Significance
This study explores the potential of a phage, PYOSa, for treating Staphylococcus aureus infections in combination with antibiotics. Population dynamic and genomic analysis identified a limitation and potential liability of using PYOSa for therapy. Due to the production of potentially pathogenic atypical small colony variants, PYOSa alone cannot eliminate S. aureus populations. However, we demonstrate that by following the administration of PYOSa with bactericidal antibiotics, this limitation and potential liability can be addressed. The methods used in this investigation to explore the efficacy of combinations of PYOSa and antibiotics for treating S. aureus infections can be employed to evaluate the clinical potential and facilitate the design of treatment protocols for any bacteria and phage that can be cultured in vitro.
Keywords: phage therapy, population dynamics, Staphylococcus aureus
Abstract
In response to increasing frequencies of antibiotic-resistant pathogens, there has been a resurrection of interest in the use of bacteriophage to treat bacterial infections: phage therapy. Here we explore the potential of a seemingly ideal phage, PYOSa, for combination phage and antibiotic treatment of Staphylococcus aureus infections. This K-like phage has a broad host range; all 83 tested clinical isolates of S.aureus tested were susceptible to PYOSa. Because of the mode of action of PYOSa, S. aureus is unlikely to generate classical receptor-site mutants resistant to PYOSa; none were observed in the 13 clinical isolates tested. PYOSa kills S. aureus at high rates. On the downside, the results of our experiments and tests of the joint action of PYOSa and antibiotics raise issues that must be addressed before PYOSa is employed clinically. Despite the maintenance of the phage, PYOSa does not clear populations of S. aureus. Due to the ascent of a phenotyically diverse array of small-colony variants following an initial demise, the bacterial populations return to densities similar to that of phage-free controls. Using a combination of mathematical modeling and in vitro experiments, we postulate and present evidence for a mechanism to account for the demise–resurrection dynamics of PYOSa and S. aureus. Critically for phage therapy, our experimental results suggest that treatment with PYOSa followed by bactericidal antibiotics can clear populations of S. aureus more effectively than the antibiotics alone.
Driven by well-warranted concerns about the increasing frequencies of infections with antibiotic-resistant pathogens, there has been a resurrection of interest in, research on, and clinical trials with a therapy that predates antibiotics by more than 15 y: bacteriophage (1–4). One direction phage therapy research has taken is to engineer lytic (virulent) phages with properties that are anticipated to maximize their efficacy for treating bacterial infections in mammals (5–8). Primary among these properties are 1) a broad host range for the target bacterial species; 2) mechanisms that prevent the generation of envelope or other kinds of high-fitness resistance in the target bacteria (9); 3) the capacity to thwart the innate and adaptive immune systems of bacteria, respectively, restriction-modification and CRISPR-Cas (7, 10, 11); 4) the ability to survive, kill, and replicate on pathogenic bacteria colonizing or infecting mammalian hosts (12, 13); and 5) little or no negative effects on the treated host (9).
To these five desired properties for therapeutic bacteriophages, there is a sixth that should be considered: the joint action of these phages and antibiotics (8, 14–17). Phages-only treatment may be reasonable for compassionate therapy, where the bacteria responsible for the infection are resistant to all available antibiotics (18–20). From a practical perspective, however, for phages to become widely employed for treating bacterial infections, they would have to be effective in combination with antibiotics. It would be unethical and unacceptable to clinicians and regulatory agencies to use phage-only therapy for infections that can be effectively treated with existing antibiotics.
Although not specifically engineered for these properties, there is a Staphylococcal phage isolated from a therapeutic phage collection from the Eliava Institute in Tbilisi, Georgia, that we call PYOSa that on first consideration appears to have all six of the properties required to be an effective agent for therapy. 1) PYOSa is likely to have a broad host range for S. aureus. The receptor of this K-like Myoviridae is N-acetylglucosamine in the wall-teichoic acid backbone of Staphylococcus aureus and is shared among most (21), if not all, S. aureus, thereby suggesting PYOSa should be able to adsorb to and potentially replicate on and kill a vast number of clinical isolates of S. aureus. 2) S. aureus does not generate classical, surface modification mutants resistant to PYOSa. Since the structure of the receptor of PYOSa is critical to the viability, replication, and virulence of these bacteria, the modifications in this receptor (22) may not be consistent with the viability or pathogenicity of S. aureus (23). 3) The replication of PYOSa is unlikely to be prevented by restriction-modification (RM) or CRISPR-Cas. Despite a genome size of 127 KB, the PYOSa phage has no GATC nucleotide restriction sites for the S. aureus restriction enzyme Sau3A1 and only one restriction site, GGNCC (guanine, guanine, any nucleotide, cytosine, cytosine), for the Sau961 restriction endonuclease (24, 25). There is no evidence for a functional CRISPR-Cas system in S. aureus or, to our knowledge, other mechanisms by which S. aureus may prevent the replication of this phage (26). 4) There is evidence that PYOSa-like phages can replicate in mammals. Early treatment with a phage with a different name but the same properties as PYOSa, Statuv, prevented mortality in otherwise lethal peritoneal infections of S. aureus in mice (27). A PYOSa-like phage has also been successfully used therapeutically in humans (28). 5) No deleterious effects of a PYOSa-like phage were observed in recent placebo-controlled trials with volunteers asymptotically colonized by S. aureus (24). 6) Finally, there is evidence to suggest synergy with antibiotics. In vitro, PYOSa increased the efficacy of low concentrations of antibiotics for the treatment of biofilm populations of S. aureus (14).
