Significance
α1-Antitrypsin deficiency is one of the most common genetic diseases. Homozygous and heterozygous carriers of the Z allele of α1-antitrypsin are susceptible to developing liver fibrosis and cirrhosis. In mouse and human samples expressing the Z allele of α1-antitrypsin, we found both miR-34b and miR-34c are up-regulated by activation of FOXO3 upon JNK phosphorylation on Ser574. Deletion of miR-34b/c resulted in early development of liver fibrosis and increased signaling of the PDGF pathway, a target of miR-34b/c. JNK-activated FOXO3 and miR-34b/c up-regulation also occur in several mouse models of liver fibrosis. Fibrosis is a major health problem and unravelling its underlying pathogenic mechanisms has potential for the development of targeted therapeutic agents.
Keywords: liver fibrosis, JNK, FOXO3, miR-34b/c, α1 antitrypsin deficiency
Abstract
α1-Antitrypsin (AAT) deficiency is a common genetic disease presenting with lung and liver diseases. AAT deficiency results from pathogenic variants in the SERPINA1 gene encoding AAT and the common mutant Z allele of SERPINA1 encodes for Z α1-antitrypsin (ATZ), a protein forming hepatotoxic polymers retained in the endoplasmic reticulum of hepatocytes. PiZ mice express the human ATZ and are a valuable model to investigate the human liver disease of AAT deficiency. In this study, we investigated differential expression of microRNAs (miRNAs) between PiZ and control mice and found that miR-34b/c was up-regulated and its levels correlated with intrahepatic ATZ. Furthermore, in PiZ mouse livers, we found that Forkhead Box O3 (FOXO3) driving microRNA-34b/c (miR‐34b/c) expression was activated and miR-34b/c expression was dependent upon c-Jun N-terminal kinase (JNK) phosphorylation on Ser574. Deletion of miR-34b/c in PiZ mice resulted in early development of liver fibrosis and increased signaling of platelet-derived growth factor (PDGF), a target of miR-34b/c. Activation of FOXO3 and increased miR-34c were confirmed in livers of humans with AAT deficiency. In addition, JNK-activated FOXO3 and miR-34b/c up-regulation were detected in several mouse models of liver fibrosis. This study reveals a pathway involved in liver fibrosis and potentially implicated in both genetic and acquired causes of hepatic fibrosis.
α1-Antitrypsin (AAT) deficiency is an inherited disorder that affects ∼1 in 3,000 individuals and is an important genetic cause of lung and liver disease (1). The most common defect is the Z variant of the SERPINA1 gene, which results in the production of misfolded and polymerogenic Z α1-antitrypsin (ATZ). ATZ-dependent liver disease has a wide spectrum of clinical manifestations ranging from liver insufficiency in newborns to chronic liver disease and hepatocellular carcinoma in adults (2, 3). Because of its misfolding and polymerization, ATZ is unable to efficiently traverse the secretory pathway. Accumulation of ATZ in the endoplasmic reticulum (ER) of hepatocytes has a proteotoxic effect.
Expression of microRNAs (miRNAs) is affected in several liver diseases with distinct profiles across diseases with different etiologies (4). Here, we investigated differentially expressed miRNAs in the liver of PiZ mice, a transgenic animal model expressing the human ATZ (5). We then confirmed the most relevant findings in liver samples from patients. Following the identification of an important miRNA cluster involved in liver fibrosis, the upstream molecules affecting its expression, and its effector, we showed this pathway is involved in various murine models of liver fibrosis.
Results
Profiling of miRNA in PiZ Mouse Livers Revealed Up-Regulation of miR-34b/c.
Transgenic PiZ mice accumulate ATZ within the ER of hepatocytes in a manner akin to that of patients affected by AAT deficiency. Therefore, these mice are a valuable experimental model for investigating the liver disease of AAT deficiency (5, 6). We evaluated differentially expressed miRNAs by next-generation sequencing in the livers of PiZ mice and strain-, age-, and sex-matched wild-type controls. Seventy miRNAs were found to be differentially expressed in PiZ livers compared with wild-type controls (SI Appendix, Fig. S1). miRNAs with the greatest fold changes in PiZ versus wild-type livers were validated by qPCR (SI Appendix, Fig. S2A). Among differentially expressed miRNAs, both miR-34b-5p and miR-34c-5p (henceforth referred to as miR-34b and miR-34c, respectively) showed the highest statistical significance and fold changes in PiZ mice compared with wild-type controls (Fig. 1A). In PiZ mice, up-regulation of miR-34b and miR-34c (miR-34b/c), which are expressed from a common primary transcript (7), was confirmed in both liver (Fig. 1B) and plasma (Fig. 1C) by targeted real-time PCR analysis whereas differences in miR-34a expression were detected in plasma but not in liver (Fig. 1 B and C). Expression of miR-16, a marker of hemolysis (8), was not significantly different between the two groups (SI Appendix, Fig. S2B), ruling out hemolysis as the factor responsible for the increased miR-34b/c levels in the blood.
Fig. 1.
Increased expression of miR-34b/c in mouse livers expressing Z α1-antitrypsin. (A) Differentially expressed miRNAs visualized by volcano plot. miRNAs with false discovery rate (FDR) <10−2 (y axis) and fold change in PiZ over wild type (WT) >|4| are shown and miR-34b/c are red-circled. (B) qPCR on PiZ versus WT for miR-34 family members on liver RNA showing significant up-regulation of miR-34b/c (at least n = 3 per group; t test: **P < 0.01 and ***P < 0.005 versus WT). (C) qPCR on PiZ versus WT for miR-34 family members on plasma RNA showing significant up-regulation of miR-34a-5p, miR-34b-3p and 5p, and miR-34c-5p (n = 4 per group; t test: *P < 0.05 and **P < 0.01 versus WT). Data are reported as average ± SE.
miR-34b/c Is Mainly Expressed by Hepatocytes and Its Levels Correlate with Hepatic ATZ Accumulation.
