Significance
Here, we used a fluorescent protein that is free in solution and is trapped in nuclear HIV-1 capsids to demonstrate that the capsids retain integrity and prevent mixing of macromolecules within the viral core and the cellular environment until just before integration. We also found that capsid integrity is maintained until just minutes before disassembly in the nucleus, revealing that uncoating proceeds rapidly after integrity loss. These valuable insights into the early stage of HIV-1 replication indicate that intact HIV-1 capsids are imported through nuclear pores, that reverse transcription is mostly completed within intact capsids, and that preintegration complex-host interactions facilitating integration and target site selection must occur within a short time frame between capsid disassembly and integration.
Keywords: HIV-1, capsid, core integrity, nuclear import, uncoating
Abstract
We recently reported that HIV-1 cores that retained >94% of their capsid (CA) protein entered the nucleus and disassembled (uncoated) near their integration site <1.5 h before integration. However, whether the nuclear capsids lost their integrity by rupturing or a small loss of CA before capsid disassembly was unclear. Here, we utilized a previously reported vector in which green fluorescent protein is inserted in HIV-1 Gag (iGFP); proteolytic processing efficiently releases GFP, some of which remains trapped inside capsids and serves as a fluid phase content marker that is released when the capsids lose their integrity. We found that nuclear capsids retained their integrity until shortly before integration and lost their GFP content marker ∼1 to 3 min before loss of capsid-associated mRuby-tagged cleavage and polyadenylation specificity factor 6 (mRuby-CPSF6). In contrast, loss of GFP fused to CA and mRuby-CPSF6 occurred simultaneously, indicating that viral cores retain their integrity until just minutes before uncoating. Our results indicate that HIV-1 evolved to retain its capsid integrity and maintain a separation between macromolecules in the viral core and the nuclear environment until uncoating occurs just before integration. These observations imply that intact HIV-1 capsids are imported through nuclear pores; that reverse transcription occurs in an intact capsid; and that interactions between the preintegration complex and LEDGF/p75, and possibly other host factors that facilitate integration, must occur during the short time period between loss of capsid integrity and integration.
HIV-1 reverse transcribes its genome, enters the nucleus, and integrates its double-stranded DNA into the host genome to form a provirus. HIV-1 uncoating, defined as disassembly of the viral capsid, a macromolecular conical structure that is formed in virions by assembly of ∼1,500 capsid (CA) proteins (1–3), is an essential step in the early stage of HIV-1 replication that is required to release newly synthesized viral DNA for integration into the host genome (4, 5). For many years, it was thought that the capsid must uncoat so that the viral reverse transcriptase (RT) inside the core can have access to the cytoplasmic deoxynucleotide triphosphates (dNTPs) needed for DNA synthesis (6, 7). Previous models of HIV-1 uncoating have suggested that the viral core uncoats immediately upon fusion near the plasma membrane (8–11), that the viral core uncoats gradually after fusion as it is transported from the plasma membrane to the nuclear envelope (NE) (12–15), or that the viral core uncoats after it is docked at the NE just before the reverse transcription complex (RTC)/preintegration complex (PIC) enters the nucleus (16–21). Consistent with the uncoating at the NE model, it has been argued that the HIV-1 capsid must remain intact so that the RT and integrase (IN) can remain associated with the viral nucleic acid until a functional PIC is formed (19). In the gradual uncoating model and the uncoating at the NE model, it remains possible that the capsid can rupture or lose its integrity and thus allow dNTPs to enter the core and support reverse transcription while a capsid lattice is maintained to prevent loss of RT and IN from the viral cores. In this scenario, the capsids would lose their integrity early on when reverse transcription is initiated but retain most of the CA until a functional PIC is formed several hours later. It is noteworthy that a ring of positively charged arginine residues in a capsid pore formed by CA hexamers was reported to facilitate the import of dNTPs into intact viral cores, allowing RT access to substrate dNTPs for reverse transcription without capsid integrity loss (22). How the same hexameric ring of arginines binds to inositol hexakisphosphate and facilitates transfer of dNTPs into viral cores is not well understood at this time (23).
The timing and cellular location of HIV-1 uncoating has been difficult to study for two main reasons. First, only one of ∼50 RTCs/PICs in infected cells leads to productive infection (24) and biochemical studies of the population of RTCs/PICs in infected cells may not reflect the properties of infectious virions. Second, methods to directly label and quantify CA in RTCs/PICs in imaging assays without significantly inhibiting virus infectivity were not available. We recently developed a method to directly label and quantify CA in RTCs/PICs in infected cells (25). Briefly, we produced virions in which a small proportion of the CA in capsids was fused to green fluorescent protein (GFP) at the N terminus of CA. The virions retained most of their infectivity, allowing us to quantify the amounts of CA associated with replication-competent RTCs/PICs. In addition, we developed a method to detect transcriptionally active proviruses, enabling us to identify infectious RTCs/PICs that led to productive infection. Our GFP-CA direct labeling method provides advantages for studies of uncoating over immunofluorescence staining for CA, which relies on epitope accessibility (17, 24, 26–28) or fluorescently labeled CA-associated host factor cyclophilin A (CypA) (16, 29), which relies on maintenance of the CypA-CA interaction.
Our studies using the GFP-CA labeling method indicated that RTCs/PICs retain >94% of the CA in intact capsids until they uncoat ∼1.5 h near the integration sites before integration and detection of HIV-1 transcription (25). We also concluded that the RTCs/PICs complete reverse transcription in the nucleus and remain sensitive to CA inhibitor PF74, indicating that the nuclear viral complexes retain assembled CA hexamers; this conclusion was supported by Dharan et al. who reported similar findings in another recent study (30). These results led us to conclude that nuclear capsids are intact (or nearly intact) until they uncoat just prior to integration. Although our results indicated that nuclear capsids retained most of the CA in intact viral cores, we could not determine whether the nuclear capsids had ruptured or lost a few CA molecules, leading to a loss of core integrity and mixing of the capsid content and the nuclear environment before disassembly of the capsid. Whether loss of core integrity and capsid disassembly are discrete steps or whether they occur simultaneously is unknown.
Cleavage and polyadenylation specificity factor 6 (CPSF6) is a nuclear host protein that interacts with incoming HIV-1 capsids (31). We recently observed that CPSF6 fused to fluorescent protein mRuby (mRuby-CPSF6) accumulated onto capsids shortly after they docked at the NE and remained associated with the capsids until they uncoated inside the nucleus just before integration (25). In addition, CPSF6 binding was shown to be essential for integration into gene-rich euchromatin located in the internal regions of the nucleus (32–34). Here, we labeled HIV-1 capsids with a GFP that is free in solution and is trapped in infectious nuclear HIV-1 capsids; the GFP served as fluid phase marker and was used to determine capsid integrity. We found that nuclear viral cores that retained their integrity led to productive infection. Furthermore, we determined that capsid integrity is lost only ∼1 to 3 min before uncoating in the nucleus, indicating that uncoating proceeds rapidly after the initial integrity loss.
Results
Labeling HIV-1 Capsids with a GFP Fluid Phase Marker.