With in vitro population and evolutionary dynamic experiments with PYOSa and S. aureus Newman in combination with three different bacteriostatic and six different bactericidal antibiotics, we explore just how well PYOSa fits the above criteria for combination antibiotic and phage therapy. Our population dynamic experiments indicate that as consequence of the ascent of potentially pathogenic PYOSa resistant small colony variants (SCVs), by itself, PYOSa does not clear S. aureus infections. After an initial decline in the density of these bacteria when confronted with PYOSa, despite the continued presence of these phage, the densities of the bacterial populations return to levels similar to those observed in their absence. Using mathematical models, we present a hypothesis to account for these demise resurrection population dynamics, and the continued maintenance of the phage following the ascent of PYOSa resistant SCVs. We test and provide evidence in support of that hypothesis with a DNA sequence analysis of the genetic basis of the SCVs. By combining PYOSa with antibiotics, the density of the S. aureus population can be markedly reduced. There are, however, significant differences in the effectiveness of this combination therapy depending on whether the antibiotics and phage are used simultaneously or sequentially and on whether the antibiotics are bacteriostatic or bactericidal. Treatment with PYOSa, followed by the administration of bactericidal antibiotics, is more effective in reducing density of these bacterial population than treatment with these antibiotics alone. The methods developed here to evaluate the clinical potential of PYOSa in combination with antibiotics and design protocols for treating S. aureus infections with these phages and antibiotics can be employed for these purposes for any phage and bacterium that can be cultured in vitro.
Results
Bacteriophage PYOSa Has a Broad Host Range for S. aureus.
We use two assays to determine the host range of PYOSa: 1) the production of zones of inhibition in soft agar lawns and 2) changes in the optical density of exponentially growing liquid cultures of S. aureus mixed with this phage. By both criteria, S. aureus Newman and 12 clinical isolates of methicillin-sensitive S. aureus from the Network on Antimicrobial Resistance (NARSA) collection (29) were all sensitive to PYOSa and appeared to be unable to generate classically resistant mutants. Additional evidence for a broad host range of PYOSa comes from a survey of 71 clinical isolates of S. aureus, including 54 methicillin-resistant S. aureus (MRSA) strains and phylogenetically similar species performed by LabCorp (SI Appendix, Table S1).
S. aureus Appears to Be Unable to Generate Classical, Surface Modification, Mutants Resistant to PYOSa.
Evidence for this comes from experiments with S. aureus Newman and 12 clinical isolates from the NARSA collection (29). Single colonies of each strain were grown in the presence of ∼106 phage particles per mL, and after 24 h of exposure to the phage in liquid culture, the optical densities of exponentially growing S. aureus were no greater than that of the media without the bacteria, and by plating, we were unable to detect colonies from these cultures.
Bacterial Population Heterogeneity and the Maintenance of Phage.
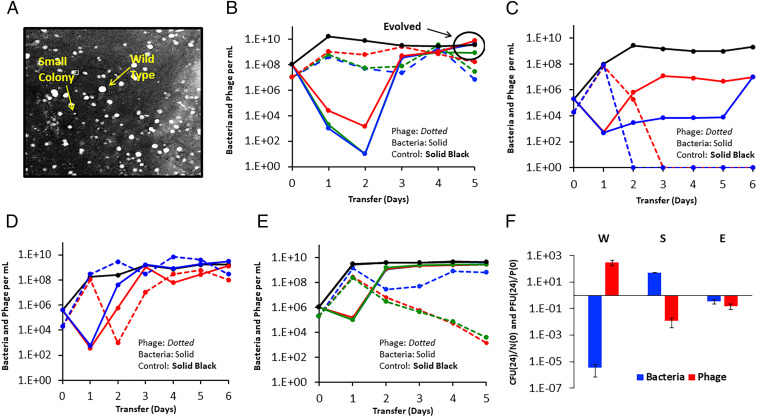
With the bacterial growth and phage infection parameters estimated for S. aureus Newman and PYOSa, a complete clearance of the bacteria in these experiments after less than 24 h of exposure to PYOSa is anticipated from a simple mass action model of the population dynamics of bacteria and phage (30). To determine whether this is the case empirically, we divided a culture with ∼104 S. aureus Newman into 28 tubes. We let these cultures replicate for 2 h, for an average density of 3 × 107 and then added ∼6 × 108 PYOSa. At 24 h, all 28 cultures were clear, and no bacterial colonies were found on the lysogeny broth (LB) plate samplings of these cultures. However, by day seven, 26 of these 28 independent cultures were turbid or somewhat turbid. These turbid cultures all had phage at densities in excess of 5 × 106. Upon plating these 7-d cultures, colonies were detected with two distinct phenotypes: colonies similar in size to the ancestral wild-type S. aureus Newman and much smaller colonies, small colony variants (Fig. 1A).
Fig. 1.
Small-colony variants and the population dynamics of PYOSa and S. aureus Newman in serial transfer culture. (A) Wild-type S. aureus and small-colony variants visualized. (B) Changes in densities of S. aureus Newman and PYOSa in three independent (red, green, blue) serial transfer experiments diluted 1/100 in fresh media daily. (C) Changes in densities of S. aureus Newman and PYOSa in serial transfer cultures initiated two small-colony variants isolated from the fifth transfer of the cultures with PYOSa and S. aureus Newman in B. (D) Changes in densities of S. aureus Newman and PYOSa in serial transfer cultures initiated with PYOSa and equal densities of cultures derived from small colonies and the ancestral S. aureus Newman. (E) Changes in densities of S. aureus and PYOSa in serial transfer cultures initiated with PYOSa and single colonies of evolved (wild-type colony morphology) bacteria isolated from the fifth transfer of the cultures in B. (F) Changes in the ratio of bacteria and phage at time 0 and 24 h. Wild-type S. aureus Newman (W), small-colony variants (S), and evolved bacteria (E). Three independent replicas.