To investigate the role of miR-34b/c in ATZ-mediated liver disease, first we measured miR-34b/c expression in parenchymal and nonparenchymal liver cells. Because mature miRNAs can be secreted and taken up by neighboring cells not expressing the miRNAs, we evaluated the miR-34b/c common primary transcript (pri-miR-34b/c) in parenchymal and nonparenchymal liver cells of PiZ mice. This precursor transcript is specific for the cells expressing the miRNAs but not for cells that take up the mature miRNAs. The pri-miR-34b/c was found to be mainly expressed in the parenchymal fraction enriched for albumin gene expression (Fig. 2A and SI Appendix, Fig. S3). ATZ accumulation visualized by periodic acid-Schiff staining after diastase digestion (PAS-D) in PiZ mouse liver is not uniform and typically regions both devoid of and containing PAS-D globules are detected on the same liver tissue section (9). To correlate ATZ accumulation with miR-34b/c levels, we performed laser-controlled microdissection (LCM) on PiZ livers to separate PAS-D–negative from PAS-D–positive regions for qPCR analysis of miR-34b/c expression. Compared with PAS-D–negative regions, PAS-D–positive regions showed increased miR-34b/c (Fig. 2 B and C). Next, we analyzed 6-wk-old PiZ mice injected with a recombinant serotype 8 adeno-associated virus (rAAV8) vector that expresses an artificial miRNA designed to target and down-regulate expression of ATZ (AAV8pCB-mir914-GFP) or injected with the same dose of an AAV8pCB-GFP expressing the green fluorescent protein (GFP) reporter gene (10). By 4 wk postinjection, livers of mice injected with AAV8pCB-mir914-GFP showed a reduction of PAS-D staining and circulating levels of ATZ (11) and decreased miR-34b/c liver content (Fig. 2D). Taken together, these findings revealed a correlation between miR-34b/c levels and ATZ accumulation in the liver.
Fig. 2.
Expression levels of miR-34b/c correlate with Z α1-antitrypsin accumulation (ATZ). (A) Expression of pri-miR-34b/c in parenchymal (Hep) and nonparenchymal cells (NPC) from PiZ mouse livers (n = 3; t test: *P < 0.05). (B) Representative image of microdissected PAS-D+ and PAS-D− areas in PAS-D–stained livers from PiZ mice. (C) qPCR for miR-34b/c on microdissected liver areas with ATZ globule accumulation (PAS-D+, red dotted lines in B) or devoid of ATZ globules (PAS-D−, blue dotted lines in B) showed enrichment of miR-34b/c in ATZ-rich areas (n = 4 per group; paired t test: *P < 0.05 and **P < 0.01 versus PAS-D−). (D) qPCR for miR-34b/c in PiZ livers injected with rAAV-miR914 or control vector showing reduction of miR-34b/c levels in rAAV-miR914–injected mice at 4 wk postinjection (n = 5 per group; t test: *P < 0.05 versus rAAV-GFP). Data are reported as average ± SE.
miR-34b/c Expression Is Up-Regulated by JNK-FOXO3.
Expression of miR-34b/c is directly regulated by Forkhead Box O3 (FOXO3) (12, 13). However, FOXO3 protein levels showed only mild and nonsignificant differences between PiZ and wild-type control mice (SI Appendix, Fig. S4 A and B). Nevertheless, PiZ mice showed increased FOXO3 nuclear signals in hepatocytes by immunohistochemistry (Fig. 3A) that was confirmed by detection of higher FOXO3 levels in the nuclear fractions of PiZ mouse livers compared with wild-type controls, suggesting increased nuclear translocation (Fig. 3B and SI Appendix, Fig. S4C). Consistent with increased nuclear FOXO3, gene set enrichment analysis (GSEA) on RNA-sequencing (RNA-seq) data revealed a significant enrichment of FOXO3 target genes among differentially expressed genes in PiZ livers compared with wild-type controls (enrichment score 0.47) (Fig. 3C and SI Appendix, Fig. S4D).
Fig. 3.
JNK-mediated FOXO3 activation and up-regulation of miR-34b/c in PiZ livers. (A) Representative FOXO3 immunohistochemistry on livers from WT and PiZ mice showing increased nuclear localization of FOXO3. Yellow arrowheads point to FOXO3-positive nuclei. Square, central vein; triangle, portal vein (n = 3 per group; magnification: Left, 20×; Middle and Right, 40×). (Scale bars, 100 μm.) (B) Western blot on liver nuclear extracts showing increased FOXO3 in PiZ compared with WT livers. RAD50 is used for nuclear protein normalization; GAPDH is shown as control for purity of nuclear extracts. CYT, cytoplasmic fraction. (C) Enrichment plot from GSEA including FOXO3 target genes showing enrichment in PiZ mice versus WT livers. (D) Western blot on whole-liver extracts showing increased phospho-Ser574-FOXO3 in PiZ compared with WT mice and reduced levels of phosphorylated FOXO3 in PiZ/Jnk1−/− mice compared with PiZ. (E) Western blot on liver nuclear extracts showing decreased FOXO3 in PiZ/Jnk1−/− nuclei compared with PiZ livers. H3 is used for nuclear protein normalization; GAPDH is shown as control for purity of nuclear extracts. (F) qPCR for miR-3b/c showing significant down-regulation of miR-34b/c levels in livers of PiZ/Jnk1−/− compared with PiZ mice (n = 5 per group; one-way ANOVA and Tukey’s post hoc test: **P < 0.01 versus PiZ). Data are reported as average ± SE.
FOXO3 is activated by JNK phosphorylation on residue Ser574 (14) and c-Jun N-terminal kinase (JNK) is activated in PiZ livers (15). PiZ mice showed increased levels of phospho-Ser574-FOXO3 whereas PiZ/Jnk1−/− mice (15) showed levels of phosphorylated FOXO3 similar to wild-type controls (Fig. 3D and SI Appendix, Fig. S4E), despite no significant changes in Foxo3 gene expression (SI Appendix, Fig. S4F), and reduced nuclear FOXO3 compared with PiZ (Fig. 3E and SI Appendix, Fig. S4G). Moreover, PiZ/Jnk1−/− livers showed reduced levels of miR-34b/c compared with PiZ control (Fig. 3F). Taken together, these results suggest that JNK-dependent activation of FOXO3 drives miR-34b/c expression in PiZ livers.