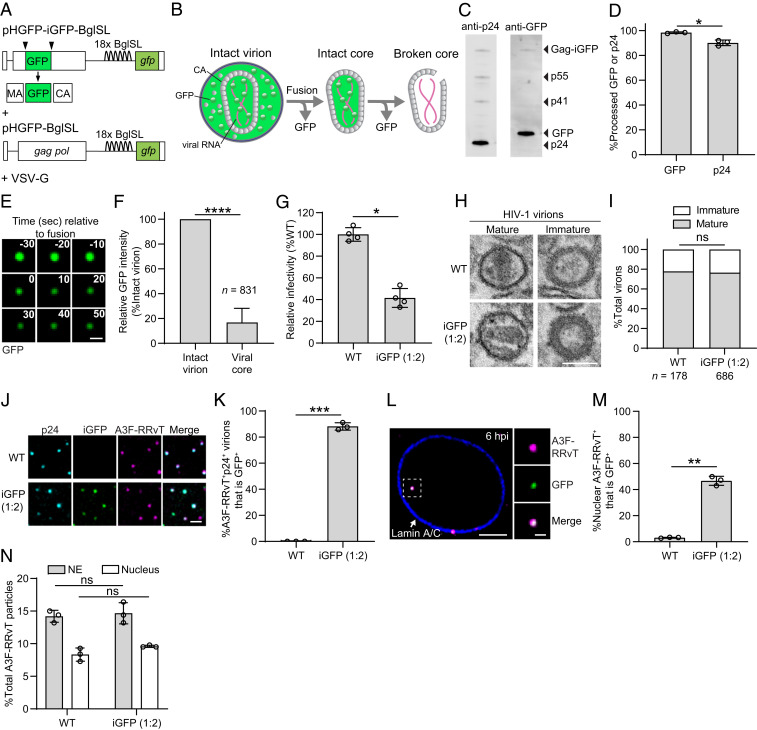
To determine whether viral cores in infected cells retained their integrity, we utilized a previously described HIV-1 vector in which GFP was inserted between matrix (MA) and CA (pNL4-3 Gag-iGFP) (35) (Fig. 1A). Proteolytic processing during virion maturation releases GFP from the Gag precursor, some of which remains trapped inside the capsid and is released upon loss of core integrity (Fig. 1B). We also utilized a previously described RNA stem-loop system in which 18 copies of BglG stem loops (BglSLs) were engineered into the HIV-1 genome in place of vif/vpr to facilitate the detection of nascent HIV-1 genomic RNA at the HIV-1 transcription site (Fig. 1A). This system can be used to identify the capsids that lead to productive infection and determine the nuclear location of integration (25). Virions were produced by cotransfection of an HIV-1 vector expressing Gag-iGFP and a vector expressing wild-type gag-pol at a 1:2 ratio (Fig. 1A). Western blot analysis of these virions indicated that nearly all GFP was fully processed from Gag-iGFP and the processing efficiency was similar to that of CA from Gag (Fig. 1 C and D). Treatment of iGFP-labeled virions with saponin detergent in vitro to disrupt the viral membrane reduced the GFP signals to 16.9 ± 11.3% compared to the GFP intensities of intact virions, indicating that nearly ∼20% of the GFP remained inside the capsids (Fig. 1 E and F). The virions retained almost half of their infectivity compared to control virions produced in the absence of iGFP (Fig. 1G). The morphology of iGFP-labeled and unlabeled virions was similar and the ratio of virions with mature and immature morphology was not significantly different (Fig. 1 H and I). To determine whether GFP-labeled capsids were detectable after nuclear import, we generated virions that were colabeled with GFP content marker and core-associated host restriction factor APOBEC3F (A3F) fused to fluorescent protein red-red vine tomato (RRvT) (25). Single virion analysis of HIV-1 particles immunostained with anti-CA antibody showed that most A3F-RRvT+ p24+ virions were labeled with GFP (∼88%, Fig. 1 J and K). In contrast to a previous report indicating the GFP content marker is lost in the cytoplasm ∼25 min after fusion (36), we observed many GFP-labeled capsids at the NE and inside the nucleus at 6 h postinfection (hpi) (Fig. 1 L and M). The percentage of A3F-RRvT-labeled RTCs/PICs in the nucleus at 6 hpi that were GFP+ was ∼47%, indicating many of the capsids that had entered the nucleus remained intact. Interestingly, ∼47% (762/1,608 × 100%) of the A3F-RRvT+ capsids also had detectable GFP content marker levels shortly after in vitro saponin treatment (SI Appendix, Fig. S1A), suggesting that the GFP content marker levels in ∼53% of the nuclear capsids were below the limit of detection. To further analyze the capsid labeling efficiency, we divided the 1,608 GFP+ intact virions into four quartiles based on their GFP intensity and determined the percentage of capsids that had detectable levels of GFP intensity for each quartile (SI Appendix, Fig. S1B). The GFP intensities of the capsids (SI Appendix, Fig. S1B) and the proportion of GFP+ capsids were correlated with the GFP intensities of the intact virions (SI Appendix, Fig. S1C), strongly suggesting that the ∼47% frequency of GFP+ nuclear capsids was expected based on their labeling and detection efficiency. The percentages of A3F-RRvT-labeled RTCs/PICs at the NE and inside the nucleus at 6 hpi were similar for GFP-labeled and unlabeled virions, indicating that GFP labeling of the capsids did not affect their efficiency of docking with the NE or their nuclear import (Fig. 1N).
Fig. 1.
Characterization of GFP content marker-labeled virions and detection of GFP-labeled capsids in nuclei of infected HeLa cells. (A) GFP content marker-labeled virions were produced by cotransfection of HIV-1 vectors pHGFP-iGFP-BglSL and pHGFP-BglSL at a 1:2 ratio. (B) Schematic of a GFP content marker-labeled HIV-1 virion and a postfusion capsid. GFP is fluid phase marker distributed throughout the virion upon virus maturation. Fusion of viral and host cell membranes releases GFP that is outside of the core, while some GFP is retained in an intact capsid until a rupture in the core or loss of CA molecules results in loss of core integrity (broken core). (C) GFP content marker-labeled virus purified through a 20% sucrose cushion was analyzed by Western blotting with anti-HIV p24 (Left lane) and anti-GFP (Right lane) antibodies. (D) Quantitation of the proportion of processed GFP or p24 CA relative to the total GFP (98%) or CA (90%) in the virions, respectively. High time-resolution images (1 frame/10 s) (E) and analyses (F) of iGFP-labeled virions upon treatment with saponin detergent. Scale bar for E, 1 μm. (G) HIV-1 virions produced by cotransfection of 293T cells with pHGFP-iGFP-BglSL and pHGFP-BglSL at a 1:2 ratio retain 40% of the infectivity in HeLa cells compared to unlabeled wild-type (WT) virions (set to 100%). (H and I) Quantitative electron microscopy to determine the proportion of HIV-1 virions containing mature capsids labeled with (77%) and without iGFP (78%). (Scale bar, 100 nm.) Statistical significance was determined using Fisher’s exact test. (J) Analysis of HIV-1 virions labeled with A3F-RRvT and iGFP or A3F-RRvT alone. The virions were centrifuged onto a slide and immunostained with α-p24 CA antibody. Scale bar, 2 μm. (K) The percentage of A3F-RRvT+p24-CA+ virions that are iGFP+. (L) Nucleus of a HeLa cell infected with virions colabeled with GFP and A3F-RRvT. The cells were fixed at 6 hpi and immunostained with anti-Lamin A/C antibody to visualize the NE. Scale bar, 5 μm; Inset, 1 μm. (M) The percentage of A3F-RRvT-labeled virus without (WT) and with GFP associated with the NE or inside the nucleus. (N) Similar proportions of viral complexes labeled with A3F-RRvT without (WT) and with GFP (1:2 ratio) that stably associated with the NE and entered the nucleus at 6 hpi. Average of three independent experiments is shown. For D, F, G, K, M, and N, error bars indicate ±SD; P values are from Welch’s t tests. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant (P > 0.05).
