loop 4 in both Raf-RBD molecules at the extreme ends of the complex. In this context, helices 4 and 5 at the dimer interface are intimately connected to the active site through the NKxD and ExSAK nucleotide-binding motifs, with helix 4 forming additional connections with loop 8 and the N-terminal end of helix 3, and helix 5 connecting to the interswitch region and the β-sheet core. Overall, the major communities in Ras separate elements associated with GTP hydrolysis that we propose is promoted by calcium in the context of the dimer (23, 30) from those across the Ras dimer and Ras/Raf interfaces important for activation of Raf kinase. It is not surprising that these two separate functions, signaling and GTP hydrolysis, would be decoupled in this system.
Two recent cryogenic electron microscopy (cryo-EM) structures of full-length Raf in complex with the scaffold protein 14-3-3 (41) and in complex with both 14-3-3 and MEK (42) reveal details of activation at the Raf kinase/MEK level but in the absence of Ras show disordered Ras binding domains, even as the kinase itself is in its active dimeric form. The fact that Raf kinase can dimerize in the absence of Ras points to a Ras function other than to simply promote kinase domain dimerization. When not in complex with Ras, the Raf cysteine-rich domain (Raf-CRD) functions to maintain autoinhibition in the absence of active Ras as revealed by the cryo-EM structure of the full-length inactive kinase, in which Raf-CRD is ordered and nestled between 14-3-3 and the Raf kinase domain (42). This autoinhibition is released when bound to Ras, allowing for activation of the kinase; however, it is not clear that Ras dimerization is necessary for this process. Here, we propose that a major function of Ras dimerization upon Raf binding is to couple with Galectin dimers to transform these protein complexes into an allosterically connected robust signaling platform (Fig. 6). The strong allosteric connection between the opposite extremes of the Ras/Raf dimer presents a unified complex that could be important for effective activation of signaling through Ras/Raf/MEK/ERK, particularly in the context of interactions with scaffold proteins such as Galectin, which is itself a homodimer critical for signaling through Ras/Raf (43, 44). Raf residue D113 is in the proposed binding site between CRaf-RBD and Galectin-1 (19) and is implicated in being allosterically involved in Ras/Raf binding by at least three research groups using independent methods (19, 31, 32). The importance of residue D113 to the signaling complex is further supported by the robust allosteric connections of this residue to critical Ras dimerization elements in helices 4 and 5. Coupling of the dimer of the Ras/Raf-RBD complex to the Galectin dimer through residue D113 in the loop 4 region could form a kinetic proofreading platform similar to that observed for the LAT/Grb2/SOS complex (27, 45–47).
Fig. 6.
Multiprotein assembly proposed for signaling through the Ras/Raf/MEK/ERK pathway. Complexes involving Ras (green), Raf-RBD (blue), and Galectin dimers (beige) at the membrane would promote protein network that result in synchronized activation for signal amplification and kinetic proofreading. The Raf-CRD and the serine/threonine rich region that follows, situated before the kinase domain (residues 132 to 340 in CRaf), are indicated by a blue line connecting the RBD to the kinase domain. The composite figure was made with structures for HRas/CRaf-RBD (PDB ID 4G0N), the Galectin-1 dimer (PDB ID 3W58), and the Raf-kinase domain dimer (PDB ID 3OMV).