Lack of miR-34b/c Accelerates the Development of Liver Fibrosis in PiZ Mice.
To investigate the role of miR-34b/c in ATZ-mediated liver disease, we generated PiZ/miR-34b/c−/− by crossing PiZ mice with miR-34b/c−/− mice (16). PiZ/miR-34b/c−/− mice showed normal fertility and sex ratio. At 13 to 15 wk of age, PiZ/miR-34b/c+/+, PiZ/miR-34b/c+/−, and PiZ/miR-34b/c−/− livers showed similar ATZ accumulation by PAS-D staining (Fig. 4 A, Left). Circulating alanine aminotransferase (ALT) levels were only mildly increased in PiZ/miR-34b/c+/+ and PiZ/miR-34b/c−/− compared with controls and not significantly different between PiZ/miR-34b/c−/− and PiZ/miR-34b/c+/+ mice (SI Appendix, Fig. S5). Nevertheless, livers of PiZ/miR-34b/c−/− and PiZ/miR-34b/c+/− mice developed more severe fibrosis than PiZ/miR-34b/c+/+, as shown by Sirius red staining (Fig. 4 A, Right). Quantification of Sirius red staining and measurement of hepatic hydroxyproline content confirmed a significant increase in liver fibrosis in PiZ/miR-34b/c−/− compared with PiZ/miR-34b/c+/+, whereas PiZ/miR-34b/c+/− showed intermediate levels of fibrosis between those of PiZ/miR-34b/c−/− and PiZ/miR-34b/c+/+ (Fig. 4 B and C).
Fig. 4.
Deletion of miR-34b/c resulted in early development of liver fibrosis in PiZ mice. (A) Representative PAS-D (left panels) and Sirius red (right panels) stainings of livers from WT, miR-34b/c−/−, and PiZ/miR-34b/c+/+ as controls, PiZ/miR-34b/c+/− and PiZ/miR-34b/c−/− showing similar ATZ accumulation by PAS-D in PiZ/miR-34b/c+/+, PiZ/miR-34b/c+/−, and PiZ/miR-34b/c−/−, and increased fibrosis in PiZ/miR-34b/c+/− and PiZ/miR-34b/c−/− compared with controls. Scale bar, 100 µm. (B) Quantification of percentages of Sirius red (SR)-positive areas showing increased stained area in PiZ/miR-34b/c−/− compared with controls (n = 5 image per animal, n = 3 to 11 animals per group; one-way ANOVA and Tukey’s post hoc test: **P < 0.01). (C) Hydroxyproline (HYP) determination on liver lysates showing increased HYP content in PiZ/miR-34b/c−/− compared with controls (n = 5 to 11 per group; one-way ANOVA and Tukey’s post hoc test: *P < 0.05). (D) Top five up-regulated clustered cellular processes (Upper) and biological components (Lower) from Gene Ontology analysis on differentially expressed genes in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ livers. (E) Expression of liver fibrosis gene signature in WT, miR-34b/c−/−, PiZ/miR-34b/c+/+, and PiZ/miR-34b/c−/−. Each column represents the average gene expression levels of n = 5 mice per group. Data are reported as average ± SE.
RNA-seq analysis showed that hepatic gene expression in PiZ/miR-34b/c−/− mice was well-clustered and separated from PiZ/miR-34b/c+/+, miR-34b/c−/−, and wild-type control mice (SI Appendix, Fig. S6). Analysis of differentially expressed genes in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ yielded 1,580 dysregulated genes (802 up- and 778 down-regulated). Functional annotation clustering analysis showed that a significant number of the differentially expressed genes are associated with biological processes of extracellular matrix components, including collagens, tissue damage, and regeneration (i.e., cell death and proliferation, angiogenesis, cell migration) (Fig. 4D and Datasets S1–S3). Moreover, GSEA using a liver fibrosis expression signature (17, 18) showed significant enrichments of fibrosis genes among the up-regulated genes in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ (Fig. 4E, SI Appendix, Fig. S7, and Dataset S4). Taken together, these findings support a protective role of miR-34b/c in the development of liver fibrosis in PiZ mice.
Activation of the Platelet-Derived Growth Factor Pathway in PiZ Mice Lacking miR-34b/c.
To investigate the antifibrotic mechanism of miR-34b/c in livers expressing ATZ, we searched for genes up-regulated in miR-34b/c−/− livers with fibrosis. By Venn diagram, we compared genes differentially expressed in miR-34b/c−/− versus wild-type and in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ livers. Next, we isolated genes that were differentially expressed in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ but not in miR-34b/c−/− versus wild-type livers (SI Appendix, Fig. S8). Within this subset of 1,418 genes, 58 up-regulated genes were miR-34b/c putative target genes (SI Appendix, Table S1). Among these genes, Pdgfra and Pdgfrb, encoding the α- and β-subunits of the platelet-derived growth factor receptor (PDGFR), respectively, were the most interesting because of the consolidated role of the PDGF pathway in liver fibrosis. PDGFA to PDGFD ligands are potent mitogens that drive hepatic stellate cell proliferation and differentiation into myofibroblasts through activation of tyrosine kinase PDGFR (19). Down-regulation of PDGFRα or PDGFRβ encoded by Pdgfra and Pdgfrb, respectively, exerts a protective role against liver fibrosis, while their overexpression is profibrotic (20–22). Interestingly, the Pdgfra 3′ untranslated region (UTR) includes two putative canonical target sites (one 8-mer and one 7-mer) for miR-34b/c. The 3′ UTR of Pdgfrb has six canonical binding sites for miR-34b/c (one 8-mer and five 6-mers) and one 8-mer site in the coding sequence. Moreover, PDGFRA and PDGFRB targeting by the miR-34 family has been validated in humans (23). Targeting by miR-34b/c was validated by luciferase assay on wild-type and mutagenized 3′ UTR from Pdgfra (Fig. 5 A and B) and Pdgfrb (Fig. 5 C and D). Up-regulation of Pdgfra and Pdgfrb was confirmed at the protein level in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ (Fig. 5E and SI Appendix, Fig. S9). Moreover, livers from PiZ/miR-34b/c−/− showed increased levels of phospho-Tyr849/857-PDGFRα/β and increased phosphorylation of its targets Janus Kinase 1 (JAK1) and AKT, consistent with released miR-34b/c repression of PDGFRα/β and activation of its downstream targets (Fig. 5E and SI Appendix, Fig. S9). Taken together, these data show that lack of miR-34b/c results in up-regulation of PDGFRα/β and activation of the PDGF pathway, thus suggesting that miR-34b/c antagonize liver fibrosis through repression of the PDGF pathway.
