Significance
Genetically modified animals are crucial for functional studies and translational biomedical research. However, the generation of genome-edited large animals is time consuming and inefficient. Here, we report the generation of transgenic pigs and chickens that ubiquitously express Cas9 nuclease. The functionality of Cas9 was demonstrated for different target genes, a variety of cell types, and in vivo for the heart and the developing brain. Genome editing can now easily be carried out in vivo in specific organs or tissues without the need to generate germline-modified animals. These Cas9 transgenic animals provide an innovative and efficient resource for in vivo genome editing in biomedical and agricultural sciences.
Keywords: Cas9 transgenic chicken, Cas9 transgenic pig, in vivo genome editing, CRISPR-Cas9
Abstract
Genetically modified animals continue to provide important insights into the molecular basis of health and disease. Research has focused mostly on genetically modified mice, although other species like pigs resemble the human physiology more closely. In addition, cross-species comparisons with phylogenetically distant species such as chickens provide powerful insights into fundamental biological and biomedical processes. One of the most versatile genetic methods applicable across species is CRISPR-Cas9. Here, we report the generation of transgenic chickens and pigs that constitutively express Cas9 in all organs. These animals are healthy and fertile. Functionality of Cas9 was confirmed in both species for a number of different target genes, for a variety of cell types and in vivo by targeted gene disruption in lymphocytes and the developing brain, and by precise excision of a 12.7-kb DNA fragment in the heart. The Cas9 transgenic animals will provide a powerful resource for in vivo genome editing for both agricultural and translational biomedical research, and will facilitate reverse genetics as well as cross-species comparisons.
Chickens and pigs are the most important livestock species worldwide. They are not only important sources of food, but also valuable models for evolutionary biology and biomedical science. Pigs share a high anatomical and physiological similarity with humans and are an important species for translational biomedical research, for example, in the areas of cancer, diabetes, neurodegenerative, and cardiovascular diseases (1–3). They also resemble the human pathophenotype more closely than rodents. For example, pig models for familial adenomatous polyposis (FAP) develop polyps in the large intestine as observed in human patients (4), whereas mouse FAP models develop them in the small intestine (5). In contrast to mammals, chickens are phylogenetically distant vertebrates from humans, but they were instrumental in the field of developmental biology due to the easy access to the embryonated egg. They are used for studying neurological and cardiovascular functions (6–8) and provided key findings in B cell development and graft versus host responses (9–11). Genetically modified livestock species also hold great promise for agriculture by offering new approaches for disease control, such as genome-edited pigs resistant to Porcine Reproductive and Respiratory Syndrome or Avian Leucosis Virus (ALV)-resistant chickens (12–15).
Due to the lack of fully functional embryonic stem cells, genetic engineering in pigs and chickens has been a laborious, inefficient, and time-consuming procedure (16). The generation of pigs with precise germline modifications required gene targeting in somatic cells followed by somatic cell nuclear transfer. This also is not practical in chickens, where precise alteration of the genome only became possible with recent improvements in the cultivation and manipulation of germline-competent primordial germ cells (PGCs) (17–19). These modified PGCs can be injected into the blood vessel system of stage 13 to 15 (Hamburger−Hamilton [HH]) embryos to produce germline chimeras and, by further breeding, genetically modified chickens.
With the advent of synthetic endonucleases such as CRISPR-Cas9 efficiency of targeted germline modification has improved in both species (20–23). It still requires the generation and breeding of new founder lines, which is time consuming in large animals. To circumvent the need for generating germline-modified animals, attempts have been made to carry out genome editing directly in specific organs or tissues (24–27). But this has been hampered by the need to deliver both Cas9 and the required guide RNA (gRNA) and by the limited cargo capacity of viral vectors. To bypass this drawback, Cas9 transgenic mice have been generated, requiring delivery of only the respective gRNAs (28).
Here, we describe the generation of both Cas9 transgenic pigs and chickens that ubiquitously express Cas9 endonuclease and provide proof of its function in vitro and in vivo. These animals provide an innovative and efficient model for in vivo genome editing to assess gene function in health and disease.
Results
Generation of Cas9 Transgenic Animals.
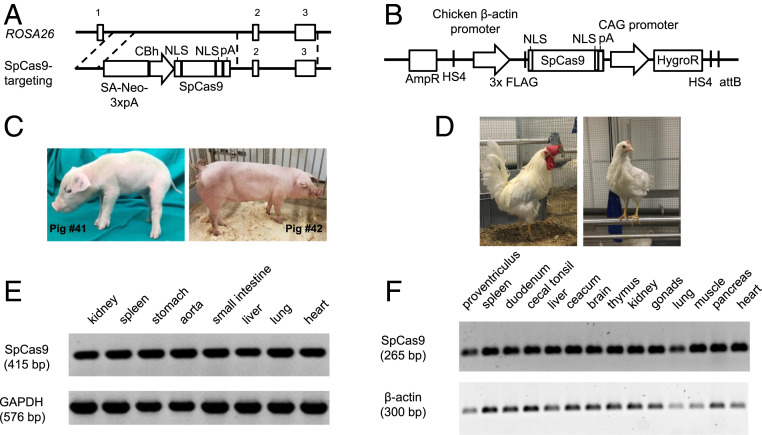
Cas9 transgenic pigs were generated by targeted placement of Streptococcus pyogenes Cas9 (SpCas9) at the ROSA26 locus (Fig. 1A), which was previously shown to support abundant ubiquitous transgene expression in pigs (29, 30). Five percent of G418-resistant cell clones showed correct gene targeting, expressed SpCas9, and were used for somatic cell nuclear transfer resulting in two liveborn piglets (#41 and #42) (Fig. 1C). For both animals, correct gene targeting was confirmed by long-range PCR across the 5′ and 3′ junctions of the targeted allele and DNA sequence analysis (SI Appendix, Fig. S1). Monoallelic insertion of SpCas9 was shown by PCR across the nontargeted ROSA26 wild-type allele (SI Appendix, Fig. S1). Sanger sequencing of the PCR product confirmed sequence integrity of the ROSA26 wild-type allele. Pig #42 served as a founder animal of an SpCas9 transgenic herd.
Fig. 1.
Generation and expression analyses of SpCas9 transgenic pigs and chickens. (A) Structure of the ROSA26-SpCas9 targeting vector and targeting strategy to introduce SpCas9 gene into the porcine ROSA26 locus. Exons are indicated by numbered boxes, and regions of homology are indicated by dotted lines. (B) Expression vector used for the generation of SpCas9-expressing PGCs. (C) SpCas9 transgenic founder pigs. (D) SpCas9 transgenic rooster and hen. (E) RT-PCR analyses of SpCas9 transgenic pig organs. SpCas9 expression is shown by a 415-bp PCR product, and porcine GAPDH (576 bp) serves as a control. (F) RT-PCR analyses of SpCas9 transgenic chicken organs. SpCas9 expression in all tissues analyzed is shown by a 265-bp PCR product, and β-actin (300 bp) serves as a control.
Cas9 transgenic chickens were generated by phiC31 integrase-mediated integration of an SpCas9 expression construct (Fig. 1B) into a chicken endogenous pseudo attP site, which has previously been reported to increase the integration frequency in PGCs (31). These PGCs already carried an enhanced green fluorescent protein (EGFP) transgene, and Cas9-EGFP-PGC clones were screened for SpCas9 expression by immunofluorescence and flow cytometry (SI Appendix, Fig. S2A). Functionality was tested by transient transfection with an expression vector carrying a gRNA directed against the EGFP gene. Analysis by flow cytometry showed that 17% of SpCas9-expressing cells lost EGFP fluorescence, while no reduction was observed for the MOCK control (SI Appendix, Fig. S2B). From 60 injections with modified PGCs (PGC clone 4) into stage 13 to 15 HH embryos, 19 male germline chimeras were obtained. Upon sexual maturity, four chimeras were mated with wild-type hens to obtain fully transgenic SpCas9 chickens (Fig. 1D). As expected, EGFP and Cas9 segregated independently in the offspring.
