Abstract
Pancreatic cancer cells expressing the surface markers CD133 have been widely reported as cancer stem cells and mainly responsible for tumor recurrence and chemoresistance in pancreatic cancer. In spite of its role as stem cell marker in Pancreatic cancer, its function remains elusive. CD133 (also known as prominin-1) is a pentaspan glycoprotein predominantly localized in Lipid rafts, a specialized membrane microdomains enriched in crucial signaling proteins. Coexistence of CD133 with these signaling proteins can modulate various signaling pathways which might be responsible for aggressive phenotype of CD133+ cells. This chapter describe detailed protocol to isolate lipid rafts from CD133+ Tumor initiating cells. Purified lipid rafts can be investigated further for protein or lipid composition by mass spectrometry which can shed some light on functional role of CD133 protein in these cancer stem cells.
Keywords: CD133, Lipid rafts, Tumor Initiating Cells, Pancreatic Cancer, Cancer Stem Cells
1. Introduction
Pancreatic cancer is the third most common cancer related cause of death in United States, with 50,000 patients being detected each year and almost as many succumbing to the disease. The major reasons for poor survival are chemoresistance and tumor recurrence. Both these phenomena have been attributed to a specialized population of cells within the tumor, the tumor initiating cells or cancer stem cells. Over the years many markers have been associated with the pancreatic tumor initiating cells (1–5). One such marker CD133, is known to be consistent in giving rise to tumors from a very low number of cells in multiple animal models of this disease (4,6). Structurally, CD133 is a cholesterol-interacting pentaspan membrane protein concentrated in plasma membrane protrusions (7,8). Its unique distribution suggests CD133 may be involved in membrane organization (8). This idea is supported by the fact that loss of CD133 from the plasma membrane of human retinal cells causes retinal degeneration, possibly due to impaired generation of evaginations and/or impaired conversion of evaginations to disks (9).
Topologically, CD133 is located in cholesterol-containing lipid rafts in membrane microdomains, where it is involved in mediating signaling cascades (10,11). Lipid rafts are small platforms, composed of sphingolipids and cholesterol in the outer exoplasmic leaflet, connected to phospholipids and cholesterol in the inner cytoplasmic leaflet of the lipid bilayer. These assemblies are fluid but more ordered and tightly packed than the surrounding bilayer. The difference in packing is due to the saturation of the hydrocarbon chains in raft sphingolipids and phospholipids as compared with the unsaturated state of fatty acids of phospholipids in the liquid-disordered phase. The organization of the lipid rafts is considered to play a significant role in regulating EMT (epithelial mesenchymal transition) which is a hallmark of metastasis. Concomitant with the acquisition of an aggressive phenotype, the EMT is marked by a profound rewiring of the cell signaling programs that affect a multitude of pathways (12,13). Many pathways are activated by extracellular ligands or receptors located at the plasma membrane (PM), suggesting that changes in the properties of the PM may facilitate the wholesale signaling network rearrangements associated with an EMT. Supporting this possibility, the lipid compositions of cells in epithelial or mesenchymal states have been shown to be distinct (14) and useful in distinguishing cells with an EMT phenotype (15).Further, alterations in the fluidity of the plasma membrane, e.g. by modulating cholesterol content, can induce or inhibit an EMT (16).
The function of CD133 is even less clear in the context of cancer. Despite its ubiquitous presence on CSCs from various solid tumors, it is unknown whether the intracellular signaling downstream of CD133 contributes to the maintenance of cellular stemness. Clinically, strong CD133 expression correlates with chemo/radio-resistance and a poor prognosis (17).
Interestingly, expression of CD133 has been closely correlated with metastasis and aggressive biology of tumors (6). Tumors having high expression of CD133 are known to metastasize more than those with lower CD133+ population. Thus, it is of utmost importance to understand the structural association of CD133 in the lipid rafts of the plasma membrane, in order to evaluate the function of this critical TIC marker in pancreatic cancer. In this article we outline the protocol for isolation of lipid rafts from CD133+ cells from pancreatic tumors. Following isolation, the lipid rafts can be analyzed further by proteomics or lipidomics methods to evaluate the composition of these membrane structures. A complete understanding of the protein and lipid components is required to elucidate the function of this protein and how it may be involved in invasion metastasis and chemoresistance.