Biocondensates, consisting of concentrated multiprotein assemblies forming distinct fluid structures that separate from surrounding areas, are ubiquitous across signal transduction networks (48). Furthermore, single-particle tracking (SPT) experiments (49, 50), most recently coupled with photoactivated localization microscopy (SPT-PALM) and detailed trajectory analysis (51), have shown that activated Ras proteins in live cells are found on the membrane in mobile and immobile phases, the latter being consistent with the predicted formation of large macromolecular assemblies that cannot freely diffuse on the membrane. Recent work with the transmembrane receptor LAT and its cytosolic binding partners Grb2 and the Ras guanine nucleotide exchange factor SOS have shown that these proteins form polymer-like assemblies on SLBs (45) with restricted mobility. By applying properties of polymer physics to this protein system, it becomes apparent that the proteins bound to several partners within the signaling competent complex form a gel-like phase at the membrane that crowding effects and Brownian motion fail to describe. Other interactions with the membrane from within these condensates, for example, involving membrane receptors (52), SOS (53–55), or PLCγ (56), further restrict molecular mobility and may influence Ras. Increasing connectivity with dimerization of the Ras/Raf complex could allosterically prime the Galectin binding site on Raf-RBD to form a multivalent protein complex on the membrane corresponding to the immobile species observed in the SPT-PALM experiments (51). This platform of synchronized activated signaling proteins (Fig. 6) could be essential for effective activation of the Ras/Raf/MEK/ERK signaling cascade, ensuring signal amplification and avoidance of random misfiring due to isolated encounters in analogy to the LAT/Grb2/SOS platform. In this scenario, isolated Ras/Raf dimers would not be effective signaling agents, explaining the requirement for Galectin in the activation of the pathway (19, 44). Scaffold proteins are known to be hubs for the control of cell signaling (57) and are often involved in biomolecular condensates associated with function either on or off membranes (48). Our proposed model puts Ras dimerization at the center of such an organizational platform promoted by the binding of Raf-RBD and recruitment of Galectin for kinetic proofreading and signal amplification, leading to activation of the critical kinases in the mitogenic signal transduction pathway. While not yet supported by experiments, this proposed model opens avenues for testing the framework that leads to activation of the MAPK pathway at the level of Ras/Raf interaction and dimerization.
Ras on the membrane forms nanoclusters consisting of oligomers with several Ras molecules interacting with each other through multiple interfaces (13, 21). Ras/Ras interactions are weak and dynamic (13, 18), and it has been shown that the KRas G-domain is found distal from the membrane (58), exposed to form the signaling active complex in the presence of Raf. Given the robust high-affinity dimerization that we observe on SLBs in the presence of Raf-RBD, we propose that binding of Raf to Ras-GTP on the membrane allosterically modulates the α4-α5 interface such that it predominates in the signaling active dimer of the complex. This results in a dramatic increase in allosteric connectivity linking the Galectin-binding D113 residue on Raf-RBD near loop 4 at the two extremes of the dimer, which we propose becomes primed to interact with the Galectin dimer to form a robust and synchronized signaling platform. In this context, the number of Ras molecules in the nanoclusters, confined to specific microdomains, may serve to spatially limit the size of the active signaling platform, and therefore the overall signal intensity, in a controlled fashion. While some aspects of the proposed model still need to be tested, the present work provides a conceptual leap forward by revealing the relationship between Ras and Raf-RBD with respect to dimerization and the high allosteric connectivity between them in the dimer, leading to the prospect that dimerization is a critical step in forming the signaling complex for synchronized activation of multiple Raf kinase molecules.
Methods
Ras and Raf-RBD Expression and Purification.
The G-domains of KRas or HRas proteins containing residues 1 to 166 or 1 to 173 were purified as previously described (59). After nucleotide exchange to replace GDP with the GTP analog GppNHp (60), protein was loaded into a 5-mL sample loop followed by injection onto a HiTrap QHP 5-mL anion exchange column (GE Life Sciences). Ras was eluted with a linear 0 to 25% gradient of Buffer B with composition described in the published protocol (59). The peak fractions were analyzed by sodium dodecyl (lauryl) sulfate-polyacrylamide gel electrophoresis, and fractions containing Ras-GppNHp were pooled and concentrated to ∼20 mg/mL and flash frozen in liquid nitrogen for storage at −80 °C until needed. CRaf-RBD containing residues 52 to 131 was expressed and purified as previously described (3).
Ras/Raf-RBD Dimerization Observed in Solution by SEC.