Fig. 5.
Increased PDGF signaling in PiZ/miR-34b/c−/− livers. Schematic representation of miR-34/c binding to 8-mer recognition sites in Pdgfra (A) and Pdgfrb (B) 3′ UTRs. miR-34b/c seed sequence pairings are depicted in blue and other base pairings are in red. Nucleotides that have been mutated for luciferase assays are indicated by asterisks. Luciferase activity assay on HeLa cells transfected with negative control (NC), miR-34b, or miR-34c mimic and with luciferase-expressing plasmids carrying WT or mutated (mut) Pdgfra (C) and Pdgfrb (D) 3′ UTR (n = 6 per group; one-way ANOVA and Tukey’s post hoc test: **P < 0.01 and ***P < 0.005). (E) Western blotting of the PDGF pathway on whole-liver extracts showing increased levels of miR-34b/c target genes PDGFRα/β, activation of PDGFRα/β, and phosphorylation of PDGFR target proteins JAK1 and AKT in PiZ/miR-34b/c−/− versus PiZ/miR-34b/c+/+ mice. Data are reported as average ± SE.
miR-34c Is Increased in Human Livers Expressing ATZ.
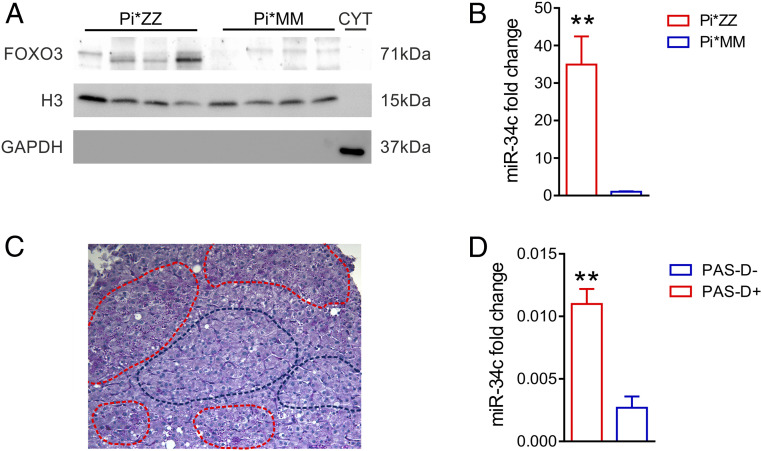
To investigate the clinical relevance of our findings, we analyzed FOXO3 and miR-34b/c in liver samples from patients with AAT deficiency. Because miR-34b and miR-34c are both expressed from the same primary transcript in humans (7), we analyzed human miR-34c levels as a proxy for miR-34b/c expression. Similar to PiZ mice, AAT-deficient patients with advanced hepatic disease requiring liver transplantation (11) showed increased FOXO3 nuclear levels and significant miR-34c up-regulation compared with control livers from patients who underwent liver transplantation for unrelated liver disorders (Fig. 6 A and B and SI Appendix, Fig. S10A). It is noteworthy that livers of patients with AAT deficiency were previously found to have activation of JNK (15). Moreover, liver specimens from four independent patients with milder liver disease (SI Appendix, Table S2) showed increased phospho-Ser574-FOXO3 levels, compared with controls with unrelated liver disease (SI Appendix, Fig. S10 B and C) and a trend toward up-regulation of miR-34c compared with controls (SI Appendix, Fig. S10D). Consistent with the data in PiZ livers, miR-34c was up-regulated in PAS-D–positive liver areas compared with globule-devoid areas obtained by LCM (Fig. 6 C and D).
Fig. 6.
FOXO3 activation and miR-34c up-regulation in livers of patients with AAT deficiency. (A) Western blot of liver nuclear extracts from AAT-deficient patients who underwent liver transplantation (Pi*ZZ) compared with control liver samples from patients undergoing liver transplantation for unrelated liver causes (Pi*MM) showing increased nuclear FOXO3 in livers from Pi*ZZ subjects. (B) Expression of miR-34c by qPCR is significantly increased in liver samples from Pi*ZZ subjects (Pi*ZZ, n = 5) compared with control liver samples (Pi*MM, n = 4) (t test: **P < 0.01). (C) Representative image of laser-microdissected PAS-D+ (red dotted lines) and PAS-D− (blue dotted lines) areas in PAS-D–stained liver from a Pi*ZZ subject. (D) qPCR for miR-34c on laser-microdissected liver areas from a single AAT-deficient patient showing enrichment in ATZ-accumulating liver areas (PAS-D+) versus liver areas devoid of ATZ globules (PAS-D−) (t test: **P < 0.01). Data are reported as average ± SE.
The JNK/FOXO3/miR-34b/c Pathway Is Activated in Liver Fibrosis of Different Etiologies.