The transgene copy number was determined in both species via droplet digital PCR (ddPCR) and revealed a single copy of SpCas9 in pigs and chickens (SI Appendix, Fig. S3 A and B). Both Cas9 transgenic chickens and pigs developed normally, showed no obvious abnormalities (i.e., in weight gain; SI Appendix, Fig. S3C), and were fertile.
Ubiquitous Expression of SpCas9 in Transgenic Animals.
SpCas9 messenger RNA (mRNA) expression was analyzed in different tissues from chickens (proventriculus, spleen, duodenum, cecal tonsil, cecum, liver, kidney, lung, pancreas, heart, brain, thymus, muscle, and gonads) and pigs (stomach, spleen, small intestine, aorta, liver, kidney, lung, and heart). Analysis of the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for pig samples and β-actin for chicken samples served to validate the quality of RNA. The RT-PCR showed SpCas9 expression in all samples of both species (Fig. 1 E and F).
Functionality of SpCas9.
To examine the functionality of SpCas9 in vitro, we isolated different primary cell types from Cas9-expressing animals and transfected those with gRNAs directed against EGFP, β2-microglobulin (B2M), a component necessary for the assembly of the MHC class I complex, or against the C-X-C chemokine receptor type 4 (CXCR4) for the avian experiments. Targets in porcine cells were the endogenous B2M gene and the α-1,3-galactosyltransferase (GGTA1) gene responsible for the presentation of alpha-gal epitopes on the cell surface of most mammals, which play a crucial role in hyperacute rejection in pig-to-primate xenotransplantation (21, 32). Successful inactivation of these target genes can be assessed by flow cytometry.
Cas9 transgenic pigs.
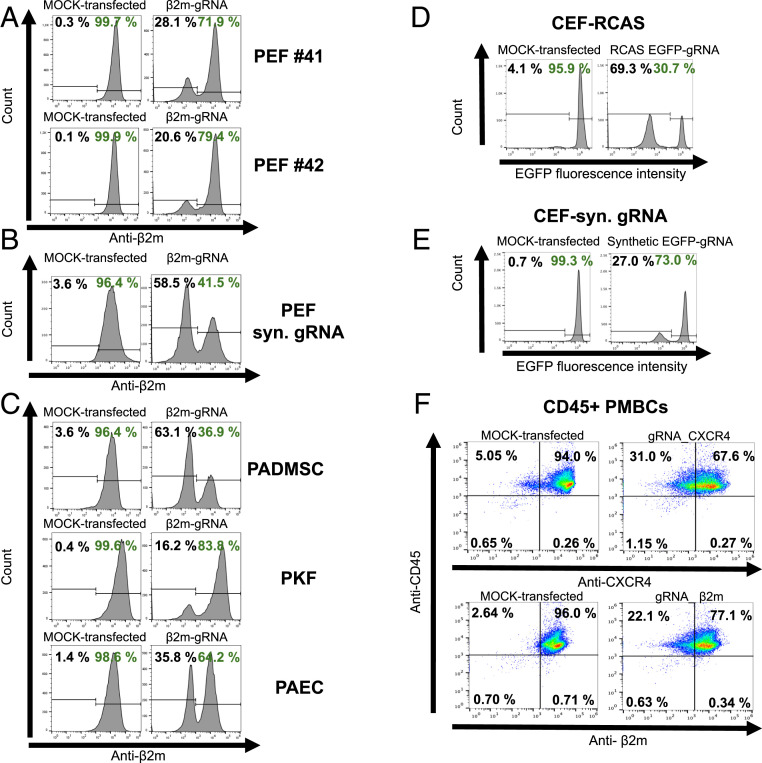
Porcine ear fibroblasts (PEFs) were isolated from the founder pigs (#41 and #42), and transiently transfected with a vector containing a gRNA against B2M (β2m-gRNA). Flow cytometry analyses revealed that 28.1% of cells from animal #41 and 20.6% of cells from pig #42 lost expression of β2m (Fig. 2A). To test whether chemically modified synthetic gRNAs can increase the editing efficiencies, as previously shown for human cells (33), PEFs were transfected with a chemically modified synthetic gRNA directed against B2M. In flow cytometry analyses, 58.5% of PEF cells were negative for β2m expression (Fig. 2B). To confirm functionality in different cell types, an SpCas9 transgenic offspring was killed, and porcine adipose-derived mesenchymal stem cells (PADMSCs), porcine aorta endothelial cells (PAECs), and porcine kidney fibroblasts (PKFs) were isolated, transiently transfected with a vector containing a gRNA against B2M, and analyzed by flow cytometry. Homozygous inactivation of the target gene was observed in all cell types (63.1% of cells in PADMSCs, 35.8% in PAECs, and 16.2% in PKFs), confirming the functionality of SpCas9 (Fig. 2C). The different efficiencies reflect the difference in transfectability of the various cell types.
Fig. 2.
SpCas9 functionality in primary cells derived from SpCas9 transgenic pigs and chickens. (A) PEFs derived from ear tissue of SpCas9 founder pigs #41 and #42 transfected with a construct carrying a gRNA against B2M or GGTA1 (MOCK-transfected control). (B) PEFs derived from an SpCas9 transgenic pig and transfected with chemically modified gRNA against porcine B2M or P16 (MOCK-transfected control). (C) PADMSCs, PKFs, and PAECs derived from an SpCas9 transgenic piglet and transfected with a construct carrying a gRNA against B2M or GGTA1 (MOCK-transfected control). (D) CEFs derived from an SpCas9-expressing chicken and transfected with RCASBP(A)-EGFP-gRNA or RCASBP(A)-INF-gRNA (MOCK-transfected control). (E) CEFs transfected with a synthetic gRNA against EGFP or B2M (MOCK-transfected control). (F) Splenic CD45+ PMBCs derived from an SpCas9-expressing chicken and transfected with chemically modified gRNA against B2M or CXCR4. Shown is one representative example of three independent replicates.
To assess SpCas9 transgene function in a more complex tissue, organoids were generated from the colonic mucosa of an SpCas9-expressing pig (SI Appendix, Fig. S4A) and transiently transfected with a vector carrying a gRNA directed against GGTA1. Tracking of Indels by Decomposition (TIDE) analysis of the sequenced PCR fragment across the gRNA target site revealed a total target cleavage efficiency of 12.0% in the organoids (SI Appendix, Fig. S4B).
Cas9 transgenic chicken.
Primary chicken embryonic fibroblasts (CEFs) were derived from an EGFP/Cas9 transgenic embryo. A gRNA against EGFP was delivered by the retroviral RCASBP(A) (Replication-Competent ALV LTR with a Splice acceptor) vector, which is a derivative of the avian Rous sarcoma virus (34, 35). Six days posttransduction, 69.3% of cells were negative for EGFP expression as measured by flow cytometry (Fig. 2D). In comparison, transfection of CEFs with a synthetic unmodified gRNA against EGFP was less efficient, with 27.0% loss of EGFP expression after 48 h (Fig. 2E). Next, genome editing was demonstrated in primary CD45+ peripheral blood mononuclear cells derived from the spleen of a Cas9-expressing chicken. Cells were electroporated with two 2′-O-methyl phosphorothioate linkage-modified synthetic gRNAs directed against CXCR4 or B2M, resulting in a significant reduction of CXCR4 (P < 0.05; 31% of cells) or β2m expression (P < 0.01; 22.1% of cells), as demonstrated in three independent replicates (Fig. 2F and SI Appendix, Fig. S5).
In Vivo Genome Editing.
The results above showed efficient gene inactivation in various primary cell types isolated from both SpCas9-expressing pigs and chickens. To confirm that genome editing would work equally well in vivo, gRNAs were directly delivered to SpCas9 transgenic animals.
Cas9 transgenic pig.