To elucidate why PYOSa does not kill all of the S. aureus Newman in these liquid cultures, we prepared three independent 10-mL cultures with ∼108 S. aureus Newman and ∼107 PYOSa and serially passaged these cultures for 5 d, transferring 1/100 of these cultures to fresh media each day. The results of these serial transfer experiments are presented in Fig. 1B. As anticipated from the experiment with the 28 tube cultures, at 24 h, all the flasks were clear. The densities of bacteria remaining in these cultures estimated by plating, ∼103 per mL, is probably an underestimate of the real density due to killing by phage on the plates. Additional support for the hypothesis that the densities of S. aureus in these liquid cultures is substantially greater than the colony-forming units (CFU) estimate is that the density of the PYOSa phage did not decline at each transfer, and therefore, the phage must have been replicating. This hypothesis was evaluated with a simple model (30) using the phage infection parameters estimated for PYOSa and S. aureus Newman. To maintain a population in a culture diluted by 100-fold the phage density must increase by at least 100-fold at each transfer and for this to occur, the product of the adsorption rated constant, δ, the burst size β, and the density of sensitive bacteria, N, has to exceed 100. With the estimated values, δ = 4.8 × 10−7 and β = 80, the density of bacteria in these cultures would have to be at least 2.6 × 106 for the phage not to be diluted out.
Most intriguingly, in all three independent serial transfer populations with the PYOSa phage present, the CFU estimates of the density of S. aureus started to increase after the second transfer. By the fourth and fifth transfer, the density of cells in these cultures was similar to that of the phage-free controls, and the phage continued to be maintained, presumably because of the continued presence of phage-susceptible populations. To test this, we spread bacterial aliquots onto agar and observed heterogeneity in colony growth with both fast-growing and small-colony variants. Using a spot test, we showed that cultures grown from fast-growing colonies, henceforth called the evolved bacteria, were susceptible to the original PYOSa as well as to the coexisting PYOSa. In contrast, the spot test showed that cultures grown from small colony variants isolated from these late transfer cultures were resistant to lysis by PYOSa.
To further evaluate the different bacterial phenotypes present in the late transfer cultures, we serially passaged two small-colony cultures in the presence of PYOSa (Fig. 1C). We observed that cultures generated from small-colony variants were unable to support the phage. These data and the spot test results suggest an explanation for the maintenance of bacteria and phages throughout the original transfer experiment (Fig. 1B). Our hypothesis is that the bacterial population is polymorphic and includes sensitive cells capable of supporting phage growth as well as a nonsensitive population. We tested this hypothesis by mixing wild-type S. aureus Newman in equal frequency with small-colony variants and observed, consistent with the hypothesis, that the phages were maintained throughout the serial transfers and that the total bacterial population density remained similar to that in phage-free controls (Fig. 1D). As a separate test of the polymorphic population hypothesis, we made serial transfer experiments initiated with mixtures of PYOSa phages and evolved bacteria isolated from the fifth transfer cultures (Fig. 1E). The results of these experiments are consistent with this reduced susceptibility hypothesis. Moreover, within a single transfer, the bacterial density returned to that of the phage-free controls and the phages declined, albeit at a rate less than that anticipated if they were not replicating at all, in two of the three cultures. In the two cultures where the phage declined significantly, the dominant bacterial population were small-colony variants, while the replicate with the coexisting phage was dominated by bacteria that generated wild-type colony morphologies. See SI Appendix, section II for a consideration of the between experiment variation in the population dynamics observed in Fig. 1 B and E.
Based on the preceding results and interpretations, we postulate that the bacteria are of three states, the ancestral wild-type, small-colony variants, and what we call the evolved state, which have wild-type or near wild-type colony growth rate and rise to high densities after serial passaging in the presence of phage. Both the wild-type and the evolved are postulated to be capable of supporting phage growth, whereas the small colonies type do not support phage growth.
The Genetic Basis of Bacterial Colony Growth Rate Variation.
To determine the genetic basis of the differences between wild-type S. aureus Newman, the small-colony variants, and evolved states, colonies with a range of sizes were isolated from early and late in serial passaging experiments similar to those shown in Fig. 1B. Whole-genome sequencing of several clones revealed a clear sequence of events during the passaging. The first event was the selection of mutations in femA which encodes a protein responsible for assembling the pentaglycine interpeptide bridges in the S. aureus cell wall (SI Appendix, Table S2). These mutants all have a small-colony variant phenotype. The range of observed colony sizes is representative of the diversity of mutations identified in femA and presumably indicates a direct correlation between the growth rate and the severity of the defect in FemA function. We observed in later transfers larger colonies that carried a femA mutation and an additional mutation (see below), suggesting that this was a compensatory mutation restoring growth rate and creating the evolved state. The small-colony variant phenotype of S. aureus is known to be very unstable and subject to rapid suppression by a wide variety of compensatory mutations (31). To investigate the nature of the evolved state, we chose several different femA mutants and used these to select faster-growing colonies. Fast growers were easily selected and were observed on agar as larger colonies growing above the slower-growing parental small-colony strains. Whole-genome sequencing revealed an array of suppressing mutations were selected, including an internal suppressor in femA, mutations in a variety of genes affecting secondary messenger metabolism, and frequent mutations in the transcriptional regulator sarA (SI Appendix, Table S3). We concluded that the initial event upon exposure of wild-type S. aureus Newman to PYOSa is the selection of small-colony variants associated with mutations in femA. These slow-growing mutants then evolve to the faster-growing evolved state by the acquisition of suppressor mutations that frequently affect global transcriptional regulators.
Phenotypic Differences between Wild-Type, Small Colony, and Evolved Clones.
The hypothesis above predicts that cultures derived from bacterial colonies of each state should respond differently when exposed to PYOSa. To test this, we cocultured high densities of each bacterial state (wild-type [W], small-colony [S], and evolved-state [E]) with PYOSa and determined the relative change in densities of both bacteria and phage, respectively, 108 and 107 cells and particles per mL, after 24 h exposure (Fig. 1F). Each of the three states exhibited a unique dynamic. The density of the wild-type and evolved bacteria declined, with the wild-type declining to a much greater extent than the evolved state. In contrast, the density of small-colony variant bacteria increased. PYOSa phage increased on the wild-type bacteria but declined on both the small-colony variant and the evolved-state bacteria. The decline in phage was greater on small-colony variants than on the evolved state.