2. Materials
Primary tumor from patients, spontaneous pancreatic cancer mouse model (KRas G12D TP53R172H PdxCre).
RPMI 1640 medium, Fetal bovine serum (FBS).
Phosphate Buffered Saline.
Collagenase IV (Worthington Biochemicals).
MACS column (Miltenyi Biotech)
anti CD133 microbeads (human) or anti-CD133/PROM1 microbeads (mouse).
Anti CD133 antibody.
OptiPrep (Sigma Aldrich).
Isolation Buffer (IB): 150 mM NaCl, 5 mM dithiothreitol (DTT), 5 mM EDTA, 25 mM Tris- HCl, pH 7.4 supplemented with a cocktail of protease inhibitors.
Triton X-100.
Ultracentrifuge with any small volume (approx. 10–13 ml) swinging bucket rotor (e.g., Beckman SW41Ti or equivalent).
Syringe with metal cannula (for underlayering) or Pasteur pipette (for overlayering).
40-micron nylon mesh, electric pipettor.
Dot blot apparatus.
3. Methods
3.1. Preparation of Single Cell Suspension
To generate single cell suspension, cut xenograft tumors from mice or primary human tumors into small pieces with sterile scissors in RPMI medium, and then mince the tissue mechanically with scissors or scalpel until the pieces are 1 mm in size (able to be pipetted without difficulty using 10-mL pipettes). Wash the tissue with RPMI twice, centrifuge samples, and discard the solution.
Resuspend the minced tissue in 20–30 mL RPMI in a 50-mL centrifuge tube depending on the amount of tissue. Large amounts of tissue may require additional tubes.
Digest minced tissue by adding ultrapure collagenase IV in medium at a final concentration of 200 units/mL.
Incubate the sample at 37°C with shaking at a speed of 120 RPM for 1.5 h for first step enzymatic dissociation.
Pipette the sample for 3 min using a 25-mL pipette to mechanically dissociate the sample and put it back to 37°C incubation.
Further mechanically dissociate the sample every 15–20 min by pipetting with a 10-mL or 5-mL pipette until whole tumor is dissociated.
Add RPMI to a total volume of 50 mL and spin down at 800 RPM for 5 minutes
Discard the supernatant and resuspend the samples with RPMI containing 2% FBS.
Filter through a 40-micron nylon mesh to eliminate clumps and debris.
Wash cells with RPMI/2% FBS twice to remove enzyme solution. Resuspend cells in RPMI/2% FBS and perform a cell count. Cells are ready for sorting (see Note 1).
3.2. MACS Sorting
Prepare MACS mini/midi column by washing them with RPMI/2%FBS while attached to magnetic stand.
While the column is being washed add anti-CD133 microbeads to single cell suspension of tumor cells (human origin) or anti-Prom1 microbeads to cells of murine origin.
Incubate cells on ice for 20 min
Add cell-microbeads mix to column attached to magnetic stand.
Collected flow through in a 15 ml tube (this is CD133- cells. Once purity has been determined, this fraction can be discarded).
Wash column with RPMI/2%FBS and pool with flow-through to remove traces of CD133- cells
Remove column from magnetic stand and place on 15ml tube. Elute CD133+ cells using RPMI/2%FBS. (This is CD133+ fraction)
Analyze 50 μl of elute of CD133+ cells and CD133- cells by flow cytometry (using anti-CD133-PE antibody to confirm purity).
3.3. Cell lysis
Wash CD133+ cell pellet from the tumor (~ 10–12 million cells from the above mentioned tumor size) twice with PBS (5 times the cell pellet volume).
Add 0.8 mL of ice cold 0.1% Triton X-100 IB to the cell pellet (see Note 2).