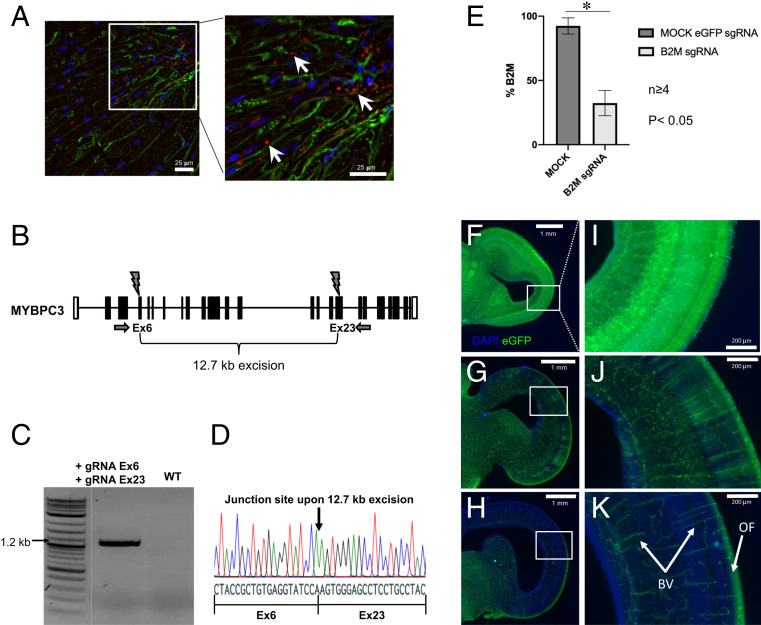
While, in the previous in vitro experiments, only individual gRNAs were used to achieve gene inactivation, the goal of the in vivo experiment was to assess whether a precise excision of a large DNA fragment (12.7 kb) could be achieved in heart tissue by delivering two gRNAs using an adeno-associated viral (AAV) vector. The target gene was MYBPC3, which encodes the cardiac myosin-binding protein C (cMyBP‐C). Mutations in MYBPC3 cause neonatal cardiomyopathy (36). To deliver gRNAs directly into the heart, polyamidoamine (PAMAM)-coated AAV2/9 vectors encoding two gRNAs targeting MYBPC3 exons 6 and 23 and an mCherry expression construct were injected into the coronary target artery (left anterior descending artery, LAD) of Cas9-expressing pigs at an age of 3.5 mo. After 3 wk, the experiment was terminated, and heart tissue sections from the distal LAD perfusion area were analyzed by fluorescence microscopy imaging. DNA was isolated from tissue sections with the highest number of mCherry-positive cells (Fig. 3A). PCR analysis and subsequent sequencing revealed precise CRISPR-Cas9−mediated deletion of the 12.7-kb DNA fragment between the two gRNA target sites located in exons 6 and 23 of the porcine MYBPC3 gene (g.4175_16906del) (Fig. 3 B–D). Indel mutations at the individual gRNA target sites alone were not observed. Quantitative PCR showed 8% efficiency for the deletion of the 12.7-kb fragment (SI Appendix, Fig. S6).
Fig. 3.
In vivo genome editing in SpCas9 transgenic pigs and chickens. (A) Porcine heart section of an SpCas9-transgenic pig transduced with an AAV virus encoding gRNAs directed against the porcine MYBPC3 gene and an mCherry reporter construct. The mCherry signal (red) is marked with white arrows. Sections were counterstained with DAPI (blue) and WGA-Alexa488 conjugate (green). (B) Structure of the porcine MYBPC3 locus. Exons are indicated by numbered boxes. Primers are shown as black arrows, and gRNA target sites are shown as black flashes. Simultaneous CRISPR-Cas9−mediated cleavage at the gRNA target sites in exons 6 and 23 leads to the excision of a 12.7-kb fragment. (C) Detection of the 1.2-kb PCR product resulting from successful CRISPR-Cas9−mediated excision of the 12.7-kb fragment. As PCR conditions were optimized for the detection of the 1.2-kb fragment, the much larger 13.9-kb wild-type fragment was not amplified. (D) Sequencing result across the junction site upon CRISPR-Cas9−mediated excision of the 12.7-kb fragment between both gRNA target sites. (E) In ovo genome editing of B2M by RCASBP(A)-mediated gRNA delivery. At ED3, embryonated eggs were injected with RCASBP(A)-β2m-gRNA−infected DF-1 cells or RCASBP(A)-EGFP-gRNA−infected DF-1 cells (MOCK control). At ED18, bursal B cells were analyzed for β2m expression by flow cytometry (n ≥ 4). *P < 0.05. Statistical analysis was done by Student´s t test, and error bars represent the SD. (F−K) In vivo genome editing via in ovo electroporation in chickens. At ED2, midbrain vesicles of EGFP-positive embryos were electroporated with pBlueScript II SK (+) vector containing a gRNA against EGFP or β2m, and embryos were collected at ED 12. DAPI-treated midbrain sections were analyzed for EGFP expression, by fluorescence microscopy. (F) Horizontal section of midbrain OT of MOCK β2m-gRNA−transfected embryo with clear EGFP signal in every cell (neurons, glia, and blood vessels). (G and H) Frontal sections of OT of EGFP-gRNA−transfected embryos showing a prominent reduction of EGFP fluorescence. (I–K) Magnified sections from F−H illustrating tectal layering and loss of EGFP in J and K. Blood vessels (BV) and optical input fibers (OF) still expressing EGFP marked with white arrows.
Cas9 transgenic chicken.
To examine the efficiency of in ovo genome editing, two different methods were tested. First, 3-d-old embryonated eggs were injected in the allantoic cavity with a DF-1 cell line producing the infectious RCASBP(A) vector encoding the B2M gRNA. The β2m expression on bursa-derived B cells was then analyzed by flow cytometry and showed a significant reduction of β2m on the cell surface compared to MOCK-transduced controls (Fig. 3E). The percentage of cells lacking β2m expression varied between the individual embryos, ranging from 30 to 80%, most likely reflecting different transduction efficiencies. Amplicon sequencing confirmed editing at the gRNA target site (SI Appendix, Fig. S9). As an alternative approach, tissue-specific in ovo electroporation was tested. Brain tissues from chicken embryos were electroporated with vectors containing either a gRNA against B2M or EGFP. In the latter case, fluorescence microscopy imaging clearly showed a loss of EGFP in Cas9/EGFP-positive embryos (Fig. 3 F–K). In contrast, no EGFP loss was observed in Cas9/EGFP embryos electroporated with a gRNA against B2M. TIDE analysis for EGFP targeting revealed a total cleavage efficiency of 11.5% (embryo #5) and 6.8% (embryo #7) (SI Appendix, Fig. S7) and, for B2M, 13.8% (embryo #6), 11.8% (embryo #12), and 11.1% (embryo #22) (SI Appendix, Fig. S8).
Discussion
Besides describing the establishment of a pig line that ubiquitously expresses SpCas9, we also reported the generation of SpCas9 transgenic chickens. Proof of principle studies showed the functionality of the SpCas9 transgene in vivo and in vitro by employing different methods for gRNA delivery, such as transfection with synthetic gRNAs, in ovo electroporation, or viral-based delivery methods. While all approaches resulted in editing of the target locus, the efficiency depended on the gRNA delivery method, ranging from 8% in vivo to 70% homozygous gene inactivation in vitro. Optimization of gRNA delivery clearly has to be a main objective for in vivo genome editing. Besides improving viral-based methods, such as AAVs or lentiviral transduction, nonviral delivery approaches should be investigated, such as delivery via polymer-based carriers or lipid nanoparticles (37–39).
The Cas9 transgenic chickens were generated by phiC31-mediated transgene integration into an endogenous pseudo attP site (40). Animals carrying a single copy of SpCas9 were healthy and fertile. However, if cell clones with more than a single copy of SpCas9 were used to produce Cas9 transgenic birds, embryo lethality between embryonic day 18 (ED18) and ED21 was observed. This is in contrast with SpCas9 homozygous mice and pigs, where two transgene copies are tolerated. As integration occurred randomly in the chicken, this could be a result of insertional mutagenesis or, more likely, due to the potential toxic effect of high Cas9 expression as observed for Cre-recombinase expressing mice (41).