The appropriate Ras protein was combined with a 20-fold molar excess of Raf in stabilization buffer (20 mM Hepes pH 7.5, 20 mM MgCl2, 50 mM NaCl, 1 mM DTT, and 2% glycerol) and concentrated to ≤100 µL at 4 °C. The solution was then spin-filtered through a 0.2-µm filter for injection onto a preequilibrated Superdex 200 Increase 10/300 GL column (GE Healthcare) for standard gel filtration analysis with a 0.5 mL/min flow rate. To reduce protein loss during injection onto the column, a 500-µL Hamilton syringe with large hub removable 22-gauge needle was used to inject the protein into a 100-µL sample loop via an INV-907 fill port. Peak integration was performed for the three major peaks, dimer KRas/Raf-RBD complex, monomer KRas/Raf-RBD, and excess Raf using a zero baseline and the Unicorn peak integration function. Integration windows were adjusted to prevent inclusion of higher order oligomers that may be present in the sample mixture. The sum of the complex dimer and monomer peaks was taken as 100% complex, and the respective percentage of dimer and monomers were calculated as the contribution of each peak to the total.
SEC In-Line with SEC-SAXS.
Samples for SEC-SAXS were prepared according to the dimerization assay protocol above and flash frozen once concentrated for storage and transportation. Data were collected at the Cornell High Energy Synchrotron Source (CHESS) at Cornell University (Ithaca, NY). Standard SEC was run in stabilization buffer as above in which the eluted sample was subjected to constant light scattering data collection using a flow cell in line with the X-ray beam (61). Because the elution profile matches exactly that of the standard Ras/Raf-RBD dimerization assay, the three peak identities for the SEC-SAXS chromatograms were known a priori. For each species (dimer Ras/Raf-RBD, monomer Ras/Raf-RBD, and Raf-RBD), five 2-s frames of data were combined (10 s total exposure time) and buffer subtracted against five 2-s frames corresponding to the column equilibrated with stabilization buffer. The ATSAS program package was used for initial SAXS data analysis, including Guinier and distance distribution analysis, and envelope generation (62, 63). SUPCOMB was used to align the molecular envelope generated using the SEC-SAXS data with the crystal structure PDB file (4G0N) with a very good normalized spatial discrepancy value of near 1 (64). Chimera was also used for alignment of the models in the SAXS-generated envelopes (65). A direct comparison of the rigid PDB model with the SAXS data yielding the reported χ2 values in SI Appendix, Figs. S5 and S7 was performed in FoXS (66). Multi-FoXS (66) was used for the two-model system associated with the frame near the midpoint between the Ras/Raf-RBD dimer and monomer peaks.
Mass Spectrometry.
Samples eluted from the SEC peak corresponding to the dimer of the Ras/Raf-RBD complex in the presence of 1 mM DTT were prepared following a previously described protocol (67). Briefly, protein from the appropriate pooled SEC fractions was precipitated using a volume ratio of 1:1:4:3 of protein:chlororform:methanol:water. The supernatant was removed and the precipitate washed with the addition and removal of another four parts methanol. Pellets were solubilized in cold (−20 °C) 80% formic acid and diluted to the original volume with high-performance liquid chromatography-grade water.
Intact protein liquid chromatography–mass spectrometry was performed using an H class Acquity Ultra-High-Pressure Liquid Chromatography (UPLC) system coupled with a Xevo G2-S Q-TOF mass spectrometer (Waters Corp, Milford, MA) as previously described (67). Briefly, reversed phase chromatography was employed for separation (Acquity UPLC protein BEH C4 300 Å pore size, 1.7-µm particle size, 100-mm bed length, 2.1-mm identification (ID) × 100 mm) with 95% water/−5% acetonitrile with 0.1% formic acid as solvent A and 95% acetonitrile/−5% water with 0.1% formic acid as solvent B.
FCS.