Use an automatic pipet to resuspend cell pellet and vortex breifly for 10 sec (see Note 3).
Pass solution through a 23-gauge needle using a 5-mL syringe 20 times (see Note 4).
Centrifuge lysed cells in solution for 10 min at 112 × g at 4 °C
Carefully retrieve post nuclear supernatant from pellet of cell debris (see note 5).
3.4. Density Gradient
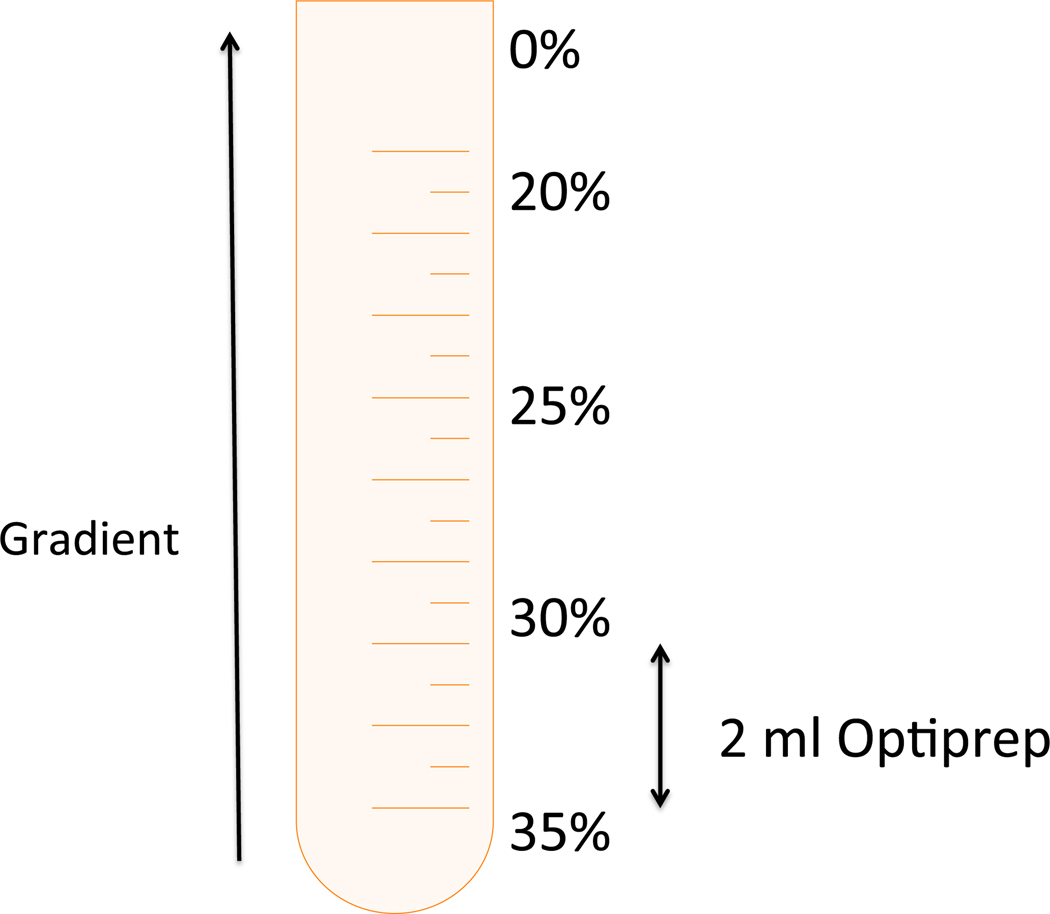
The density gradient is made of 5 layers of OptiPrep with different concentrations: 35%, 30%, 25%, 20% and 0%. The lower layer (35% OptiPrep) contains the cell lysate (see Table 1). Work with pre-cooled Lysis Buffer, OptiPrep (60% w/v), OptiPrep gradient layers, and 2 ml microcentrifuge tubes.
Table 1.