For the pig, the SpCas9 transgene was inserted in the ROSA26 locus. This locus is known to be a “safe harbor” for transgene expression without interrupting the function of essential endogenous genes (42). Moreover, previous reports showed that it is readily targeted, supports abundant ubiquitous expression, and is dispensable for normal physiology and development (29, 30). Unlike previous work, where a Cre-inducible Cas9-expressing line has been generated (43), our pig line expresses a single copy of SpCas9 ubiquitously under the control of the CBh promoter. This eliminates the need for two consecutive recombination events to occur and should increase editing efficiency. Ubiquitously Cas9-expressing pigs are healthy and fertile and could be bred to homozygosity, consistent with findings in ROSA26-Cas9−expressing mice (28).
The functionality of the SpCas9 transgene was tested not only in a series of porcine primary cells but also in porcine colonic organoids, which form a more complex three-dimensional structure. Our group has an interest in modeling human cancers in pigs, such as colorectal cancer (4). Modification of colonic organoids from SpCas9-expressing pigs enables in vitro local inactivation of single or multiple tumor-relevant genes and the analysis of oncogenic transformation prior to subsequent autologous reimplantation, as shown for mice (44). This procedure as well as the option to inactivate multiple tumor-suppressor genes directly in vivo will eliminate the need to generate germline-modified pigs and will allow closer simulation of human oncogenesis.
In vivo genome editing in Cas9-expressing pigs and chickens was confirmed using three different approaches. Previous studies reported successful in vivo genome editing in avian species by direct injection of adenoviral delivered CRISPR-Cas9 components into the blastoderm of a newly laid egg (45), but the need to deliver the Cas9 endonuclease has generally been the limiting factor, due to its large size. Here, a retroviral RCAS vector expressing only the gRNA to efficiently inactivate B2M in avian B cells was used. In addition, tissue-specific in vivo genome editing without the need for viral vectors was demonstrated in chicken embryos via in ovo-electroporation of brain tissue. For the targeted delivery of two gRNAs to the porcine heart, an AAV vector was used, resulting in the precise excision of a 12.7-kb DNA fragment. Overall, there are very few reports on direct in vivo genome editing in pigs. We previously used AAV vectors carrying an intein-split Cas9 system and a pair of gRNAs to mediate genome editing in muscles (46), while Wang et al. (43) used a lentiviral-mediated approach to deliver gRNAs into lungs of inducible Cas9-expressing pigs. Even though both approaches are successful, one required the transduction of cells with two viruses simultaneously, whereas the other required the delivery not only of the gRNA but also of Cre-recombinase to first activate the latent Cas9 gene, potentially reducing efficiency and risking Cre-toxicity in the target cells. In contrast to these reports, our model requires the delivery of only gRNAs and is not dependent on cotransduction of multiple viruses or Cas9 activation.
In conclusion, we generated a versatile tool and showed efficient genome editing of five different target genes in a number of cell types and in vivo, confirming SpCas9 functionality in both pig and chicken. We are confident that the SpCas9 transgenic pigs and chickens provide a powerful platform for in vitro and in vivo genome editing in mammalian and nonmammalian livestock species, eliminating the need for Cas9 delivery in vivo and in vitro.
Methods
Generation of SpCas9 and gRNA Constructs.
For the generation of SpCas9 expression constructs, we used the sequence of a human codon-optimized SpCas9 obtained from pX330-U6-Chimeric_BB-CBh-SpCas9 from Feng Zhang (Addgene plasmid # 42230; http://n2t.net/addgene:42230; RRID:Addgene_42230). Oligonucleotides were synthesized by Eurofins Genomics.
For stable integration in chicken PGCs, the SpCas9 construct contained an attB site to ensure the insertion of the transgene using phiC31 integrase (40). SpCas9 was amplified by PCR, and the PCR product was assembled with a −8.7-kb backbone construct using NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs) according to manufacturer’s instructions. The SpCas9 expression cassette contained a 1.3-kb chicken β-actin promoter, followed by an SV40 nuclear localization signal (NLS), a 4.1-kb SpCas9 gene, a nucleoplasmin NLS, and an SV40 poly(A). The generated construct contained a CAG-hygromycin resistance cassette, and the SpCas9 gene was flanked by duplicated copies of the core 300-bp HS4 insulator from the chicken β-globin gene to ensure proper transgene expression (Fig. 1B).
For precise placement of SpCas9 at the porcine ROSA26 locus, we generated a promoter trap vector consisting of a 2.2-kb 5′ homology arm corresponding to a region of porcine ROSA26 intron 1; a 1.6-kb fragment composed of splice acceptor, Kozak sequence, promoterless neomycin resistance gene, and a triple-poly-A signal; a 5.3-kb SpCas9 expression cassette; and a 4.7-kb 3′ homology arm corresponding to a region of porcine ROSA26 introns 1 to 3. The SpCas9 cassette contained a 0.8-kb CBh promoter (47), followed by an SV40 NLS, a 4.1-kb SpCas9 gene, a nucleoplasmin NLS, and a 0.2-kb BGHpA (Fig. 1A).
The gRNA constructs directed against EGFP and chicken B2M were generated by cloning the respective gRNA oligonucleotides (EGFP: 5′-CCGGCAAGCTGCCCGTGCCC-3′; chicken B2M: 5′-GAACGTCCTCAACTGCTTCG-3′) with additional BbsI overhangs into a BbsI-digested pBluescript II KS (+) vector carrying a 0.2-kb U6 promoter and a single-guide RNA (sgRNA) scaffold sequence. The gRNA constructs directed against porcine GGTA1 and B2M were generated by cloning the respective gRNA oligonucleotides (GGTA1 exon 7: 5′-GTCGTGACCATAACCAGA-3′; porcine B2M exon 1: 5′-TAGCGATGGCTCCCCTCG-3′), with additional BbsI overhangs into a BbsI-digested vector carrying a 0.2-kb U6 promoter, an sgRNA scaffold sequence, and, additionally, a 1.6-kb puromycin resistance cassette composed of a 0.8-kb CBh promoter, a 0.6-kb puromycin resistance gene, and a 0.2-kb BGHpA. Oligonucleotides were synthesized by Eurofins Genomics. In chickens, gRNAs were also delivered by the RCAS system. The sequences coding for gRNAs and a human U6 promoter were introduced into the ClaI restriction site of the RCASBP(A) vector as previously described (48, 49).
For AAV-mediated in vivo delivery of gRNAs directed against target sites in porcine MYBPC3 exon 6 and exon 23, a construct was generated consisting of 1) intact and ∆trs inverted terminal repeats (ITRs) of AAV2 (0.1 kb each), 2) expression cassettes (0.4 kb each) for gRNAs directed against MYBPC3 exon 6 (5′-GCTGTGAGGTATCCACCA-3′) and exon 23 (5′-GACTCCTGCACGGTGCAGT-3′) driven by human and murine U6 promoters, respectively, and 3) a 1.2-kb mCherry cassette composed of a truncated CMV promoter, an mCherry coding sequence, and a synthetic polyA.
Generation of SpCas9-Expressing Cells.
Gene targeting in porcine cells was performed in porcine kidney fibroblasts that were isolated and cultured by standard methods (50). Cells were transfected with 4 μg to 6 μg of linearized targeting vector DNA (ROSA26-SpCas9) using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific), selected with 1,400 μg/mL to 1,600 μg/mL G418 (Genaxxon Bioscience) for 7 d to 10 d, and targeted clones identified by 5′- and 3′-junction PCR and DNA sequence analysis.
For the generation of transgenic chickens, EGFP-PGCs were derived from the germinal crescent as previously described (51); 5 × 106 of EGFP-PGCs were resuspended in a total volume of 100 µL of Nucleofector Solution V (Lonza) premixed with 10 µg of Cas9 plasmid and the same amount of phiC31 integrase (52). Electroporation was conducted by ECM 830 Square Wave Electroporation System (BTX) using eight square wave pulses (350 V, 100 µs). After transfection, cells were plated on 48-well plates and selected with 50 µg/mL hygromycin.
Animals.