We hypothesized that miR-34b/c and its upstream regulators JNK and FOXO3 may be involved in other forms of liver fibrosis besides AAT deficiency. To investigate this hypothesis, we evaluated JNK, FOXO3, and miR-34b/c in various models of liver fibrosis, including biliary fibrosis in Abcb4−/− mice (24) and in mice with bile duct ligation (25), and pharmacologically induced pan-lobular fibrosis in mice administered carbon tetrachloride (CCl4) or thioacetamide (TAA) (26). Consistent with previous studies (27, 28), JNK activation and concomitant increased phospho-Ser574-FOXO3 were detected in fibrotic livers compared with controls (Fig. 7 A, C, E, and G and SI Appendix, Fig. S11). Moreover, miR-34b/c was up-regulated in fibrotic livers compared with controls (Fig. 7 B, D, F, and H), confirming activation of the JNK/FOXO3/miR-34b/c pathway in liver fibrosis induced by various etiologies.
Fig. 7.
JNK/FOXO3/miR-34b/c pathway is activated in liver fibrosis. Western blot analysis for total and phosphorylated JNK and FOXO3, and qPCR analysis for miR-34b/c on whole-liver extracts from Abcb4−/− mice (A and B), mice with bile duct ligation (BDL) (C and D), and mice treated with thioacetamide (TAA) (E and F) or carbon tetrachloride (CCl4) (G and H) versus vehicle-treated controls (n = 4 for each group; t test: *P < 0.05 and ***P < 0.005). Data are reported as average ± SE.
Discussion
In the present study, we found JNK phosphorylation on Ser574, FOXO3 activation, and miR-34b/c up-regulation in murine and human livers with ATZ accumulation. PiZ mice deleted for miR-34b/c showed greater liver fibrosis and increased signaling of PDGF, a profibrotic pathway that is a target of miR-34b/c. Interestingly, JNK-activated FOXO3 and miR-34b/c up-regulation were also found in several mouse models of liver fibrosis, suggesting that this pathway is broadly implicated in hepatic diseases.
JNK signaling is associated with cell death, survival, differentiation, proliferation, and tumorigenesis in hepatocytes. Moreover, it is involved in inflammation and fibrosis (29). JNK is activated in livers expressing ATZ (15) and can phosphorylate FOXO3, thus promoting its nuclear translocation (30). As previously observed with hepatitis C virus infection (14), JNK can phosphorylate FOXO3 on the Ser574 residue in livers expressing ATZ. Ser574 phosphorylation drives FOXO3-dependent apoptosis (14) and, consistently, a correlation between apoptosis and the amount of ATZ has been previously shown (31). Previous studies have reported that FOXO3 binds to the miR-34b/c promoter up-regulating its expression (12, 13) and, accordingly, we found that up-regulation of miR-34b/c in PiZ mice was dependent on FOXO3 activation by JNK. Interestingly, our findings suggest an antifibrotic role for FOXO3-mediated up-regulation of miR-34b/c in livers expressing ATZ, as well as in livers of mice subjected to various types of injuries resulting in fibrosis. Up-regulation of miR-34 family members was previously found in animal models of pharmacologically induced liver fibrosis and miR-34b up-regulation was associated with fibrosis due to viral hepatitis in humans (32). While a growing body of evidence supports a profibrotic role for miR-34a (33–36), the role of miR-34b/c has been less clear and both antifibrotic (37, 38) and profibrotic activities have been observed (33). However, most of these studies were performed in vitro without coculturing of hepatocytes with hepatic stellate cells. In contrast, we evaluated the consequences of miR-34b/c deletion in vivo in the PiZ mouse model that spontaneously develops liver fibrosis by 16 to 24 wk of age (39).
The findings of this study also suggested that miR-34b/c reduces liver fibrosis by repressing PDGF signaling. Activation of the PDGF pathway mainly occurs in hepatic stellate cells and portal fibroblasts, promoting their proliferation and transdifferentiation toward myofibroblasts, which drive the development and progression of fibrosis (19). We detected FOXO3 activation and miR-34b/c expression mainly in hepatocytes. Therefore, it could be argued that repression of PDGF signaling by miR-34b/c in hepatocytes protects from liver fibrosis. However, miRNAs can be secreted and the paracrine activity of secreted miR-34b/c on other liver cell types cannot be excluded. Accordingly, miR-34b/c levels were increased in PiZ plasma, supporting their secretion by hepatocytes. On the other hand, PDGFRα expression is induced in damaged hepatocytes and hepatocyte-restricted deletion of PDGFRα decreased liver fibrosis (40), supporting a hepatocyte-specific antifibrotic effect of miR-34b/c. Nevertheless, the involvement of additional genes unrelated to the PDGF pathway that are still targets of miR-34b/c cannot be excluded.
Interestingly, FOXO3 plays a crucial role in fibrogenesis occurring in idiopathic pulmonary fibrosis, a lethal, progressive fibrosing parenchymal lung disease (41). Therefore, the results of the present studies raise the attractive hypothesis that miR-34b/c up-regulation might also be involved in lung fibrosis.
Liver fibrosis is not very common in young individuals who are homozygotes for the Z allele of SERPINA1, but its incidence increases significantly with age. According to recent studies, clinically relevant fibrosis occurs in 20 to 35% of adult Pi*ZZ and the degree of fibrosis correlates with the mutant protein burden (42, 43). The mechanisms underlying the variability in liver fibrosis development among homozygotes for the Z allele are unknown. Polymorphisms in the promoter region affecting miR-34b/c levels (44, 45) might protect or increase individuals’ susceptibility to develop liver fibrosis. The heterozygote Z allele has recently emerged as the strongest single-nucleotide polymorphism-based risk factor for cirrhosis in nonalcoholic fatty liver disease and alcohol misuse (46). Therefore, it can be speculated that miR-34b/c polymorphisms might increase the risk of liver fibrosis also in individuals who are heterozygotes for the Z allele.