Preparation of OptiPrep Density Gradient Layers.
| Gradient Layer | Final OptiPrep (%) | Cell lysate (vol) | IB (ml) | OptiPrep (ml) | Total volume (ml) |
|---|---|---|---|---|---|
| 1 (bottom) | 35 | 0.84 | 0 | 1.16 | 2 |
| 2 | 30 | - | 1 | 1 | 2 |
| 3 | 25 | - | 1.16 | 0.84 | 2 |
| 4 | 20 | - | 1.3 | 0.7 | 2 |
| 5 (top) | 0 | - | 1 | 0 | 1 |
Prepare the 5 solutions that will form the OptiPrep gradient layers according to Table 1. Mix each one well by vortexing. Keep the prepared solutions on ice (see Note 6).
Put 2 ml of gradient layer 1 (35% OptiPrep containing the cell lysate) at the bottom of the precooled ultracentrifuge tube.
Place each OptiPrep gradient layer over the other in order (see Figure 1) using a Pasteur pipette. It is recommended to use an electric pipettor. Gradient is visible to the naked eye.
Spin the density gradient in ultracentrifuge at 200,000 × g for 4 h at 4 °C.
Take the tubes carefully out of the ultracentrifuge and put them on ice.
Mark 9 microcentrifuge tubes from 1–9. Tube number 1 will be used for the lowest % of the gradient (the top fraction of the ultracentrifuge tube).
CD133 lipid rafts are recovered as a fine dense band at the border of 20% (w/v) and 30% (w/v) OptiPrep layers after ultracentrifugation at ~200,000 × g for 4 hours. Proteins that are part of the rafts or bound to these structures are present in the rafts enriched fraction.
Figure 1:
Demonstration of setting up an OptiPrep gradient for lipid raft isolation.
3.5. Fractions Collection
Mark a line on a Pasteur pipette to indicate a 1 ml volume. Connect the pipette to an electric pipettor.
Carefully collect 1 ml fractions from top to bottom of the ultracentrifuge tube and transfer each fraction to a marked microcentrifuge tube. The number of total collected fractions can vary between 7–9. Number the topmost fraction 1 and so on.
Keep the fractions on ice for later use.
Fractions can be stored in −80⁰C for up to 6 months.
3.6. Gradient Fraction Analysis
Perform dot-blot with 2–3 μl of each gradient fraction as well as original lysate on nitrocellulose membrane.
Hybridize with CD133 antibody to detect fraction containing lipid raft.
4. Notes
Digestion of a 1-cm3 xenograft tumor will typically result in 10–20 million cells. Excess cells may be frozen for future use by placing cells in a solution of FBS with 10% DMSO.
Lipid rafts have a unique feature of relative resistance to solubilization in an ice cold TRITON X-100 solution. This feature is used for their isolation. Since the procedure is highly temperature dependent, the work should be performed in the cold room. Note that at 8°C the Caveolae/Rafts proteins may already be soluble in the TRITON X-100 solution and will not float up in the gradient.
Solution should appear homogeneous with no clumps of cells in solution.
Drawing solution into the syringe and then forcing it out counts as 2X.
Cell debris pellet will be soft and longer spin time may be needed (up to 20 min). Do NOT spin at any higher speed or membrane lipids will be lost.
In order to create an OptiPrep layer, which contains precisely 35% OptiPrep, the volume of cell lysate in tube number 1 should be exactly 0.84 ml.