Permission for the generation of transgenic Cas9-expressing chickens and pigs, and for the animal experiments, was issued by the government of Upper Bavaria, Germany (ROB-55.2-2532.Vet_02-17-101 for chickens; ROB-55.2-2532.Vet_02-18-33 and AZ 55.2-2532.Vet_02-18-79 for pigs). Experiments were performed according to the German Welfare Act and European Union Normative for Care and Use of Experimental Animals. All animals received standard diet and water ad libitum.
Transgenic pigs were generated as previously described (53). Donor cells were arrested at G0/G1 phase by serum deprivation. Oocytes, isolated from prepubertal gilts, were maturated in vitro and enucleated, and single donor cells were inserted into the perivitelline space. Cells were fused, and oocytes activation was induced via electric pulse. Reconstructed embryos were then transferred into the oviducts of hormonally synchronized recipient sows.
Transgenic chickens were generated as previously described (54). Briefly, 3,000 Cas9-PGCs were injected into the vasculature of 65-h-old embryos (Lohmann selected White Leghorn line, Lohmann-Tierzucht GmbH), subsequently transferred into a turkey surrogate shell, and incubated until hatching of chimeric roosters. Upon sexual maturity, sperm was collected for DNA isolation and genotyping. After breeding with wild-type hens, fully Cas9 transgenic chickens were obtained.
Derivation and Functional Assays in SpCas9-Expressing Cells.
PEFs, PADMSCs, PAECs, and PKFs were isolated, according to standard protocols (50, 55, 56), from an SpCas9-expressing pig and cultured in Dulbecco´s Modified Eagle Medium (DMEM) (PEFs and PKFs) or Advanced DMEM (PADMSCs and PAECs) supplemented with 20% fetal bovine serum (FBS), 2 mM Ala-Gln, 1× MEM nonessential amino acid solution, and 1 mM sodium pyruvate (Corning) at 37 °C and 5% CO2. Cells were then transfected with 1 μg of gRNA constructs against porcine B2M or GGTA1 using Lipofectamine 2000 transfection reagent, and, 24 h posttransfection, transiently transfected cells were selected using 1.5 μg/mL puromycin (InvivoGen) for 2 d. The remaining cell pool was analyzed by flow cytometry. PEFs were further transfected with 25 pmol of chemically modified gRNAs directed against porcine B2M (G*U*A*GCGAUGGCUCCCCUCG) or porcine P16 (G*A*G*GCUAGCCAGUCGGCCGA) (Synthego) using Stemfect RNA Transfection Kit (Stemgent) according to the manufacturer’s instruction.
To test SpCas9-expressing PGCs before injection into the embryo, 4 × 106 cells were electroporated with 10 µg of DNA of pBluescript II SK(+) DNA vector containing a gRNA directed against EGFP using the Nucleofector V Kit (Lonza). Subsequently, cells were analyzed for loss of EGFP expression by flow cytometry.
Chicken embryonic fibroblasts (CEFs) were prepared from Cas9-positive embryos as previously described (57) and cultured in Iscove’s medium supplemented with 5% FBS, 2% chicken serum, and 1% Penicillin/Streptomycin at 40 °C and 5% CO2. Transfection with RCAS-gRNAs was performed using ViaFect Transfection Reagent (Promega) according to the manufacturer’s instructions, with slight modifications. Briefly, 2.5 × 105 cells were seeded in six-well plates; 24 h later, transfected with 500 ng of DNA (transfection reagent to DNA ratio of 6:1); and incubated for 20 min at room temperature.
Transfection with synthetic gRNAs was conducted using Xfect RNA Transfection Reagent Protocol-At-A-Glance (Takara) following the manufacturer’s instructions. Twenty-four hours after seeding 100,000 EGFP-Cas9 CEF cells in a 24-well plate, these were transfected with 25 pmol of synthetic EGFP-gRNA premixed with Xfect RNA transfection polymer and incubated for 4 h, and the transfection mixture was then replaced by 500 µL of cell culture medium. Cells were analyzed at different time points for indels and loss of EGFP expression.
Splenic CD45+ cells were derived from a Cas9-expressing animal and isolated as previously described (58). The spleen was removed aseptically and homogenized by passing it through a cell strainer. Cells were isolated by Ficoll density gradient separation and directly electroporated with 25 pmol of chemically modified gRNAs directed against CXCR4 (CXCR4_1434: A*A*A*UUCAAUGAGUAUGCCAG) or B2M (β2m_1444: U*C*U*UGGUGCCCGCAGAGGCG) (Synthego) using the human T Cell Nucleofector Kit (Lonza) according to manufacturer’s instructions. Cells were resuspended in RPMI, 10% FBS, 1% Penicillin/Streptomycin, and 1% Glutamax and seeded on a 12-well plate, and medium was replaced after 6 h. Forty-eight hours postelectroporation, cells were analyzed by flow cytometry.
In Ovo Functionality of SpCas9 in Chicken Bursal B Cells.
Transduction of embryonic B cells was carried out using the RCAS retroviral gene transfer system as previously described (35, 59–61) (Fig. 3B). Briefly, DF-1 cells were transfected with RCASBP(A)-β2m-gRNA or MOCK RCASBP(A)-EGFP-gRNA using ViaFect Transfection Reagent (Promega) according to the manufacturer’s instructions. The transfected cells were cultured for 10 d to ensure completed infection. Then 1 × 106 cells in 100 μL of DMEM were injected into the allantoic cavity of ED3-old embryos using a sterile 1-mL syringe. At ED10, blood was taken from the embryos, and genotyping was performed to identify SpCas9-positive embryos. At ED18, heart tissue was collected for RCAS detection by PCR with primers RCAS_B2M_F (5′-CGAAGCAGTTGAGGACGTTC-3′) and RCAS_B2M_R (5′-CATATTTGCATATACGATACAAGGC-3′) resulting in a 245-bp amplicon. The bursa was dissected from the embryo to obtain B cells isolated by density gradient centrifugation. Cells were then stained for β2m and analyzed by flow cytometry.
Functionality of SpCas9 in Porcine Colonic Organoids.
Colonic organoids were isolated as follows: Colonic mucosa was incubated in dissociation buffer (phosphate-buffered saline [PBS], 30 mM EDTA, and 10 mM dithiothreitol) for 20 min at 4 °C; colonic crypts were then mechanically separated under a microscope and embedded in Matrigel (Corning). Organoids were cultivated in growth medium as previously described (62), with minor changes (50% mouse Wnt3a-conditioned medium, 50 ng/mL mouse Wnt3a recombinant protein [Biolegend], 15% mouse R-spondin1-conditioned medium, 1.3% porcine Noggin-conditioned medium, 2.5 µM CHIR99021 [Calbiochem/Merck], 100 µg/mL primocin [Invitrogen]). Colonic organoids were electroporated with 2 µg of gRNA construct as previously described (63) using the BTX ECM 830 with the following instrument settings: 180 V, two pulses of 5-ms length, and 100-ms pulse interval. After 24 h, cells were selected using 1 µg/mL puromycin (InvivoGen) for 48 h, and cell pools were analyzed for the presence of indels.
AAV Preparation and G2-PAMAM Coating.
Recombinant self-complementary AAVs of pseudotype 2/9 were produced using the triple transfection method described previously (64). Briefly, HEK 293T cells were transfected with a vector encoding mCherry and U6-driven gRNAs targeting MYBPC3 exons 6 and 23 (pscAAV-hU6.gRNA-mU6.gRNA-trCMV.mCherry), a vector encoding AAV2 rep and AAV9 cap sequences, and the helper plasmid pAdDeltaF6 (Puresyn) using PEI Max (Polysciences). After 72 h, cells were harvested, and the virus was purified by iodixanol gradient centrifugation. The virus was further purified by gravity flow size-exclusion purification using Sephadex G100 SF resin (Sigma-Aldrich) in Econo-Pac columns (Bio-Rad). The virus was concentrated in phosphate-buffered saline using Amicon Ultra-15 centrifugal filter units (Merck, Germany) and stored at 4 °C. Viral titers were quantified by qPCR using a probe binding the inverted terminal repeat. For G2-PAMAM coating, 1 × 1014 virus genomes were mixed with 18 µg of G2-PAMAM nanoparticles (Andrews ChemServices) in Opti-MEM I (Thermo Fisher Scientific) by gentle pipetting and incubated for 30 min at room temperature before further use.