Capsid-Associated GFP Is Efficiently Released from Gag by Proteolytic Processing.
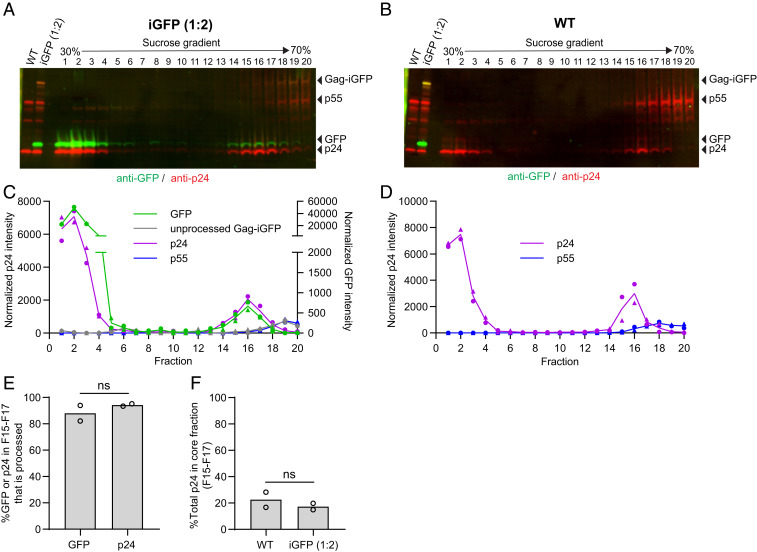
To establish that the GFP associated with capsids can serve as a reporter of viral core integrity, we determined whether the GFP associated with viral cores was efficiently cleaved from the Gag polyprotein and did not remain covalently attached to Gag. To characterize the GFP associated with capsids, the GFP-labeled and unlabeled virions were subjected to detergent lysis and sucrose gradient fractionation following a previously described protocol (37) (Fig. 2). In these gradients, free CA remained at the top of the 30 to 70% sucrose gradient (fractions 1 through 4) while the GFP-labeled and unlabeled viral cores sedimented and concentrated in fractions 15 through 17 (Fig. 2 A and B, respectively). The GFP-labeled and unlabeled capsids were concentrated in the same fractions, indicating that GFP labeling did not significantly alter their sedimentation. Western blot analysis of the fractions using anti-GFP antibody (Fig. 2 A and C) indicated that most of the GFP associated with fractions 15 through 17 was fully proteolytically cleaved, and no Gag-iGFP or partially cleaved products were detectable. Most of the Gag-iGFP fusion protein was detected in fractions 18 through 20, consistent with previous reports indicating that immature viral cores sediment to the bottom of the gradient and concentrate in these fractions (Fig. 2 A and C) (38). As expected, Western blot of fractions containing unlabeled virions did not detect any GFP proteins (Fig. 2B). Most of the unprocessed Gag (p55) was also present in fractions 18 through 20, consistent with immature viral cores concentrating in these fractions near the bottom of the gradient (Fig. 2 B and D). Importantly, most of the GFP and CA in fractions 15 through 17 was fully processed (Fig. 2E, 88 ± 8.4% and 94 ± 1.3%, respectively), indicating that the GFP signals associated with viral cores mostly represent free GFP and not Gag-iGFP fusion proteins. A similar proportion of CA from GFP-labeled and unlabeled virions was associated with the viral cores in fractions 15 through 17, indicating that GFP labeling did not significantly influence the in vitro stability of the viral cores (Fig. 2F).
Fig. 2.
Intact capsids contain processed GFP. (A and B) Western blots of 20 fractions obtained from detergent treatment and sucrose-gradient fractionation (30 to 70% sucrose) of HIV-1 virions labeled with (A) and without (B) GFP. GFP was detected using anti-GFP antibody followed by IRDye 800CW-labeled secondary antibody (green) and gag was detected using anti-p24 antibody followed by IRDye 680-labeled secondary antibody (red). Capsid-associated GFP and p24 sediment to fractions 15 through 17, whereas GFP and p24 between the viral membrane and capsid remains at the top of the gradient (fractions 1 through 4). Most of the unprocessed Gag-iGFP and p55 Gag sediment to the bottom of the gradient (fractions 18 through 20). (C and D) Quantitation of processed GFP, p24, unprocessed Gag-iGFP, and p55 for virions labeled with (C) and without (D) iGFP from two independent experiments, indicated by circles and triangles. (E) The percentage of processed GFP (88%) or p24 CA (94%) relative to unprocessed Gag-iGFP and p55, respectively, in the core fractions (15 through 17) for virions labeled with GFP. (F) The percentage of total p24 CA signal in the gradient (fractions 1 through 20) that is in the core fractions (15 through 17) for HIV-1 virions labeled with (17%) and without (23%) GFP. P values are from Welch’s t tests. ns, not significant (P > 0.05).
Postfusion Capsids in the Cytoplasm and in the Nucleus Retain Similar Levels of GFP.
As illustrated in Fig. 1B, we expect the GFP between the viral membrane and the capsid to diffuse away upon fusion of the viral and host membranes, and the capsid-associated GFP to remain until there is a loss of core integrity. To determine the proportion of GFP in the virions that is trapped inside viral cores, we performed a high-time resolution (1 frame/min) analysis of infected living cells to capture fusion events and compared the GFP signal intensities before and after fusion (Fig. 3 A and B and Movie S1). We infected cells at very low multiplicities of infection (MOI ≤ 0.01; <2 GFP+ virions/cell) so that individual virions could be tracked. We captured 43 virions that exhibited a substantial loss of the GFP signal in a single step, indicative of fusion, followed by stable GFP signals during the remaining observation period (∼25 min; Fig. 3B). Next, we compared the GFP intensities of the postfusion capsids identified in these movies to the intensities of the GFP-labeled viral cores in the nuclei of HeLa cells and CEM-SS T cells (Fig. 3 C and D). These results showed that nuclear viral cores in HeLa and CEM-SS T cells have similar GFP levels compared to the postfusion cytoplasmic viral cores, indicating that the nuclear capsids in HeLa and CEM-SS T cells have retained their integrity through nuclear import and inside the nucleus.
Fig. 3.
Postfusion capsids in the cytoplasm and capsids in the nucleus retain similar levels of GFP. (A) Representative live-cell microscopy images showing the fusion of GFP-labeled virions and release of the capsid in the cytoplasm of a HeLa cell expressing Bgl-mCherry, which was used to visualize the nucleus. (B) The GFP intensities of 43 GFP-labeled virions before and after fusion. Intensities were normalized to the average intensities from the three frames before fusion (set to 100). (C) Representative images of a GFP-labeled capsid in the nucleus of a HeLa cell (Left) and a CEM-SS T cell (Upper Right) expressing mRuby-Lamin B at 6 hpi, and a primary CD4+ T cell (Lower Right) stained with DAPI (blue) and anti-CD4 antibody (red) at 5 hpi. (D) Comparison of the GFP intensities of intact virions and postfusion capsids in the cytoplasm of HeLa cells, and capsids in the nucleus of HeLa, CEM-SS, and primary CD4+ T cells. Lines are mean ± SD; P values are from Welch’s t tests; ns, not significant (P > 0.05). (Scale bars for A and C, 5 µm; for Inset, 1 µm.)