References
- 1.Bednar F, Simeone DM (2009) Pancreatic cancer stem cells and relevance to cancer treatments. Journal of cellular biochemistry 107 (1):40–45. doi: 10.1002/jcb.22093 [DOI] [PubMed] [Google Scholar]
- 2.Bhagwandin VJ, Shay JW (2009) Pancreatic cancer stem cells: fact or fiction? Biochim Biophys Acta 1792 (4):248–259. doi: 10.1016/j.bbadis.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donahue TR, Dawson DW (2011) Nodal/Activin signaling: a novel target for pancreatic cancer stem cell therapy. Cell stem cell 9 (5):383–384. doi: 10.1016/j.stem.2011.10.006; 10.1016/j.stem.2011.10.006 [DOI] [PubMed] [Google Scholar]
- 4.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C (2007) Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell stem cell 1 (3):313–323. doi: 10.1016/j.stem.2007.06.002 [DOI] [PubMed] [Google Scholar]
- 5.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM (2007) Identification of pancreatic cancer stem cells. Cancer research 67 (3):1030–1037. doi: 10.1158/0008-5472.can-06-2030 [DOI] [PubMed] [Google Scholar]
- 6.Banerjee S, Nomura A, Sangwan V, Chugh R, Dudeja V, Vickers S, Saluja AK (2014) CD133+ tumor initiating cells (TIC) in a syngenic murine model of pancreatic cancer respond to Minnelide. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-13-2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbeil D, Marzesco AM, Wilsch-Brauninger M, Huttner WB (2010) The intriguing links between prominin-1 (CD133), cholesterol-based membrane microdomains, remodeling of apical plasma membrane protrusions, extracellular membrane particles, and (neuro)epithelial cell differentiation. FEBS letters 584 (9):1659–1664. doi: 10.1016/j.febslet.2010.01.050; 10.1016/j.febslet.2010.01.050 [DOI] [PubMed] [Google Scholar]
- 8.Marzesco AM (2013) Prominin-1-containing membrane vesicles: origins, formation, and utility. Advances in Experimental Medicine and Biology 777 (Journal Article):41–54. doi: 10.1007/978-1-4614-5894-4_3; 10.1007/978-1-4614-5894-4_3 [DOI] [PubMed] [Google Scholar]
- 9.Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, Kumaramanickavel G, John S, Nancarrow D, Roper K, Weigmann A, Huttner WB, Denton MJ (2000) A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Human molecular genetics 9 (1):27–34 [DOI] [PubMed] [Google Scholar]
- 10.Roper K, Corbeil D, Huttner WB (2000) Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat Cell Biol 2 (9):582–592. doi: 10.1038/35023524 [DOI] [PubMed] [Google Scholar]
- 11.Giebel B, Corbeil D, Beckmann J, Hohn J, Freund D, Giesen K, Fischer J, Kogler G, Wernet P (2004) Segregation of lipid raft markers including CD133 in polarized human hematopoietic stem and progenitor cells. Blood 104 (8):2332–2338. doi: 10.1182/blood-2004-02-0511 [DOI] [PubMed] [Google Scholar]
- 12.McCaffrey LM, Macara IG (2011) Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol 21 (12):727–735. doi: 10.1016/j.tcb.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 13.Martin-Belmonte F, Perez-Moreno M (2012) Epithelial cell polarity, stem cells and cancer. Nature reviews Cancer 12 (1):23–38. doi: 10.1038/nrc3169 [DOI] [PubMed] [Google Scholar]
- 14.Bose R, Wrana JL (2006) Regulation of Par6 by extracellular signals. Curr Opin Cell Biol 18 (2):206–212. doi: 10.1016/j.ceb.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Lopez S, Lerner RG, Petritsch C (2014) Asymmetric cell division of stem and progenitor cells during homeostasis and cancer. Cell Mol Life Sci 71 (4):575–597. doi: 10.1007/s00018-013-1386-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, Minhas S, Soeda A, Hoeppner DJ, Ravin R, McKay RD, McLendon RE, Corbeil D, Chenn A, Hjelmeland AB, Park DM, Rich JN (2011) Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis 2:e200. doi: 10.1038/cddis.2011.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su YJ, Lin WH, Chang YW, Wei KC, Liang CL, Chen SC, Lee JL (2015) Polarized cell migration induces cancer type-specific CD133/integrin/Src/Akt/GSK3beta/beta-catenin signaling required for maintenance of cancer stem cell properties. Oncotarget 6 (35):38029–38045. doi: 10.18632/oncotarget.5703 [DOI] [PMC free article] [PubMed] [Google Scholar]