FCS measurements were performed on a home-built confocal system integrated into an inverted microscope. The experimental methods have been published previously (8). The light source was a pulsed (100 ps at 20 MHz repetition rate) supercontinuum laser (NKT Photonics). The average excitation power for a typical FCS measurement was 0.5 µW for 488 ± 5 nm. The fluorescent signals were collected by the objective and passed a 50-µm pinhole detected by avalanche photodiode detectors (Hamamatsu) and processed by a hardware correlator (http://Correlator.com). To calibrate the spot size of the focus, a bilayer with a known surface density of fluorescent lipids, BODIPY FL DHPE for 488 nm, was measured, which consistently yielded the radii of 0.20 ± 0.01 µm. Each signal was focused into 0.15 × 0.15 µm avalanche photodiode elements (Hamamatsu) and subsequently processed by a hardware correlator (http://Correlator.com). The resulting autocorrelation G(τ) traces were fit to two-dimensional Gaussian diffusion model,
All FCS experiments were performed in imaging buffer to suppress oxidative cross-linking of Ras. The imaging buffer was of the following composition: 10 mM BME, 20 mM glucose, 2 mM Trolox, 320 μg/mL glucose oxidase, and 50 μg/mL catalase in the Hepes buffer. Trolox stock solution was prepared at a concentration of 200 mM in solvent by mixing DMSO and 1 M Tris base at a 4:1 volume ratio followed by 305 nm ultraviolet treatment for 15 min.
SMT.
The experimental methods for single molecule total internal reflection fluorescence (smTIRF) have been described previously (55). TIRF images were acquired using a Nikon Eclipse Ti inverted microscope equipped with a 100x 1.49 NA oil immersion TIRF objective and an Andor iXon EMCCD camera. Both 488-nm and 637-nm diode lasers (Coherent Inc.) were used as illumination sources for TIRF imaging. The laser intensity was set to 0.5 and 15 mW at the objective for a 488- and 637-nm laser, respectively. In order to track KRas at the single molecule level in a wide range of surface density, 100 pM of Alexa 647 GppNHp-loaded KRas was mixed with various concentrations of eGFP-KRas loaded with nonfluorescent GppNHp (typically up to 50 nM). TIRF intensity of eGFP channel was used to estimate the overall density of KRas, and Alexa 647 GppNHp-loaded KRas were imaged for SMT. All single-particle experiments were performed with the same imaging buffer used for FCS experiments. The images were acquired at 20 ms exposure with no delay time. Particle localization and trajectory liking were done with TrackMate (ImageJ plugins) (68). The step size distribution was calculated in Igor Pro (WaveMetrics).
MD Simulations.
MD simulations of 90 ns production run were performed for HRas/CRaf-RBD monomer and HRas/CRaf-RBD dimer in which both structures were started from coordinates with PDB ID 4G0N (23). The simulations were conducted at the Northeastern Discovery Cluster (http://www.northeastern.edu/rc). In each of the PDB files, the GTP analog, GppNHp, was modified to form GTP by replacing the β-γ–bridging nitrogen atom with oxygen. Calcium and acetate molecules were left at the allosteric site. All the crystallographic water molecules were included in the simulation. Each protein construct was additionally solvated by TIP3P waters with 150 mM NaCl (plus net charge neutralizing ions) for a total of 28,808 and 61,101 atoms for the monomer and dimer, respectively. Each system was minimized for 5,000 frames then equilibrated to 300 K. A 1-fs time step was used for the first 30 ns then increased to 2 fs for the remaining 60 ns. Standard parameters were used with Particle Mesh Ewald (PME) method for long range electrostatic interaction, a cutoff of 1.1 nm for the van der Waals (vdW) potential and short electrostatic interaction, SHAKE algorithm for constraint of all covalent bonds to hydrogen, and a Langevin thermo- and barostat at 300 K and 1 bar, respectively. The simulations were prepared using Visual MD and performed on Nanoscale MD software (69). The Chemistry at Harvard Macromolecular Mechanics (CHARMM) force field was used for the simulations (70).