We also infected primary CD4+ T cells, the primary target of HIV-1 infection, with GFP content marker-labeled virions (Fig. 3 C, Lower Right). We readily observed GFP signals in the nuclei of infected cells at 5 hpi, indicating that capsids that retained their core integrity were imported into the nucleus of primary CD4+ T cells. The GFP signal intensities of the nuclear capsids in the primary CD4+ T cells were not significantly different from the postfusion cytoplasmic capsids in HeLa cells (Fig. 3D), indicating that the capsids in the nuclei of primary CD4+ T cells retained their integrity through nuclear import.
Loss of Capsid Integrity in the Nucleus near the Integration Site.
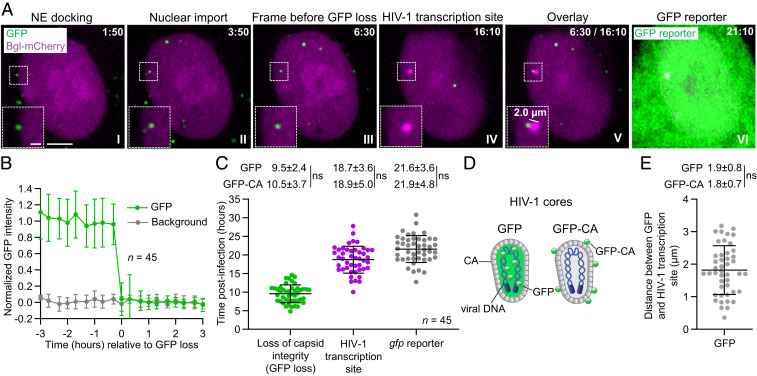
To determine if the capsids labeled with the GFP content marker inside the nucleus are infectious, we utilized our previously described live-cell imaging strategy to identify viruses that lead to productive infection as well as determine the nuclear location of integration (25). Briefly, 18 copies of BglSLs were inserted into the vif/vpr region of the HIV-1 genome (Fig. 1A). HeLa cells that stably expressed bacterial protein Bgl fused to mCherry (Bgl-mCherry), a protein which specifically recognizes the BglSL, were infected at a low MOI (<0.1 GFP-expressing proviruses/cell) with GFP-labeled virions. Integration of the viral DNA into the host genome, followed by transcription of the integrated provirus, resulted in detection of transcriptionally active proviruses as bright mCherry+ puncta in the nucleus that appeared several hours later near the sites where the GFP content marker signal disappeared (Movie S2). Fig. 4A shows an example of the capsid docked at the NE at 1:50 h:min postinfection (I) and imported into the nucleus at 3:50 h:min postinfection (II); the GFP signal disappeared at 6:30 h:min postinfection (III), and an HIV-1 transcription site became detectable at 16:10 h:min postinfection (IV). An overlay of the frame before the GFP signal loss and the first frame of HIV-1 transcription site detection showed that the HIV-1 transcription site appeared 2 µm from the site of the GFP signal loss (V). The GFP reporter expression became detectable at 21:10 h:min postinfection (VI).
Fig. 4.
GFP-labeled capsids lose their integrity in the nucleus prior to detection of HIV-1 transcription site. (A) Representative live-cell microscopy images of a HeLa:Bgl-mCherry cell infected with GFP-labeled virions. An GFP-labeled nuclear viral core docked at NE 1:50 h:min postinfection (I), entered the nucleus at 3:50 h:min postinfection (II), and lost the GFP signal between 6:30 h:min postinfection and 6:50 h:min postinfection, indicative of loss of core integrity (III); HIV-1 transcription site appeared at 16:10 h:min postinfection (IV). Overlay of last frame in which iGFP signal was detectable and first frame of HIV-1 transcription site appearance (after correcting for cell movement) indicates that the HIV-1 transcription site appeared ∼2.0 µm from the site of GFP disappearance (V). GFP reporter expression was detected 21:10 h:min postinfection (VI). (Scale bar, 5 µm; Inset, 2 µm.) (B) Average normalized GFP intensities of 45 infectious capsids were stable in the nucleus for several hours before abrupt GFP signal loss within a single frame (<20 min). (C) The time of GFP loss, HIV-1 transcription site detection, and gfp reporter detection for 45 infectious RTCs/PICs. (D) Illustrations of GFP- and GFP-CA-labeled capsids. (Left) A capsid labeled with GFP, which is not covalently attached to any viral protein and is expected to behave as a fluid phase marker. (Right) A capsid that is labeled with a small amount of CA protein fused to GFP. (E) The distance between GFP signal detected in the frame prior to loss and associated HIV-1 transcription site (after correcting for cell movement). For C and E, values were compared to those previously determined for 59 infectious GFP-CA-labeled viruses (25). Lines are mean ± SD; P values are from Welch’s t tests; ns, not significant (P > 0.05).
We identified 45 infectious capsids labeled with the GFP content marker inside the nucleus that resulted in productive infection and appearance of an HIV-1 transcription site (Fig. 4B). The GFP signals for these 45 capsids remained steady for several hours inside the nucleus until abrupt loss of the GFP content marker signal within a single 20-min time period between consecutive frames in movies captured at 20 min/frame, indicating a loss of core integrity inside the nucleus (Fig. 4B). The average time of GFP content marker signal loss was ∼9.5 hpi (Fig. 4C) followed by detection of the HIV-1 transcription sites at ∼18.7 hpi and gfp reporter expression at ∼21.6 hpi (Fig. 4C). The average time of the GFP content marker signal disappearance, HIV-1 transcription site appearance, and GFP reporter detection were not significantly different from the previously determined average time of GFP-CA signal disappearance (∼10.5 hpi), HIV-1 transcription site detection (∼18.9 hpi), and GFP reporter detection (∼21.9 hpi), respectively (Fig. 4C) (25). As illustrated in Fig. 4D, the GFP content marker signal loss indicates loss of core integrity, whereas loss of GFP-CA signal indicates uncoating. These results demonstrated that the capsids inside the nucleus retain their core integrity for several hours following nuclear import, and core integrity loss and uncoating occur within the same 20-min period between frames. The average distance between the site of GFP content marker signal loss and HIV-1 transcription site appearance was ∼1.9 µm, which was not significantly different from the average distance between GFP-CA signal loss and HIV-1 transcription site appearance (∼1.8 µm; Fig. 4E) (25). We previously determined that the average distance between GFP-CA signal loss and HIV-1 transcription site appearance was similar to the average distance HIV-1 transcription sites moved within the same time frame; furthermore, the average distance was in general agreement with the previously reported constrained diffusion of genes with a 1.5-µm radius (39). Thus, these observations indicate that the loss of HIV-1 capsid integrity occurred at or very close to the chromosomal sites of integration.
High Concentration of Inhibitor PF-3450074 (PF74) Results in Nuclear Capsid Integrity Loss and Uncoating.