Measurement of the growth rates in liquid culture of the three bacterial states confirmed the growth parameters previously inferred from colony sizes (SI Appendix, Table S4). Mutations causing a small-colony phenotype on agar also caused severe growth defects in liquid culture, while the evolved clones with secondary suppressing mutations showed partial restoration of growth rate toward wild-type levels.
The pentapeptide crosslinks that FemA synthesizes are notably the target of lysostaphin. FemA mutants resistant to lysostaphin have previously been observed to become hypersensitive to penicillin antibiotics (32, 33). To test for evidence of these phenotypes, the minimal inhibitory concentration (MIC) of the three states were measured against lysostaphin and oxacillin (SI Appendix, Table S4). We observed that all of the strains carrying femA mutations (both small colony and evolved state) became resistant to lysostaphin and hypersensitive to oxacillin. Accordingly, the compensatory evolution from small-colony variant to evolved state does not phenotypically recreate a wild-type phenotype: wild-type and evolved are genetically and phenotypically distinct states. Since FemA is responsible for adding the second and third glycines to the interpeptide bridge, and FemB is responsible for adding the fourth and fifth residues, mutations in femA are expected to result in single-glycine bridges between the cell-wall peptides. To the degree that the femA mutants identified have different growth and susceptibility phenotypes, the cell-wall defect in strains carrying these mutations is likely not complete but rather represents a balance between full-length pentapeptide crosslinks and single-glycine crosslinks. FemA was recently reported to be essential (34), and it is notable that in our experiments no unequivocally femA null mutations were identified. Based on the number of mutants we have screened, this indicates that femA null mutants are either nonviable or are counterselected in these experiments.
PYOSa Does Not Replicate on Stationary Phase S. aureus.
In infected hosts, many of the bacteria will not be replicating, and thereby, phage would be particularly effective for treatment if they, like some antibiotics (35), could kill nonreplicating S. aureus. To determine whether PYOSa can kill and replicate on nongrowing bacteria, three independent 48-h stationary phase cultures were mixed with PYOSa for average initial densities of ∼4 × 109 S. aureus and 106 PYOSa. The bacteria and phages were incubated with shaking for 24 h, and the viable cell and phage densities were estimated and compared with the initial densities. There was no evidence for the stationary phase bacteria being killed; the mean and SE of the N(24)/N(0) ratio was 0.96 ± 0.03. Moreover and critically, there was a significant decline in the density of phage, N(24)/N(0) = 0.043 ± 0.006. Thus, not only does PYOSa not kill stationary phase S. aureus, these nonreplicating bacteria act as a sink and could reduce the density of PYOSa from treated hosts.
The results of experiments to determine whether the decline in the density of phage can be attributed to changes in the medium, such as a high pH, were negative. The density of PYOSa did not decline in sterile filtrates of 48-h stationary phase cultures. There is, however, the suggestion that the bacteria must be viable to lead to the reduction in the viable density. When the S. aureus in the 48-h stationary phase culture are killed with chloroform, the density of PYOSa does not decline
Accounting for the Population Dynamics Presented in Fig. 1B.
In the SI Appendix, section IV and Fig. S2, using a mathematical model and numerical solutions, we present a hypothesis for the kill-recovery dynamics observed for the bacteria, and the maintenance of the phage in the serial transfer cultures depicted in Fig. 1B. In accord with this hypothesis, 1) the bacterial population recovers from its initial demise due to predation by PYOSa phage by the generation and ascent of small-colony variants which are immune to and selected by the phage, and 2) the phage are maintained because of the instability of the small-colony variants, which continuously generate the evolved-state bacteria upon which the phage can replicate.
The Joint Action of PYOSa and Antibiotics.
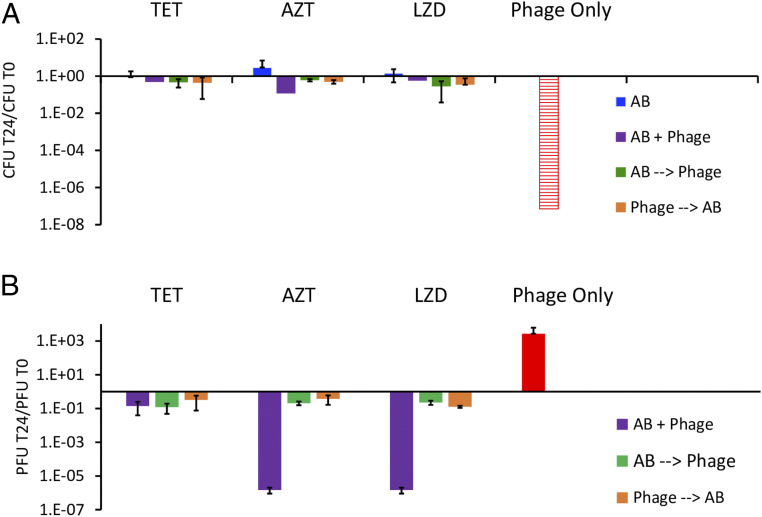
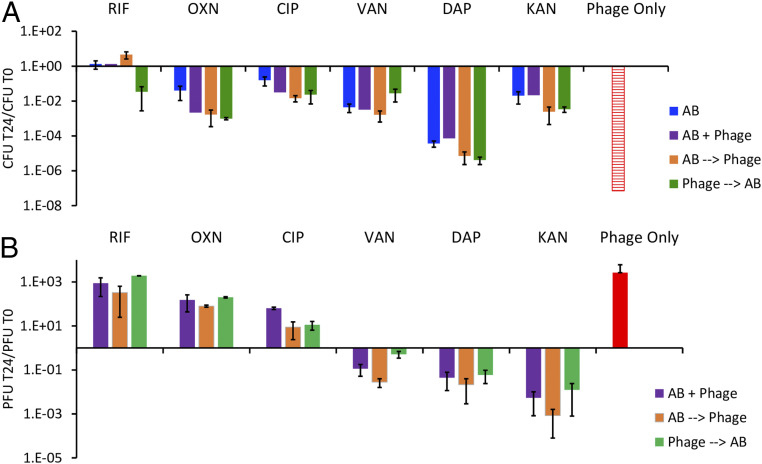