In conclusion, we identified miR‐34b/c up-regulation and JNK-FOXO3 activation in livers injured by expression of ATZ or other types of insults resulting in fibrosis. In PiZ mice, lack of miR-34b/c resulted in more severe liver fibrosis, likely as a consequence of increased PDGF signaling. Taken together, these results unravel an important pathway implicated in pathogenesis of liver diseases. Fibrosis is a major global health problem and the elucidation of its pathogenic mechanism and hence of key therapeutic targets is a research priority (47). Elucidation of molecular mechanisms underlying liver fibrosis is fundamentally important for establishing antifibrotic therapies (48) and this study revealed a pathway that might become a target for therapeutic interventions.
Materials and Methods
Mouse Studies.
Mouse procedures were approved by the Italian Ministry of Health. Male 4- to 15-wk-old C57BL/6 (Charles River Laboratories), PiZ (6), PiZ/Jnk1−/−, miR-34b/c−/− (16), and PiZ/miR-34b/c−/−, Abcb4−/− (49) mice were used. Synthetic miR-914 has been described elsewhere (10). rAAV8pCB-mir914-GFP and rAAV8pCB-GFP were generated, purified, and titered by the UMass Gene Therapy Vector Core as previously described (50). Bile duct ligation was performed in C57BL/6 mice as previously described (26) and mice were killed 1 wk postsurgery. TAA (Sigma-Aldrich) was dissolved in phosphate-buffered saline (PBS) and administered to C57BL/6 mice by intraperitoneal injection three times a week for 4 wk with escalating doses, starting from 50 to 200 mg/kg, as previously described (27). CCl4 (Sigma-Aldrich) was dissolved in corn oil (Sigma-Aldrich) and administered to C57BL/6 mice by gavage three times a week for 4 wk with escalating doses, starting from 0.875 to 2.5 mL/kg, as previously described (27). When killed, mice were perfused with PBS. ALT levels were measured by scil VitroVet analyzer (Scil vet).
Human Liver and Serum Samples.
Human liver samples were collected, snap-frozen, and anonymized according to human study approvals obtained from the Institute of Pathology, University Hospital of Basel and Cardinal Glennon Children’s Medical Center, St. Louis University School of Medicine. Liver specimens from patients with milder liver disease and AAT deficiency confirmed on pathology and the corresponding controls were anonymously obtained from the Institute of Pathology, University Hospital of Basel (SI Appendix, Table S2). Liver samples with severe disease were anonymously obtained from Cardinal Glennon Children’s Medical Center, St. Louis University School of Medicine from patients under 18 y of age homozygous for the Z allele of SERPINA1 who underwent hepatic transplantation for liver failure. Control liver specimens were obtained from age-matched patients homozygous for the wild-type allele of AAT who also underwent liver transplantation. Pathology features of these specimens were previously reported (11).
miRNA Analyses.
Small RNA libraries were constructed using a TruSeq Small RNA Sample Preparation Kit (Illumina) following the manufacturer’s protocol. Methods are described in greater detail in SI Appendix. The data were deposited in GEO with accession number GSE85413.
For targeted miRNA expression analysis in mice and in human livers with mild liver disease, miRNA-enriched total RNA was extracted from liver tissues using the miRNeasy Mini Kit and from plasma using the miRNeasy Serum/Plasma Kit (Qiagen). Ten to 20 ng of total RNA was reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit and TaqMan miRNA assay (SI Appendix, Table S3) (Applied Biosystems). qPCR was performed in triplicate using 1 to 3 μL complementary DNA (cDNA), the TaqMan MicroRNA assay, and TaqMan Universal Master Mix II no UNG (Applied Biosystems) on a Light Cycler 480 System (Roche). The running program was as follows: preheating, 10 min at 95 °C; 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Small nucleolar RNA234, miR-23a, and miR-152 were used as housekeeping for mouse livers and plasma and human livers, respectively. For pri-miR-34b/c analysis, isolation of hepatocytes and nonparenchymal liver cells from PiZ mouse livers was performed as previously described (11). Two micrograms of total RNA was retrotranscribed using the High Capacity cDNA Reverse Transcription Kit according to the manufacturer’s protocol (Applied Biosystems). qPCR was performed using the TaqMan pri-miRNA assay (SI Appendix, Table S3) as for mature miRNAs. 18S was used as housekeeping. Data analysis was performed using LightCycler 480 software version 1.5 (Roche).
To validate Pdgfra and Pdgfrb as targets of miR-34b/c, murine Pdgfra and Pdgfrb 3′ UTRs were amplified by PCR from genomic DNA of C57BL/6 wild-type mice and cloned downstream of the firefly luciferase gene into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). The miR-34b/c 8-mer recognition sites in the pmirGLO Pdgfra.3′ UTR and pmirGLO Pdgfrb.3′ UTR plasmids were mutagenized by the QuikChange Site‐Directed Mutagenesis Kit (Agilent) according to the manufacturer’s instructions. Primers used for construct generation are shown in SI Appendix, Table S4. HeLa cells were cultured in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum and 5% penicillin/streptomycin. Cells were cotransfected with the plasmid containing the wild-type or mutagenized pmirGLO Pdgfra.3′ UTR and pmirGLO Pdgfrb.3′ UTR and with negative control, miRIDIAN Mimic miR-34b-5p, or miRIDIAN Mimic miR-34c-5p using Interferin transfection reagent (Polyplus). Cells were harvested 48 h after transfection and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Data were expressed relative to renilla luciferase activity to normalize for transfection efficiency. Two sets of experiments were performed with n = 3 in each experiment. The firefly-to-renilla activity ratio for each replicate was normalized to the average of negative control-transfected samples.
For analyses of miRNA in human livers with end-stage liver disease, total RNA was isolated from liver tissue with TRIzol (Life Technologies) and diluted to 10 ng/µL. miR-34c and U47 small RNAs were jointly retrotranscribed using small RNA-specific stem-loop primers (Life Technologies) and the TaqMan miRNA Reverse Transcription Kit (Life Technologies). Droplets were generated using a QX-200 Droplet Generator (Bio-Rad) and end-point droplet digital PCR (ddPCR) was performed using small RNA-specific primers and FAM-labeled probes (Life Technologies) and 2× ddPCR Supermix for Probes no dUTP (Bio-Rad). Positive and negative droplets were quantified (Bio-Rad). Only samples with at least 10,000 accepted droplets and at least 100 negative droplets were included in the data analysis. Samples were run in triplicate, the replicates were averaged, and the ratio of miR-34c–positive to U47-positive droplets was calculated.