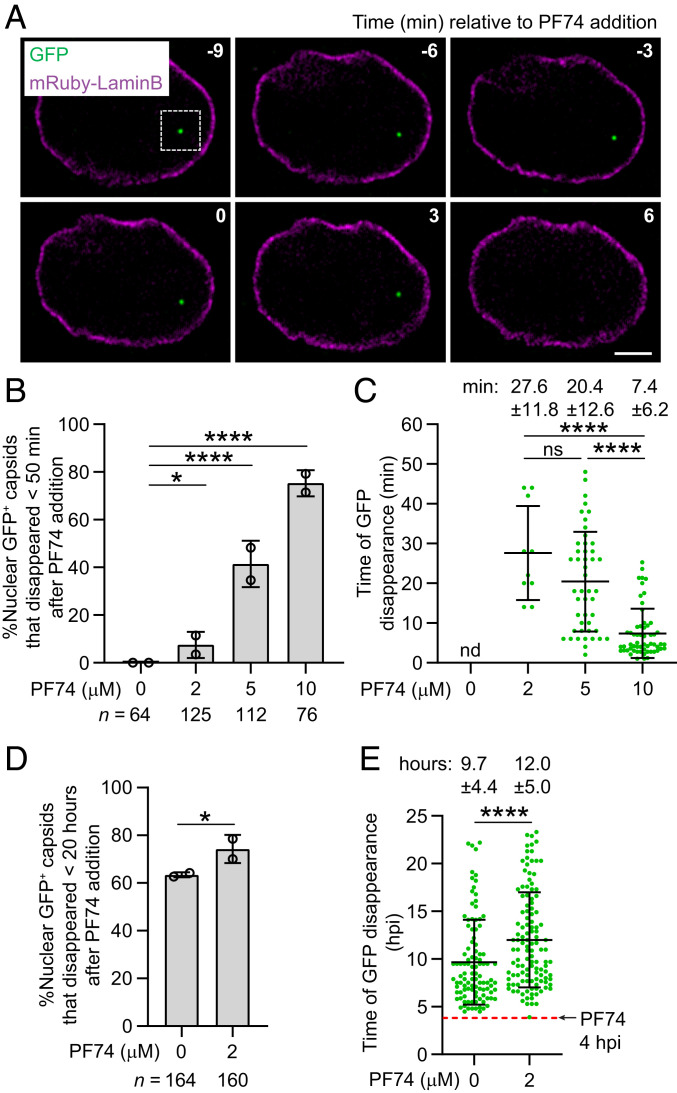
The CA-binding inhibitor PF74 binds to the interface between capsid monomers in assembled capsids and destabilizes that capsid lattice (40–42). PF74 susceptibility is therefore dependent on the assembled capsid. We previously observed that treatment of infected cells with 10 µM PF74 resulted in capsid destabilization and disassembly (25). Consistent with this observation, treatment of infected cells after nuclear import of GFP-labeled viral cores with 10 µM PF74 resulted in rapid disappearance of the GFP fluid phase marker signals from the nuclei (Fig. 5A), indicating that most nuclear capsids lost integrity upon PF74 binding. There was a dose-dependent increase in the percentage of the GFP signals that disappeared inside the nucleus within 50 min of PF74 treatment (Fig. 5B) and a dose-dependent decrease in the time of GFP disappearance after PF74 treatment (Fig. 5C). These results indicate that higher concentrations of PF74 (>5 µM) lead to a rapid loss of integrity for most capsids. The GFP fluid phase marker was lost from viral cores within a single frame (<2 min) in cells treated with either 2 or 10 µM PF74, indicating that the GFP fluid phase marker diffused away rapidly from the viral cores that lost their integrity even at a low PF74 concentration when most viral cores remained intact (SI Appendix, Fig. S2). To further evaluate the effect of low PF74 concentration on capsid stability, we acquired time-lapse images every 20 min for 20 h immediately following treatment of the infected cells with 2 µM PF74 at 4 hpi. Treatment with 2 µM PF74 led to a slight increase in the percentage of GFP-labeled capsids that disappeared inside the nucleus <20 h after PF74 addition compared to GFP-labeled capsids in control cells (Fig. 5D). However, 2 µM PF74 treatment led to a 2.3-h delay in the time of GFP disappearance relative to GFP-labeled capsids in control cells, indicating that low amounts of PF74 can transiently stabilize capsids and delay the loss of the GFP content marker (Fig. 5E). We also observed the rapid disappearance of GFP signal from most GFP-CA-labeled nuclear capsids (81/82) after 10 µM PF74 treatment, but not 2 µM PF74 treatment (0/51) (SI Appendix, Fig. S3A). Overall, most viral cores lost the GFP-CA signal or the GFP content marker at 10 µM PF74 but not at 2 µM PF74, suggesting that GFP-CA labeling of capsids did not substantially influence their sensitivity to PF74; however, a subtle effect of GFP-CA labeling on PF74 sensitivity cannot be excluded. Notably, the time of disappearance of the GFP content marker-labeled capsids in cells treated with 10 µM PF74 was the same as that of GFP-CA-labeled capsids (SI Appendix, Fig. S3B), suggesting that PF74 susceptibility was not altered by GFP-CA labeling of the capsids.
Fig. 5.
Disruption of nuclear capsids labeled with GFP with CA-binding inhibitor PF74. (A) Representative live-cell microscopy images of a nuclear GFP-labeled capsid in a HeLa cell expressing mRuby-Lamin B before and after addition of 10 µM PF74. Numbers in white indicate time (minutes) relative to the time of PF74 addition. (Scale bar, 5 µm.) (B and C) The percentage of nuclear capsids labeled with GFP content marker that disappeared within <50 min of treatment with different concentrations of PF74 (B) and the time of GFP disappearance relative to the time of PF74 addition (C). (D and E) The percentage of nuclear capsids labeled with GFP content marker that disappeared within 20 h of treatment with 2 µM PF74 at 4 hpi or mock treatment at 4 hpi (D) and the time of GFP disappearance relative to the time of infection (E). For B and D, P values are from Fisher’s exact tests; ****P < 0.0001; *P < 0.05. For C and E, lines are mean ± SD; P values are from Mann–Whitney test; ****P < 0.0001; ns, not significant (P > 0.05).
Capsid Integrity Is Maintained Until Just Minutes before Disassembly.
We found that the GFP content marker and GFP-CA signals were lost within the same 20-min time period between consecutive movie frames, indicating that core integrity loss and uncoating occurred within 20 min of each other. To determine the relative kinetics of core integrity loss and uncoating more precisely, we infected HeLa cells that stably expressed mRuby-CPSF6, which directly binds to CA in viral capsids and can be used to indirectly determine the level of CA associated with viral cores (25). We then performed high time-resolution analysis (1 frame/5 min) of the infected cells to determine the timing of GFP content marker and GFP-CA signal loss relative to mRuby-CPSF6 signal loss to determine the kinetics of core integrity loss and capsid disassembly, respectively.
We infected HeLa cells expressing mRuby-CPSF6 with GFP content marker-labeled virions and determined the kinetics of loss of GFP and mRuby-CPSF6 signals for 25 GFP-labeled nuclear viral cores (Fig. 6). Sixteen of the 25 nuclear GFP-labeled viral cores exhibited loss of the GFP and mRuby-CPSF6 signal within the same 5-min time period between consecutive frames (Fig. 6 A and B and Movie S3). However, 9 of the 25 nuclear viral cores lost the GFP signal in the 5-min time period before the loss of the mRuby-CPSF6 signal (Fig. 6C and SI Appendix, Fig. S4A and Movie S4). In contrast, all 26 GFP-CA-labeled viral cores lost both GFP-CA and mRuby-CPSF6 signals within the same 5-min time period between consecutive frames (Fig. 6D and SI Appendix, Fig. S4B and Movie S5). Overall, 64% of the GFP-labeled viral cores lost the GFP and the mRuby-CPSF6 signals in the same 5-min time period, whereas 100% of the GFP-CA-labeled viral cores lost the GFP-CA and the mRuby-CPSF6 signals in the 5-min time period (Fig. 6E). These results suggested that viral core uncoating, as determined by GFP-CA signal loss, and mRuby-CPSF6 signal loss have very similar kinetics. However, capsid integrity loss, as determined by GFP content marker signal loss, may occur a few minutes before uncoating.