AAV Infusion into Cas9-Expressing Pigs.
Pigs (n = 2) were anesthetized, intubated, and ventilated. Two 6F sheaths were inserted into the carotid artery and the external jugular vein. For antegrade AAV9-gRNA infusion (1 × 1014 virus genomes), the coronary target artery (LAD) was blocked by an over-the-wire (OTW) balloon of 2.5 mm diameter, distal to the second diagonal branch. Through the lumen of the OTW balloon, 10 mL of saline solution containing the virus load was injected in fractions of 1 mL/min. During that period, the venous outflow was blocked by a Swan Ganz catheter inserted deeply into the anterior interventricular vein, accompanying the LAD. With this technique, the transition time of fluid was decelerated significantly to about 30 s. After injection of 10 mL, the balloons were deflated and removed. After a final demonstration of open coronary vessels and normal heart function, the catheters and sheaths were removed, and wounds were closed surgically. Anesthesia was stopped, and animals were extubated in the operating room. The animals were killed after 3 wk, and the hearts were excised, sampled in a systematic manner (65), and snap frozen in liquid nitrogen-cooled 2-methylbutane for further investigation.
Analysis of In Vivo Functionality of SpCas9 in Porcine Hearts.
Snap-frozen heart tissue was sectioned by cryotome and counterstained with DAPI and WGA-Alexa488 conjugate (Thermo Fisher Scientific). MCherry signal, indicating successful transduction, was visualized with the Thunder imaging system (Leica). Genomic DNA from heart tissue was isolated by Proteinase K (Carl-Roth) digestion and phenol-chloroform extraction. Deletion of the fragment between the MYBPC3_Ex6 and MYBPC3_Ex23 gRNA target sites was screened by PCR using Q5 polymerase (New England Biolabs) and flanking primers ∼0.6 kb distal to the gRNA target sites (Table 1). Correctness of amplified bands was confirmed by Sanger sequencing. Efficiency of editing was determined by qPCR using Viia7 Real-time PCR System (Applied Biosystems). Briefly, MYBPC3 wild-type exon 6 and the deletion variant (g41575_16906 del) were amplified by PCR using the primers MYBPC3_Ex6_F (5′-GCCCCAAGTCTCCTCTAACA-3′) and MYBPC3_Ex6_R (5′-TCACCTATTCACCTGTGCCC-3′) for exon 6 and MYBPC3_Ex6_F and MYBPC3_Del_R (5′-CAATGGGCATGAAGGGCTG-3′) for the deletion variant. Amplicons were quantified photospectometrically, standard curves were generated by serial dilution, and initial quantities of both amplicons in the tissue sample were determined by qPCR using the primers above. The frequency of the deletion variant was obtained by calculating the ratio of wild-type exon 6 to the deletion variant.
Table 1.
Primers used for indel detection
| sgRNA target site | Forward primer (5′−3′) | Tm (°C) | Reverse primer (5′−3′) | Temperature (°C) | PCR size |
| GGTA1 exon 7 | GCCAGTCACCACAAGCCATG | 63.5 | TGGCCCTGTGACACCATTCT | 63.5 | 362 bp |
| pB2M exon 1 | CCACCCAGTCCAACCTTTGCC | 65.5 | CCAGAGTTAGCGCCCGGAGT | 65.8 | 377 bp |
| MYBPC3 Δ6 to 23 | CTTGAGAGGCTGTTGTTGCTGG | 66.7 | GCACACCCCATCTCTGATTCCT | 67.2 | 1,225 bp |
| EGFP | ATGGTGAGCAAGGGCGAGGAGCT | 70.0 | CGTCCTCGATGTTGTGGCGGATC | 66.9 | 523 bp |
| chB2M | CAAGGTGCAGGTGTACTCC | 59.4 | ACTTGTAGACCTGCGGCTC | 61.1 | 269 bp |
In Vivo Functionality of SpCas9 in the Chicken Central Nervous System.
Central nervous tissue (neurons and glial cells) was transfected using in ovo electroporation (66–70). Briefly, Cas9xWT and Cas9xEGFP embryos at developmental stages HH 10 to 13 (after ∼44 h of incubation) were transfected with a pBlueScript II SK (+) vector containing a gRNA directed against B2M or EGFP. Adhesive tape was attached to the eggshell to prevent cracks, and 2 mL of albumin was extracted from the egg using a syringe. A window of about 1 cm2 to 2 cm2 was cut into the upper side to expose the blastodisc with the embryo and illuminated with blue light to increase contrast. The plasmid solutions were mixed 4:1 with Fast Green Dye resulting in 667 ng/μL pBlueScript II SK (+) vector containing a gRNA against B2M and 1,255 ng/μL pBlueScript II SK (+) vector containing a gRNA against EGFP. A small volume of plasmid solution filling only the tip of a 100-μm glass micropipette was then injected into the mesencephalic vesicle (second brain vesicle). Next, a gold electrode (positive) was positioned at about 1 mm distance to the left of the embryo, and a tungsten microelectrode (negative) was inserted into the second brain vesicle. For electroporation, three rectangular pulses at 2 Hz with 15 V each and 25-ms duration were applied (Grass S48 stimulator, Medical Instruments with Genetrodes 45-0115, Harvard Apparatus Inc.). Electrodes were removed, and 1 mL to 2 mL of 4 °C chicken ringer solution (150 mM NaCl, 5.4 mM KCl, 2.2 mM CaCl2, and 2.4 mM NaHCO3) was dripped onto the embryo for liquid resupply and to reduce damage caused by heating. The eggshells were sealed with adhesive tape, and the embryos were incubated until ED12, when the embryonic brains were dissected for genotyping, TIDE analysis, and histological preparation of midbrain slices of EGFP-positive embryos. For histological analysis of EGFP-positive embryos, the ED12 midbrains including the optic tectum (OT) were fixed in 4% paraformaldehyde for at least 24 h, transferred into 30% sucrose solution for 4 h for cryoprotection, cryosectioned to 50 µm, and mounted with DAPI mounting medium (0.1 mg DAPI in 100 mL n-propyl gallate). All images were taken with an epifluorescence microscope (Olympus BX63 with XM10 digital camera, Olympus) using the same exposure time and gain.
Indel Detection.
Cell pools were analyzed for the presence of indels by PCR spanning the gRNA target site using primers presented in Table 1. The resulting amplicons were sequenced (Eurofins Genomics) and analyzed with TIDE (http://shinyapps.datacurators.nl/tide/) (71). Amplicon sequencing over the B2M target region was carried out by Genewiz.
Genotyping PCRs.
The following primers were used to identify correct placement of SpCas9 at the porcine ROSA26 locus: 5′-junction PCR used primers ROSA26_5′F (5′-TATGGGCGGGATTCTTTTGC-3′) and Neo_5′R (5′-AGCCCCTGATGCTCTTCGTC-3′) to amplify a 3.1-kb fragment; 3′-junction PCR used primers Cas9_3′F (5′-GCAGATCAGCGAGTTCTCCA-3′) and ROSA26_3′R (5′-CAGGTGGAAAGCTACCCTAGCC-3′) to amplify a 5.6-kb fragment. The wild-type ROSA26 allele was amplified with primers ROSA26_5′F (sequence above) and ROSA26_I1R (5′-GTTTGCACAGGAAACCCAAG-3′), producing a 3.1-kb fragment. PCRs were carried out using AccuStart Taq DNA polymerase HiFi (QuantaBio) and thermal cycling conditions: 94 °C for 1 min, followed by 40 cycles at 94 °C for 20 s, 60 °C for 30 s, 78 °C for 1 min per kb, and a final extension at 68 °C for 5 min.
The following primers were designed for the specific detection of SpCas9 in transgenic chickens Cas9-593-For (5′-GAGAGAATGAAGCGGATCGAAGAG-3′) and Cas-440-Rev (5′–TTGCTGTAGAAGAAGTACTTGGCG-3′). PCRs were carried out using FIREPol DNA Polymerase (Solis Biodyne) and thermal cycling conditions: 95 °C for 5 min, followed by 40 cycles at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min per kb, and a final extension at 72 °C for 5 min.