Liver Staining, Laser-Controlled Microdissection, and Western Blotting.
Livers from PBS-perfused mice were fixed in 4% paraformaldehyde for 12 h, stored in 70% ethanol, and embedded in paraffin blocks. PAS-D staining was performed on 5-μm-thick paraffin sections of livers. Sections were rehydrated and treated with 0.5% α-amylase type VI-B (Sigma-Aldrich) for 20 min and stained with PAS reagent according to the manufacturer’s instructions (Bio-Optica). Methods for Sirius red staining and immunohistochemistry are provided in SI Appendix.
For laser-controlled microdissection, 10-μm sections from formalin-fixed paraffin-embedded PiZ livers were quickly rehydrated (xylene 2 min twice, 100% ethanol 1 min, 95% ethanol 1 min, 75% ethanol 1 min), stained with Mayer’s hematoxylin (Bio-Optica) and eosin Y (Sigma), and quickly dehydrated (75% ethanol 1 min, 95% ethanol 1 min, 100% ethanol 1 min). Solutions were all prepared in diethyl pyrocarbonate-treated water and kept at 4 °C to minimize RNA degradation. Dried sections underwent laser-controlled microdissection using PALM Microbeam (Zeiss). A total area of 5 × 105 μm2 for each sample was dissected. Serial PAS-D–stained sections were used to identify areas for dissection. Total RNA was extracted using the RNeasy FFPE Kit (Qiagen) according to the manufacturer’s protocol.
For Western blotting, proteins from tissues were extracted in RIPA buffer according to standard procedures. Nuclear protein extracts were prepared using the CelLytic NuCLEAR Extraction Kit (Sigma-Aldrich). Primary antibodies were diluted in TBS-T (0.8% NaCl, 0.02% KCl, 0.3% Tris-base-0.1% Tween 20)/5% milk (Bio-Rad) (SI Appendix, Table S5). Secondary antibodies were enhanced chemiluminescence (ECL) anti-rabbit horseradish peroxidase (HRP) and ECL anti-mouse HRP (GE Healthcare). Peroxidase substrate was provided by the ECL Western Blotting Substrate Kit (Pierce). Analysis of band intensities was performed using Quantity One 1-D Analysis software version 4.6.7 (Bio-Rad).
RNA-Seq and GSEA.
Library preparation was performed with a total of 100 ng of RNA from each sample using the QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen) according to the manufacturer’s instructions. Detailed information is provided in SI Appendix. Data were deposited in the Gene Expression Omnibus (GEO) with accession number GSE141593. Methods for identification of putative miR-34b/c target genes and GSEA are provided in SI Appendix.
Statistical Analyses.
Two-tailed Student’s t test and ANOVA plus Tukey’s honestly significant difference post hoc were used as statistical tests for mean comparisons. Experimental group sizes are reported in the figure legends. Data are reported as average ± SE.
Supplementary Material
Acknowledgments
We are grateful to Milton Finegold and Sandro Banfi for helpful discussions and critical review of the manuscript. We thank the Telethon Institute of Genetics and Medicine (TIGEM) Next-Generation Sequencing core for miRNA sequencing, and Margherita Mutarelli from the TIGEM Bioinformatics core for miRNA-sequencing analysis. We are grateful to Steven Weinman for providing the anti–phospho-Ser574-FOXO3 antibody and to Antonella De Matteis for the anti-P115 antibody. We acknowledge the University of Massachusetts Medical School Center for Clinical and Translational Science (UMCCTS Grant UL1TR001453). This work has been supported by the Alpha-1 Foundation (Gordon L. Snider Award 2016 and Research Grant 2018 to P.P. and Research Grant 2019 to N.B.-P.) and the “Federico II” University of Naples (STAR Program to P.P.). R.F. is a recipient of a “Prof. Mario Coppo” Fellowship Award. S.A. has been supported by an Alpha-1 Antitrypsin Laurell’s Training Award.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025242118/-/DCSupplemental.
Data Availability
miRNA-seq and RNA-seq data are available in GEO with the accession numbers GSE85413 and GSE141593, respectively. All study data are included in the article and/or supporting information.