Fig. 6.
High time-resolution analysis of the loss of GFP, GFP-CA, and capsid-associated mRuby-CPSF6. (A) Representative live-cell microscopy images (1 frame/5 min) of infected HeLa:mRuby-CPSF6 cells showing disappearance of both GFP and mRuby-CPSF6 signals in the same frame. Scale bar, 5 μm; Inset, 1 μm. (B–D) The GFP and mRuby-CPSF6 intensities for 16 viral cores dual-labeled with GFP and mRuby-CPSF6 from which both signals disappear in the same 5-min period between consecutive frames (B); 9 viral cores dual-labeled with GFP and mRuby-CPSF6 from which GFP disappears one frame before mRuby-CSPF6 disappearance (C); and 26 viral cores dual-labeled with GFP-CA and mRuby-CPSF6 from which both signals disappear in the same 5-min period between consecutive frames (D). (E) The percentage of GFP-labeled (Left) or GFP-CA-labeled (Right) viral cores in the nucleus from which the GFP signal and the mRuby-CPSF6 signal disappeared in the same frame or the GFP signal was lost one frame before mRuby-CPSF6. (F) Comparison of the observed values with the expected values derived by modeling the probability of capturing GFP and mRuby-CPSF6 signals disappearing in the same 5-min period between consecutive frames or the GFP signal disappearing 1 to 4 min before mRuby-CPSF6 disappearance. P values are from Fisher’s exact tests. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant (P > 0.05).
To estimate the average time of GFP content marker signal loss relative to the mRuby-CPSF6 signal loss we modeled the probability of capturing the loss of both GFP and mRuby signals within the same 5-min period or between adjacent 5-min periods (Fig. 6F). We observed that 9 of 25 viral cores lost the GFP content marker signal in the frame before the mRuby signal was lost, and 0 of 26 GFP-CA signals were lost before the mRuby signal were lost. If the GFP content marker signal was lost an average of 1 min before the mRuby signal loss, we would expect 5 of 25 particles to lose the GFP content marker signal in the 5-min period before the mRuby signal loss; similarly, 10 of 25, 15 of 25, and 20 of 25 viral cores would be expected to lose the GFP signal in the 5-min period before the mRuby signal was lost, if the GFP signal was lost an average of 2, 3, or 4 min before the mRuby signal loss, respectively. The expected proportion of GFP-labeled viral cores that lost the GFP signal in the frame before the mRuby signal loss was not significantly different from the observed proportion (9 of 25), when the average time of GFP signal loss was assumed to be 1, 2, or 3 min before the mRuby signal loss (5 of 25, 10 of 25, and 15 of 25, respectively; P > 0.05); however, it was significantly different when the average time of GFP signal loss was assumed to be 0 or 4 min before the mRuby signal loss (0 of 25 and 20 of 25, respectively; P < 0.01). These results and the modeling analysis suggest that the nuclear viral capsids lose their integrity, as indicated by loss of the GFP signal, ∼1 to 3 min before disassembly, as indicated by loss of the mRuby-CPSF6 signal (SI Appendix, Fig. S4 C, Upper). In contrast, the GFP-CA signal loss occurs <1 min before the mRuby-CPSF6 signal loss; consequently, all 26 particles observed lost both signals within the same 5-min period (SI Appendix, Fig. S4 C, Lower).
Discussion
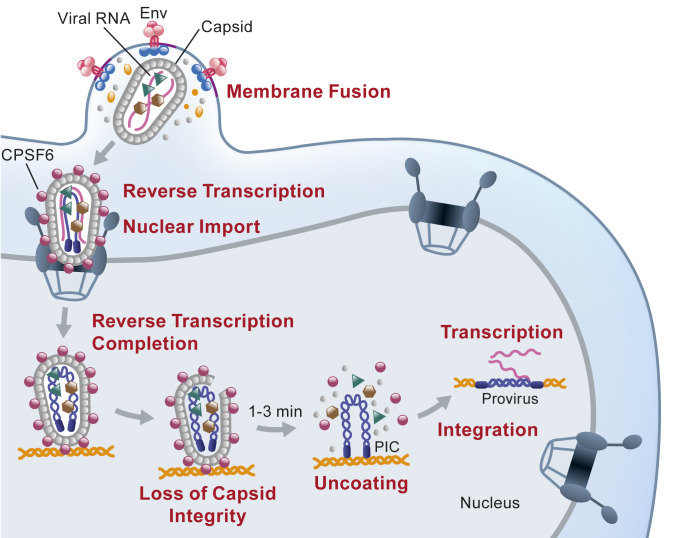
The results of our studies show that the HIV-1 viral cores that lead to productive infection retain their integrity inside the nucleus of infected cells until ∼1 to 3 min before uncoating. A model for the early stage of HIV-1 replication based on the current and recently published studies (25) is shown in Fig. 7. The intact HIV-1 capsid docks at a nuclear pore complex and recruits CPSF6. The intact capsid and associated CPSF6 enter the nucleus and quickly transport to the site of integration, where they remain for several hours. The capsids lose their integrity, either by developing holes (loss of CA) or rupturing (no loss of CA), just ∼1 to 3 min before uncoating, resulting in a dramatic and rapid loss of CA. Upon uncoating, the PIC is released from the capsid, after which it can interact with LEDGF/p75 (43–45) and potentially other host factors, and integrate into the chromatin. These recent studies (25, 30) challenge previous models suggesting that capsid disassembly primarily occurs in the cytoplasm (36) or at the nuclear pore complex prior to nuclear import (16, 18, 46, 47). Our finding that capsids retain their integrity until shortly before viral core uncoating imply that host factors that promote integration must engage the RTC/PICs in the nucleus during a short time period (<1.5 h) between capsid integrity loss and integration. Overall, these observations provide insights into nuclear import, reverse transcription, formation of the PIC, and integration.
Fig. 7.
Model for nuclear import and HIV-1 uncoating. Intact viral cores are released in cytoplasm upon fusion and transported to the nuclear envelope. Reverse transcription is initiated in the cytoplasm. After docking to the nuclear pore complex, the viral cores recruit CPSF6, which facilitates their entry into the nucleus. Reverse transcription is completed inside the intact viral core. Loss of capsid integrity occurs ∼10 h after infection, followed ∼1 to 3 min later by rapid (<1 min) disassembly of the viral core. This results in the release of the preintegration complex (PIC) and the viral DNA integrates into the host chromatin near the site of uncoating to form a provirus.