Determination of the Transgene Copy Number by Droplet Digital PCR.
Droplet digital PCR (ddPCR) was performed as described previously (30). The transgene copy number was determined using a fluorescence-labeled probe for SpCas9 ([5′FAM-GCACGCCATTCTGCGGCGGC-BHQ3′]; primers used: ddSpCas9_F [5′-AGTTCATCAAGCCCATCCTG-3′] and ddSpCas9_R [5′-TCTTTTCCCGGTTGTCCTTC-3′]) or hygromycin ([5′FAM-TCGTGCACGCGGATTTCGGCTCCAA-3′]; primers used: ddHygro_F [5′-CATATGGCGCGATTGCTGATC-3′] and ddHygro_R [5′-GTCAATGACCGCTGTTATGC-3′]). GAPDH ([5′HEX-TGTGATCAAGTCTGGTGCCC-BHQ3′]; primers used: ddGAPDH_F [5′-CTCAACGACCACTTCGTCAA-3′] and ddGAPDH_R [5′-CCCTGTTGCTGTAGCCAAAT-3′]) or β-actin ([5′HEX-GTGGGTGGAGGAGGCTGAGC-BHQ3′]; primers used: ddBeta_actin_F: [5′-CAGGATGCAGAAGGAGATCA-3′] and ddBeta_actin_R: [5′-TCCACCACTAAGACAAAGCA- 3′]) were used as references. The target gene was quantified using the QX100 system (Bio-Rad Laboratories).
Transgene Expression Analysis.
RNA from porcine samples was isolated using Sure Prep RNA/DNA/Protein Purification Kit (Fisher Scientific) for cells, or the innuSPEED tissue RNA kit (Analytic Jena) for tissues, and complementary DNA (cDNA) was synthesized using the FastGene Scriptase (Nippon Genetics). SpCas9 mRNA was detected with primers pCas9_F1 (5′-GCAGATCAGCGAGTTCTCCA-3′) and pCas9_R1 (5′-GGGAGGGGCAAACAACAGAT-3′), resulting in a 415-bp amplicon. Porcine GAPDH mRNA was detected with primers GAPDHF (5′-TTCCACGGCACAGTCAAGGC-3′) and GAPDHR (5′-GCAGGTCAGGTCCACAAC-3′), resulting in a 576-bp amplicon. PCR was performed using GoTaq G2 DNA Polymerase (Promega) according to the manufacturer’s instructions.
RNA from chicken samples was isolated by Reliaprep RNA Tissue Miniprep System according to manufacturer instructions (Promega), followed by cDNA synthesis using GoScript Reverse transcription mix (Promega). SpCas9 mRNA was detected with primers chCas9_F1 (5′-GAGAGAATGAAGCGGATCGAAGAG-3′) and chCas9_R2 (5′-CAGTTCCTGGTCCACGTACATATC-3′), resulting in a 150-bp amplicon. β-actin mRNA was detected with primers Beta_actin_F (5′-TACCACAATGTACCCTGGC-3′) and Beta_actin_R (5′-CTCGTCTTGTTTTATGCGC-3′), resulting in a 300-bp amplicon. PCR was performed using FIREPol DNA Polymerase (Solis Biodyne) according to the manufacturer’s instructions.
Immunofluorescence.
Cytoplasmic detection of Cas9 in the generated PGC clones was done by FLAG-Tag staining. Briefly, a total number of 5 × 105 cells were centrifuged, washed with PBS, and incubated with 100 µL of fixation buffer (eBioscience) for 10 min. Cells were washed with 2% bovine serum albumin (BSA) in PBS (FLUO-Buffer) followed by permeabilization with 0.1% Triton X-100 in PBS for 10 min. Cells were incubated for 1 h with mouse anti-FLAG M2 antibodies at a dilution of 1:500 in FLUO-Buffer. Subsequently, cells were washed in PBS and incubated with an Alexa 568 secondary anti-mouse IgG for 1 h in the dark. Cells were washed in PBS and resuspended in 20 µL of mounting medium supplemented with DAPI, and immunofluorescence was detected under a fluorescence microscope (ApoTome, Zeiss).
Flow Cytometry.
Extracellular staining was carried out to detect β2m and CXCR4. Briefly, 3 × 105 to 1 × 106 cells were washed with 2% BSA in PBS (FLUO-Buffer). To determine the living cell population, cells were incubated with Fixable Viability Dye eFluor 780 (eBioscience) at a dilution of 1:1,000. After washing with FLUO-Buffer, primary antibodies (concentration shown in Table 2) were applied for 20 min. Cells were washed in FLUO-Buffer to remove unbound antibodies and incubated with conjugated secondary antibodies for 20 min. Subsequently, cells were again washed and analyzed using an Attune flow cytometer (Thermo Fisher Scientific). To detect CD45+ cells, a direct fluorescein isothiocyanate (FITC)-labeled primary antibody was used, and the cells were washed with FLUO-Buffer before analysis. Antibodies and concentrations used are shown in Table 2.
Table 2.
Antibodies used for flow cytometry
| Antibody | Company/origin | Concentration |
| Anti chicken β2m F21-21(75) | Cell culture supernatant | 1:5 dilution |
| Anti pig-B2M-Biotin clone B2M-02 | Sigma-Aldrich (SAB4700015) | 20 µg/mL |
| Streptavidin-PE | BD (554061) | 2.5 µg/mL |
| CD45-FITC | Biozol (SBA-8270-02) | 5 µg/mL |
| CXCR4 | Bio-Rad (MCA6012GA) | 0.2 µg/mL |
| H+L-APC | Biozol (MBS-MBS4151013) | 0.625 µg/mL |
For intracellular staining, cells were fixed and permeabilized with 0.1% Triton X-100 in PBS for 10 min. The subsequent staining process was carried out as described above. Detection of EGFP signal loss in different cells was examined by quantifying the loss of EGFP signal. For all flow cytometry analyses, an Attune flow cytometer (Thermo Fisher Scientific) was used. Data were analyzed with FlowJo 10.4.1 software (FlowJo, LLC 2006-2017).
Statistical Analysis.
Statistical analysis was carried out using SPSS24 statistics (version 24.0.0.0) software (IBM). Normally distributed data (Shapiro−Wilk test P > 0.05) were analyzed by student’s t test. The Mann−Whitney U Test was applied for not normally distributed data. P values < 0.05 were considered as significant. Graphs were constructed using GraphPad Prism (version 8.0.1 145) (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Daniela Huber for the generation and evaluation of gRNAs directed against the MYBPC3 gene. This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants Schu2446/4-1, KA3492/6-1, and FL948/1-1 awarded to B.S., B.B.K., and T.F. respectively; by the DFG Collaborative Research Center/Transregio 267 and the DFG Collaborative Research Center 1321; and by the German Center for Cardiovascular Research on “Gene therapy for neonatal sarcomeric cardiomyopathies: Towards first-in-patient” (High Risk High Volume Grant: 81X2600413) awarded to L.C., A.S., and E.W.