Molecular model of the 2:2 KRas/CRaf-RBD dimer was constructed using available crystal structures (PDB 4DSO for KRas4B and 4G0N for the RBD; 4G0N is a complex of HRas/CRaf-RBD which showed the dimer in the crystal lattice, onto which KRas4B was modeled. The RBD was then homology docked onto each side of the KRas4B dimer). The complex was anchored into a membrane of 280 POPC and 70 POPS (80%:20%). Two POPC lipid molecules were removed on the leaflet where KRas is anchored in order to relax the membrane tension. The system was charged at pH 7.0 (using amino acid protonation states, and HSD, hydrogen on delta position for a neutral histidine) and solvated by TIP3P water with 150 mM NaCl (plus net charge neutralizing ions) for a total of 160,358 atoms. We used Mg2+ and GTP parameters as described previously and the CHARMM36m force field (70). The initial equilibration simulation was performed for 30 ns with a 2-fs time step using NAMD/2.12 package (69). Standard parameters were used with PME method for long range electrostatic interaction, a cutoff of 1.2 nm for the vdW potential and short electrostatic interaction, SHAKE algorithm for constraint of all covalent bonds to hydrogen, and a Langevin thermo- and barostat at 310 K and 1 bar, respectively. The equilibrium simulations were then transferred to the Anton 2 supercomputer for production simulations of 1 μs (71).
Dynamical Network Analysis.
Dynamical network analysis is a general method used to obtain an accurate picture of network topology and long-range signaling in protein complexes derived from MD simulations (28). Each amino acid residue in the complex is assigned a node centered on its Cα atom and used as a base to construct significant regions of amino acid interactions and pathways of allosteric modulation that connect them. Edges are placed to connect the nodes between residues that remain within a 4.5Å distance for at least 75% of the simulation time. The edges are weighted using pairwise correlation data calculated by the program Carma (29). This information can then be mined to define community networks using the Girvan–Newman algorithm (72) and the diversity of paths that connect sites of functional significance in the complex. Nodes in the same community network can communicate with each other easily through multiple paths, whereas those in distinct community networks either do not communicate well or communicate through one or a small number of nodes essential for allosteric modulation. Once “source” and “sink” residues are defined as those whose allosteric connections are being evaluated, the shortest allosteric path between them is termed the optimal path. All others are labeled as suboptimal paths. A length offset of 20 edges that prevents the suboptimal paths from being more than 20 edges longer than the optimal path was used to prevent the program from identifying uneccesarily long paths connecting the residues.
Supplementary Material
Acknowledgments
We thank Andrew Stephen and the Ras initiative team at the Frederick National Laboratory for Cancer Research, National Cancer Institute, for providing the farnesylated and methylated full-length KRas4B used in the SLB experiments. We thank Richard Gillilan (CHESS) for assistance with collection of the SEC-SAXS data. The protein structure coordinates used in this work were previously published in the PDB and are accompanied in the text by their respective accession codes. The simulations of KRas/CRaf-RBD on the membrane were performed by Z.L. on Anton 2 computer, with time provided by the Pittsburgh Supercomputing Center through NIH R01GM116961 and supported by NIH Grant R01EY029169 to M.B., preceded by R01GM112491. Support for FCS and SMT work at UC Berkeley was provided by NIH National Cancer Institute Physical Sciences in Oncology Network project 1-U01CA202241 and by the Novo Nordisk Foundation Challenge Program under the Center for Geometrically Engineered Cellular Systems. This project was supported by NSF Grant MCB-1517295 awarded to C.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.S.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015648118/-/DCSupplemental.
Data Availability
Coordinates with PDB ID 4G0N, 6W4E, 3K8Y, and 4DSO were used for analysis or model preparation for the simulations in this study. The MD simulation trajectories have been deposited on GitHub and can be accessed at https://github.com/mattoslab/mattoslabsite/tree/PNAS2021.