The studies presented here provide valuable insights into the dynamics of capsid integrity loss and uncoating in infected cells. Capsids that retain the GFP fluid phase marker must be intact enough to prevent the release of GFP, which could only result from holes in the capsid lattice larger than the dimensions of GFP (∼4.2 nm × ∼2.4 nm) (48) or a rupturing of the capsid lattice without loss of CA protein. By comparison, the exterior diameter of a CA hexamer is ∼9.0 nm (49), suggesting that loss of a single hexamer, and perhaps even a single CA molecule (assuming a ∼4.5-nm diameter that is half the size of the hexamer diameter), would be sufficient to allow loss of the GFP content marker from the capsid. However, the minimum size of the holes in the capsid lattice needed to permit rapid loss of the GFP content marker is unknown.
Our high-time resolution (1 frame/5 min) analyses of the timing of GFP, GFP-CA, and core-associated mRuby-CPSF6 signal loss demonstrating capsid integrity loss ∼1 to 3 min before uncoating support a model in which uncoating is initiated by loss of core integrity and is completed when most or all of the CA is dissociated from the viral nucleic acid-protein complex, a process which occurs inside the nucleus of an infected cell in <3 min. Interestingly, the kinetics of the loss of capsid integrity and uncoating were in close agreement with the disassembly kinetics after the opening of capsids in vitro (half-life of 70 to 100 s) (50).
A previous analysis of HIV-1 capsids in intact virions by cryoelectron tomography (cryoET) reported that most capsids had regions of CA lattice that were disrupted or missing, and only a small proportion of the capsids (6 of 107) formed intact conical fullerene cones (2). As the authors noted, these imperfections in the capsids may represent assembly defects or disruptions during virus purification. It is likely that most of the viral cores with assembly defects lose the GFP content marker upon fusion and lead to abortive infection. Recently, Christensen et al. established an efficient in vitro reverse transcription/integration assay (51). They visualized capsids by cryoET that had disassembled to variable degrees and had loops of DNA strands protruding from the capsids, suggesting that intermediates in the uncoating process were captured. In view of the rapid kinetics of uncoating that we observed in vivo, we hypothesize that the process of uncoating proceeds more slowly in vitro, enabling the visualization of the partially disassembled capsids. A previous in vitro study that visualized capsid uncoating using time-lapse atomic force microscopy reported that the capsid undergoes a partial or complete rupture near the narrow end of the conical structure (52). We did not observe a significant loss of GFP-CA signal in the 5-min time period before uncoating (Fig. 6D and SI Appendix, Fig. S4B), suggesting that most of the CA remains associated with the capsids until uncoating. However, based on the sensitivity of the imaging and quantitation, loss of several CA hexamers at the time of integrity loss cannot be excluded.
A previous study also tracked HIV-1 labeled with GFP content marker by live-cell imaging and used a combination of low MOI infection and gfp reporter expression to identify the virions that led to productive infection (36). However, they observed the disappearance of the GFP content marker in the cytoplasm ∼25 min after fusion of the viral and host membranes; based on the loss of GFP signal disappearance soon after fusion it was concluded that uncoating occurs in the cytoplasm. It is not clear what biological and/or technical differences could have resulted in these differing observations and conclusions. We and others observed that only a small fraction (∼20%) of the GFP content marker signal inside a virion is contained within the capsid whereas the majority (∼80%) of the GFP content marker in the virion is located between the viral membrane and capsid and is lost at the time of fusion (36). Consistent with these observations, the authors did observe reduction of the GFP content marker signal in most viral complexes after fusion. A minority viral complexes (6/19) in their study retained some residual GFP signals after fusion until the end of the observation period (120 min); based on our observations, these particles could have led to productive infection. However, it was concluded that the particles that lost the residual GFP signals ∼25 to 30 min after fusion led to productive infection. We speculate that a combination of low detection efficiency and/or photobleaching resulted in an inability to detect the GFP content marker-labeled cores in the nucleus. In addition, in their system, the cells may have been infected with one or more unlabeled capsids that led to productive infection, along with a labeled capsid that lost the GFP signal soon after fusion.
It is unclear why HIV-1 capsids have evolved to remain intact until they disassemble in the nucleus. Consistent with a previous study indicating that capsids with altered stability are defective in reverse transcription (53), and recent studies indicating that HIV-1 completes reverse transcription in the nucleus (25, 30), we propose that the intact capsid provides a protective microenvironment that ensures high concentrations of RT and nucleocapsid near the viral nucleic acid to ensure completion of viral DNA synthesis. Similarly, an intact core might be necessary to maintain a high concentration of integrase near the viral DNA to ensure formation of a functional intasome. We and others have recently reported that time-of-addition assays with capsid inhibitors PF74 and GS-CA1 (25, 30, 54) inhibited viral replication and resulted in disassembly of nuclear viral cores (25), supporting the view that premature uncoating of the viral core prevents formation of a functional PIC that can integrate and form a provirus. Furthermore, our previous time-of-addition assays showed that the sensitivity to RT inhibitors is lost before sensitivity to PF74, suggesting that assembled capsids composed of CA hexamers are retained until after completion of reverse transcription (25).
Recent studies indicating that dNTPs can be transported into capsids through central pores in CA hexamers support the idea that reverse transcription can be completed inside an intact capsid (22). An intact capsid may provide additional functions, including protection from host restriction factors, facilitation of nuclear import, and evasion of innate sensing by cytoplasmic (55) and/or nuclear DNA sensors to suppress cellular immune responses (56). It was proposed that reverse transcription of a flexible RNA into a rigid double-stranded DNA increases the internal pressure on the capsid, leading to initiation of capsid disassembly (52). This hypothesis suggests that progression of reverse transcription and stiffness of the capsid, both of which may be affected by viral and cellular factors, play a role in initiating uncoating, but further studies will be needed to determine the molecular events that initiate and control the timing of uncoating.
The results of these studies indicate that intact viral cores that retain the GFP content marker are imported through the nuclear pores into the nucleus. A direct interaction of CPSF6 with the core at the nuclear pore complex is required for nuclear import (25, 28), although the exact mechanism by which CPSF6 facilitates the nuclear import of an intact core is unclear. Some degree of flexibility of the capsid and/or the nuclear pore may be required for the passage of intact capsids (width of ∼61 nm) (57) through nuclear pores (inner diameter of ∼39 nm) (58). In this regard, pearl necklace-like viral complexes containing CA and nucleic acid were observed in the nuclei of CD4+ T cells by electron microscopy (59). Although we did not determine the shape or flexibility of the capsids, our results suggest that if the intact capsids are imported through nuclear pores by altering their shape, such deformations do not result in a loss of the GFP content marker and core integrity. Electron microscopy studies will be needed to determine the shape of viral cores at the NE and in the nucleus. However, electron microscopy studies alone cannot determine if individual capsids are completely intact or identify the capsids that lead to productive infection. It will also be difficult to capture the different stages of uncoating in infected cells by electron microscopy methods, given that the loss of capsid integrity and uncoating occurs in <3 min.
Overall, our results indicate that the infectious HIV-1 viral cores retain capsid integrity and maintain a separation between the viral core content and the cellular environment until just before integration. In addition, we observed that capsids lose their integrity just a few minutes before uncoating, suggesting that capsid disassembly proceeds rapidly after the loss of core integrity. The observation that capsids remain intact until <1.5 h before integration has important implications for most postentry replication events, including nuclear import, reverse transcription, uncoating, and integration. The fact that nuclear capsids retain their integrity indicates that the mechanism by which capsids are imported through nuclear pores cannot induce major deformations that might disrupt the viral core structure. Furthermore, the results indicate that reverse transcription mostly occurs in an intact viral core and raise the possibility that a functional intasome may form within an intact capsid before uncoating occurs. Finally, the results indicate that interactions between the PIC and LEDGF/p75 and other potential host factors that facilitate integration must occur within a short time frame between uncoating and integration. Thus, these studies provide unprecedented insights into the kinetics of nuclear uncoating of viral cores and most of the essential steps in the early stage of HIV-1 replication.