Footnotes
Competing interest statement: L.C. holds a patent on gene therapy vectors for treating cardiomyopathy that was licensed to DiNAQOR, is a member of the Scientific Advisory Board, and has shares in DiNAQOR.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022562118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Flisikowska T., Kind A., Schnieke A., Genetically modified pigs to model human diseases. J. Appl. Genet. 55, 53–64 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Swindle M., Smith A. C., Comparative anatomy and physiology of the pig. Scand. J. Lab. Anim. Sci. 25, 11–21 (1998). [Google Scholar]
- 3.Wolf E., Braun-Reichhart C., Streckel E., Renner S., Genetically engineered pig models for diabetes research. Transgenic Res. 23, 27–38 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Flisikowska T., et al., A porcine model of familial adenomatous polyposis. Gastroenterology 143, 1173–1175.e7 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Karim B. O., Huso D. L., Mouse models for colorectal cancer. Am. J. Cancer Res. 3, 240–250 (2013). [PMC free article] [PubMed] [Google Scholar]
- 6.Van Herck S. L., Geysens S., Delbaere J., Darras V. M., Regulators of thyroid hormone availability and action in embryonic chicken brain development. Gen. Comp. Endocrinol. 190, 96–104 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Goodfellow F. T., et al., Zika virus induced mortality and microcephaly in chicken embryos. Stem Cells Dev. 25, 1691–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiecker C., The chick embryo as a model for the effects of prenatal exposure to alcohol on craniofacial development. Dev. Biol. 415, 314–325 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Murphy J. B.. The effect of adult chicken organ grafts on the chick embryo. J. Exp. Med. 24, 1–5 (1916). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glick B., Chang T. S., Jaap R. G., The bursa of Fabricius and antibody production. Poult. Sci. 35, 224–225 (1956). [Google Scholar]
- 11.Xiao Y., et al., A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: Implications for the origin and evolution of mammalian defensins. BMC Genomics 5, 56 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkard C., et al., Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 13, e1006206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmich R.et al., Acquiring resistance against a retroviral infection via CRISPR/Cas9 targeted genome editing in a commercial chicken line. Front. Genome Ed. 2, 3, 10.3389/fgeed.2020.00003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koslová A., et al., Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus. Proc. Natl. Acad. Sci. U.S.A. 117, 2108–2112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitworth K. M., et al., Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22 (2016). [DOI] [PubMed] [Google Scholar]
- 16.van de Lavoir M. C., et al., High-grade transgenic somatic chimeras from chicken embryonic stem cells. Mech. Dev. 123, 31–41 (2006). [DOI] [PubMed] [Google Scholar]
- 17.van de Lavoir M. C., et al., Germline transmission of genetically modified primordial germ cells. Nature 441, 766–769 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Schusser B., et al., Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur. J. Immunol. 46, 2137–2148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whyte J., et al., FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Rep. 5, 1171–1182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer K., et al., Viable pigs after simultaneous inactivation of porcine MHC class I and three xenoreactive antigen genes GGTA1, CMAH and B4GALNT2. Xenotransplantation 27, e12560 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Petersen B., et al., Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation 23, 338–346 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov L., et al., Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One 11, e0154303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oishi I., Yoshii K., Miyahara D., Kagami H., Tagami T., Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 6, 23980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiou S. H., et al., Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 29, 1576–1585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie C., et al., Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 26, 1099–1111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue W., et al., CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddalo D., et al., In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platt R. J., et al., CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., et al., Dual fluorescent reporter pig for Cre recombination: Transgene placement at the ROSA26 locus. PLoS One 9, e102455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieblinger B., et al., Strong xenoprotective function by single-copy transgenes placed sequentially at a permissive locus. Xenotransplantation 25, e12382 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Leighton P. A., et al., Generation of chickens expressing Cre recombinase. Transgenic Res. 25, 609–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwaki K., et al., Heart transplantation in baboons using alpha1,3-galactosyltransferase gene-knockout pigs as donors: Initial experience. Nat. Med. 11, 29–31 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Hendel A., et al., Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes S. H., Greenhouse J. J., Petropoulos C. J., Sutrave P., Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61, 3004–3012 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahronian L. G., Lewis B. C., Generation of high-titer RCAS virus from DF1 chicken fibroblasts. Cold Spring Harb. Protoc. 2014, 1161–1166 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Carrier L., Mearini G., Stathopoulou K., Cuello F., Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 573, 188–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn J. D., et al., A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z., et al., Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid Commun. 40, e1800068 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Wilbie D., Walther J., Mastrobattista E., Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res. 52, 1555–1564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leighton P. A., van de Lavoir M. C., Diamond J. H., Xia C., Etches R. J., Genetic modification of primordial germ cells by gene trapping, gene targeting, and phiC31 integrase. Mol. Reprod. Dev. 75, 1163–1175 (2008). [DOI] [PubMed] [Google Scholar]
- 41.The Company of Biologists Ltd. , Cre-recombinase-associated toxicity highlights limitations of genome editing. Dis. Model. Mech. 6, 1299–1300 (2013). [Google Scholar]
- 42.Li X., et al., Rosa26-targeted swine models for stable gene over-expression and Cre-mediated lineage tracing. Cell Res. 24, 501–504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K., et al., Cre-dependent Cas9-expressing pigs enable efficient in vivo genome editing. Genome Res. 27, 2061–2071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roper J., et al., In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J., Ma J., Lee K., Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail. Proc. Natl. Acad. Sci. U.S.A. 116, 13288–13292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moretti A., et al., Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 26, 207–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray S. J., et al., Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 22, 1143–1153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes S. H., The RCAS vector system. Folia Biol. (Praha) 50, 107–119 (2004). [PubMed] [Google Scholar]
- 49.Reuter A., et al., Antiviral activity of lambda interferon in chickens. J. Virol. 88, 2835–2843 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richter A., et al., Potential of primary kidney cells for somatic cell nuclear transfer mediated transgenesis in pig. BMC Biotechnol. 12, 84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song Y, et al. Characteristics of long-term cultures of avian primordial germ cells and gonocytes. Biol. Reprod. 90, 15 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Trefil P., et al., Male fertility restored by transplanting primordial germ cells into testes: A new way towards efficient transgenesis in chicken. Sci. Rep. 7, 14246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurome M., Kessler B., Wuensch A., Nagashima H., Wolf E., Nuclear transfer and transgenesis in the pig. Methods Mol. Biol. 1222, 37–59 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Schusser B., et al., Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. U.S.A. 110, 20170–20175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leuchs S., et al., Inactivation and inducible oncogenic mutation of p53 in gene targeted pigs. PLoS One 7, e43323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oropeza M., et al., Transgenic expression of the human A20 gene in cloned pigs provides protection against apoptotic and inflammatory stimuli. Xenotransplantation 16, 522–534 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Bai C., et al., Establishment and biological characteristics of a Jingning chicken embryonic fibroblast bank. Eur. J. Histochem. 55, e4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pieper J., Methner U., Berndt A., Heterogeneity of avian gammadelta T cells. Vet. Immunol. Immunopathol. 124, 241–252 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Fekete D. M., Cepko C. L., Replication-competent retroviral vectors encoding alkaline phosphatase reveal spatial restriction of viral gene expression/transduction in the chick embryo. Mol. Cell. Biol. 13, 2604–2613 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davani D., Pancer Z., Ratcliffe M. J. H., Ligation of surface Ig by gut-derived antigen positively selects chicken bursal and peripheral B cells. J. Immunol. 192, 3218–3227 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Sayegh C. E., Demaries S. L., Iacampo S., Ratcliffe M. J., Development of B cells expressing surface immunoglobulin molecules that lack V(D)J-encoded determinants in the avian embryo bursa of fabricius. Proc. Natl. Acad. Sci. U.S.A. 96, 10806–10811 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato T., et al., Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Fujii M., Matano M., Nanki K., Sato T., Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 10, 1474–1485 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Kupatt C., et al., Cotransfection of vascular endothelial growth factor-A and platelet-derived growth factor-B via recombinant adeno-associated virus resolves chronic ischemic malperfusion role of vessel maturation. J. Am. Coll. Cardiol. 56, 414–422 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Hinkel R., et al., Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation 128, 1066–1075 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Itasaki N., Bel-Vialar S., Krumlauf R., ‘Shocking’ developments in chick embryology: Electroporation and in ovo gene expression. Nat. Cell Biol. 1, E203–E207 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Nakamura H., Funahashi J., Introduction of DNA into chick embryos by in ovo electroporation. Methods 24, 43–48 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Yang B., Geary L. B., Ma Y.-C., In ovo electroporation in chick midbrain for studying gene function in dopaminergic neuron development. J. Vis. Exp. e4017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blank M. C., Chizhikov V., Millen K. J., In ovo electroporations of HH stage 10 chicken embryos. J. Vis. Exp. e408 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weigel S., Flisikowska T., Schnieke A., Luksch H., Hybrid voltage sensor imaging of eGFP-F expressing neurons in chicken midbrain slices. J. Neurosci. Methods 233, 28–33 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.