References
- 1.Sveger T., The natural history of liver disease in alpha 1-antitrypsin deficient children. Acta Paediatr. Scand. 77, 847–851 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Piitulainen E., Carlson J., Ohlsson K., Sveger T., Alpha1-antitrypsin deficiency in 26-year-old subjects: Lung, liver, and protease/protease inhibitor studies. Chest 128, 2076–2081 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Eriksson S., Carlson J., Velez R., Risk of cirrhosis and primary liver cancer in alpha 1-antitrypsin deficiency. N. Engl. J. Med. 314, 736–739 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Szabo G., Bala S., MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 10, 542–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson J. A., et al., Accumulation of PiZ alpha 1-antitrypsin causes liver damage in transgenic mice. J. Clin. Invest. 83, 1183–1190 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson J. A., et al., Multiple tissues express alpha 1-antitrypsin in transgenic mice and man. J. Clin. Invest. 82, 26–36 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L., et al., A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134, 10.1038/nature05939 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirschner M. B., et al., Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 6, e24145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geller S. A., Nichols W. S., Dycaico M. J., Felts K. A., Sorge J. A., Histopathology of alpha 1-antitrypsin liver disease in a transgenic mouse model. Hepatology 12, 40–47 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Mueller C., et al., Sustained miRNA-mediated knockdown of mutant AAT with simultaneous augmentation of wild-type AAT has minimal effect on global liver miRNA profiles. Mol. Ther. 20, 590–600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attanasio S., et al., CHOP and c-JUN up-regulate the mutant Z α1-antitrypsin, exacerbating its aggregation and liver proteotoxicity. J. Biol. Chem. 295, 13213–13223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kress T. R., et al., The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell 41, 445–457 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Masui K., et al., mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 18, 726–739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tikhanovich I., et al., Regulation of FOXO3 by phosphorylation and methylation in hepatitis C virus infection and alcohol exposure. Hepatology 59, 58–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastore N., et al., Activation of the c-Jun N-terminal kinase pathway aggravates proteotoxicity of hepatic mutant Z alpha1-antitrypsin. Hepatology 65, 1865–1874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Concepcion C. P., et al., Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 8, e1002797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Koppen A., et al., Uncovering a predictive molecular signature for the onset of NASH-related fibrosis in a translational NASH mouse model. Cell. Mol. Gastroenterol. Hepatol. 5, 83–98.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau-Corona D., Bae W. K., Hennighausen L., Waxman D. J., Sex-biased genetic programs in liver metabolism and liver fibrosis are controlled by EZH1 and EZH2. PLoS Genet. 16, e1008796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borkham-Kamphorst E., Weiskirchen R., The PDGF system and its antagonists in liver fibrosis. Cytokine Growth Factor Rev. 28, 53–61 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Kocabayoglu P., et al., β-PDGF receptor expressed by hepatic stellate cells regulates fibrosis in murine liver injury, but not carcinogenesis. J. Hepatol. 63, 141–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi A., et al., Platelet-derived growth factor receptor α contributes to human hepatic stellate cell proliferation and migration. Am. J. Pathol. 187, 2273–2287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell J. S., et al., Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 102, 3389–3394 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garofalo M., et al., miR-34a/c-dependent PDGFR-α/β downregulation inhibits tumorigenesis and enhances TRAIL-induced apoptosis in lung cancer. PLoS One 8, e67581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauad T. H., et al., Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am. J. Pathol. 145, 1237–1245 (1994). [PMC free article] [PubMed] [Google Scholar]
- 25.Tag C. G., et al., Bile duct ligation in mice: Induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J. Vis. Exp. (96), 52438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y. O., Popov Y., Schuppan D., Optimized mouse models for liver fibrosis. Methods Mol. Biol. 1559, 279–296 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Kluwe J., et al., Modulation of hepatic fibrosis by c-Jun-N-terminal kinase inhibition. Gastroenterology 138, 347–359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwak B. J., et al., The role of phospho-c-Jun N-terminal kinase expression on hepatocyte necrosis and autophagy in the cholestatic liver. J. Surg. Res. 241, 254–263 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Seki E., Brenner D. A., Karin M., A liver full of JNK: Signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 143, 307–320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho K. K., et al., Phosphorylation of FOXO3a on Ser-7 by p38 promotes its nuclear localization in response to doxorubicin. J. Biol. Chem. 287, 1545–1555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindblad D., Blomenkamp K., Teckman J., Alpha-1-antitrypsin mutant Z protein content in individual hepatocytes correlates with cell death in a mouse model. Hepatology 46, 1228–1235 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Singh A. K., et al., Global microRNA expression profiling in the liver biopsies of hepatitis B virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology 67, 1695–1709 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Li X., et al., MicroRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor γ. Mol. Med. Rep. 11, 1017–1024 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Tian X. F., Ji F. J., Zang H. L., Cao H., Activation of the miR-34a/SIRT1/p53 signaling pathway contributes to the progress of liver fibrosis via inducing apoptosis in hepatocytes but not in HSCs. PLoS One 11, e0158657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan Y., et al., Regulation of cellular senescence by miR-34a in alcoholic liver injury. Am. J. Pathol. 187, 2788–2798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan G., et al., MicroRNA-34a promotes hepatic stellate cell activation via targeting ACSL1. Med. Sci. Monit. 21, 3008–3015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F., Lu Y., Zheng S., Peroxisome proliferator-activated receptor-γ cross-regulation of signaling events implicated in liver fibrogenesis. Cell. Signal. 24, 596–605 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Chen L., et al., Therapeutic effects of serum extracellular vesicles in liver fibrosis. J. Extracell. Vesicles 7, 1461505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunt E. M., et al., Hepatic progenitor cell proliferation and liver injury in α-1-antitrypsin deficiency. J. Pediatr. Gastroenterol. Nutr. 51, 626–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim B. J., et al., Selective deletion of hepatocyte platelet-derived growth factor receptor α and development of liver fibrosis in mice. Cell Commun. Signal. 16, 93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Tamari H. M., et al., FoxO3 an important player in fibrogenesis and therapeutic target for idiopathic pulmonary fibrosis. EMBO Mol. Med. 10, 276–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark V. C., et al., Clinical and histologic features of adults with alpha-1 antitrypsin deficiency in a non-cirrhotic cohort. J. Hepatol. 69, 1357–1364 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Hamesch K., et al., Liver fibrosis and metabolic alterations in adults with alpha-1-antitrypsin deficiency caused by the Pi*ZZ mutation. Gastroenterology 157, 705–719.e18 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Xu Y., et al., A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int. J. Cancer 128, 412–417 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Son M. S., et al., Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene 524, 156–160 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Strnad P., et al., Heterozygous carriage of the alpha1-antitrypsin Pi*Z variant increases the risk to develop liver cirrhosis. Gut 68, 1099–1107 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Henderson N. C., Rieder F., Wynn T. A., Fibrosis: From mechanisms to medicines. Nature 587, 555–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trautwein C., Friedman S. L., Schuppan D., Pinzani M., Hepatic fibrosis: Concept to treatment. J. Hepatol. 62 (suppl. 1), S15–S24 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Smit J. J., et al., Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 75, 451–462 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Gao G. P., et al., Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U.S.A. 99, 11854–11859 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
miRNA-seq and RNA-seq data are available in GEO with the accession numbers GSE85413 and GSE141593, respectively. All study data are included in the article and/or supporting information.