The high-throughput method to evaluate the simultaneous action of antibiotics and phage developed by Gu Liu and colleagues (17) is based on changes in optical densities and thereby provides no information about the effect of the antibiotics on the replication of the phage. The low throughput method employed in this investigation is based on changes in colony- and plaque-forming units, CFU and PFU, and provides information about the synergy or antagonism of the antibiotics on the replication of the PYOSa phage as well as the bacteria. In these experiments, we mixed growing cultures of S. aureus Newman and PYOSa in Mueller-Hinton, cation-adjusted broth (MHII) at densities of 4 × 106 with 4 × 105, respectively, and super MIC concentrations of the antibiotics. The densities of bacteria and phages were estimated just before the antibiotics were added and 24 h later, respectively, N(0) and N(24), where N is either CFU or PFU. In these experiments, the antibiotics and phages were introduced into the growing cultures of S. aureus Newman in three different ways: 1) simultaneously, AB+PYOSa, 2) antibiotics first and phage 30 min later, AB→PYOSa, and 3) phage first and antibiotics 30 min later, PYOSa→AB. As controls, we also treated parallel cultures of S. aureus Newman with antibiotics only, AB, and with phage alone. In Fig. 2 (bacteriostatic antibiotics) and Fig. 3 (bacteriocidal antibiotics), we present N(24)/N(0) ratios for the bacteria and phages for three independent experiments with each antibiotic and phage combination.
Fig. 2.
Joint action of bacteriostatic antibiotics and PYOSa. The concentrations of these antibiotics are 10 μg/mL. (A) The ratio of the change in density of S. aureus after 24 h of exposure to antibiotics (blue), antibiotics plus phage (purple, green, orange, refers to order of addition as explained in the text) or phage alone (red). Hash red, the density of S. aureus recovered was below the detection limit, ∼102 cells per mL. (B) The ratio of the change in the density of PYOSa after 24 h of confronting wild-type S. aureus in combination with antibiotics or alone.
Fig. 3.
The joint action of bactericidal antibiotics and PYOSa. Concentrations of the different antibiotics in μg/mL (RIF 0.02, OXA 3, CIP 0.5, VAN 8, DAP 64, KAN 46), corresponding to minimum bacteriocidal concentrations. (A) The ratio of the change in density of S. aureus after 24 h of exposure to antibiotics, antibiotics and phage, or phage alone. Hash red, the density of S. aureus recovered was below the detection limit, ∼102 per mL. (B) The ratio of the change in the density of PYOSa after 24 h of confronting S. aureus Newman in combination with antibiotics or alone.
With the bacteriostatic antibiotics, tetracycline (TET), azithromycin (AZM), and linezolid (LZD), the greatest decline in the density of bacteria and increase in the density of phages obtained in the experiment where phages are used alone. There is clearly a negative synergy between these bacteriostatic antibiotics and the phage. Whether administered simultaneously or sequentially, at the concentrations used, these antibiotics prevent PYOSa from killing S. aureus Newman. When the phages are administered simultaneously with AZM and LZD, the phage density declines.
In Fig. 3, we present the results of experiments with bactericidal antibiotics, rifampin (RIF), oxacillin (OXA), ciprofloxacin (CIP), vancomycin (VAN), daptomycin (DAP), and kanamycin (KAN). The data suggest that the simultaneous or sequential administration of PYOSa may modestly increase the rate at which bactericidal antibiotics kill S. aureus at the concentrations employed. However, as was observed for the parallel experiment with the bacteriostatic drugs (Fig. 2), the phages kill more S. aureus in the absence of antibiotics than with these drugs, an antagonistic interaction once again. The failure of RIF to reduce the viable density of S. aureus can be attributed to RIF resistance emerging. It should be noted, however, that in one of the three treatments where PYOSa was used before adding RIF, it prevented the ascent of resistance. Most interestingly, while treatment with RIF, OXA, and CIP allowed PYOSa to replicate, this was not the case for VAN, DAP, and KAN, which appeared to suppress the replication of PYOSa.
Sequential Treatment, Phage Followed by Antibiotics.
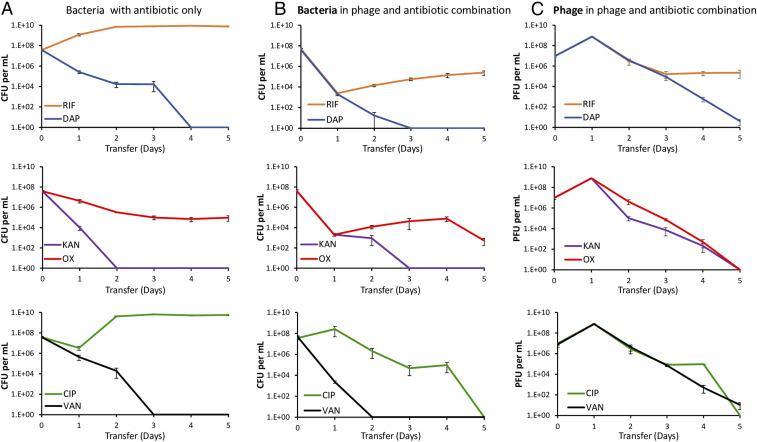
The serial transfer results presented in Fig. 1B indicate that, as a consequence of the emergence and ascent of small colony variants, PYOSa by itself will not be able to control an S. aureus population for an extended time. Although the phage continues to be present, the density of the bacteria returns to levels similar to that of the phage-free control (Fig. 1B). We asked what result would be obtained if antibiotics were administered to these populations? To address this question, we performed the serial transfer experiments with the addition of bactericidal antibiotics following the phage-mediated reduction in the density of wild-type S. aureus observed during the first 24 h. The results of these experiments are presented in Fig. 4.