References
- 1.Prior I. A., Lewis P. D., Mattos C., A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior I. A., Hancock J. F., Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 23, 145–153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson C. W., et al., The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J. Biol. Chem. 292, 12981–12993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muratcioglu S., et al., GTP-dependent K-ras dimerization. Structure 23, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovrigina E. A., Galiakhmetov A. R., Kovrigin E. L., The ras G domain lacks the intrinsic propensity to form dimers. Biophys. J. 109, 1000–1008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Güldenhaupt J., et al., N-Ras forms dimers at POPC membranes. Biophys. J. 103, 1585–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inouye K., Mizutani S., Koide H., Kaziro Y., Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 275, 3737–3740 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Chung J. K., Lee Y. K., Lam H. Y., Groves J. T., Covalent ras dimerization on membrane surfaces through photosensitized oxidation. J. Am. Chem. Soc. 138, 1800–1803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung J. K., et al., K-Ras4B remains monomeric on membranes over a wide range of surface densities and lipid compositions. Biophys. J. 114, 137–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrogio C., et al., KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Nan X., et al., Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayyed-Ahmad A., Cho K. J., Hancock J. F., Gorfe A. A., Computational equilibrium thermodynamic and kinetic analysis of K-ras dimerization through an effector binding surface suggests limited functional role. J. Phys. Chem. B 120, 8547–8556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash P., et al., Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci. Rep. 7, 40109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang H., Zhang M., Nussinov R., The quaternary assembly of KRas4B with Raf-1 at the membrane. Comput. Struct. Biotechnol. J. 18, 737–748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar-Banerjee S., et al., Spatiotemporal analysis of K-ras plasma membrane interactions reveals multiple high order homo-oligomeric complexes. J. Am. Chem. Soc. 139, 13466–13475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y., et al., Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 34, 862–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barklis E., Stephen A. G., Staubus A. O., Barklis R. L., Alfadhli A., Organization of farnesylated, carboxymethylated KRAS4B on membranes. J. Mol. Biol. 431, 3706–3717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K. Y., et al., Two distinct structures of membrane-associated homodimers of GTP- and GDP-bound KRAS4B revealed by paramagnetic relaxation enhancement. Angew. Chem. Int. Ed. Engl. 59, 11037–11045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaževitš O., et al., Galectin-1 dimers can scaffold Raf-effectors to increase H-ras nanoclustering. Sci. Rep. 6, 24165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzmán C., et al., The efficacy of Raf kinase recruitment to the GTPase H-ras depends on H-ras membrane conformer-specific nanoclustering. J. Biol. Chem. 289, 9519–9533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., et al., Lipid-sorting specificity encoded in K-ras membrane anchor regulates signal output. Cell 168, 239–251.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathew E., Mirza A., Menhart N., Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchrotron Radiat. 11, 314–318 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Fetics S. K., et al., Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spencer-Smith R., et al., Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W. C., et al., H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. U.S.A. 111, 2996–3001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillette W. K., et al., Farnesylated and methylated KRAS4b: High yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 5, 15916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W. Y. C., et al., A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethi A., Eargle J., Black A. A., Luthey-Schulten Z., Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U.S.A. 106, 6620–6625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glykos N. M., Software news and updates. Carma: A molecular dynamics analysis program. J. Comput. Chem. 27, 1765–1768 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Buhrman G., Holzapfel G., Fetics S., Mattos C., Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc. Natl. Acad. Sci. U.S.A. 107, 4931–4936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gohlke H., Kiel C., Case D. A., Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 330, 891–913 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Sikić M., Tomić S., Vlahovicek K., Prediction of protein-protein interaction sites in sequences and 3D structures by random forests. PLoS Comput. Biol. 5, e1000278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z. L., Prakash P., Buck M., A “tug of war” maintains a dynamic protein-membrane complex: Molecular dynamics simulations of C-Raf RBD-CRD bound to K-Ras4B at an anionic membrane. ACS Cent. Sci. 4, 298–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker J. A., Volmar A. Y., Pavlopoulos S., Mattos C., K-ras populates conformational states differently from its isoform H-ras and oncogenic mutant K-RasG12D. Structure 26, 810–820.