Materials and Methods
Experimental details and methods can be found in SI Appendix, including sources of cell lines and procedures for their maintenance, description of HeLa:Bgl-mCherry, HeLa:mRuby-CPSF6, and CEM:SS-mRuby-Lamin B cell lines; description of lentiviral vectors pHGFP-GFPCA-BglSL, pHGFP-BglSL; construction of lentiviral vector pHGFP-iGFP-BglSL; and procedures for virus production and infection. Details of microscopy and image processing, live-cell imaging of fusion of GFP content marker-labeled virus, GFP content marker loss, nuclear capsids colabeled with core-associated mRuby-CPSF6 and GFP content marker or GFP-CA, image analysis using custom written MATLAB programs, and fixed-cell imaging and image analysis are described in SI Appendix. Methods for single virion analysis, in vitro analysis of intact virions and viral cores, transmission electron microscope analysis of virus pellets, fractionation of viral cores using sucrose gradients, and data analysis and statistics are also described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. John Coffin and Eric Freed for valuable discussions and suggestions during manuscript preparation. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by Intramural AIDS Targeted Antiviral Program grant funding (to V.K.P. and to W.-S.H.), and under contract HHSN26120080001E.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019467118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Gres A. T., et al., Structural virology. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 349, 99–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattei S., Glass B., Hagen W. J., Kräusslich H. G., Briggs J. A., The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354, 1434–1437 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Zhao G., et al., Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell E. M., Hope T. J., HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 13, 471–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arhel N., Revisiting HIV-1 uncoating. Retrovirology 7, 96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff S. P., Intracellular trafficking of retroviral genomes during the early phase of infection: Viral exploitation of cellular pathways. J. Gene Med. 3, 517–528 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Xu H., et al., Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology 10, 70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M., A hard way to the nucleus. Mol. Med. 10, 1–5 (2004). [PMC free article] [PubMed] [Google Scholar]
- 9.Dvorin J. D., Malim M. H., Intracellular trafficking of HIV-1 cores: Journey to the center of the cell. Curr. Top. Microbiol. Immunol. 281, 179–208 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Lehmann-Che J., Saïb A., Early stages of HIV replication: How to hijack cellular functions for a successful infection. AIDS Rev. 6, 199–207 (2004). [PubMed] [Google Scholar]
- 11.Suzuki Y., Craigie R., The road to chromatin–Nuclear entry of retroviruses. Nat. Rev. Microbiol. 5, 187–196 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Fassati A., Goff S. P., Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75, 3626–3635 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald D., et al., Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159, 441–452 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nermut M. V., Fassati A., Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J. Virol. 77, 8196–8206 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warrilow D., Tachedjian G., Harrich D., Maturation of the HIV reverse transcription complex: Putting the jigsaw together. Rev. Med. Virol. 19, 324–337 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Francis A. C., Melikyan G. B., Single HIV-1 imaging reveals progression of infection through CA-dependent steps of docking at the nuclear pore, uncoating, and nuclear transport. Cell Host Microbe 23, 536–548.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burdick R. C., et al., Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLoS Pathog. 13, e1006570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dismuke D. J., Aiken C., Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80, 3712–3720 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arhel N. J., et al., HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 26, 3025–3037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita M., Perez O., Hope T. J., Emerman M., Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 3, 1502–1510 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iordanskiy S., Berro R., Altieri M., Kashanchi F., Bukrinsky M., Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology 3, 4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacques D. A., et al., HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 536, 349–353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dick R. A., Mallery D. L., Vogt V. M., James L. C., IP6 regulation of HIV capsid assembly, stability, and uncoating. Viruses 10, 640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdick R. C., Hu W. S., Pathak V. K., Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proc. Natl. Acad. Sci. U.S.A. 110, E4780–E4789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burdick R. C., et al., HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. U.S.A. 117, 5486–5493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulme A. E., Kelley Z., Foley D., Hope T. J., Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-Infected cells. J. Virol. 89, 5350–5361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng K., et al., Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. eLife 3, e04114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bejarano D. A., et al., HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. eLife 8, e41800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francis A. C., Marin M., Shi J., Aiken C., Melikyan G. B., Time-resolved imaging of single HIV-1 uncoating in vitro and in living cells. PLoS Pathog. 12, e1005709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dharan A., Bachmann N., Talley S., Zwikelmaier V., Campbell E. M., Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 5, 1088–1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K., et al., Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito A., et al., Capsid-CPSF6 interaction is dispensable for HIV-1 replication in primary cells but is selected during virus passage in vivo. J. Virol. 90, 6918–6935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francis A. C., et al., HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Nat. Commun. 11, 3505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Achuthan V., et al., Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 24, 392–404.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hübner W., et al., Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81, 12596–12607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mamede J. I., Cianci G. C., Anderson M. R., Hope T. J., Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc. Natl. Acad. Sci. U.S.A. 114, E7169–E7178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah V. B., Aiken C., In vitro uncoating of HIV-1 cores. J. Vis. Exp. 3384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warrilow D., Stenzel D., Harrich D., Isolated HIV-1 core is active for reverse transcription. Retrovirology 4, 77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn E. H., et al., Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya A., et al., Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. U.S.A. 111, 18625–18630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito A., et al., Roles of capsid-interacting host factors in multimodal inhibition of HIV-1 by PF74. J. Virol. 90, 5808–5823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balasubramaniam M., et al., PF74 inhibits HIV-1 integration by altering the composition of the preintegration complex. J. Virol. 93, e01741-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherepanov P., et al., HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Cherepanov P., Devroe E., Silver P. A., Engelman A., Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279, 48883–48892 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Llano M., Delgado S., Vanegas M., Poeschla E. M., Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279, 55570–55577 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Krishnan L., et al., The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 84, 397–406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christ F., et al., Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18, 1192–1202 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Hink M. A., et al., Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. J. Biol. Chem. 275, 17556–17560 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Ganser-Pornillos B. K., Cheng A., Yeager M., Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell 131, 70–79 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Márquez C. L., et al., Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. eLife 7, e34772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christensen D. E., Ganser-Pornillos B. K., Johnson J. S., Pornillos O., Sundquist W. I., Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science 370, eabc8420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rankovic S., Varadarajan J., Ramalho R., Aiken C., Rousso I., Reverse transcription mechanically initiates HIV-1 capsid disassembly. J. Virol. 91, e00289-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forshey B. M., von Schwedler U., Sundquist W. I., Aiken C., Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76, 5667–5677 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yant S. R., et al., A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 25, 1377–1384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monroe K. M., et al., IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diner B. A., Lum K. K., Cristea I. M., The emerging role of nuclear viral DNA sensors. J. Biol. Chem. 290, 26412–26421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Briggs J. A., Wilk T., Welker R., Kräusslich H. G., Fuller S. D., Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22, 1707–1715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knockenhauer K. E., Schwartz T. U., The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanco-Rodriguez G., et al., Remodeling of the core leads HIV-1 preintegration complex into the nucleus of human lymphocytes. J. Virol. 94, e00135-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.