Fig. 4.
Changes in the densities of bacteria and phage in serial transfer cultures treated with antibiotics alone or in combination with phages. (A) Bacteria treated with antibiotics alone. (B) Bacterial densities in cultures containing both the antibiotic and the phage. (C) Phage densities in cultures containing both the antibiotic and the phage. Mean and SEs of three independent experiments. Row 1: Treatment with RIF and DAP, 0.02 and 64 µg/mL, respectively. Row 2: Treatment with KAN and OXA, 46 and 3 µg/mL, respectively. Row 3: Treatment with CIP and VAN, 0.5 and 8 µg/mL, respectively.
Three of the antibiotics, DAP, KAN, and VAN, were alone sufficient to eliminate the S. aureus population (Fig. 4A). In contrast, with RIF by the second transfer the density of S. aureus in treated cultures reached a density observed for antibiotic and phage free cultures (∼2 × 109 bacterial per mL). The reason for the failure of RIF to clear the culture was the emergence of mutants resistant to this drug. All six colonies tested from the fifth transfer were resistant to RIF. However, the viable cells recovered from the phage and RIF combination cultures were as sensitive to RIF as were the antibiotic-free controls. Neither OXA nor CIP alone cleared the cultures. In the case of OXA, the viable cell density declined but continued to persist at a density of ∼105 cells per mL. The bacteria recovered from these cultures were sensitive to OXA. We postulate that this leveling off can be attributed to persistence (36); refer to SI Appendix, section VI and Fig. S5. In the case of CIP, the initial exposure led to a substantial decline in the viable cell density of S. aureus; however, following the second transfer, the bacterial population recovered and was sustained at densities similar to that in the antibiotic-free controls. The colonies of S. aureus recovered at the end of this experiment were susceptible to CIP. We postulate that these resurrection dynamics could be attributed to heteroresistance (37), refer to SI Appendix, section VI and Fig. S5.
Discussion and Conclusions
On first consideration, PYOSa seems to be an ideal phage for treating S. aureus infections. The results of this study provide evidence in support of three virtues of PYOSa as a therapeutic phage. 1) PYOSa is likely to kill virtually all methicillin-resistant as well as methicillin-sensitive S. aureus. 2) S. aureus are unable to generate classical surface-resistant mutants to PYOSa; thus, mixtures of multiple phages would not be needed to ensure coverage or prevent resistance. 3) S. aureus Newman has a high adsorption rate and burst size with PYOSa and, when first confronting growing populations of S. aureus, the bacteria are killed, and the phages replicate at a high rate.
On the downside, our experiments raise caveats about the use of PYOSa for treating S. aureus infections alone and suggest a possible liability as well. Not only is PYOSa unable to clear cultures of S. aureus Newman, it also selects for potentially pathogenic small-colony variants (38–41) that are refractory to this phage. Through a “leaky resistance” mechanism (42), the phage population continues to be maintained, and the bacterial population continues to persist at densities not much less than they do in the absence of PYOSa. Although not observed for the small-colony variants tested here, at least some small colonies are more resistant to antibiotics than the bacteria from which they are derived (31, 43, 44).
For phage therapy to be a practical and acceptable enterprise, these viruses would have to be used in combination with antibiotics. Our results indicate that when administered simultaneously, or nearly simultaneously with antibiotics, PYOSa does worse in killing S. aureus than it does when used alone. The ribosome-targeting bacteriostatic antibiotics, TET, AZM, and LZD, suppress the ability of PYOSa to kill S. aureus Newman. This observation is consistent with the failure of PYOSa to replicate on stationary phase populations of S. aureus. It is also consistent with the Numbers Game hypothesis for the action of bacteriostatic antibiotics (45), according to which, the number of free ribosomes is too low to support the protein synthesis needed for replication of the phage.
When administered simultaneously with bactericidal antibiotics, PYOSa is also less effective in killing S. aureus than it is in the absence of these drugs. We postulate that this can be attributed to pharmaco- and population dynamics of the joint action of antibiotics and phage; the antibiotics reduce the densities of the bacteria and thereby lower the capacity of the phage to replicate (SI Appendix, section IV). More broadly, these results raise another element that should be considered in the design and evaluation of joint antibiotic and phage treatment protocols: the importance of sequential therapy to maximize efficacy. Accordingly, by reducing the density of bacteria, phage may increase the efficacy of antibiotics (46). Quite the opposite obtains for the phage when antibiotics are used simultaneously with, or prior to, the administration of these viruses. For more considerations of the role of population dynamic processes for the therapeutic use of phages, see ref. 47.
Sequential Treatment with PYOSa and Antibiotics.
Our experiments suggest a way to deal with the major caveat and potential liability of treatment with PYOSa alone, the recovery of the bacterial population due to the ascent of small-colony variants. The administration of bactericidal antibiotics following the initial decline in the density of bacteria due to PYOSa prevents bacterial population recovery and eliminates or prevents the selection of small-colony variants. The latter is not the case when the antibiotics are used alone. One interpretation of this is that sequential treatment, initially with phages, then with a bactericidal antibiotic may be more effective than treatment with antibiotics alone or phages alone.
Conclusion and a Recommendation.
We interpret the results of this in silico and in vitro study to suggest that PYOSa will be effective for treatment of S. aureus infections but only if the administration of bactericidal antibiotics follows that of phage. The next step will, of course, be to test this sequential phage and antibiotic treatment hypothesis with S. aureus infections in experimental animal models.