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Werkmüller A., Triola G., Waldmann H., Winter R., Rotational and translational dynamics of ras proteins upon binding to model membrane systems. ChemPhysChem 14, 3698–3705 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Smith E. M., Macdonald P. J., Chen Y., Mueller J. D., Quantifying protein-protein interactions of peripheral membrane proteins by fluorescence brightness analysis. Biophys. J. 107, 66–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muratcioglu S., et al., Oncogenic K-Ras4B dimerization enhances downstream mitogen-activated protein kinase signaling. J. Mol. Biol. 432, 1199–1215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aramini J. M., et al., The RAS-binding domain of human BRAF protein serine/threonine kinase exhibits allosteric conformational changes upon binding HRAS. Structure 23, 1382–1393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorfe A. A., Grant B. J., McCammon J. A., Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure 16, 885–896 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor A., Travesset A., Differential dynamics of RAS isoforms in GDP- and GTP-bound states. Proteins 83, 1091–1106 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Kondo Y., et al., Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases. Science 366, 109–115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park E., et al., Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abankwa D., Gorfe A. A., Hancock J. F., Mechanisms of Ras membrane organization and signalling: Ras on a rocker. Cell Cycle 7, 2667–2673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elad-Sfadia G., Haklai R., Ballan E., Gabius H. J., Kloog Y., Galectin-1 augments Ras activation and diverts Ras signals to Raf-1 at the expense of phosphoinositide 3-kinase. J. Biol. Chem. 277, 37169–37175 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Huang W. Y. C., Chiang H.-K., Groves J. T., Dynamic scaling analysis of molecular motion within the LAT:Grb2:SOS protein network on membranes. Biophys. J. 113, 1807–1813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang W. Y., et al., Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc. Natl. Acad. Sci. U.S.A. 113, 8218–8223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su X., et al., Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lommerse P. H., et al., Single-molecule diffusion reveals similar mobility for the Lck, H-ras, and K-ras membrane anchors. Biophys. J. 91, 1090–1097 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakoshi H., et al., Single-molecule imaging analysis of Ras activation in living cells. Proc. Natl. Acad. Sci. U.S.A. 101, 7317–7322 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y., et al., High-throughput, single-particle tracking reveals nested membrane domains that dictate KRas(G12D) diffusion and trafficking. eLife 8, e46393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J. J. Y., et al., Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 12, eaat8715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iversen L., et al., Molecular kinetics. Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics. Science 345, 50–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christensen S. M., et al., One-way membrane trafficking of SOS in receptor-triggered Ras activation. Nat. Struct. Mol. Biol. 23, 838–846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y. K., et al., Mechanism of SOS PR-domain autoinhibition revealed by single-molecule assays on native protein from lysate. Nat. Commun. 8, 15061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi J., Balagopalan L., Nguyen T., McIntire K. M., Samelson L. E., TCR microclusters form spatially segregated domains and sequentially assemble in calcium-dependent kinetic steps. Nat. Commun. 10, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Good M. C., Zalatan J. G., Lim W. A., Scaffold proteins: Hubs for controlling the flow of cellular information. Science 332, 680–686 (2011).21551057 [Google Scholar]
- 58.Van Q. N., et al., Uncovering a membrane-distal conformation of KRAS available to recruit RAF to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 117, 24258–24268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson C. W., Buhrman G., Ting P. Y., Colicelli J., Mattos C., Expression, purification, crystallization and X-ray data collection for RAS and its mutants. Data Brief 6, 423–427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holzapfel G., Buhrman G., Mattos C., Shift in the equilibrium between on and off states of the allosteric switch in Ras-GppNHp affected by small molecules and bulk solvent composition. Biochemistry 51, 6114–6126 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Acerbo A. S., Cook M. J., Gillilan R. E., Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J. Synchrotron Radiat. 22, 180–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petoukhov M. V., et al., New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franke D., et al., ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 50, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozin M. B., Svergun D. I., Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 (2001). [Google Scholar]
- 65.Yang Z., et al., UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 179, 269–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donnelly D. P., et al., Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat. Methods 16, 587–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tinevez J. Y., et al., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Phillips J. C., et al., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J., et al., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaw D. E.et al., “Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer” in Internaltional Conference for High Performance Computing, Networking, Storage and Analysis (Institute of Electrical and Electronic Engineers (IEEE), Piscataway, NJ, 2014). [Google Scholar]
- 72.Girvan M., Newman M. E., Community structure in social and biological networks. Proc. Natl. Acad. Sci. U.S.A. 99, 7821–7826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates with PDB ID 4G0N, 6W4E, 3K8Y, and 4DSO were used for analysis or model preparation for the simulations in this study. The MD simulation trajectories have been deposited on GitHub and can be accessed at https://github.com/mattoslab/mattoslabsite/tree/PNAS2021.