The methods used in this study with PYOSa in combination with antibiotics could also be employed to evaluate the potential clinical efficacy and facilitate the design of treatment protocols for any phages and bacteria that can be cultured in vitro. We recommend that this be done prior to the evaluation of efficacy and treatment protocols with experimental animals. Existing data suggested that PYOSa met all of the criteria desired for a phage to be effective for therapy. The in vitro experiments performed in this investigation uncovered a limitation and potential liability of using this phage for therapy that would not have been anticipated. This jointly theoretical and experimental study also revealed a way to deal with this limitation and liability.
Materials and Methods
Strains and Growth Media.
Unless otherwise noted, all experiments were performed from derivatives of the parent strain S. aureus Newman (ATCC 25904). The parent S. aureus Newman was obtained from Bill Schafer of Emory University. The small-colony variant and evolved strains were obtained from PYOSa challenged S. aureus Newman by experiments performed in our laboratories. The investigation for classical resistance was performed in the following MSSA strains obtained from Abraham Moller in the Reid Lab at Emory University: NRS52, NRS102, NRS180, NRS110, NRS252, NRS253, NRS266, NRS109, NRS148, and NRS205.
Bacterial cultures were grown at 37 °C in Mueller-Hinter II (MHII) Broth (275710, BD) and on Luria-Bertani Agar (LB) Plates (244510, BD). PYOSa lysates were prepared from single plaques at 37 °C in MHII broth alongside wild-type S. aureus Newman by plate lysis. Specifically, individual phage plaques were picked with a sterile stick, resuspended in 4 mL of soft agar with 0.1 mL of overnight bacterial culture and plated on top of phage plates. The plates were then incubated at 37 °C overnight. The soft agar was scraped with a sterile iron scoop, resuspended in 10 mL MHII with ∼0.5 mL of chloroform to kill the surviving bacteria. The lysates were then centrifuged to remove the agar, sterilized by filtration (0.2 μm), and stored at 4 °C.
Sampling Bacterial and Phage Densities.
Bacteria and phage densities were estimated by serial dilutions in 0.85% NaCl solution followed by plating. The total density of bacteria was estimated on LB (1.6%) agar plates. To estimate the densities of free phage, chloroform was added to suspensions before serial dilutions. These suspensions were mixed with 0.1 mL of overnight MHII grown cultures of wild-type S. aureus Newman (about 5 × 108 cells per mL) in 4 mL of LB soft (0.65%) agar and poured onto semihard (1%) LB agar plates.
Parameter Estimations.
The parameters critical for the interaction of the PYOSa phage and S. aureus Newman used in this study were estimated in independent experiments MHII broth. The maximum growth rate of different clones of S. aureus Newman was measured by Bioscreen, as described in ref. 48. Phage burst sizes (β) were estimated with one-step growth experiments similar to ref. 49. Adsorption of PYOSa to S. aureus was estimated as described in ref. 49.
Serial Transfer Experiments.
All serial transfer experiments were carried out in 10 mL MHII cultures grown at 37 °C with vigorous shaking. The cultures were initiated by 1:100 dilution from 10-mL overnight cultures grown from single colonies. Phage was added to these cultures to reach the initial density of ∼106 PFU/mL. At the end of each transfer, 0.1 mL of each culture was transferred into flasks with fresh medium (1:100 dilution). Simultaneously, 0.1-mL samples were taken for estimating the densities of CFUs and PFUs by serial dilution and plating on solid agar.
Antibiotics and Their Sources.
TET, OXA, VAN, KAN, and Streptomycin are from Sigma Aldrich, AZM is from Tocris, DAP is from MP Biochemicals, RIF is from Applichem, and LZD is from Chem-Impex International.
Whole-Genome Sequencing.
For sequencing individual clones of S. aureus, genomic DNA was prepared using the MasterPure Gram-Positive kit, following the manufacturer's instructions (Epicentre, Illumina Inc.). Final DNA was resuspended in EB buffer. Genomic DNA concentrations were measured in a Qubit 2.0 Fluorometer (Invitrogen via Thermo Fisher Scientific). DNA was diluted to 0.2 ng/µL in water (Sigma-Aldrich), and the samples were prepared for whole-genome sequencing according to Nextera XT DNA Library Preparation Guide (Illumina Inc.). After the PCR cleanup step, samples were validated for DNA fragment size distribution using the Agilent High Sensitivity D1000 ScreenTape System (Agilent Technologies). Sequencing was performed using a MiSeq desktop sequencer, according to the manufacturer's instructions (Illumina Inc.). The sequencing data were aligned and analyzed in CLC Genomics Workbench version 11.0 (CLCbio).
Supplementary Material
Acknowledgments
We thank Melony Ivey and Esther Lee for superb technical help. We are grateful to our Staphylococcus maven, Abraham (Jon) Moller, for providing strains and sage advice and to Waqas Chaudhry, Andrew Smith, and the reviewers of an earlier incarnation of this report for helpful comments and suggestions. Funds for this research were provided by grants from the US National Institutes of General Medical Sciences, R01 GM091875 and R35 GM 136407 (B.R.L.); Vetenskapsrådet (Swedish Research Council), 2017-0359; and the Scandinavian Society for Antimicrobial Chemotherapy, SLS-693211 and SLS-876451 (D.H.). These funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008007118/-/DCSupplemental.
Data Availability
The paired-end sequence reads of the strains described in the text have been deposited in NCBI Bioproject database with accession number PRJNA688212. All other data discussed in the paper are available in the main text and SI Appendix.
References
- 1.D’Herelle F., The Bacteriophage and Its Behavior (The Williams & Wilkins Company, Baltimore, MD, 1926), pp. 578. [Google Scholar]
- 2.Ansaldi M., et al., “French phage network”-Third meeting report. Viruses 10, 123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watts G., Phage Therapy: Revival of the Bygone Antimicrobial (Elsevier, 2017). [DOI] [PubMed] [Google Scholar]
- 4.Kortright K. E., Chan B. K., Koff J. L., Turner P. E., Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Schmidt C., Phage therapy’s latest makeover. Nat. Biotechnol. 37, 581–586 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Martel B., Moineau S., CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 42, 9504–9513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu T. K., Collins J. J., Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. U.S.A. 106, 4629–4634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson S. B., et al., Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front. Microbiol. 10, 2537 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krylov V. N., [Phagotherapy in terms of bacteriophage genetics: Hopes, perspectives, safety, limitations]. Genetika 37, 869–887 (2001). [PubMed] [Google Scholar]
- 10.Labrie S. J., Samson J. E., Moineau S., Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Moradpour Z., Ghasemian A., Modified phages: Novel antimicrobial agents to combat infectious diseases. Biotechnol. Adv. 29, 732–738 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Dubos R. J., Straus J. H., Pierce C., The multiplication of bacteriophage in vivo and its protective effect against an experimental infection with Shigella dysenteriae. J. Exp. Med. 78, 161–168 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bull J. J., Levin B. R., DeRouin T., Walker N., Bloch C. A., Dynamics of success and failure in phage and antibiotic therapy in experimental infections. BMC Microbiol. 2, 35 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickey J., Perrot V., Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PLoS One 14, e0209390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkhilaishvili T., Winkler T., Müller M., Perka C., Trampuz A., Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64, e00924-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tagliaferri T. L., Jansen M., Horz H. P., Fighting pathogenic bacteria on two fronts: Phages and antibiotics as combined strategy. Front. Cell. Infect. Microbiol. 9, 22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Liu C., et al., Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. mBio 11, e01462-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schooley R. T., et al., Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan B. K., et al., Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedrick R. M., et al., Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia G., et al., Wall teichoic Acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 193, 4006–4009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki T., Swoboda J. G., Campbell J., Walker S., Gilmore M. S., In vitro antimicrobial activity of wall teichoic acid biosynthesis inhibitors against Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55, 767–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia G., Kohler T., Peschel A., The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int. J. Med. Microbiol. 300, 148–154 (2010). [DOI] [PubMed] [Google Scholar]
- 24.McCallin S., Sarker S. A., Sultana S., Oechslin F., Brüssow H., Metagenome analysis of Russian and Georgian Pyophage cocktails and a placebo-controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ. Microbiol. 20, 3278–3293 (2018). [DOI] [PubMed] [Google Scholar]
- 25.O’Flaherty S., et al., Genome of staphylococcal phage K: A new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186, 2862–2871 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato’o Y., et al., Tailor-made gene silencing of Staphylococcus aureus clinical isolates by CRISPR interference. PLoS One 13, e0185987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh S. E., Lo H. H., Chen S. T., Lee M. C., Tseng Y. H., Wide host range and strong lytic activity of Staphylococcus aureus lytic phage Stau2. Appl. Environ. Microbiol. 77, 756–761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutter E., et al., Phage therapy in clinical practice: Treatment of human infections. Curr. Pharm. Biotechnol. 11, 69–86 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Fey P. D., et al., A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin B. R., Stewart F. M., Chao L., Resource-limited growth, competition, and predation: A model and experimental studies with bacteria and bacteriophage. Am. Nat. 977, 3–24 (1977). [Google Scholar]
- 31.Cao S., Huseby D. L., Brandis G., Hughes D., Alternative evolutionary pathways for drug-resistant small colony variant mutants in Staphylococcus aureus. mBio 8, e00358-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strandén A. M., Ehlert K., Labischinski H., Berger-Bächi B., Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179, 9–16 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Climo M. W., Ehlert K., Archer G. L., Mechanism and suppression of lysostaphin resistance in oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1431–1437 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monteiro J. M., et al., Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCall I. C., Shah N., Govindan A., Baquero F., Levin B. R., Antibiotic killing of diversely generated populations of nonreplicating bacteria. Antimicrob. Agents Chemother. 63, e02360-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balaban N. Q., et al., Publisher correction: Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicoloff H., Hjort K., Levin B. R., Andersson D. I., The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Kahl B., et al., Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177, 1023–1029 (1998). [DOI] [PubMed] [Google Scholar]
- 39.von Eiff C., et al., A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179, 4706–4712 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sendi P., Proctor R. A., Staphylococcus aureus as an intracellular pathogen: The role of small colony variants. Trends Microbiol. 17, 54–58 (2009). [DOI] [PubMed] [Google Scholar]
- 41.von Eiff C., Staphylococcus aureus small colony variants: A challenge to microbiologists and clinicians. Int. J. Antimicrob. Agents 31, 507–510 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Chaudhry W. N., et al., Leaky resistance and the conditions for the existence of lytic bacteriophage. PLoS Biol. 16, e2005971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baumert N., et al., Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8, 253–260 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Norström T., Lannergård J., Hughes D., Genetic and phenotypic identification of fusidic acid-resistant mutants with the small-colony-variant phenotype in Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 4438–4446 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin B. R., et al., A numbers game: Ribosome densities, bacterial growth, and antibiotic-mediated stasis and death. mBio 8, e02253-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Udekwu K. I., Parrish N., Ankomah P., Baquero F., Levin B. R., Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63, 745–757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bull J. J., Levin B. R., Molineux I. J., Promises and pitfalls of in vivo evolution to improve phage therapy. Viruses 11, 1083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Concepción-Acevedo J., Weiss H. N., Chaudhry W. N., Levin B. R., Malthusian parameters as estimators of the fitness of microbes: A cautionary tale about the low side of high throughput. PLoS One 10, e0126915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellis E. L., Delbrück M., The growth of bacteriophage. J. Gen. Physiol. 22, 365–384 (1939). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The paired-end sequence reads of the strains described in the text have been deposited in NCBI Bioproject database with accession number PRJNA688212. All other data discussed in the paper are available in the main text and SI Appendix.